Abstract
We have identified a pineal night-specific ATPase (PINA), a novel splice variant of the ATP7B gene disrupted in Wilson disease (WD). PINA expression exhibits a dramatic diurnal rhythm in both pineal gland and retina with 100-fold greater expression at night than at day. PINA is expressed in pinealocytes and a subset of photoreceptors in adult rats and is transiently expressed in the retinal pigment epithelium and the ciliary body during retinal development. Nocturnal pineal expression of PINA is under the control of a suprachiasmatic nucleus clock mediated by superior cervical ganglion innervation of the pineal. In vitro, PINA expression in pineal cells can be stimulated by agents activating the cAMP signal transduction pathway. PINA is able to restore copper transport activity in Saccharomyces cerevisiae deficient in the homologous copper-transporting ATPase CCC2, suggesting that this protein may function as a copper transporter in rat pinealocytes. These studies suggest a potential role of rhythmic copper metabolism in pineal and/or retina circadian function.
Keywords: pineal, P-type ATPase, Wilson disease, circadian rhythms, photoreceptor, development
The pineal gland is a functional component of the circadian timing system that measures and translates the duration of environmental light into rhythmic neuronal signals. The central component of the system, the suprachiasmatic nucleus (SCN), contains a self-sustained clock (Sassone-Corsi, 1998) that is entrained by light input from the retina and by the hormone melatonin secreted from the pineal. In nonmammalian vertebrates, such as birds, reptiles, and fish, the retina and the pineal both possess a light-sensing apparatus and endogenous clock machinery and share many common molecular functions including nocturnal melatonin synthesis. In contrast, adult mammalian pineals have lost their light-sensing capability and their clock function, although the pineal gland still possesses many components of phototransduction (Lolley et al., 1992;Blackshaw and Snyder, 1997) that are used by photoreceptors of the retina. The diurnal rhythm of melatonin formation in the mammalian pineal is driven instead by a complicated neuronal network controlled by the SCN that is modulated by light impacting the retina at night (Foulkes et al., 1997).
Molecular insights into pineal circadian physiology have been facilitated by the cDNA cloning of the rate-limiting enzyme of melatonin synthesis, serotonin N-acetyltransferase (NAT) (Borjigin et al., 1995; Coon et al., 1995). NAT mRNA rhythm parallels circadian NAT catalytic activity (Borjigin et al., 1995) and NAT protein rhythm (Gastel et al., 1998). A cAMP response element (CRE) has been identified in the promoter of the NAT gene and is able to interact with an inducible cAMP early repressor (ICER) both in vitro and in vivo (Foulkes et al., 1996). Furthermore,in vivo study of NAT regulation using ICER mutant mice has demonstrated that NAT transcription is suppressed in vivoby ICER (Foulkes et al., 1996). Thus cAMP signaling seems to be essential for the temporal regulation of melatonin synthesis. While searching for transcription factors responsible for spatial control of the enzymes required for melatonin synthesis, we identified a unique nucleotide sequence designated PIRE (pineal regulatory element) in the promoter regions of several pineal-specific genes including NAT and hydroxyindole-O-methyltransferase (HIOMT) (Li et al., 1998). PIRE interacts with cone rod homeobox (CRX) that is a photoreceptor and pinealocyte-specific transcription factor (Chen et al., 1997; Freund et al., 1997; Furukawa et al., 1997; Li et al., 1998).
Our initial identification of the NAT gene used subtractive hybridization between day and night pineal gland RNA (Borjigin et al., 1995). We now report molecular cloning and characterization of a novel transcript expressed selectively at night and encoding a pineal night-specific ATPase (PINA). PINA and NAT share identical temporal expression patterns and tissue distributions. PINA is generated from alternative splicing of the Wilson disease (WD) gene (Bull et al., 1993; Petrukhin et al., 1993; Tanzi et al., 1993;Yamaguchi et al., 1993) and can function as a copper transporter.
MATERIALS AND METHODS
Animals. Sprague Dawley rats were purchased from Charles River Laboratories (Wilmington, MA) and housed in 14:10 hr light/dark lighting conditions with lights off at 21:00 hr for >1 week before the experiments. Sprague Dawley rats in which the superior cervical ganglia were bilaterally removed by surgery [superior cervical ganglionectomized (SCGX)] were purchased from Zivic-Miller Laboratories (Allison Park, PA). During the dark periods, animals were killed under safe red lights (cutoff, 600 nm) with rapid decapitation by guillotine.
Cloning of PINA cDNAs. The subtractive hybridization used to obtain a partial PINA cDNA (clone 5) was as described (Borjigin et al., 1995). Clone 5 was then used to screen a rat pineal night cDNA library (Borjigin et al., 1995) to obtain full-length PINA cDNAs. The longest clones for both transcripts (PINA5.1 and PINA5.2) were sequenced in their entirety on both strands.
Northern, Western, and RNase protection analyses. RNA samples subjected to Northern analysis and RNase protection study were prepared using an RNeasy kit (Qiagen, Hilden, Germany). RNase protection was performed with an RNase protection analysis kit (Ambion). Northern blots were hybridized with a labeled full-length PINA5.1 cDNA or full-length NAT and GAPDH cDNAs. PINA antibody used in Western analysis was generated against the N-terminal soluble domain of PINAm1 (as described below), which corresponds to the polypeptide between residues 758 and 882 of rat ATP7B protein (Wu et al., 1994).
In situ analysis. PINA probes used for in situ hybridization are derived from the PINA5.1 coding region (2 kb) as well as the pineal-specific 5′-untranslated region. The full-length rat NAT cDNA (Borjigin et al., 1995) and mouse CRX cDNA (a kind gift from Shiming Chen and Don Zack) were used for the in situ studies. The in situ hybridization technique was as described (Blackshaw and Snyder, 1997).
Yeast strains, growth conditions, and manipulations. Saccharomyces cerevisiae strains used in this study were as follows: ccc2Δ(IHY5): MATα, his3 (his3–200 or his3-Δ1), trp1 (trp1-D or trp1–289), ura3–52, leu1; CCC2: LEU2, GAL; IHY4: MATα, his3-Δ1, trp1–289, ura3–52, leu2, GAL. Yeast strains were transformed by the lithium acetate method, and uracil-based selection was used to screen for transformants. All strains were grown at 30°C in appropriate dropout media (BIO 101, La Jolla, CA) supplemented with 2% glucose. Yeast lysates were prepared as described (Yaffe and Schatz, 1984), except that the samples were not heated before analysis.64Copper incorporation into Fet3p was performed as reported previously (Yuan et al., 1995; Klomp et al., 1997).
