Abstract
The period (per) gene is an essential component of the circadian timekeeping mechanism inDrosophila. This gene is expressed in a circadian manner, giving rise to a protein that feeds-back to regulate its own transcription. A 69 bp clock regulatory sequence (CRS) has been identified previously upstream of the period gene. The CRS confers wild-type mRNA cycling when used to drive alacZ reporter gene in transgenic flies. To determine whether the CRS also mediates proper developmental and spatial expression and behavioral rescue, we used the CRS to drive eitherlacZ or per in transgenic flies. The results show that the CRS is able to activate expression in pacemaker neuron precursors in larvae and essentially all tissues that normally express per in pupae and adults. The CRS is sufficient to rescue circadian feedback loop function and behavioral rhythms inper01 flies. However, the period of locomotor activity rhythms shortens if a stronger basal promoter is used. This study shows that regulatory elements sufficient for clock-dependent and tissue-specific per expression in larvae, pupae, and adults are present in the CRS and that the period of adult locomotor activity rhythms is dependent, in part, on the overall level of per transcripts.
Keywords: Drosophila, circadian clock, transcriptional regulation, behavior, period gene, developmental expression
In Drosophila melanogaster, an autoregulatory feedback loop in gene expression is a central feature of the circadian timekeeping mechanism. In this feedback loop, the period (per) andtimeless (tim) genes are rhythmically expressed such that circadian fluctuations in per and timmRNA levels are controlled by fluctuating levels of PER and TIM proteins (Rosato et al., 1997; Hardin and Sehgal, 1998). As PER and TIM accumulate, they bind to each other and move into the nucleus (Vosshall et al., 1994; Curtin et al., 1995; Gekakis et al., 1995; Saez and Young, 1996; Zeng et al., 1996), where they act to repress the transcription of their own genes (Hardin et al., 1992; Zeng et al., 1994; Sehgal et al., 1995; So and Rosbash, 1997; Darlington et al., 1998).
To understand how PER and TIM regulate cyclic transcription, we have identified the sequences that control circadian transcription of theper gene. A 69 bp circadian regulatory sequence (CRS), situated ∼500 bp upstream of the per transcriptional initiation site, mediates mRNA cycling with an amplitude and phase similar to that of the wild-type per transcript (Hao et al., 1997). Within the CRS a consensus “E-box” transcription factor binding site is required for high-level per transcription (Hao et al., 1997).
E-box–dependent transcriptional activation is mediated by two members of the basic helix-loop-helix-PAS (bHLH-PAS) family of transcription factors, Drosophila CLOCK (dCLK) and BMAL1 (Darlington et al., 1998), also known as CYCLE (CYC) (Rutila et al., 1998). Mutations that impair the activity of either dCLK or CYC result in very low levels of per mRNA and behavioral arrhythmicity, showing that these proteins are essential for circadian clock function inDrosophila (Allada et al., 1998; Rutila et al., 1998). Consistent with the role of PER and TIM as transcriptional repressors, dCLK- or CYC-dependent activation is inhibited by the presence of PER and TIM in Drosophila tissue culture (Darlington et al., 1998). A similar regulatory circuit may also occur in mammals because orthologs of dCLK and CYC, called CLOCK (King et al., 1997) and BMAL1 (Ikeda and Nomura, 1997; Gekakis et al., 1998), respectively, activate transcription via E-boxes upstream of the mouse PER1 (mPER1) gene (Gekakis et al., 1998).
Although the per CRS is sufficient for circadian transcription in adult Drosophila, it is unclear whether the CRS also controls tissue- and developmental stage–specific expression. The spatial expression pattern of per has been well characterized and includes neuronal and non-neuronal tissues in the head and body (Hall, 1995). Among these tissues, a set of neurons in the lateral brain (LNs) appears to be the pacemaker cells for locomotor activity rhythms (Frisch et al., 1994). per is active in late embryos through adults (Young et al., 1985), and expression in LN precursors during development may be important for mediating the “time memory” of adults that were entrained as larvae (Sehgal et al., 1992; Kaneko et al., 1997).
In this study we have tested whether the per CRS mediates normal spatial and developmental expression and rescues behavioral rhythms in per01 mutants. These studies show that the CRS confers accurate (i.e., per-like) spatial expression in larvae, pupae, and adults. CRS-dependent perexpression rescues behavioral rhythms, resulting in shorter or longer periods depending on whether strong or weak promoters are used, respectively. Thus, the per CRS is a target for transcription factors that regulate circadian, spatial, and developmental expression.
MATERIALS AND METHODS
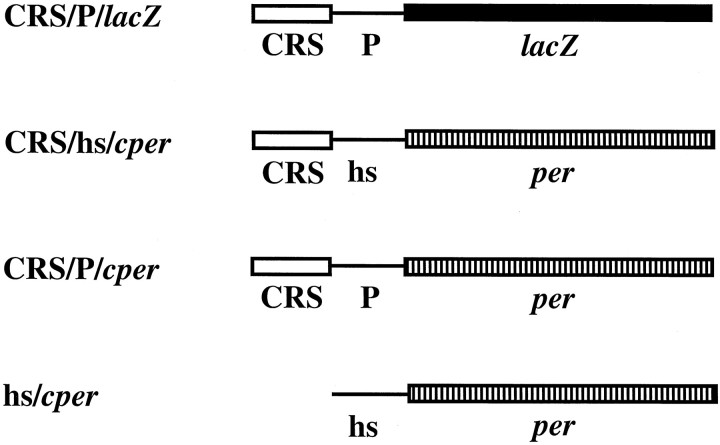
Construction of transformation plasmids. The CRS/P/lacZ transgene was constructed as follows. The CRS was amplified from the −563 to −494/hs/lacZ construct template with a sense (5′-GAGAATTCGAGAAACCGTAGG-3′) and an antisense (5′-GTGGATCCGATTTTGCTGGCC-3′) primer pair. This PCR fragment was inserted into the CPLZ vector (Wharton and Crews, 1993) at theEcoRI and BamHI sites.
