Abstract
Our understanding of myelination has been greatly enhanced via the study of spontaneous mutants that harbor a defect in a gene encoding one of the major myelin proteins (myelin mutants). In this study, we describe a unique genetic defect in a new myelin mutant called the Long Evans shaker (les) rat that causes severe dysmyelination of the CNS. Myelin deficits result from disruption of the myelin basic protein (Mbp) gene caused by the insertion of an endogenous retrotransposon [early transposons (ETn) element] into a noncoding region (intron 3) of the gene. The ETn element alters the normal splicing dynamics of MBP mRNA, leading to a dramatic reduction in the levels of full-length isoforms (<5% of normal) and the appearance of improperly spliced, chimeric transcripts. Although these aberrant transcripts contain proximal coding regions of the MBP gene (exons 1–3), they are unable to encode functional proteins required to maintain the structural integrity of the myelin sheath. These chimeric transcripts seem capable, however, of producing the necessary signal to initiate and coordinate myelin gene expression because normal numbers of mature oligodendrocytes synthesizing abundant levels of other myelin proteins are present in the mutant CNS. Theles rat is thus an excellent model to study alternative functions of MBP beyond its well characterized role in myelin compaction.
Keywords: myelin mutants, dysmyelination, MBP, intron insertion, retrotransposon, ETn element
The ability of a neuron to conduct an action potential rapidly along its axon is facilitated by the insulating properties provided by the surrounding myelin sheath. The myelin sheath is an extension of the glial cell plasma membrane that has wrapped repeatedly around a portion of an axon to form a condensed, multilamellar structure. Within the CNS, myelin is synthesized exclusively by oligodendrocytes. Although myelin is composed primarily of lipids, it also contains a significant complement of novel proteins that serve to maintain the structural integrity of the myelin sheath (Hudson, 1990; Lemke, 1992; Holz and Schwab, 1997). The most abundant proteins identified in CNS myelin include proteolipid protein (PLP) and myelin basic protein (MBP), whereas less prominent proteins include 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP), myelin oligodendrocyte basic protein, myelin-associated glycoprotein, and myelin oligodendrocyte glycoprotein.
Myelination of the rodent CNS occurs primarily during postnatal development and requires the coordinated activation of genes synthesizing various structural proteins and enzymes. In rats and mice, the most active period of CNS myelination takes place between 2 and 4 weeks and is complete by 2 months. Myelin protein gene expression is regulated principally at the transcriptional level with changes in relative amounts of protein reflecting corresponding changes in mRNA levels (Campagnoni and Macklin, 1988). For example, MBP mRNA is initially detected in mouse brain at 2–10 d postnatally, reaches maximal levels at 15–22 d, and decreases to 20% of maximal levels after 30 d (Campagnoni, 1988; Lemke, 1988). Expression of certain myelin proteins, such as MBP, is also regulated at the post-transcriptional level (Kumar, 1989; Verdi and Campagnoni, 1990;Brophy et al., 1993), although these influences induce only modest changes in expression levels (2- to 3-fold effect) compared with those that control transcription initiation (20- to 30-fold effect). In addition, because MBP as well as many of the myelin proteins exists as multiple isoforms that appear at different times during development (Barbarese and Barry, 1988; Mathisen et al., 1993), myelin gene expression can also be influenced by events that affect alternative splicing (De Ferra et al., 1985; Roach and Takahashi, 1985; Newman et al., 1987; Okano et al., 1991). The numerous regulatory mechanisms identified in oligodendrocytes to manipulate myelin protein expression affirm the importance of precise regulation of spatial and temporal expression of each myelin protein to ensure proper formation of the myelin sheath (Trapp et al., 1987; Pfeiffer et al., 1993).
In spite of our understanding of the organization and overall protein composition of the myelin sheath, the exact role many of these proteins play in oligodendrocyte development and myelination remains unknown. Many of the insights we have gained into the function of the different myelin proteins have come from the study of spontaneous animal mutants. These mutants, collectively referred to as the myelin mutants, exhibit varying degrees of myelin deficiencies that result from a mutation in one of the major myelin protein genes (Duncan, 1995; Griffiths, 1996;Scherer, 1997). These mutants are divided into two groups based on the manner in which the dys- or hypomyelinating phenotype is inherited. The X-linked mutants share a common characteristic in that they harbor a defect in the Plp gene. Extensive analysis of the various X-linked mutants has been instrumental in defining the structural role PLP plays in facilitating the close apposition of the external surfaces of adjacent membrane layers in compacted myelin (Boison et al., 1995). In addition, these mutants have led to an understanding of how an alternatively spliced form of PLP, called DM20, is important in oligodendrocyte development (Griffiths, 1996).