RESULTS
PINA is an alternatively spliced product of the Wilson disease gene ATP7B
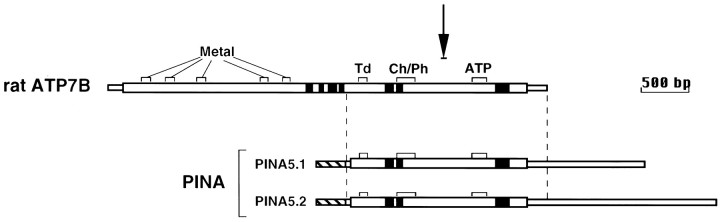
In a pilot analysis of 24 randomly selected clones from the night-subtracted rat pineal cDNA library aimed at identifying molecules responsible for melatonin synthesis, two novel genes were isolated. One of these genes (NAT) has been described elsewhere (Borjigin et al., 1995). A second novel gene, designated PINA, is the focus of this report. Several full-length PINA cDNAs were isolated from a rat pineal night cDNA library and were sequenced in their entirety. Sequence analysis revealed a complete open reading frame that is identical to the C-terminal half of ATP7B (Wu et al., 1994) (Fig.1). A stretch of 300 bp preceding the open reading frame is absent from the ATP7B sequence and is pineal specific. To confirm that PINA is a splice variant of ATP7B and not a duplicate copy of a part of the ATP7B gene, we isolated rat genomic DNA encoding PINA. We found that the pineal-specific 5′ end is a part of the intron sequence immediately upstream of exon 9 of the ATP7B gene (data not shown) (see Li et al., 1998), indicating that PINA is a result of the usage of an alternative intronic promoter (Li et al., 1998). PINA consists of 3.5 kb (PINA5.1) and 4.3 kb (PINA5.2) transcripts that diverge 30 bp 5′ to the polyA sequence in PINA5.1. PINA5.2 contains an additional 822 bp-untranslated sequence further downstream of PINA5.1 and is most likely generated by an alternative polyadenylation. With the first ATG, the deduced open reading frame encodes a 665 amino acid polypeptide with a predicted mass of 74 kDa. In 293S cells transfected with the full-length PINA constructs, however, two monomer bands (PINAm1 and PINAm2) are observed (data not shown). The lower molecular weight product (PINAm2) reflects the usage of the second ATG, which is 26 amino acid residues downstream from the first ATG and contains a better Kozak consensus sequence for translation initiation (data not shown).
Fig. 1.
Structure of the PINA cDNA in comparison with ATP7B. PINA consists of 3.5 kb (PINA5.1) and 4.3 kb (PINA5.2) transcripts that contain a coding region identical in each transcript and in the C-terminal half of rat ATP7B, the rat homolog of the Wilson disease gene. Thick open boxes represent the coding region, and thinner open boxes represent the 5′- and 3′-untranslated regions. Functional domains that are conserved among copper transporters are marked as follows: Metal, metal-binding domains (CXXC motifs); Td, transduction domain (TGEA); Ch/Ph, channel (CPC) and phosphorylation domain (DKTGT); ATP, ATP-binding domain (GDGXNDXP). Hydrophobic regions that are proposed to form transmembrane domains (Bull and Cox, 1994) are indicated by solid black boxes. The region between the vertical dashed lines is identical in each clone, except at amino acid residue 1315 of rat ATP7B where there is an insertion of two amino acids (methionine and alanine) in the correspondingPINA5.1 and PINA5.2 sequences. OurPINA sequence agrees with both the mouseATP7B sequence (GenBank accession number U38477) and the human ATP7B sequence (GenBank accession number P35670) at the corresponding positions. In the 5′-untranslated region ofPINA, there is a 300 bp sequence (cross-hatched box) not found in ATP7B cDNA and specific to the pineal. The 3′ end of the PINA transcripts contains additional untranslated sequences (1550 bp for the 4.3 kb and 780 bp for the 3.5 kb message) that were not found in the reported ratATP7B sequence. The arrow above the map points to the region where the proximal deletion breakpoint maps in Long–Evans Cinnamon (LEC) rats (Wu et al., 1994).
In contrast to that of ATP7B, the predicted amino acid sequence of PINA lacks the multiple copper-binding sequences in the N-terminal half of the protein and the four putative transmembrane segments. PINA and ATP7B share a number of conserved elements including transduction, channel and phosphorylation, and ATP-binding domains, signature elements of P-type ATPase proteins (Pedersen and Carafoli, 1987). In addition, a CPC motif is found in the second putative transmembrane segment of PINA, which is conserved in all heavy metal-transporting ATPases and is thought to be crucial for heavy metal transport (Solioz and Vulpe, 1996). To date, all of the heavy metal transporters identified contain at least one heavy metal-binding motif upstream of the ATPase domain (Bull and Cox, 1994; Solioz and Vulpe, 1996). PINA represents a novel subclass of the putative metal transporters because of the absence of metal-binding sequences (see below).
The product of the PINA-coding ATP7B gene is a copper transporter disrupted in WD (Vulpe and Packman, 1995). A large number of mutations in ATP7B have been identified in WD patients with >80% clustered in the PINA-coding ATPase region (Thomas et al., 1995). Affected WD patients present with liver disease, movement disorder, neuropsychiatric manifestations, or combinations of the three. Circadian defects such as sleep disorders have not been reported in WD.
PINA expression in the pineal is diurnally and developmentally regulated
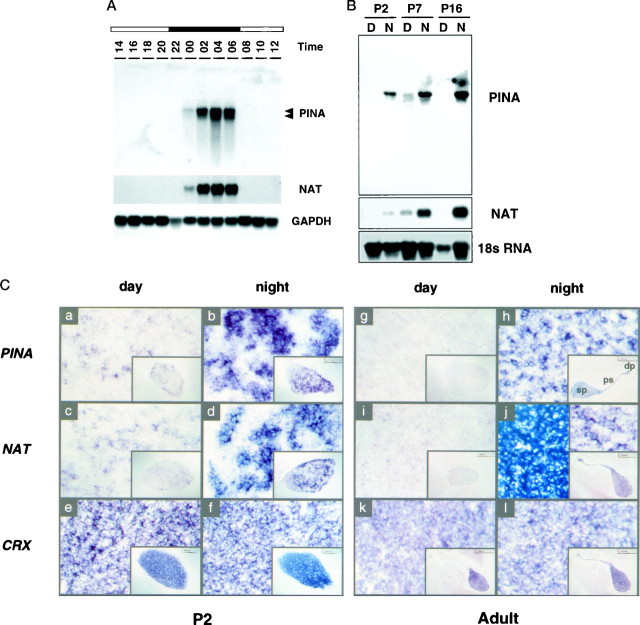
To examine the tissue specificity of PINA expression, we analyzed RNA from day and night adult pineal, eye, and several other tissues on a Northern blot. Abundant transcripts of 4.3 and 3.5 kb are detected in the night pineal, and a weak signal is observed in night eye. We have not detected PINA RNA in any other tissues examined in the night or day (Li et al., 1998). To study the circadian pattern of PINA expression, we isolated pineal RNA at 2 hr intervals from adult rats (housed in 14:10 hr light/dark lighting conditions with lights off at 21:00 hr) and analyzed the RNA by Northern blot (Fig.2A). PINA transcripts are first detected at midnight, 3 hr into the dark period; reach their maximum levels during the second half of the night; and disappear within 1 hr after lights are turned on at 08:00 hr. This pattern of PINA rhythm is indistinguishable from that of NAT studied in the same set of experiments.