The hs/cper transformation vector was constructed as follows. A 5.9 kb per cDNA fragment spanning from theSalI site of exon 3 to the EcoRI site at the 3′ downstream sequence was cut out from the hspcper (Edery et al., 1994a) and cloned into pBluescript KS− to form Rec2. The remainder of the per cDNA and the heat-shock basal promoter were generated by PCR using the hspcper as a template, a sense primer (5′-GGCTCGAGGAGCGCCGGAGTATAAATAG-3′), and an antisense primer (5′-GGCTCGTCGACGCCGAG-3′). This PCR product was cloned into the XhoI and SalI restriction sites of Rec2 to form the hs/cper fusion Rec5. The hs/cperfusion gene was then cloned into the KpnI- andXbaI-cut polylinker sequences of a modified (i.e.,XhoI sites deleted) CaSpeR-4 transformation vector (Thummel and Pirotta, 1991).
The CRS DNA fragment was generated using the −563 to −494/hs/lacZ construct (Hao et al., 1997) (also called CRS/hs/lacZ) as a template for PCR with sense (5′-GAGGTACCTACGGTTTCTCGG-3′) and antisense (5′-CACTCGAGGCGGATTTGCTGGCC-3′) primers. The PCR product was inserted into the hs/cper transformation vector at theKpnI and XhoI sites to form the CRS/hs/cper construct.
Construction of the CRS/P/cper transgene was as follows. A 550 bp XhoI/SalI fragment containing thehsp70 basal promoter fused to the 5′ portion of theper cDNA was subcloned into pBluescript KS−, forming X/S-550. The hsp70 promoter region was removed by digestion with XhoI andNcoI and replaced with a 100 bp PCR fragment containing the P-element transposase basal promoter generated using CaSpeR-4 as a template for sense (5′-GTCTCGAGAAGCTTACCGAAG-3′) and antisense (5′-GACCATGGTAAGGGTTAATG-3′) primers, ultimately forming X/S-P550. The P-element transposase basal promoter containing theXhoI/SalI fragment from X/S-P550 was then removed and used to replace the hsp70 basal promoter-containing fragment from Rec4, which contains the 3′ portion of the percDNA plus 2.1 kb of downstream per sequences, forming Rec4-P. The CRS was removed from the CRS/hs/cper construct by digestion with KpnI and XhoI and was inserted into Rec4-P, forming CRS-Rec4-P. A KpnI/EcoRI fragment containing the CRS, the P-element basal promoter, and theper cDNA and 3′-flanking sequences was ligated into CaSpeR-4, forming CRS/P/cper.
The nucleotide sequence of all constructs was confirmed using octamer sequencing (Hardin et al., 1996).
Fly stocks and germ-line transformation. D. melanogaster strains were raised on a cornmeal, sugar, agar, yeast, and Tegosept (a mold inhibitor) medium at 25°C. The wild-typeD. melanogaster strain was Canton-S. P-element–mediated germ-line transformation was performed as described previously (Hao et al., 1997). At least four independent transformant lines with inserts on the second or third chromosomes were generated for each construct and balanced with In(2LR)CyO and In(3LR)TM2, respectively. CRS/P/lacZ transformants were crossed into a y per01 w genetic background for β-galactosidase staining as adults and into a w genetic background for β-galactosidase staining as larvae and pupae. The BG6 transgene (Dembinska et al., 1997) was used as a positive control for larval staining (Kaneko et al., 1997). To test whether larval staining was dependent on dClk or Cyc, we crossed BG6 transformants into a homozygous dClkJrk(Allada et al., 1998) or Cyc (Rutila et al., 1998) genetic background. CRS/P/cper transformants were crossed into ay per01 w genetic background to test for molecular and behavioral rhythms.
β-Galactosidase staining. Each of five independent CRS/P/lacZ transgenic lines was dissected at Zeitgeber time 1 (ZT1) as L1, L2, or L3 larvae and as early, mid, or late pupae and then assayed for β-galactosidase activity using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) histochemistry as described (Smith and Shepherd, 1996). CRS/P/lacZ adults were sectioned and stained using X-gal histochemistry as described (Liu et al., 1988). At least eight individuals were assayed from each independent CRS/P/lacZline at each developmental stage. Twenty BG6 larvae were assayed at the L3 stage, and each gave the reported staining pattern (Kaneko et al., 1997). Twenty BG6;dClkJrk and six BG6;Cyc larvae were assayed at the L3 stage, and none showed CNS staining.
Locomotor activity analysis. Locomotor activity of adult male Canton-S, y per01 w,y per01w;CRS/P/cper, y per01 w;CRS/hs/cper, andy per01 w;hs/cpertransgenic flies were monitored and analyzed as described (Hamblen et al., 1986). Briefly, flies were entrained in 12:12 hr light/dark (LD) cycles at 25°C for 3 d and then were transferred into constant darkness (DD). Locomotor behavior was monitored continuously starting from the entrainment, and data collected during DD were analyzed using periodogram analysis (Hamblen et al., 1986). Flies with powers >15 and a width greater than two in periodogram analysis were designated rhythmic.
RNase protection assays. Flies used for RNase protection analyses were entrained in 12:12 hr LD cycles at 25°C for 3 d and then transferred into DD and collected during the first day. For each time point, RNA was extracted from the heads and used for RNase protection assays as described (Hardin et al., 1990). The probe used in these studies was Rec5 (used to detect endogenousper01 transcript and transgenic hs/cper transcript). The Rec5 probe contains a 329 nucleotide (nt) antisense RNA from +208 bp of the heat-shock leader sequences to the SalI site in per exon 3. The probe protects a 329 nt fragment from the transgenic pertranscript (hs/cper) and a 283 nt fragment from the endogenous per01 transcript. As a control for the amount of RNA in each lane, an antisense ribosomal protein probe (RP49) was included in each RNase protection assay (Hardin et al., 1990).
Immunohistochemistry. Flies were entrained in 12:12 hr LD cycles at 25°C for at least 3 d and then transferred into DD and collected during the first day. Sectioning and staining were performed as described (Siwicki et al., 1988). Polyclonal anti-PER antibody raised in rabbits (a gift from J. Hall and R. Stanewsky) was used with a biotinylated goat anti-rabbit IgG secondary antibody to detect PER via immunostaining at a 1:4000 and a 1:2000 dilution, respectively.