Mutants that inherit a defect in myelination in an autosomal recessive manner form the second group and are much more heterogeneous than the X-linked mutants in terms of both genotypes and phenotypes. Mutants in this group include the shiverer (shi) mouse (Biddle et al., 1973), its allelic variant shimld(Doolittle and Schweikart, 1977), the taiep rat (Holmgren et al., 1989; Duncan et al., 1992), and the quaking (qk) mouse (Hardy, 1998). The most thoroughly characterized of these mutants isshi in which CNS dysmyelination results from a lack of MBP caused by a large deletion in the Mbp gene (Roach and Takahashi, 1985; Molineaux et al., 1986). This mutant provided the first direct evidence of the critical role MBP plays in myelinogenesis (Bird et al., 1978; Chernoff, 1981) and myelin sheath compaction (Dupouey et al., 1979; Privat et al., 1979). In addition,shi demonstrated that MBP is not required to maintain the structural integrity of the myelin sheath in the peripheral nervous system (PNS), indicating that this protein may have multiple functions that play different roles in the PNS and CNS (Kirschner and Ganser, 1980; Rosenbluth, 1980; Peterson and Bray, 1984).
We recently identified a new myelin mutant that developed spontaneously in a colony of Long Evans rats (Delaney et al., 1995). This mutant, referred to as the Long Evans shaker (les) rat, exhibits classical clinical symptoms of CNS dysmyelination, including whole-body tremors by 2 weeks of age followed by seizures, progressive ataxia, and hindlimb paresis. The CNS appears devoid of any myelin, whereas myelin in the PNS appears normal. Some discernible myelin sheaths are evident in the CNS during early postnatal development (<2 weeks), although these appear to be eventually lost because only rare, thinly myelinated axons are observed in animals beyond 4 weeks of age. Although the genetic defect responsible for dysmyelination in this mutant was unknown, the autosomal recessive nature of the phenotype coupled with a lack of detectable MBP immunolabeling in either the CNS or PNS implicated the MBP gene as the likely source of the defect (Kwiecien et al., 1998). In this study, we identify the genetic mutation in theles Mbp gene as a large retroviral-like insertion that alters the normal processing of MBP mRNA. Novel aspects of dysmyelination in this mutant provide further evidence in support of the hypothesis that MBP plays a much broader role in myelination than its well characterized function in myelin sheath compaction.
MATERIALS AND METHODS
Immunohistochemistry. Rats were anesthetized with sodium pentobarbital and perfused transcardially with saline followed by 4% paraformaldehyde in 0.1 m phosphate, pH 7.4. Brains were removed, post-fixed in perfusate, cryoprotected in PBS containing 30% sucrose, and sectioned on a freezing sliding microtome. Twenty-micrometer-thick free-floating sections were washed in PBS and permeabilized with 1% Triton X-100 containing 1% H2O2 (to quench endogenous peroxidase activity). After three washes in PBS, sections were incubated with 3% normal goat serum in PBS to prevent nonspecific binding followed by incubation at 4°C overnight with primary antibody diluted in PBS. Antibodies used in this study included a mouse monoclonal anti-MBP (Boehringer Mannheim, Indianapolis, IN) at a dilution of 1:4000 and a rabbit polyclonal anti-PLP/DM20 IgG (gift from Dr. Ian Griffiths) at a dilution of 1:10,000. After the overnight incubation, tissue sections were washed in PBS and incubated with a 1:500 dilution of biotinylated secondary antibody for 30 min at room temperature. Secondary antibodies included a donkey anti-rabbit IgG (The Jackson Laboratory, Bar Harbor, ME) or a goat anti-mouse IgG (Vector Laboratories, Burlingame, CA). After an additional PBS wash, secondary antibodies were detected using the avidin–biotin complex reaction (ABC Elite kit; Vector Laboratories) and developed with a solution of 6.7 mg/ml aqueous diaminobenzidine (DAB) and 0.04% H2O2. The reaction product was enhanced and stabilized by incubation in a 0.04% aqueous osmium tetroxide solution (Electron Microscopy Sciences) and wet mounted on slides using glycerol. Where appropriate, a simplified myelin stain (Kaatz et al., 1992) with nitroblue tetrazolium and DAB was used to visualize myelin in adjacent brain sections (40 μm).