Fig. 2.
Circadian PINA expression in the developing and mature pineals. A, Temporal expression pattern of PINA mRNA in adult pineals. Northern blot of pineal RNA collected from rats at 2 hr intervals is shown. The blot was hybridized sequentially with full-length PINA5.1,NAT, and GAPDH probes. Eachlane contains 5 μg of total RNA. The positions of the RNA bands for PINA5.1 (top) and PINA5.2 transcripts (bottom) are marked on the right.B, Developmental pattern of PINA mRNA rhythms in the pineal. Northern blot of pineal RNA collected at 04:00 hr (N for night) and 16:00 hr (D for day) from rats of P2, P7, andP16 is shown. Total RNA from one single pineal was loaded in each lane, and blots were hybridized sequentially with PINA5.1 and NATfull-length probes. An 18 S RNA probe was used for control of equal loading between day and night samples. The absence of P16 NAT and PINA expression during the daytime was confirmed in two other independent experiments (data not shown).C, Midsagittal brain sections at day (16:00 hr) and night (04:00 hr) of P2 and adult rats processed forin situ hybridization with digoxigenin-labeled riboprobes using rat PINA5.1 (see Fig. 1), ratNAT (Borjigin et al., 1995), and mouseCRX (Chen et al., 1997) full-length cDNAs as templates. Daytime expression of both PINA(a) and NAT(c) is clearly visible at P2compared with those of adult sections (g,i, respectively). Compared withCRX-expressing cells, PINA- andNAT-expressing cells display a punctuate pattern atP2 and adult with a greater variation atP2. The day sections (a,c, e, g, i,k) as well as the night sections (b,d, f, h, j,l) are consecutive sections from the same animals at the specified times. Color reactions of all sections were developed overnight with the exception of those shown in theright top and bottom insets inj, which were developed for 4 hr to revealNAT-positive signals comparable with that of other sections. The entire pineal structure including the deep pineal gland (dp), the pineal stalk (ps), and the superficial pineal (sp) is positive for all three probes (exemplified in inset inh). Sense probes revealed no positive signals (data not shown).
The developmental pattern of day and night PINA expression was also studied using Northern blot analysis (Fig. 2B). Although undetectable in day pineals of adult rats, PINA and NAT transcripts are present in postnatal day 2 (P2) and P7 rat pineals during the day. This daytime PINA and NAT expression disappears at P16, coinciding with the maturation of the sympathetic innervation to the pineal (Hakanson et al., 1967). Interestingly, there is already a marked diurnal rhythm of both PINA and NAT expression at P2, the earliest time point examined. To analyze the patterns of PINA expression in pineal further, we performed in situhybridization on developing and adult brain midsagittal sections of day (16:00 hr) and night (04:00 hr) rats (Fig. 2C). Consistent with the Northern blot data, at P2 PINA and NAT transcripts display prominent diurnal rhythms with nighttime levels much higher than daytime values (Fig. 2Ca–d). This pattern of expression differs from that reported by Pfeffer and Stehle (1998) and perhaps reflects differing sensitivity of the detection methods used. A constitutively expressed pineal- and retina-specific transcript CRX (Li et al., 1998) does not show detectable diurnal variation (Fig.2Ce,f). There is a marked heterogeneity among night pineal cells expressing PINA (and NAT) compared with the CRX-expressing cells at P2. The punctate-appearing PINA and NAT-expressing cells in night pineals of P2 rats may represent areas of pineal innervated by developing sympathetic nerves (Hakanson et al., 1967).
In adult pineals, PINA and NAT are not expressed during the day, whereas CRX expression is similar during the day and night. At night, expression of PINA, NAT, and CRX is observed throughout the pineal structure including the superficial pineal, the pineal stalk, and the deep pineal gland (Fig. 2C, insets at thelower right corners ofh,j,l). The deep pineal, closely associated with both the habenular and the posterior commissures (Fig. 2C), is functionally connected to the superficial pineal gland because the surgical removal of the superior cervical ganglion (SCG) or constant light treatment abolishes the expression of PINA and NAT in the deep pineal (data not shown). In adult pineals, the intensity of labeling is heterogeneous among different populations of pineal cells for PINA and NAT (Fig.2Ch,j); in contrast, CRX labeling is more homogeneous at this stage (Fig. 2Cl).
PINA is expressed in a subset of retina photoreceptors at night
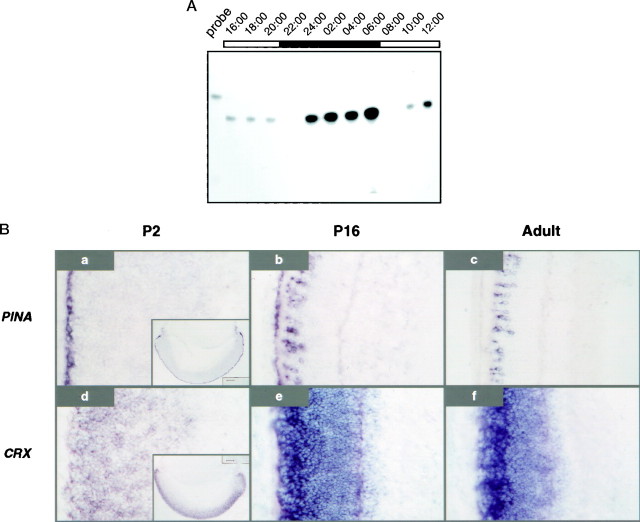
To examine the expression profile of PINA in the eye, we analyzed RNA from adult rat eyes at 2 hr intervals by RNase protection using the pineal-specific 5′-untranslated region of PINA cDNA as a probe (Fig.3A). PINA transcripts are expressed most abundantly at night after midnight and decline significantly when lights are turned on. This pattern resembles the PINA rhythm in pineal (Fig. 2A) and is similar to that of NAT in rat eyes (data not shown). In situhybridization analysis of PINA expression at night was performed on developing and adult eyes of night rats (Fig. 3B). PINA transcripts are abundantly expressed in the developing retinal pigment epithelium (RPE) layer at embryonic day 14.5 (data not shown). During early postnatal retinal development until P16, PINA expression is still found in RPE cells with increasing expression in a subset of photoreceptor cells (Fig. 3Ba,b) that are situated in the layer typically occupied by cone photoreceptor cells. PINA expressions in RPE cells become undetectable in the adult retina of day or night animals (Fig. 3Bc). In addition to photoreceptor and RPE cells, PINA is also found transiently in the developing ciliary body at P2 (Fig. 3B, inset ina) and P16 (data not shown) but is undetectable in the adult ciliary body. In contrast, the retina/pineal-specific, constitutively expressed transcript CRX is not detected in RPE or the ciliary body at any stage examined (Fig. 3Bd). There is no day and night difference in PINA expression in the eye until after P16 (data not shown), which corresponds to the completion of retinal development (Cepko et al., 1996). The onset of the development of the PINA diurnal rhythm in the eye is later than that in the pineal (see Fig.2C). There is no detectable daytime (16:00 hr) expression of PINA in any region of the adult eyes examined (data not shown), and the night expression of PINA in rat photoreceptors does not display the typical gradient observed for the blue cone opsin (Chiu and Nathans, 1994) or the red/green cone opsin (Wikler et al., 1996).