Western blotting. Western-blotting analyses were performed as described (Edery et al., 1994b) with the following modifications. The polyclonal anti-PER antibody was diluted to 1:20,000, and the horseradish peroxidase–linked anti-rabbit IgG was diluted to 1:5000 in blocking solution; the blots were incubated with the primary antibody at 4°C overnight and with the secondary antibody at room temperature for 1 hr.
RESULTS
The CRS mediates per-like spatial expression in adults
Our previous reporter gene studies showed that the CRS is capable of driving rhythmic transcription (Hao et al., 1997). The transgenes used in these studies produced cytoplasmic β-galactosidase that was difficult to resolve as individual cells. To improve cellular resolution, we inserted the CRS into the CPLZ transformation vector (Wharton and Crews, 1993), which produces a β-galactosidase product that is localized to the nucleus because it is fused to the N terminal of the P-element transposase that contains a nuclear localization signal (O’Kane and Gehring, 1987). This vector also uses the P-element transposase basal promoter to drive lacZ (Fig.1).
Fig. 1.
Schematic drawings of transgenic constructs.CRS, per circadian regulatory sequence;hs, Drosophila heat-shock protein 70 gene basal promoter plus leader sequences; lacZ, fusion of the N terminal of the P-element transposase (including the nuclear localization signal) and the Escherichia coli lacZ-coding sequences; P,Drosophila P-element transposase gene basal promoter plus leader sequences; and per, per cDNA plus 2.1 kb of 3′-flanking sequences.
In adults, X-gal staining of each CRS/P/lacZ transformant line reveals nuclear β-galactosidase activity in photoreceptors, glial cells of the optic lobe, most LNs and dorsal neurons (DNs), the ventriculus, cardia, fat bodies, and Malpighian tubules (Fig.2). We have observed that a few cells, including glia in the lamina, dorsal LNs, and the first group of dorsal neurons (DN1s), show little or no staining in CRS/P/lacZtransformants compared with that shown by lacZ driven by ∼4 kb of per upstream sequence (Liu et al., 1988, 1991;Stanewsky et al., 1997) and of PER in wild-type flies (Zerr et al., 1990). Staining throughout the entire cell is seen in the abdomen and represents endogenous β-galactosidase activity (Liu et al., 1988). Thus, the CRS contains not only regulatory sequences capable of driving rhythmic transcription but also regulatory sequences that mediate essentially normal per expression in adults.
Fig. 2.
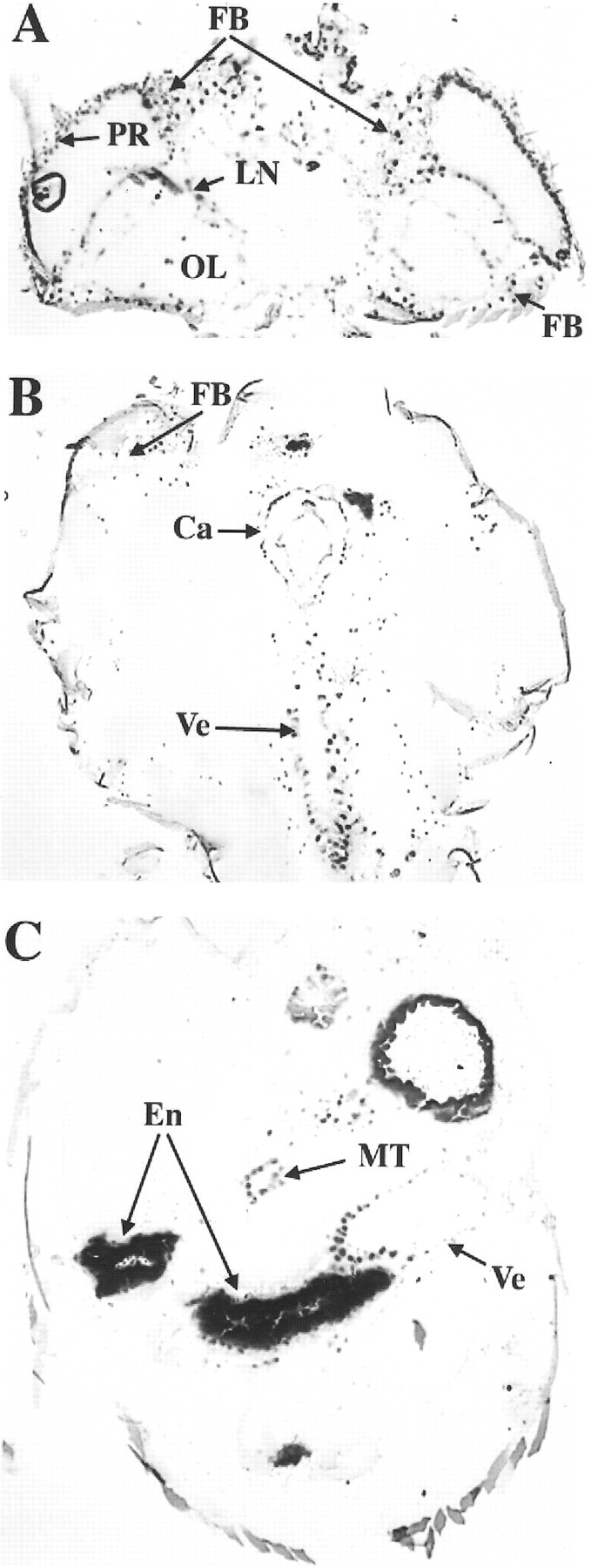
Spatial expression of the CRS/P/lacZ transgene in adults. A, X-gal–stained head of a male transgenic CRS/P/lacZ fly.B, X-gal–stained thorax of a male transgenic CRS/P/lacZ fly. C, X-gal–stained abdomen of a male transgenic CRS/P/lacZ fly. Ca, Cardia; En, endogenous ventricular staining;FB, fat bodies; LN, lateral neurons;MT, Malpighian tubules; OL, optic lobe glia; PR, photoreceptor cells; and Ve, ventriculus.