Northern blot analysis. Total RNA (20 μg) extracted from brains, spinal cords, and sciatic nerves of mutant and age-matched controls at 2, 4, 8, 12, and 16 weeks of age was separated on 1.2% agarose–formaldehyde gels and transferred to a Zeta-Probe GT nylon membrane (Bio-Rad, Hercules, CA). Membranes were hybridized according to the manufacturer’s recommendations for 18–24 hr at 65°C with a32P-labeled cDNA probe synthesized by random primer extension (Feinberg and Vogelstein, 1983). The MBP probe used in this analysis was generated from a 1.4 kbEcoRI–HindIII restriction fragment from pMBPSP72 (Mentaberry and Adesnik, 1986), the PLP/DM20 probe was from a 0.98 kbEcoRI–SacI restriction fragment from pPLPG3 (Hudson et al., 1987), the CNP probe was from a 0.85 kb PstI restriction fragment from pCNP7 (Bernier et al., 1987), and the cyclophilin probe was from a 0.68 kb BamHI–PstI fragment from pIBI5 (Danielson et al., 1988). Quantitation of hybridization levels on blots was performed using a Scanalytics Ambis Radioanalytical Imaging System (Scanalytics/CSPI, Billerica, MA). The individual mean for each RNA sample was normalized to cyclophilin expression and compared with age-matched control samples run in the same gel (expressed as percent of control). The means of three separate brains for each age (n = 3) were evaluated by Student’s t test. Results are presented in histograms as mean ± SEM.
To identify chimeric MBP transcripts containing sequences of the early transposons (ETn) element, probes synthesized from PCR products generated from primers located in either Mbp exon 1 or 3 and the long terminal repeat (LTR) of the ETn element (see description of primers below) were used in Northern blots. In other experiments, probes specific for either a 362 bp EcoRI or a 355 bp PstI fragment located ∼1.7 or 4.2 kb, respectively, from the 3′ end of the LTR primer (see below) were used.
In situ hybridization. Cellular localization of mRNA encoding MBP and PLP/DM20 was determined using a free-floating in situ hybridization protocol. Animals were anesthetized with halothane and perfused transcardially with saline followed by a 0.4% paraformaldehyde solution in 0.1 m phosphate buffer (PB), pH 7.4. Brains were removed and post-fixed in perfusate overnight at 4°C, washed three times in PB saline solution (PBS), and cryoprotected in PBS containing 30% sucrose overnight. Tissue was sectioned sagittally at 25 μm on a freezing microtome and washed three times (for 5 min each) in PBS at room temperature. Tissue sections were incubated subsequently in PBS containing 0.1m glycine (two washes for 3 min each at room temperature), in PBS (15 min at room temperature), in a solution of 0.1 mTris-HCl, pH 8, containing 0.05 m EDTA and 1 μg/ml proteinase K (30 min at 37°C), and in a solution of 0.1 mtriethanolamine and 0.25% acetic anhydride, pH 8 (10 min at room temperature). Sections were then washed twice in 2× SSC (1× is 0.15m NaCl and 0.3 m sodium citrate, pH 7) for 15 min each at room temperature and incubated for 60 min at 60°C in hybridization buffer that consisted of 35× Denhardt’s solution (50× is 10 mg/ml Ficoll, 10 mg/ml BSA, and 10 mg/ml polyvinyl-pyrrolidone), 10% sodium dextran sulfate, 50% formamide, 20 mmdithiothreitol (DTT), 0.15 mg/ml yeast tRNA, and 0.3 mg/ml salmon sperm DNA.
Radiolabeled cRNA probes specific for MBP and PLP/DM20 mRNAs were transcribed from linearized plasmids (described above in Northern blots) using SP6 or T7 RNA polymerase in the presence of35S-UTP. Tissue sections were incubated in hybridization buffer containing the appropriate probe (2 × 104 cpm per microliter of buffer) at 60°C for 18–48 hr. After hybridization, tissue sections were washed twice in 4× SSC (30 min each at 60°C) and incubated in a solution of 0.5m NaCl, 10 mm Tris-Cl, pH 8, and 1 mm EDTA containing 20 μg/ml RNase A for 30 min at 45°C. Tissue sections were then washed in SSC buffers under increasingly stringent conditions including 2× SSC (three washes for 30 min each at room temperature and an additional wash for 60 min at room temperature), 0.5× SSC (two washes for 30 min each at 60°C), 0.1× SSC (30 min wash at 60°C), and a final wash in 0.1× SSC at room temperature. All SSC washes contained 5 mm DTT to stabilize the radiolabeled probe. Tissue sections were mounted on coverslips coated with poly-l-lysine in 0.05 m phosphate buffer, pH 7.4, defatted in chloroform, and dipped in Kodak NTB2 emulsion (Eastman Kodak, Rochester, NY). Sections were exposed at 4°C for 3–6 d, developed, and counterstained with hematoxylin.