Fig. 3.
Circadian PINA expression in developing and mature eyes. A, Temporal expression pattern of PINA mRNA in adult eyes. RNase protection analysis was performed with total eye RNA collected from rats at 2 hr intervals. Each lane contains 10 μg of total RNA.B, Night eye sections from rats at P2,P16, and day 60 (adult) hybridized withPINA and CRX antisense probes. Thein situ sections from day and night eyes were also hybridized with a probe derived from the pineal-specific 5′-untranslated region of PINA cDNA, and the same results were obtained (data not shown). Sections were collected from rats at 04:00 hr. PINA expression at P2is confined within the retinal pigment epithelium layer (a) and the developing ciliary body (see theinset in a) where the CRXsignal (d) is absent. At P16,PINA signal is found in RPE cells, a subset of photoreceptor cells (b), and the ciliary body (data not shown), whereas the CRX signal is found in the entire photoreceptor cell layers (e) and again is absent in RPE and the ciliary body (data not shown). In adult eyes,PINA is restricted to a subset of photoreceptor cells located in the upper outer nuclear layer where cone cells normally reside (c). Daytime expression ofPINA is present at a level comparable with those at night at P2 and is undetectable in adult eyes (data not shown).
Diurnal rhythm of PINA is regulated by clock-controlled cAMP signaling in the night pineal
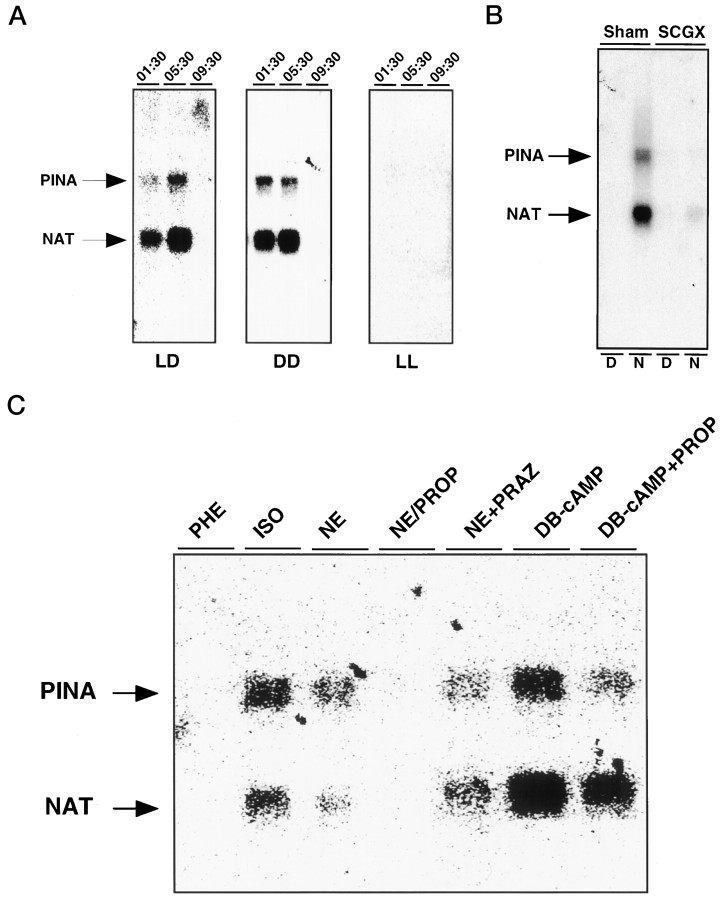
To examine neural and light regulation of PINA expression, Northern blot analysis was performed with RNA isolated from the pineals of rats after exposure to constant light or dark for 3 d or rats subjected to surgical removal of the SCG. The PINA rhythm persists in constant dark and is absent in constant light (Fig.4A), indicating that the PINA rhythm is under control of a circadian clock that is suppressed by light. Removal of SCG, the sympathetic ganglion responsible for delivery of information from the SCN to the pineal, abolishes the PINA rhythm (Fig. 4B).
Fig. 4.
Regulation of pineal PINAtranscription. A, Clock control of PINAexpression. PINA expression in the pineal under normal lighting (LD), constant dark (DD), and constant light (LL) conditions was assayed by Northern blot analysis. Adult male Sprague Dawley rats were housed in 14:10 hr light/dark cycles (LD), complete darkness for 3 d (DD), or constant room lighting for 3 d (LL) before use. Each lane contains 2 μg of total pineal RNA isolated at the indicated time points.B, Role of SCG in clock-controlledPINA expression. Bilateral SCGX male adult Sprague Dawley rats and sham control rats (sham) were purchased from commercial sources. Two micrograms of total pineal RNA isolated from rats at 16:00 (D) and 04:00 hr (N) was loaded as indicated in eachlane, and the blot was hybridized with bothPINA and NAT probes. C, Regulation of PINA expression in vitro. Northern analysis of RNAs from pineals cultured in vitroin the presence of various drugs is shown. The concentrations of the drugs used are as follows: dibutyryl cAMP (DB-cAMP), 1 μm; isoproterenol (ISO), 1 μm; norepinephrine (NE), 100 nm; phenylephrine (PHE), 1 μm; prazosin (PRAZ), 1 μm; and propranolol (PROP), 1 μm.
The sympathetic innervation of the pineal gland influences its biochemical properties via the release of norepinephrine acting primarily at β-adrenergic receptors that stimulate cAMP formation. To ascertain whether PINA is similarly regulated, we used organ culture of whole-rat pineal glands (Fig. 4C). Exposure to norepinephrine and the β-adrenergic receptor agonist isoproterenol augments expression of PINA. By contrast, phenylephrine, which selectively activates α-receptors, has no effect. Further evidence of an involvement of β-receptors is our finding that the β-adrenergic receptor antagonist propranolol blocks the stimulatory effect of norepinephrine, whereas the α-adrenergic receptor antagonist prazosin is ineffective. Treatment of the culture with a nonhydrolyzable analog of cAMP, dibutyryl-cAMP, stimulates PINA expression, an action that is not affected by propranolol or prazosin. Thus, both PINA and NAT are regulated by β-adrenergic stimulation via cAMP.