The CRS drives expression in LN precursors
The Drosophila circadian timekeeping system operates from the first larval instar (L1) onward (Sehgal et al., 1992). During the L1 stage, per begins to be expressed in one cluster of lateral neurons and in two clusters of dorsal neurons (DN1 and DN2) (Kaneko et al., 1997). Although per is rhythmically expressed in all three clusters of larval neurons, the observation thatper expression is only maintained in the LNs into adulthood suggests that this cluster of neurons conveys circadian phase to adults (Kaneko et al., 1997). Because per expression in larval neurons is correlated with the onset of circadian timekeeping, we wanted to determine whether the CRS could drive per-like expression in larvae.
Larvae containing the CRS/P/lacZ or BG transgenes were stained at the mid-L1, -L2, and -L3 stages. The BG transgene, which serves as a positive control, is a per–lacZ fusion gene that is expressed in larval LNs, DN1s, and DN2s (Kaneko et al., 1997). The CRS/P/lacZ-staining patterns were similar for each larval instar (data not shown); thus we will only show the results obtained from L3 larvae. In BG and two representative CRS/P/lacZ lines, β-galactosidase staining was readily detected in the nuclei of four to five larval LNs (Fig.3). In contrast to BG, CRS/P/lacZ larvae showed no staining in DN1s, and staining in DN2s was detected in only 10% of the brain hemispheres (data not shown). Because the CRS/P/lacZ transgene was expressed in larval and adult LNs (Figs. 2, 3), we suspected that this transgene would also be expressed in pupal LNs. When brains from early (12–24 hr), mid (48 hr), and late (72–96 hr) CRS/P/lacZ pupae were stained, LN staining was observed (data not shown). These data demonstrate that the CRS mediates expression in LNs from the first larval instar to adults.
Fig. 3.
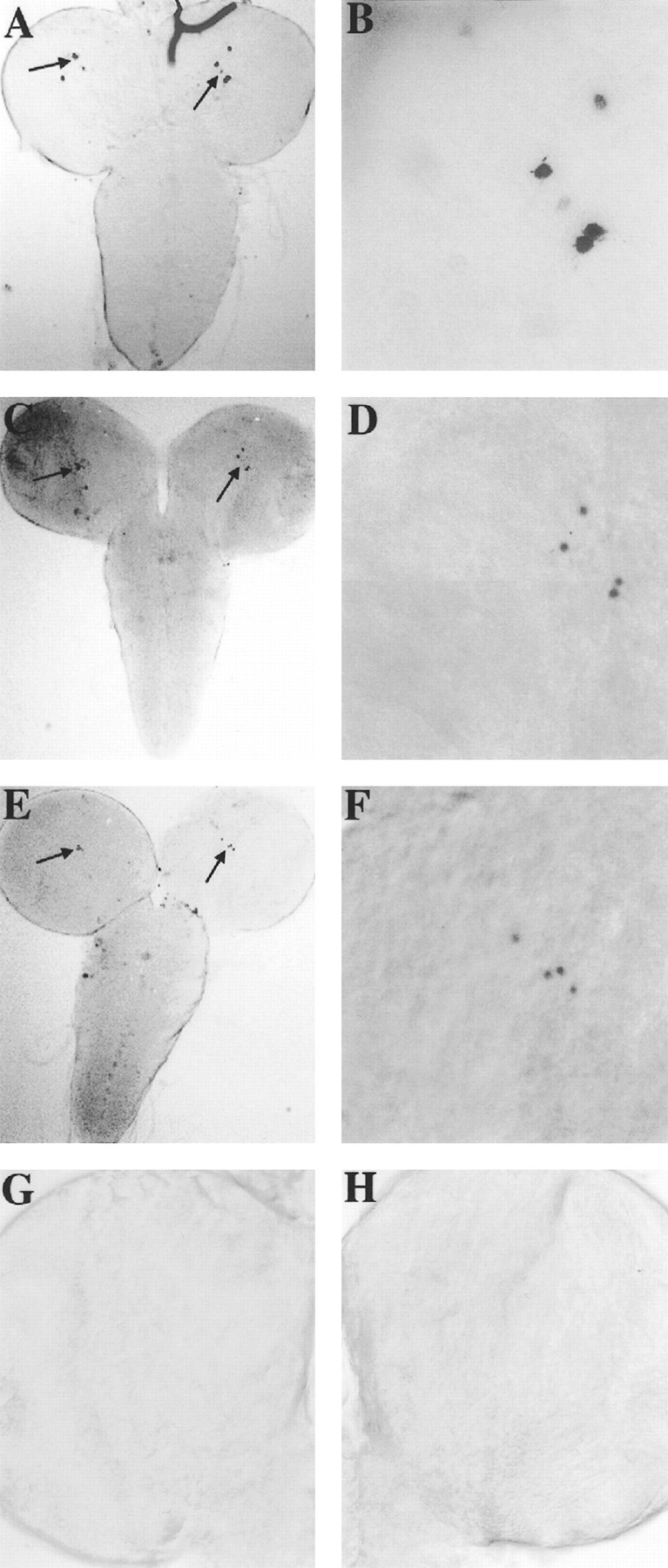
Spatial expression of the CRS/P/lacZ transgene in L3 larvae. A–F, Anterior on top. A, CNS of a BG6 third instar larva dissected at ZT1. In this focal plane, staining is restricted to a cluster of four or five LNs (arrows) in each hemisphere. B, Higher magnification of LN staining from the right hemisphere in A. C, CNS of a CRS/P/lacZ-3 third instar larva dissected at ZT1. Staining is restricted to a cluster of four or five LNs (arrows) in each hemisphere. D, Higher magnification of LN staining from the right hemisphere inC. E, CNS of a CRS/P/lacZ-8 third instar larva dissected at ZT1. Staining is restricted to a cluster of four or five LNs (arrows) in each hemisphere.F, Higher magnification of LN staining from the right hemisphere in E. G, Left optic lobe of a representative BG6;dClkJrk third instar larva dissected at ZT1. H, Right optic lobe of a representative BG6;Cyc third instar larva dissected at ZT1.