PCR analysis. Alternatively spliced isoforms of MBP mRNA were detected by PCR amplification of RNA isolated from brain, spinal cord, and sciatic nerve of 2-, 4-, 8-, 12-, and 16-week-old rats using primers complementary to sequences in MBP exons 1 (5′-CCAAGAAGATCCCACAGCAG-3′) and 7 (5′-CATGGGAGATCCAGAGCG-3′). First-strand cDNA was synthesized using Moloney’s murine leukemia virus reverse transcriptase and random hexanucleotide primers as described previously (Kawasaki, 1990). cDNA generated from 100 ng of total RNA was amplified in a buffer containing 50 mm KCl, 10 mm Tris-HCl, pH 9, 1.5 mmMgCl2, 0.2 mm dNTPs, 20 pmol of each primer, and 2.5 U of Taq polymerase in a final volume of 50 μl. Samples were incubated in a thermocycler (Perkin-Elmer, Norwalk, CT) for 25–40 cycles (94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec) followed by a final extension at 72°C for 7 min. PCR products were separated by electrophoresis on an 8% polyacrylamide gel and visualized by staining with ethidium bromide (EtBr). To confirm the identity of PCR products, we cloned DNA fragments into the pGEM-T vector (Promega, Madison, WI) and sequenced the fragments at the University of Wisconsin Biotechnology Center Sequencing Facility.
To identify chimeric transcripts containing Mbp exons fused with ETn sequences, we subjected RNA samples to a reverse transcription (RT)-PCR analysis similar to that described above except primers were used that were complementary to sequences in either Mbpexon 1 (see above) or exon 3 (5′-CAAGAACTACCCACTACGGC-3′) and sequences within the LTR of the ETn element (5′-CATGATCTCATCAGGCAC-3′) at an annealing temperature of 59°C. Purified PCR products were subsequently used as probes in Northern blots to estimate the size of chimeric mRNA expressed in nervous tissue from les rats.
To determine whether there were any large inserts or deletions inMbp introns 2 and 3, a modified PCR procedure was used that had been optimized for amplifying long DNA target sequences usingrTth DNA polymerase (XL PCR Kit; Perkin-Elmer). Genomic DNA (250 ng) isolated from livers or spleens of normal andles rats was amplified according to the manufacturer’s instructions using primers in Mbp exon 2 (5′-CTGTGCCACATGTACAAG-3′) and exon 3 (5′-GAAGAAGTGGACTACTGGG-3′) or primers in exon 3 (5′-CAAGAACTACCCACTACGGC-3′) and exon 4 (5′-ATGGAGGGGGTGTACGAG-3′) at an annealing temperature of 60°C. PCR products were separated on 1% agarose gels and visualized by staining with EtBr. To identify the 6 kb insert present inMbp intron 3 from the les mutant, we cloned the 2 and 8 kb PCR products generated from control and lessamples, respectively, into pGEM-T and sequenced the products at the University of Wisconsin Biotechnology Center Sequencing Facility. Various restriction fragments from the ETn sequence were isolated and used as probes in Northern blots to identify chimeric mRNA expressed in nervous tissue from the les rat.
Levels of MBP mRNA in mutant brains of 4- and 12-week-old rats (n = 3) were estimated by quantitative autoradiography of MBP PCR products that had been normalized to actin mRNA levels. MBP and actin PCR products were synthesized from cDNA in a single reaction using Mbp exon 1 and 7 primers (described above) and commercially available actin primers (Promega) that had been end-labeled with 32P by T4 polynucleotide kinase. All PCR reaction conditions were the same as described previously except synthesis was limited to 27 cycles because this was the minimum number needed to generate a sufficient amount of MBP PCR products that could be visualized by autoradiography.
RESULTS
Expression of myelin basic protein is severely reduced in theles rat
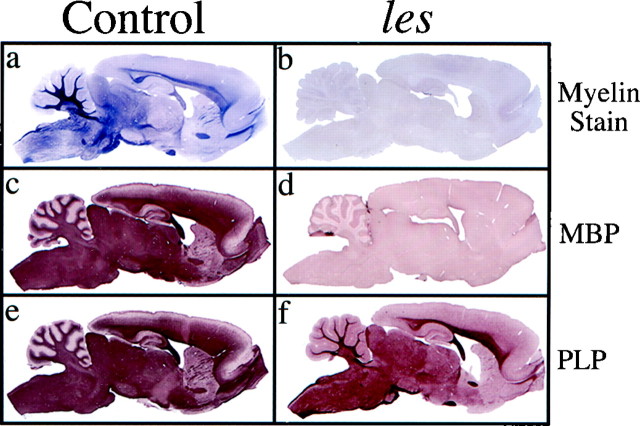
The les rat is a recently identified myelin mutant that exhibits severe dysmyelination of the CNS while the PNS remains normally myelinated (Kwiecien et al., 1998). We have shown previously that axons throughout the spinal cord and optic nerve of this mutant are either devoid of discernible myelin or surrounded by thin, poorly compacted myelin sheaths (Kwiecien et al., 1998). The severity of myelin deficits observed in les is reminiscent of those described in other CNS myelin mutants that result from a defect in a gene encoding one of the myelin structural proteins (Lunn et al., 1995;Griffiths, 1996). To gain a better understanding of the genetic defect causing CNS dysmyelination in les rats, we analyzed expression levels of the major CNS myelin proteins in the brains of affected and age-matched control animals at various ages (2–16 weeks). Consistent with our previous observations in mutant spinal cords and optic nerves, a paucity of myelin was readily apparent throughout the brains of les rats at all ages analyzed (Fig.1a,b). Immunohistochemical analysis of adjacent brain sections revealed no detectable MBP immunoreactivity (Fig. 1c,d), whereas levels of PLP appeared to be only marginally reduced compared with that in age-matched controls (Fig.1e,f).