PINA is a putative copper transporter
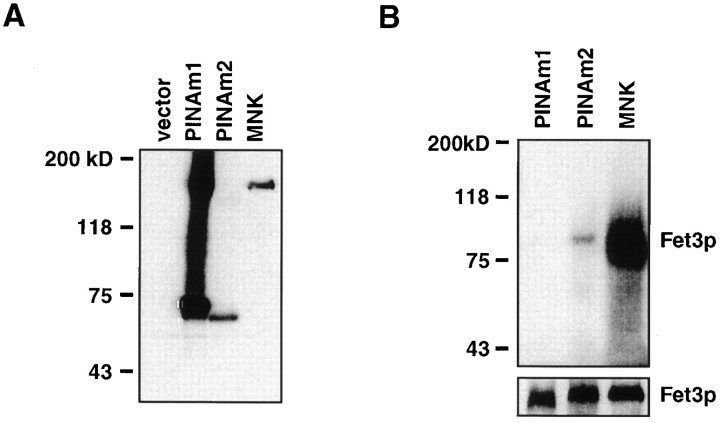
To ascertain whether PINA can function as a copper transporter, we expressed PINA in the ccc2 mutant of S. cerevisiae. Previous studies have demonstrated that yeast strains lacking the Wilson/Menkes gene homolog CCC2 are deficient in high-affinity iron uptake because of a failure to incorporate copper into the ceruloplasmin homolog Fet3p (Yuan et al., 1995). To examine expression of the PINA proteins in these yeast strains, immunoblot analysis was performed on equivalent amounts of total protein lysates from PINAm1- and PINAm2-transformed yeast cells. To create a positive control, we also transformed yeast with a known copper transporter ATP7A (Payne and Gitlin, 1998), the gene product defective in Menkes (Chelly et al., 1993; Mercer et al., 1993; Vulpe et al., 1993). Both PINA proteins (PINAm1 and PINAm2) and the ATP7A protein (Menkes, MNK) are expressed in these yeast with PINAm1 more highly expressed than PINAm2 (Fig. 5A). PINAm2 protein migrates faster than PINAm1 protein in the SDS gel, as expected from its smaller size. The PINA antibody cross-reacts with the human Menkes protein and not with the endogenous CCC2 protein in wild-type IHY4 cells (data not shown).
Fig. 5.
PINA is a putative copper transporter. A, Western blot analysis ofPINA proteins expressed in a strain of yeast (Saccharomyces cerevisiae) deficient in CCC2, the homolog of the human Menkes/Wilson gene. Membrane protein lysates (40 μg each) were separated by SDS-PAGE, transferred to nitrocellulose, and reacted with a rabbit polyclonal antibody againstPINA. B, Radioactive copper (64Cu) incorporation into Fet3p. Yeast cells were pulse labeled with 64Cu, and 200 μg of protein from membrane fractions was separated on nonreducing SDS-PAGE followed by autoradiography. Fet3p protein levels in each sample were estimated by Western blot as shown in the bottom panel.
The function of the PINA protein in these yeast transformants was examined by analyzing copper incorporation by the Fet3p protein. Consistent with previous studies (Payne and Gitlin, 1998), theccc2 mutant expressing the wild-type Menkes protein contains high levels of copper-bound Fet3p (Fig. 5B), whereas the mutant cells transformed with vector alone and the fet3pdeletion mutant do not display copper binding to Fet3p proteins (data not shown). Expression of PINAm2 leads to copper incorporation into Fet3p, indicating that PINA protein is capable of transporting copper in yeast cells, although at a much lower efficiency than does ATP7A (Fig. 5B). The capability of the copper transport action of PINAm2 protein was confirmed in an independent experiment (data not shown).
It is not clear why PINAm1 is entirely devoid of copper transport activity compared with that of PINAm2. In a preliminary experiment, an ATP7B mutant, whose N-terminal copper-binding domain is deleted while the first four putative transmembrane segments are intact, was shown to be a nonfunctional copper transporter (J. D. Gitlin, unpublished observations). These results suggest that the region upstream of the second in-frame ATG in the PINA coding sequence as well as the region upstream of this ATG in the ATP7B protein may serve as an inhibitory domain for copper transport. The inhibition may be released only after a conformational change induced by copper binding to the heavy metal-associated (HMA) (Bull and Cox, 1994) elements in ATP7B.
A novel HXXM motif is present in the C terminal of all ATP7B proteins
In the absence of the CXXC copper-binding motifs, how do PINA proteins recognize copper to transport? Copper binding is achieved mainly by coordinated interactions of three amino acid residues within a three-dimensional structure of a given protein: cysteine, histidine, and methionine (Adman, 1991; Arnold and Haymore, 1991). The importance of the CXXC motif [also termed HMA (see Bull and Cox, 1994)] in heavy metal binding and transport has been well described (Solioz and Vulpe, 1996). In place of CXXC motifs, many copper-related proteins contain methionine- and histidine-rich sequences that can also be metal-binding domains. A MXXM motif found in multiple copies in the yeast copper transporter Ctr1 (Dancis et al., 1994) and in bacterial copper-resistance proteins of Pseudomonas syringae (CopA) (Cha and Cooksey, 1991) and Enterococcus hirae (CopB) (Odermatt et al., 1993) has been implicated in copper binding. We thus searched for similar motifs in the PINA protein.
We observe three copies of an HXXM sequence in the C-terminal cytoplasmic tail of the PINA protein (Table1). This motif is present in at least one copy in all known ATP7B proteins and is absent in all known ATP7A proteins at the corresponding region. In ATP7A proteins, however, a conserved HXXM element is found at the end of the N-terminal soluble domain. Multiple HXXM sequences occur in bacterial and plant proteins that handle copper such as the copper transporter CopB (Solioz and Odermatt, 1995) of E. hirae, the copper-resistance proteins CopA and CopB of P. syringae, and COPT1 ofArabidopsis thaliana (Kampfenkel et al., 1995) (Table 1). Interestingly, the HXXM motif is not found anywhere in proteins that transport or bind metals other than copper, such as all of the Cd2+ transporters (GenBank data) and the mercury proteins MerAs and MerP (Silver et al., 1989), and thus may be a distinguishing feature of copper transporters.
Table 1.
An HXXM motif present in PINA/ATP7B and proteins involved in copper trafficking
| Source | Protein | *HXXM motifs | GenBank accession number |
|---|---|---|---|
| Rat | PINA/ATP7B | 589/1375HGRM | Q64535 |
| 603/1389HVGM | |||
| 641/1427HGGM | |||
| Human | ATP7B | 1389HGHM | P35670 |
| 1403HIGM | |||
| Sheep | ATP7B | 1368HGHM | AF032881 |
| Mouse | ATP7B | 1386HGRM | U38477 |
| 1400HIGM | |||
| Human | ATP7A | 696HSSM | Q04656 |
| Mouse | ATP7A | 687HSAM | Q64430 |
| Chinese hamster | ATP/A | 686HSSM | P49015 |
| Enterococcus hirae | CopB | 62HGHM | P05425 |
| 70HSHM | |||
| 75HEDM | |||
| 83HSHM | |||
| 88HENM | |||
| Pseudomonas syringae | CopA | 376HGSM | P12374 |
| 409HSKM | |||
| 470HGAM | |||
| 420HGAM | |||
| 596HMEM | |||
| Pseudomonas syringae | CopB | 37HSQM | P12375 |
| 45HSKM | |||
| 53HSQM | |||
| 61HSKM | |||
| 69HSQM | |||
| Arabidopsis thaliana | COPT1 | 3HDHM | Z49859 |
| 34HHHM | |||
| 36HMKM |
*The number represents the position of the histidine residues.