The CRS regulates correct per mRNA and protein expression in adults
Because the CRS is capable of mediating per-like circadian, spatial, and developmental expression, we hypothesized thatper expression driven by this regulatory element would rescue molecular and behavioral rhythms inper01 flies. To test this hypothesis, we inserted the CRS upstream of a Drosophila heat-shock protein 70 (hsp70) basal promoter/per cDNA (hs/cper) fusion gene that contains per coding sequences plus 2.1 kb of noncoding genomic sequences (Fig. 1). P-element–mediated germ-line transformation was used to generate four transgenic lines, which were then crossed into aper01 background.
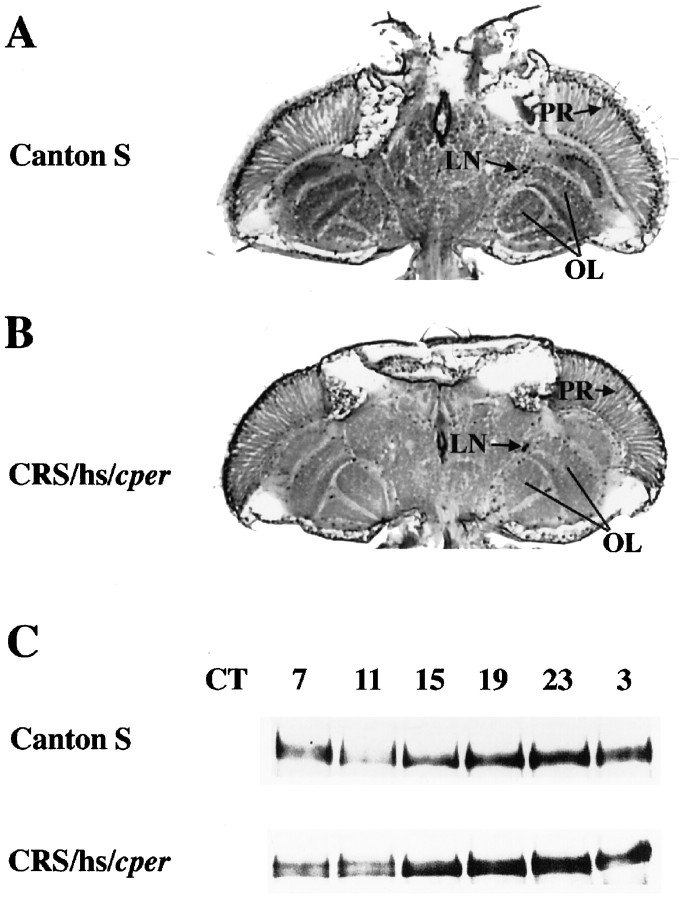
The PER spatial expression pattern in transformant flies containing CRS/hs/cper resembles that of wild-type flies; CRS/hs/cper flies collected at circadian time 22 (CT22) and sectioned and stained with anti-PER antibody show PER in photoreceptors, brain glia, and LNs (Fig.4A,B). The abundance of head-specific PER generated from the transgene also shows daily fluctuations during the first day of DD by Western blot analysis (Fig. 4C). The levels of PER in these transformants peak between CT19 and CT23 and fall to their lowest levels between CT7 and CT11. This cycling appears to phase lead PER cycling in wild-type flies, which peaks at CT23 and is least abundant at CT11, consistent with previous observations (Edery et al., 1994b; Zeng et al., 1996).
Fig. 4.
Spatial and circadian expression of PER inper01;CRS/hs/cpertransformants. A, Spatial distribution of PER staining within the head of a wild-type (Canton-S) fly collected at ZT22.B, Spatial distribution of PER staining within the head of a per01;CRS/hs/cpertransformant collected at ZT22. C, Western blot of PER abundance during constant darkness. Wild-type (Canton-S) andper01;CRS/hs/cperflies were collected every 4 hr during the first day in DD after 3 d of entrainment. The circadian time (CT) of each time point is noted above the lane. For abbreviations, see Figures 1 and 2.
CRS/hs/cper-derived transcripts from heads cycle with an approximately fivefold amplitude during the first day of DD (Fig.5). Consistent with previous observations, circadian cycling of endogenousper01 transcripts is also rescued (Hardin et al., 1990). The peak level for both the endogenous and transformant-derived transcripts is between CT11 and CT15, consistent with that of wild-type transcripts (Hardin et al., 1990). The overall level of CRS/hs/cper-derived transcripts is more than twofold higher than that of rescued per01transcripts at their peak levels, which may be attributable to the relatively strong hsp70 basal promoter (Hao et al., 1997). Thus, the histochemical staining, Western blot, and RNase protection results show that the CRS is capable of regulating the transcriptional aspects of per expression needed for feedback loop function in the appropriate cells of the head.
Fig. 5.