Fig. 1.
No detectable MBP immunostaining was evident throughout the CNS of the les rat, whereas levels of PLP appear to be only modestly reduced compared with that in controls.a–f, Bright-field photomicrographs of the immunohistochemical analysis of sagittal brain sections taken from 44-d-old les (b, d,f) and control (a,c, e) rats stained for MBP (c, d) and PLP (e,f). a, b, Adjacent brain sections stained for myelin.
To determine whether a lack of MBP immunostaining reflected a corresponding decrease in mRNA levels, we measured MBP mRNA inles rats during the period of most active CNS myelination (4 weeks of age) by Northern blot analyses. No MBP transcripts were detected in RNA extracted from mutant brains, spinal cords, or sciatic nerves, whereas full-length MBP mRNA (2.2 kb) was observed in age-matched controls (Fig.2A). The lack of MBP mRNA in les could not be attributed to RNA degradation or differences in RNA loading because hybridization levels for cyclophilin, an mRNA expressed abundantly throughout the CNS (Danielson et al., 1988), were similar in both mutant and control samples. Additional samples of RNA extracted from brains of animals at 2, 4, 8, and 12 weeks of age were subsequently analyzed by Northern blots to ascertain whether detectable levels of MBP mRNA were expressed in the CNS at any point during postnatal development. Again, no MBP mRNA was observed in mutants compared with robust expression in age-matched controls (Fig. 2B). By contrast, expression levels of other major myelin genes, including Plp and Cnp, appeared similar in les and controls (Fig.2B). Quantitative analyses of Northern blots confirmed that although MBP mRNA levels were severely diminished in this mutant (<5% of normal), expression of Plp was only moderately decreased (50–80% of normal) and Cnp was not significantly different (p < 0.5) from that in age-matched controls (Fig. 2C).
Fig. 2.
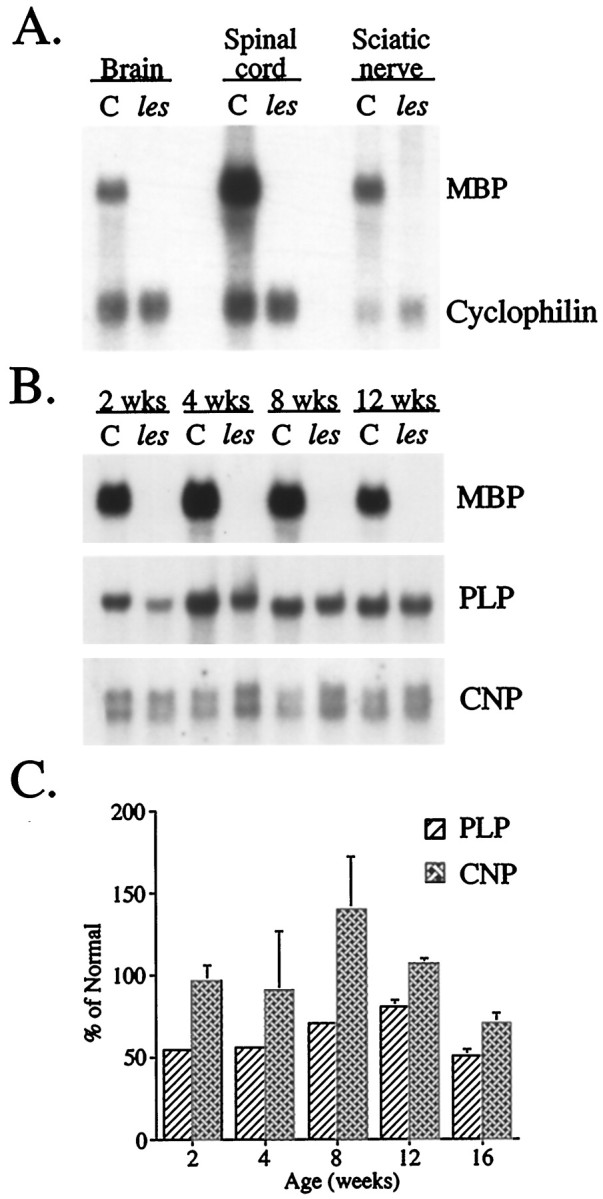
MBP mRNA cannot be detected in either the CNS or PNS of les rats, whereas levels of other major myelin mRNAs remain indistinguishable from that of controls. A, Autoradiogram of a Northern blot in which RNA samples (20 μg) extracted from brain, spinal cord, and sciatic nerve of 4-week-old mutant and control (C) rats were hybridized with32P-labeled probes for MBP and cyclophilin mRNAs.B, Autoradiograms of Northern blots in which RNA samples extracted from 2-, 4-, 8-, and 12-week-old mutant and control brains were hybridized with 32P-probes specific for either MBP, PLP, or CNP mRNAs. C, Histogram summarizing the quantitation of Northern blots of PLP and CNP mRNA levels in different brain RNA samples (n = 3) of mutant and age-matched controls at different ages. Results are expressed as the percent of control (mean ± SEM).