DISCUSSION
PINA and copper transport
Copper is required for all living organisms for growth and development because of its essential role in numerous physiological processes. However, copper in excess is toxic to the cells, resulting in cell death. A critical balance must therefore be maintained by specialized cellular transport mechanisms that regulate intracellular copper level. The process of copper transport involves copper uptake at the plasma membrane, intracellular copper distribution and use mediated by various copper-binding proteins, and copper export. Defects in copper transport result in two human inherited disorders, Menkes disease and Wilson disease. Two diseases result from defects in homologous genes with similar function (Hung et al., 1997; Payne and Gitlin, 1998) expressed in different tissues. Both genes encode copper-transporting P-type ATPases, a class of ATP-dependent integral membrane proteins that transport cations in diverse organisms. This large family of proteins includes Ca2+-, Cu+-, Cu2+-, Cd2+-, H+-, K+-, Mg2+-, H+-/K+-, and Na+-/K+-transporting ATPases (Carafoli, 1992). P-type ATPases form subfamilies with distinct cation transport specificity, and heavy metal-transporting ATPases are more closely related to each other than to other P-type ATPases.
All known metal transporters contain in their N terminals (Silver et al., 1993) invariant metal-binding cysteine motifs (CXXC) shown to be important in metal binding in a mercury protein (Sahlman and Skarfstad, 1993). They also possess a cluster of histidine- and methionine-rich domains (Bull and Cox, 1994; Solioz and Vulpe, 1996). Surprisingly, PINA lacks the entire N terminal region and the flanking transmembrane segments. In spite of this, the PINA protein is capable of transporting copper in a yeast system, although at a lower efficiency than the Menkes transporter ATPase. Accordingly, the N-terminal metal-binding region may primarily serve a regulatory role for metal transporters expressed constitutively and may not be essential for the transport activity. According to this formulation, PINA would lack the copper recognition domain because its function is regulated by diurnal variations in transcription and not by allosteric changes based on copper detection. The reduced copper-transport activity of PINA protein compared with the Menkes product may be attributable to the presence in night pineals of additional factor(s) needed for its full action. Alternatively, PINA may transport metals other than copper. Consistent with its proposed role in metal transport, PINA possesses the conserved intramembranous CPC element (Solioz and Vulpe, 1996) that is important in cation transport (Vilsen et al., 1989) and the conserved HXXM motif that provides a potential site for PINA to interact directly with metal.
PINA and circadian rhythms
The pineal gland contains a number of oscillating transcripts including ICER (Stehle et al., 1993), Fra2 (Baler and Klein, 1995), and NAT (Borjigin et al., 1995). PINA is the only pineal/retina-specific and night-specific molecule other than NAT. The rhythmic induction of pineal night genes reflects a clock-driven adrenergic input and cAMP signaling at night (Stehle et al., 1993; Baler and Klein, 1995;Roseboom et al., 1996). Consistent with the role of clock and cAMP signaling in the temporal regulation of pineal rhythms, PINA transcription is under clock control and inducible in vivoand in vitro using agents activating the β-adrenergic receptor and cAMP signaling pathway. A consensus CRE element in the promoter of PINA is able to interact with ICER in vitro(data not shown) and may be the in vivo determinant of the night specificity of PINA expression.
Although sympathetic input is thought to be the primary driving force behind the diurnal rhythm of pineal night-specific genes, PINA and NAT mRNA rhythms are already evident at P2, when the SCG innervation is still immature and no NAT activity (Ellison et al., 1972) or melatonin rhythms are detected (Stehle et al., 1995). Our findings suggest that a clock-driven adrenergic innervation may be present early in development. Alternatively there may be other inputs contributing to the generation of pineal rhythms besides the sympathetic innervation.
In addition to the pineal, PINA occurs in the adult retina where it is also diurnally controlled. Rhythmic melatonin synthesis driven by diurnal NAT activity is well documented in the retina of various species. Recently, NAT mRNA has also been found rhythmically expressed in the retinal photoreceptors of several species (Klein et al., 1997; J. Borjigin, unpublished observations). Because the NAT activity rhythm in retina is stimulated by agents activating the cAMP signal transduction pathway, the nocturnal increase in PINA transcription may be similarly controlled. In the retina, the PINA rhythm is not present at P2, contrasting with the pineal PINA rhythm evident at P2. The diurnal PINA rhythm begins after P16, corresponding to the completion of retinal development. This difference may be caused by a delay in the maturation of the retinal clock (Tosini and Menaker, 1996) compared with that of the SCN clock (Reppert et al., 1988), which is evident at birth.
The similar phylogenetic origins of the pineal gland and retina and the existence of a number of common molecules such as PINA and NAT in these tissues stimulated our search for common transcriptional machinery regulating spatial expressions of both pineal- and retina-specific genes. The presence of a number of homeobox-binding sequences, pineal response elements (PIRE) in the promoter regions of pineal genes such as PINA and NAT, and the ability of CRX, the newly discovered retina- and pineal-specific homeodomain-containing transcription factor, to transactivate PIRE-reporter constructs indicate that CRX is a functional component of such machinery (Li et al., 1998). In the present study, we have compared expression profiles for PINA, NAT, and CRX in the pineal and eye of both adult and developing rats. Consistent with the possible involvement of CRX in regulating PINA expressionin vivo, PINA and NAT expression seems to be confined within the domains in which CRX is expressed in pineal and adult retina. In the developing eyes, however, PINA is transiently detected in cell types (RPE and ciliary body) devoid of CRX expression. Thus, besides CRX, other homeodomain-containing proteins may coordinate the expression of pineal- and retina-specific genes including PINA.
Unlike the reported expression pattern of NAT transcripts in night retina (Klein et al., 1997), which is diffusely present in the entire photoreceptor layer, PINA expression is uniquely detected in a subset of photoreceptor cells in a location normally occupied by cone photoreceptor cells. If the PINA rhythm is under endogenous retinal clock control, the PINA-containing photoreceptors may be one of the cellular sites of the retinal clock machinery. It is not clear yet, however, whether the PINA-expressing cells belong to a known class of photoreceptors or reflect a distinct type of new photoreceptor cells (Foster, 1998).