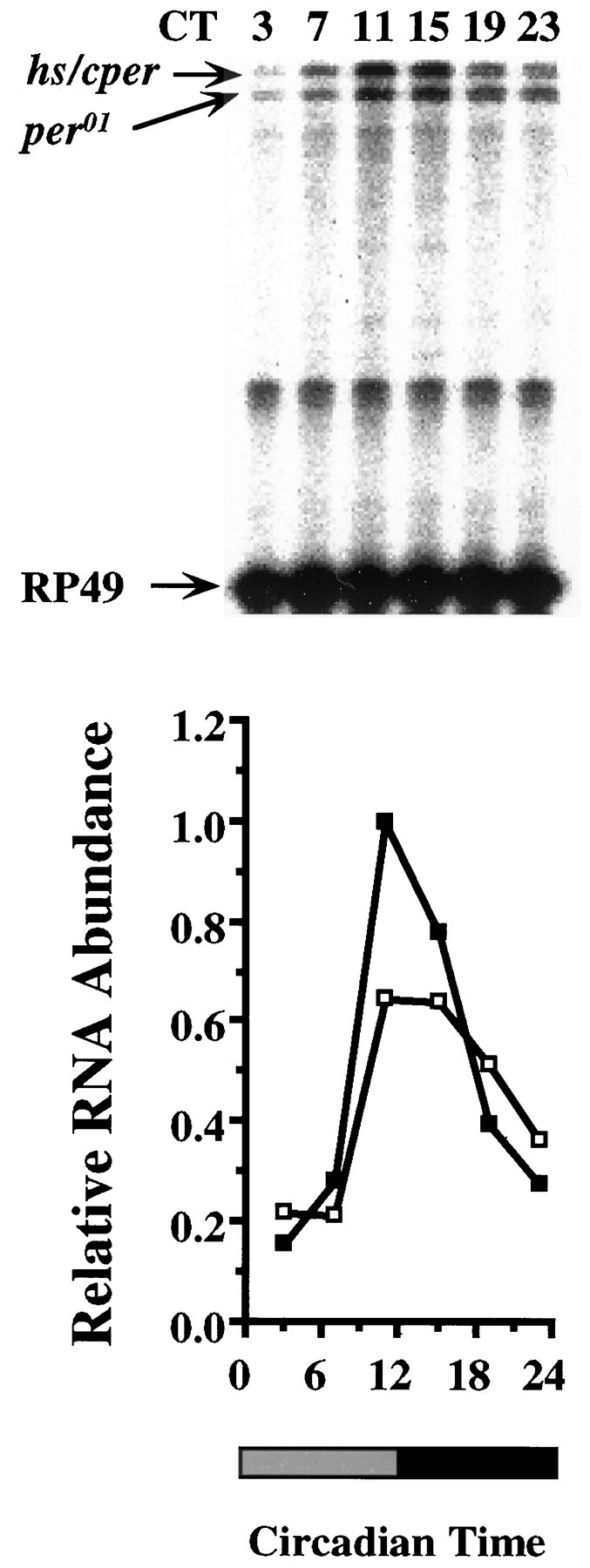
CRS/hs/cper drives mRNA cycling in DD. Top, RNase protection gels ofper01;CRS/hs/cpertransformants taken at CT 3, 7, 11, 15, 19, and 23.Bottom, The quantitation of the protection gel. Data are normalized to the peak in hs/cper mRNA abundance, which is set to 1.0. The hs/cper RNA is shown withfilled squares, and the endogenousper01 RNA is shown with open squares. The shaded and filled boxes below the x-axis represent subjective day or night under DD conditions, respectively. This experiment was repeated five times with similar results. Ribosomal protein 49 (RP49), Antisense ribosomal protein 49 RNA probe. For other abbreviations, see Figures 1 and 4.
CRS-driven PER expression rescues locomotor activity rhythms
The ability of the CRS/hs/cper construct to rescue molecular feedback loop function suggests that it would also rescue behavioral rhythms. Indeed, the CRS/hs/cper transgene rescues rhythmic locomotor activity in 50–93% (average, ∼77.8%) of the per01 flies tested with periods of ∼22.5 hr (Table 1). The penetrance of CRS/hs/cper flies is strong compared with that of previous transformant flies containing per genomic sequences (penetrance ranging from 25 to 100%), and they have shorter periods than other per transformant types (periods range from ∼23 to ∼37 hr) (Bargiello et al., 1984; Hamblen et al., 1986; Baylies et al., 1987, 1992; Citri et al., 1987; Liu et al., 1991). Rhythms in CRS/hs/cper flies have an average power of 40 ± 3.5 (Table 1), similar to the 48.5 ± 1.8 value forper01 flies transformed with a 13.2 kbper genomic DNA fragment containing ∼4 kb of upstream sequence, the entire per transcribed sequence, and ∼2 kb of downstream sequence (Cheng et al., 1998).
Table 1.
Activity rhythms of per01; CRS/hs/cper and per01; CRS/P/cper flies
| Genotype | Line | Number tested | Percent rhythmic (%) | Average period (hr ± SEM) | Average power (±SEM) |
|---|---|---|---|---|---|
| CRS/hs/cper | 4 | 20 | 90 | 23.1 ± 0.1 | 53.3 ± 7.5 |
| 5 | 16 | 81.8 | 22.7 ± 0.2 | 40.2 ± 7.6 | |
| 10 | 16 | 93.8 | 22.3 ± 0.3 | 29.8 ± 3.8 | |
| 19 | 20 | 50 | 22.5 ± 0.2 | 30.9 ± 4.9 | |
| pooled | 72 | 77.8 | 22.7 ± 0.1 | 40.0 ± 3.5 | |
| CRS/P/cper | 1a | 10 | 60 | 23.9 ± 0.3 | 38.9 ± 3.4 |
| 2a | 10 | 100 | 23.3 ± 0.3 | 54.5 ± 6.9 | |
| 3 | 10 | 80 | 26.0 ± 0.2 | 65.3 ± 14.2 | |
| 1 | 10 | 90 | 25.4 ± 0.1 | 66.6 ± 9.1 | |
| 2 | 10 | 100 | 24.2 ± 0.1 | 43.4 ± 5.1 | |
| pooled | 50 | 86 | 24.5 ± 0.2 | 54.3 ± 4.0 | |
| Canton-S | 18 | 94.7 | 24.2 ± 0.1 | 78.2 ± 4.2 |
Young male Canton-S, per01; CRS/hs/cper and per01; CRS/P/cper flies were entrained in 12 hr light/dark cycles for 3/d, and locomotor activity was monitored in constant darkness at 25°C for 7/d. Periodogram analysis was done as previously described (Hamblen et al., 1986). Power and width are defined in Frisch et al. (1994) and were used to distinguish between rhythmic and arrhythmic flies (Cheng et al., 1998).
Because the CRS/hs/cper flies have ∼1.5 hr shorter periods than wild-type flies, we postulated that this difference was attributable to the relatively strong hsp70 basal promoter. Because the P-element basal promoter produces approximately fivefold lower levels of transcript than the hsp70 basal promoter (Hao et al., 1997), we expected that if the CRS were driving this promoter, the period would be closer to that of wild type. Such a result would agree with previous observations that lower permRNA titers correlate with longer periods (Baylies et al., 1987). Indeed, the CRS/P/cper construct rescued locomotor activity rhythms in per01 flies (Table 1). Overall, 86% of the CRS/P/cper flies were rhythmic with a period of 24.5 ± 0.2 hr and an average power of 54.3 ± 4.0. The penetrance and power values are in line with that of other transformants that mediate per behavioral rescue (Bargiello et al., 1984; Hamblen et al., 1986; Baylies et al., 1987, 1992; Citri et al., 1987; Liu et al., 1991), and the period is in line with our expectation given the weakness of the P-element promoter.