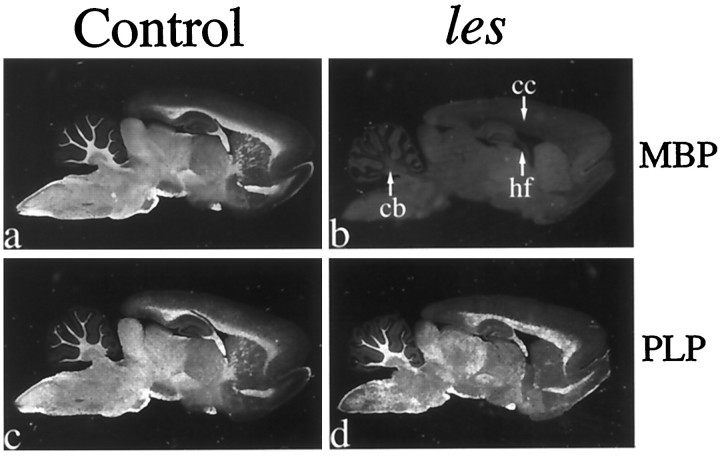
To provide further evidence of a lack of MBP mRNA expression inles oligodendrocytes, we evaluated cellular localization of MBP mRNA in sagittal brain sections by in situhybridization. Consistent with results from Northern blots, hybridization levels observed over prominent white matter tracts (e.g., corpus callosum, hippocampal fimbria, and cerebellar white matter) were dramatically reduced in mutant brains compared with controls (Fig.3a,b) and similar to the nonspecific hybridization observed in control sections incubated with the sense probe (data not shown). Results from Northern blots andin situ hybridization analyses indicated that myelin deficits in les resulted from a defect in the Mbpgene that disrupted expression at the transcriptional level. Alternatively, drastically reduced levels of MBP could also be caused by a decrease in oligodendrocyte number or a failure of oligodendrocytes to mature properly to the stage in which they can express MBP. Near normal levels of PLP detected by immunohistochemistry and Northern blots argued strongly against these latter possibilities, however. To provide more definitive evidence that normal numbers of mature oligodendrocytes were present in the CNS of this mutant, we analyzed the cellular distribution of PLP mRNA in adjacent brain sections by in situ hybridization. In marked contrast to MBP mRNA, the intensity and distribution of PLP hybridization in mutant brains were virtually identical to that observed in age-matched controls (Fig. 3c,d), confirming thatles oligodendrocytes were indistinguishable from normal in terms of numbers, distribution, and maturity.
Fig. 3.
No MBP mRNA was detected throughout theles brain, whereas the expression and distribution of PLP mRNA appear similar to that in the control. a–d, Dark-field photomicrographs of 44-d-old sagittal brain sections fromles (b, d) and control (a, c) rats hybridized with35S-labeled probe for MBP (a,b) and PLP (c, d) mRNA. Robust expression of PLP mRNA in les verifies that normal numbers of mature oligodendrocytes are present in the mutant CNS. cb, Cerebellum; cc, corpus callosum;hf, hippocampal fimbria.