PINA and Wilson disease
Copper accumulation in the livers of WD patients results from defective copper export because of malfunction of the copper-transporting ATPase gene ATP7B. There is extensive heterogeneity of symptoms within WD, some patients presenting with primarily hepatic problems, some with primarily neurological impairment, and some with both types of symptoms. Age of onset is variable, from childhood to early teens for hepatic presentation and usually late teens to adult for neurological problems. The abnormalities in the CNS in WD patients have been postulated to result from copper spillover from the liver (Bellary et al., 1995). Although the defect in the WD gene ATP7B leads to copper accumulation in the liver, brain, and eye, the only confirmed expression of ATP7B with identified cell types is found in the liver. PINA is the first novel WD transcript expressed both in brain and eye that possesses copper-transporting activity. To date, defects in the circadian timing system have always been associated with rhythmic physiology such as circadian sleep disorders or seasonal affective disorders and have never been linked to any nonrhythmic disorders in humans. This tradition, however, does not exclude the exciting possibilities of direct contribution of PINA in some aspects of the WD pathogenesis.
Because PINA expression patterns and regulation in vivo andin vitro parallel that of NAT, we tested the role of PINA in melatonin synthesis by analyzing melatonin formation in LEC rats, the WD animal model in which PINA is deleted (data not shown). Although LEC pineals display a defect in NAT protein and activity, linkage analysis demonstrated that the NAT defect in LEC rats is independent of the PINA mutation and is caused instead by a germ-line mutation in the NAT gene. Identification of the NAT mutation in LEC rats enabled us to separate the PINA mutation from the NAT mutation in breeding experiments; availability of PINA−/NAT+ rats should allow further clarification of the role of PINA in circadian biology. Identification of NAT defects independent of the PINA mutation indicates that PINA serves a yet unrecognized rhythmic physiological function in the pineal gland that presumably involves copper or another metal and may not be related to the regulation of melatonin synthesis.
Footnotes
This work was supported by United States Public Health Service Grant DA-00266 (S.H.S.) and by National Institute of Mental Health Grant MH57299 (J.B.). J.B. is a Merck fellow of the Life Science Research Foundation. We thank Dr. V. L. Culotta for use of the atomic absorption spectroscopy apparatus for copper measurement and for help with yeast studies, Drs. S. Chen and D. L. Zack for the mouse CRX cDNA, and Drs. J. Nathans and M. Takahashi for helpful comments on this manuscript.
Correspondence should be addressed to Dr. Solomon H. Snyder, Departments of Neuroscience, Pharmacology and Molecular Science, and Psychiatry, The Johns Hopkins University School of Medicine, Baltimore, MD 21205.
Dr. Borjigin’s present address: Department of Embryology, Carnegie Institution of Washington, 115 West University Parkway, Baltimore, MD 21210.
Dr. Ovodenko’s present address: State University of New York Health Science Center at Brooklyn, 450 Clarkson Avenue, Brooklyn, NY 11203.
REFERENCES
- 1.Adman ET. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 2.Arnold F, Haymore B. Engineered metal-binding proteins: purification to protein folding. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 3.Baler R, Klein DC. Circadian expression of transcription factor Fra-2 in the rat pineal gland. J Biol Chem. 1995;270:27319–27325. doi: 10.1074/jbc.270.45.27319. [DOI] [PubMed] [Google Scholar]
- 4.Bellary S, Hassanein T, Van Thiel D. Liver transplantation for Wilson’s disease. J Hepatol. 1995;23:373–381. doi: 10.1016/0168-8278(95)80194-4. [DOI] [PubMed] [Google Scholar]
- 5.Blackshaw S, Snyder SH. Developmental expression pattern of phototransduction components in mammalian pineal implies a light-sensing function. J Neurosci. 1997;17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borjigin J, Wang MM, Snyder SH. Diurnal variation in mRNA encoding serotonin N-acetyltransferase in pineal gland. Nature. 1995;378:783–785. doi: 10.1038/378783a0. [DOI] [PubMed] [Google Scholar]
- 7.Bull PC, Cox DW. Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 8. Bull PC, Thomas GR, Rommens JM, Forbes JR, Cox DW. 1993. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet [Erratum(1994)6:214] 5:327–337 [DOI] [PubMed]
- 9.Carafoli E. P-type ATPases. Introduction. J Bioenerg Biomembr. 1992;24:245–247. [PubMed] [Google Scholar]
- 10.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha J, Cooksey D. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelly J, Tumer Z, Tonnesen T, Petterson A, Ishikawa-Brush Y, Tommerup N, Horn N, Monaco AP. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat Genet. 1993;3:14–19. doi: 10.1038/ng0193-14. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 14.Chiu MI, Nathans J. A sequence upstream of the mouse blue visual pigment gene directs blue cone-specific transgene expression in mouse retinas. Vis Neurosci. 1994;11:773–780. doi: 10.1017/s0952523800003072. [DOI] [PubMed] [Google Scholar]
- 15.Coon SL, Roseboom PH, Baler R, Weller JL, Namboodiri MA, Koonin EV, Klein DC. Pineal serotonin N-acetyltransferase: expression cloning and molecular analysis. Science. 1995;270:1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- 16.Dancis A, Yuan DS, Haile D, Askwith C, Elde D, Moehle C, Kaplan J, Klausner RD. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 17.Ellison N, Weller JL, Klein DC. Development of a circadian rhythm in the activity of pineal serotonin N-acetyltransferase. J Neurochem. 1972;19:1335–1341. doi: 10.1111/j.1471-4159.1972.tb01458.x. [DOI] [PubMed] [Google Scholar]
- 18.Foster R. Shedding light on the biological clock. Neuron. 1998;20:829–832. doi: 10.1016/s0896-6273(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 19.Foulkes NS, Borjigin J, Snyder SH, Sassone-Corsi P. Transcriptional control of circadian hormone synthesis via the CREM feedback loop. Proc Natl Acad Sci USA. 1996;93:14140–14145. doi: 10.1073/pnas.93.24.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foulkes NS, Borjigin J, Snyder SH, Sassone-Corsi P. Rhythmic transcription: the molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997;20:487–492. doi: 10.1016/s0166-2236(97)01109-0. [DOI] [PubMed] [Google Scholar]
- 21.Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 23.Gastel JA, Roseboom PH, Rinaldi PA, Weller JL, Klein DC. Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science. 1998;279:1358–1360. doi: 10.1126/science.279.5355.1358. [DOI] [PubMed] [Google Scholar]