To insure that behavioral rescue is CRS dependent, we tested the ability of an hs/cper gene lacking any perupstream regulatory sequences to rescue locomotor activity rhythms inper01 flies. All transgenic lines were arrhythmic (data not shown), showing that behavioral rescue is CRS dependent and that neither the per coding region nor the 2.1 kb of downstream sequences contain regulatory elements capable of driving expression in the LNs. These results show that theper CRS is sufficient for strong, high penetrance rescue of behavioral rhythms with ∼24 hr circadian periods and that the period shortens with a stronger basal promoter.
DISCUSSION
In Drosophila, a rapidly expanding list of genes is required for circadian feedback loop function including per(Hardin et al., 1990), tim (Sehgal et al., 1994),dClock (Allada et al., 1998; Darlington et al., 1998),Cycle (Darlington et al., 1998; Rutila et al., 1998), anddouble-time (Kloss et al., 1998; Price et al., 1998). These genes act at the transcriptional or post-transcriptional levels to regulate circadian feedback loop function (Hardin et al., 1990, 1992;Vosshall et al., 1994; Price et al., 1995, 1998; So and Rosbash, 1997;Allada et al., 1998; Cheng et al., 1998; Darlington et al., 1998; Kloss et al., 1998; Rutila et al., 1998). Two of these genes, dClkand Cyc, encode proteins that activate per andtim transcription via E-boxes located in their respective upstream sequences (Darlington et al., 1998; Gekakis et al., 1998). One of the E-boxes targeted by dCLK and CYC is located within the 69 bp CRS from per, which is required for rhythmic transcription (Hao et al., 1997).
In this study, we show that the per CRS also mediatesper-like spatial and developmental expression and that CRS-dependent per expression rescues feedback loop function and behavioral rhythms. The fact that the CRS contains all of the regulatory information required for per-like developmental, spatial, and circadian expression suggests that dCLK and CYC might mediate all aspects of per expression. If true, we might expect dClk and/or Cyc to be expressed in the same cells as per, thereby restricting peractivation to the proper cell types. In favor of this possibility is the observation that there appear to be no obvious pleiotropic effects from mutations in dClk or Cyc (Allada et al., 1998; Rutila et al., 1998), thereby tentatively limiting the function of dCLK and CYC to clock gene activation. If the dCLK and CYC proteins are responsible for per and tim spatial expression, then the minimal E-box within the CRS may well be the only regulatory sequence that is needed to drive correct spatial expression. Although the CRS is capable of mediating many, if not all, aspects ofper expression, transgenes lacking the CRS can also rescue behavioral rhythms and drive expression in LNs (Ewer et al., 1990; Liu et al., 1991; Frisch et al., 1994), indicating that importantper regulatory elements are not exclusive to the CRS.
There is evidence that PER- and/or TIM-dependent transcriptional repression also occurs via dCLK and CYC binding at the E-box (Darlington et al., 1998). This repression could occur directly via an interaction between PER and CYC and/or dCLK that disrupts activation (perhaps via PAS domains) or indirectly by PER and/or TIM activating a transcriptional repressor or another factor that acts to disrupt dCLK and/or CYC activation. If repression were caused by the direct disruption of dCLK and/or CYC activation by PER and/or TIM, then the minimal E-box needed for activation would also be sufficient for repression and therefore circadian cycling. There is precedent for such a small DNA binding target being sufficient for correct spatial and developmental activation; the SINGLEMINDED and TANGO bHLH-PAS proteins activate expression along the CNS midline in stage 10Drosophila embryos using four repeats of the 18 bp midline enhancer (Wharton et al., 1994; Sonnenfeld et al., 1997; Darlington et al., 1998).
Several clusters of neurons express per in larvae, including putative precursors to the adult LNs (Kaneko et al., 1997). By the use of antibodies to PER and TIM, this larval expression was shown to be rhythmic, but with different phases depending on the neuronal cluster (Kaneko et al., 1997). The CRS/P/lacZ transgene is expressed normally in larval, pupal, and adult LNs (Figs. 2, 3) (data not shown). Because the CRS is a target for dCLK and CYC, perhaps these transcription factors also drive per expression in larvae. To determine whether this is the case, we tested homozygous BG6;dClkJrk and BG6;Cyc larvae for β-galactosidase expression at the L3 stage and found no staining in the CNS (Fig. 3G,H). This result demonstrates that dCLK and CYC are required for perexpression in the larval CNS. Because PER and TIM levels cycle in larval and pupal LNs (Kaneko et al., 1997) and perexpression in larval LNs is dependent on dClk andCyc, it is likely that the circadian feedback loop is operating in larval and pupal LNs as it does in adult LNs. If so, the feedback loop could convey circadian phase from larvae to adults, thereby accounting for larval time memory.
The period of locomotor activity rhythms is sensitive to the number ofper gene copies in that half the dosage (one copy of this X-linked gene in females) results in 0.5–1 hr longer periods and two to five times the dosage (two to five per copies in males) results in 1–1.5 hr shorter periods (Smith and Konopka, 1982). Dosage-dependent alterations are also seen when the dosage of thedClk and Cyc genes is lowered, resulting in longer period rhythms (Allada et al., 1998; Rutila et al., 1998). Altering the dosage of per or its transcriptional activators presumably alters per mRNA titer because lower levels ofper mRNA have been shown to correlate with longer period locomotor activity rhythms (Baylies et al., 1987). CRS-drivenper expression results in rhythms that are close to 24 hr, indicating that this regulatory sequence contains all the information necessary for feedback loop function in LNs. The period of this rhythm in CRS-driven per flies, however, is sensitive to the strength of the basal promoter, with longer periods from the weaker P-element transposase basal promoter and shorter periods from the stronger hsp70 basal promoter. The hsp70 basal promoter produces approximately fivefold more RNA than the P-element transposase basal promoter (Hao et al., 1997), which results in an ∼2 hr difference in behavioral period. This magnitude of difference is consistent with the dosage studies mentioned above and shows that factors other than the number of per gene copies or the levels of per gene activators affect the period of behavioral rhythms.