The les mutation causes aberrant splicing of MBP mRNA
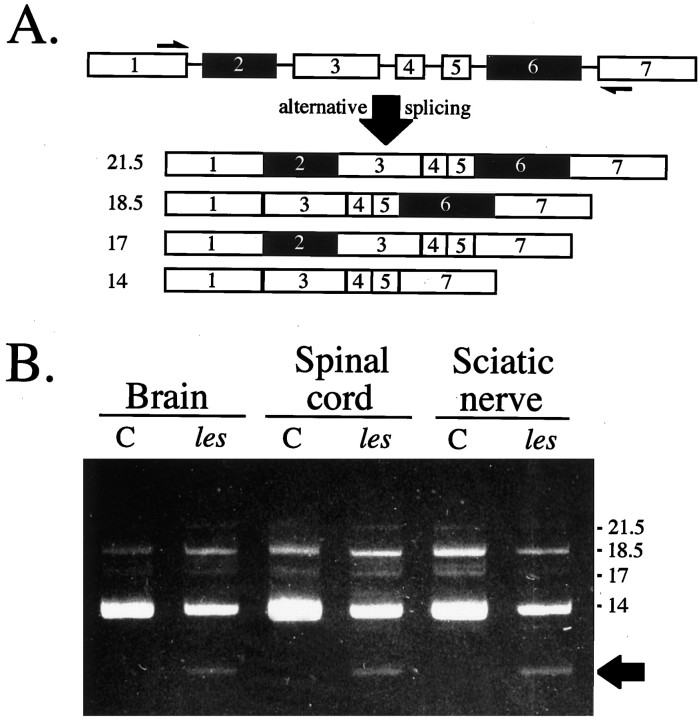
The ability to detect normal numbers of mature oligodendrocytes throughout the CNS of this mutant indicated that these cells either were incapable of synthesizing MBP mRNA or produced such low levels that they were undetectable by the molecular techniques used. To discriminate between these two possibilities, RNA examined previously in Northern blots was further analyzed by RT-PCR using primers complementary to sequences in exons 1 and 7 of the rat cDNA (Roach et al., 1983). The rodent Mbp gene consists of several exons that are alternatively spliced to generate the four major isoforms (21.5, 18.5, 17, and 14 kDa peptides) expressed in the adult CNS (Campagnoni, 1988). Primers were designed that were specific to exons 1 and 7 because they are present in all MBP isoforms and would enable us to identify the major alternatively spliced transcripts (Fig.4A). As expected, four PCR products were amplified from control RNA samples that were of the appropriate lengths to encode the major MBP isoforms (Fig.4B). Interestingly, these four transcripts were also detected in RNA extracted from the CNS and PNS of les rats. Subsequent cloning and sequencing of three of these mutant PCR products confirmed they were identical to normal transcripts (those encoding the 21.5, 18.5, and 14 kDa isoforms) in respect to both exon composition and sequence. In addition to the normal MBP transcripts, a fifth PCR product was routinely amplified from RNA samples extracted from the CNS and PNS of les rats. Cloning and sequence analysis of this transcript revealed that it was similar to that encoding the 14 kDa isoform (exons 1, 3–5, and 7) except it was lacking the normally invariant exon 3. Additional PCR analysis of les RNA further demonstrated that transcripts encoding other MBP isoforms (21.5 and 18.5 kDa) lacking exon 3 could also be detected in the CNS and PNS of this mutant (data not shown). Thus, the les mutation appears not only to affect the normal transcription of the Mbp gene but also alters post-transcriptional splicing dynamics, enabling novel, improperly processed MBP transcripts to be generated.
Fig. 4.
A novel isoform of MBP was detected in the CNS and PNS of the les rat in addition to the low levels of properly spliced MBP transcripts. A, Schematic of the rodent Mbp gene and the predominant CNS isoforms generated from alternative splicing. Half arrows in exons 1 and 7 indicate the approximate positions of primers used in RT-PCR analysis. Numbers to the leftindicate the mass (kDa) of the MBP isoform generated from each transcript. B, EtBr-stained gel of RT-PCR analysis of RNA extracted from 4-week-old brains, spinal cord, and optic nerves using primers detecting all four major adult MBP isoforms. Thearrow to the right of thegel indicates the novel isoform generated inles. Sequence analysis of this PCR product revealed that it was similar to that encoding the 14 kDa isoform except it was missing exon 3. C, Control.
The relative abundance of MBP transcripts in les appeared to be extremely low compared with that in controls because an extended number of amplification cycles (35–40 for les compared with 25 for control) was required before PCR products could be visualized by EtBr staining in polyacrylamide gels. To estimate MBP mRNA levels inles more accurately, quantitative analysis of the most abundant MBP transcript generated in these PCR reactions, the 14 kDa isoform, was performed and normalized to actin mRNA levels. MBP mRNA levels in mutant brains from 1- and 3-month-old animals were ∼2% of that observed in age-matched control brains (data not shown).