- 24.Hakanson R, Lombard des Gouttes MN, Owman C. Activities of tryptophan hydroxylase, dopa decarboxylase, and monoamine oxidase as correlated with the appearance of monoamines in developing rat pineal gland. Life Sci. 1967;6:2577–2585. doi: 10.1016/0024-3205(67)90107-5. [DOI] [PubMed] [Google Scholar]
- 25.Hung IH, Suzuki M, Yamaguchi Y, Yuan DS, Klausner RD, Gitlin JD. Biochemical characterization of the Wilson disease protein and functional expression in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:21461–21466. doi: 10.1074/jbc.272.34.21461. [DOI] [PubMed] [Google Scholar]
- 26.Kampfenkel K, Kushnir S, Babiychuk E, Inze D, Van Montagu M. Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J Biol Chem. 1995;270:28479–28486. doi: 10.1074/jbc.270.47.28479. [DOI] [PubMed] [Google Scholar]
- 27.Klein DC, Coon SL, Roseboom PH, Weller JL, Bernard M, Gastel JA, Zatz M, Iuvone PM, Rodriguez IR, Begay V, Falcon J, Cahill GM, Cassone VM, Baler R. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–357. [PubMed] [Google Scholar]
- 28.Klomp L, Lin S, Yuan D, Klausner R, Culotta V, Gitlin J. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J Biol Chem. 1997;272:9221–9226. doi: 10.1074/jbc.272.14.9221. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Chen S, Wang Q, Zack DJ, Snyder SH, Borjigin J. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc Natl Acad Sci USA. 1998;95:1876–1881. doi: 10.1073/pnas.95.4.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lolley RN, Craft CM, Lee RH. Photoreceptors of the retina and pinealocytes of the pineal gland share common components of signal transduction. Neurochem Res. 1992;17:81–89. doi: 10.1007/BF00966868. [DOI] [PubMed] [Google Scholar]
- 31.Mercer J, Livingston J, Hall B, Paynter J, Begy C, Chandrasekharappa S, Lockhart P, Grimes A, Bhave M, Siemieniak D, Glover TW. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat Genet. 1993;3:20–25. doi: 10.1038/ng0193-20. [DOI] [PubMed] [Google Scholar]
- 32.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 33.Payne AS, Gitlin JD. Functional expression of the Menkes disease protein reveals common biochemical mechanisms among the copper-transporting P-type ATPases. J Biol Chem. 1998;273:3765–3770. doi: 10.1074/jbc.273.6.3765. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen PL, Carafoli E. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem Sci. 1987;12:314–318. [Google Scholar]
- 35.Petrukhin K, Fischer S, Pirastu M, Tanzi R, Chernov I, Devoto M, Brzustowicz L, Cayanis E, Vitale E, Russo J. Mapping, cloning and genetic characterization of the region containing the Wilson disease gene. Nat Genet. 1993;5:338–343. doi: 10.1038/ng1293-338. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer M, Stehle JH. Ontogeny of a diurnal rhythm in arylalkylamine-N-acetyltransferase mRNA in rat pineal gland. Neurosci Lett. 1998;248:163–166. doi: 10.1016/s0304-3940(98)00356-5. [DOI] [PubMed] [Google Scholar]
- 37.Reppert S, Weaver D, Rivkees S. Maternal communication of circadian phase to the developing mammal. Psychoneuroendocrinology. 1988;13:63–78. doi: 10.1016/0306-4530(88)90007-8. [DOI] [PubMed] [Google Scholar]
- 38.Roseboom PH, Coon SL, Baler R, McCune SK, Weller JL, Klein DC. Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology. 1996;137:3033–3045. doi: 10.1210/endo.137.7.8770929. [DOI] [PubMed] [Google Scholar]
- 39.Sahlman L, Skarfstad E. Mercuric ion binding abilities of MerP variants containing only one cysteine. Biochem Biophys Res Commun. 1993;196:583–588. doi: 10.1006/bbrc.1993.2289. [DOI] [PubMed] [Google Scholar]
- 40.Sassone-Corsi P. Molecular clocks: mastering time by gene regulation. Nature. 1998;392:871–874. doi: 10.1038/31821. [DOI] [PubMed] [Google Scholar]
- 41.Silver S, Nucifora G, Chu L, Misra TK. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989;14:76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- 42.Silver S, Nucifora G, Phung L. Human Menkes X-chromosome disease and the staphylococcal cadmium-resistance ATPase: a remarkable similarity in protein sequences. Mol Microbiol. 1993;10:7–12. doi: 10.1111/j.1365-2958.1993.tb00898.x. [DOI] [PubMed] [Google Scholar]
- 43.Solioz M, Odermatt A. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem. 1995;270:9217–9221. doi: 10.1074/jbc.270.16.9217. [DOI] [PubMed] [Google Scholar]
- 44.Solioz M, Vulpe C. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci. 1996;21:237–241. [PubMed] [Google Scholar]
- 45.Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pevet P, Sassone-Corsi P. Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland. Nature. 1993;365:314–320. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- 46.Stehle JH, Foulkes NS, Pevet P, Sassone-Corsi P. Developmental maturation of pineal gland function: synchronized CREM inducibility and adrenergic stimulation. Mol Endocrinol. 1995;9:706–716. doi: 10.1210/mend.9.6.8592516. [DOI] [PubMed] [Google Scholar]
- 47.Tanzi RE, Petrukhin K, Chernov I, Pellequer JL, Wasco W, Ross B, Romano DM, Parano E, Pavone L, Brzustowicz LM, Devoto M, Peppercorn J, Bush AI, Sternlieb I, Pirastu M, Gusella JF, Evgratov O, Penchaszadeh GK, Honig B, Edelman IS, Soares MB, Scheinberg IH, Gilliam TC. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 48. Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. 1995. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet [Erratum(1995)9:451] 9:210–217 [DOI] [PubMed]
- 49.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 50.Vilsen B, Andersen J, Clarke D, MacLennan D. Functional consequences of proline mutations in the cytoplasmic and transmembrane sectors of the Ca2(+)-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264:21024–21030. [PubMed] [Google Scholar]
- 51. Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. 1993. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat Genet [Erratum(1993)3:273] 3:7–13 [DOI] [PubMed]
- 52.Vulpe CD, Packman S. Cellular copper transport. Annu Rev Nutr. 1995;15:293–322. doi: 10.1146/annurev.nu.15.070195.001453. [DOI] [PubMed] [Google Scholar]
- 53.Wikler K, Szel A, Jacobsen A. Positional information and opsin identity in retinal cones. J Comp Neurol. 1996;374:96–107. doi: 10.1002/(SICI)1096-9861(19961007)374:1<96::AID-CNE7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 54.Wu J, Forbes J, Chen H, Cox D. The LEC rat has a deletion in the copper transporting ATPase gene homologous to the Wilson disease gene. Nat Genet. 1994;7:541–545. doi: 10.1038/ng0894-541. [DOI] [PubMed] [Google Scholar]
- 55.Yaffe M, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi Y, Heiny M, Gitlin J. Isolation and characterization of a human liver cDNA as a candidate gene for Wilson disease. Biochem Biophys Res Commun. 1993;197:271–277. doi: 10.1006/bbrc.1993.2471. [DOI] [PubMed] [Google Scholar]
- 57.Yuan D, Stearman R, Dancis A, Dunn T, Beeler T, Klausner R. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc Natl Acad Sci USA. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]