In this study we show that a single copy of the CRS is sufficient to drive per expression in its normal spatial pattern and to mediate robust behavioral rescue. The per coding and downstream-flanking sequences do not mediate behavioral rescue (Table1), showing that the CRS is a necessary transcriptional element. Among the clock regulatory sequences identified (Anderson and Kay, 1995;Bell-Pederson et al., 1996; Liu et al., 1996; Hao et al., 1997), this is the first case in which a minimal circadian regulatory sequence has been shown to support both clock gene expression and phenotypic rescue. By dissecting the CRS further, we will determine whether the E-box is the key element involved in both circadian and spatial expression or whether other sequences are required for these functions. The discovery of an E-box upstream of tim that functions to activate expression suggests that other rhythmically transcribed feedback loop components and perhaps clock output genes will be regulated by the same mechanism in Drosophila (Darlington et al., 1998). This work may also provide insight into mammalian clock function because CLOCK and BMAL1 activate mPER1 expression via E-boxes upstream of its putative transcription start site (Gekakis et al., 1998).
Footnotes
This study was supported by National Institutes of Health Grant NS31214. We thank Ralf Stanewsky for providing anti-PER antibody, Isaac Edery for providing the hspcper DNA construct, and Jeff Hall for providing the BG6 transformant line. We also thank Cai Wu and Jerry Houl for behavioral analysis and Jerry Houl for his assistance with experiments. We thank Balaji Krishnan for comments on this manuscript.
Correspondence should be addressed to Dr. Paul Hardin, Department of Biology, University of Houston, Houston, TX 77204-5513.
REFERENCES
- 1.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 2.Anderson SL, Kay SA. Functional dissection of circadian clock and phytochrome regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA. 1995;92:1500–1504. doi: 10.1073/pnas.92.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 4.Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance and structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;328:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 5.Baylies MK, Vosshall LB, Sehgal A, Young MW. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. doi: 10.1016/0896-6273(92)90194-i. [DOI] [PubMed] [Google Scholar]
- 6.Bell-Pederson D, Dunlap JC, Loros JJ. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol Cell Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Y, Gvakharia B, Hardin PE. Two alternatively spliced transcripts from the Drosophila period gene rescue behavioral rhythms with different periods. Mol Cell Biol. 1998;18:6505–6514. doi: 10.1128/mcb.18.11.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 9.Curtin KD, Huang ZJ, Rosbash M. Temporally regulated entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 10.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian feedback loop: CLOCK induced transcription of its own inhibitors, period and timeless. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 11.Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a period-lacZ fusion protein in Drosophila: evidence for cyclical degradation. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 12.Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994a;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- 13.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994b;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewer J, Hamblen-Coyle M, Rosbash M, Hall JC. Requirement for period gene expression in the adult and not during development for locomotor activity rhythms of imaginal Drosophila melanogaster. J Neurogenet. 1990;7:31–73. doi: 10.3109/01677069009084151. [DOI] [PubMed] [Google Scholar]
- 15.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash MR, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 16.Gekakis N, Saez L, Delahaye-Brown A-M, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 17.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 18.Hall JC. Tripping along the trail to the molecular mechanisms of biological clocks. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 19.Hamblen M, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zwiebel LJ, Konopka RJ, Rosbash M, Hall JC. Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per− mutants. J Neurogenet. 1986;3:249–291. doi: 10.3109/01677068609106855. [DOI] [PubMed] [Google Scholar]
- 20.Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardin PE, Sehgal A. Molecular components of a model circadian clock: lessons from Drosophila. In: Lydic R, Baghdoyan H, editors. Handbook of behavioral state control: molecular and physiological mechanisms. CRC; Boca Raton, FL: 1998. pp. 61–74. [Google Scholar]
- 22.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;342:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 23.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardin SH, Jones LB, Homayouni R, McCollum JC. Octamer primed cycle sequencing: design of an optimized primer library. Genome Res. 1996;6:545–550. doi: 10.1101/gr.6.6.545. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic- helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko M, Helfrich-Forster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloss B, Price JL, Saez L, Balu J, Rothenfluh A, Wesley C, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Lorenz LJ, Yu Q, Hall JC, Rosbash M. Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 1988;2:228–238. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Yu Q, Huang Z, Zwiebel LJ, Hall JC, Rosbash M. The strength and periodicity of D. melanogaster circadian rhythms are differentially affected by alterations in period gene expression. Neuron. 1991;6:753–766. doi: 10.1016/0896-6273(91)90172-v. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Taub CC, McClung CR. Identification of an Arabidopsis thaliana ribulose-1,5-bisphosphate caboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiol. 1996;112:43–51. doi: 10.1104/pp.112.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Kane C, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price JL, Dembinska ME, Young MW, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 35.Rosato E, Piccin A, Kyriacou CP. Circadian rhythms: from behavior to molecules. Bioessays. 1997;19:1075–1082. doi: 10.1002/bies.950191206. [DOI] [PubMed] [Google Scholar]
- 36.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 37.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 38.Sehgal A, Price J, Young MW. Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:1423–1427. doi: 10.1073/pnas.89.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 41.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 42.Smith RF, Konopka RJ. Effects of dosage alterations at the per locus on the period of the circadian clock of Drosophila. Mol Gen Genet. 1982;189:30–36. [Google Scholar]
- 43.Smith SA, Shepherd D. Central afferent projections of proprioceptive sensory neurons in Drosophila revealed with the enhancer-trap technique. J Comp Neurol. 1996;364:311–323. doi: 10.1002/(SICI)1096-9861(19960108)364:2<311::AID-CNE9>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 44.So VW, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- 46.Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thummel CS, Pirotta V. New pCaSpeR P-element vectors. Drosoph Inf Serv. 1991;71:150. [Google Scholar]
- 48.Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 49.Wharton KA, Crews ST. CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- 50.Wharton KA, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 51.Young MW, Jackson FR, Shin H-S, Bargiello TA. A biological clock in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:865–875. doi: 10.1101/sqb.1985.050.01.104. [DOI] [PubMed] [Google Scholar]
- 52.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 54.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]