Insertion of retrotransposon detected in intron 3 of theles MBP gene
Although results from molecular studies were consistent with a defect in Mbp, the question remained as to the nature of the mutation and where it resided within the gene. Differences in hybridization patterns observed in Southern blots of mutant and control genomic DNA digested with different restriction enzymes provided the first indication that a major rearrangement had occurred within theles Mbp gene (data not shown). Previous detection of aberrantly spliced MBP transcripts lacking exon 3 further suggested that the les mutation altered sequences flanking this exon that may be important in alternative splicing. Because sequencing of MBP PCR products from les revealed no detectable mutations in exon 3, we focused our attention on the surrounding introns as the most probable location of the les mutation.
A PCR strategy was used to identify any large deletions or insertions in either intron 2 or 3 of the les Mbp gene. Sequence analyses of the various Mbp introns have not been published, although the length of intron 2 has been estimated to be ∼11 kb whereas intron 3 appears to be much shorter (Molineaux et al., 1986). To amplify efficiently across such extended distances, a PCR protocol adapted for amplifying long DNA fragments was performed onles and control genomic DNA using primers complementary to sequences in exons 2 and 3 (to amplify across intron 2) and exons 3 and 4 (to amplify across intron 3). The PCR products generated from amplification across intron 2 were of similar lengths in lesand controls (data not shown), whereas those generated across intron 3 were markedly different in size (Fig.5A). The wild-type (wt) form of intron 3 observed in controls was ∼2 kb, whereas that generated from les was ∼8 kb.
Fig. 5.
Intron 3 of the les Mbp gene contains a 6 kb insertion that is related to the ETn family of retrotransposons. A, EtBr-stained gel of PCR products generated from amplification of genomic DNA isolated fromles and control (C) rats using primers located in Mbp exons 3 and 4. B, Nucleotide sequence of MBP intron 3 from the les rat containing the ETn insertion. The insertion sites are indicated bydownward arrows, and the LTR of the ETn element is shown in bold. The 6 bp duplicated sequence (gtgatg) isboxed, and the polyadenylation sites in the LTR areunderlined. The 9 bp sequence detected inles intron 3 but not in wt isdoubly underlined.
Cloning and sequencing of the wt and mutant forms of Mbpintron 3 confirmed that an additional 5.7 kb sequence was present in the les gene. This insert was located ∼0.4 kb downstream from the exon 3–intron 3 junction (Fig. 5B). The insert was unrelated to known MBP sequences, indicating that it did not arise from duplication of adjacent regions. Interestingly, the size of the insert was similar to that reported for a specific family of retrovirus-like elements (retrotransposons) that have been described extensively in several spontaneous murine mutants (Herrmann et al., 1990; Steinmeyer et al., 1991; Adachi et al., 1993; Mitreiter et al., 1994; Moon and Friedman, 1997). This family of retrotransposons was first identified in undifferentiated embryonic carcinoma cell lines and referred to as an ETn element (Brulet et al., 1983). Comparison of the lesinsert sequence with previously published murine ETn elements (Shell et al., 1990) revealed that although there was <42% overall sequence similarity, the les insert contained certain sequence domains that are characteristic of this family of retrovirus-like elements. The most prominent of these domains was a 0.36 kb directed LTR located at each end of the insert (Fig. 5B). In addition, the intervening sequence between these LTRs in theles insert was similar to those described in murine ETn elements in that it contained only short open-reading frames that were incapable of synthesizing essential retroviral proteins (i.e.,gag and pol) required for transposition. Moreover, the dinucleotide repeats found at each terminal of theles insert (5′-TG … CA-3′) are the same as those described in the majority of retroviruses. Although the exact function of these repeats is unknown, their prevalence indicates a critical role in the integration process. The les MBP gene was also found to contain a short direct-repeat sequence flanking the insert, a phenomenon observed in all genes that have been disrupted by retroviruses. The fact that this sequence (GTGATG) was also found in intron 3 of the wt Mbp gene was consistent with it resulting from the duplication of the integration target site. Taken together, these similarities strongly suggest that the insert we have identified in the MBP gene of the les rat is actually an endogenous ETn element that has been introduced by transposition. To our knowledge, this is the first ETn element described in rats. Finally, when intron 3 sequences from the les MBP gene were compared with the wt sequence, we noticed a unique 9 bp sequence located just upstream of the ETn element in les that was not observed in the wt MBP gene (Fig. 5B). This sequence was not related to or otherwise associated with the ETn element and thus may have resulted from a independent duplication event.
The ETn element promotes aberrant splicing of MBP
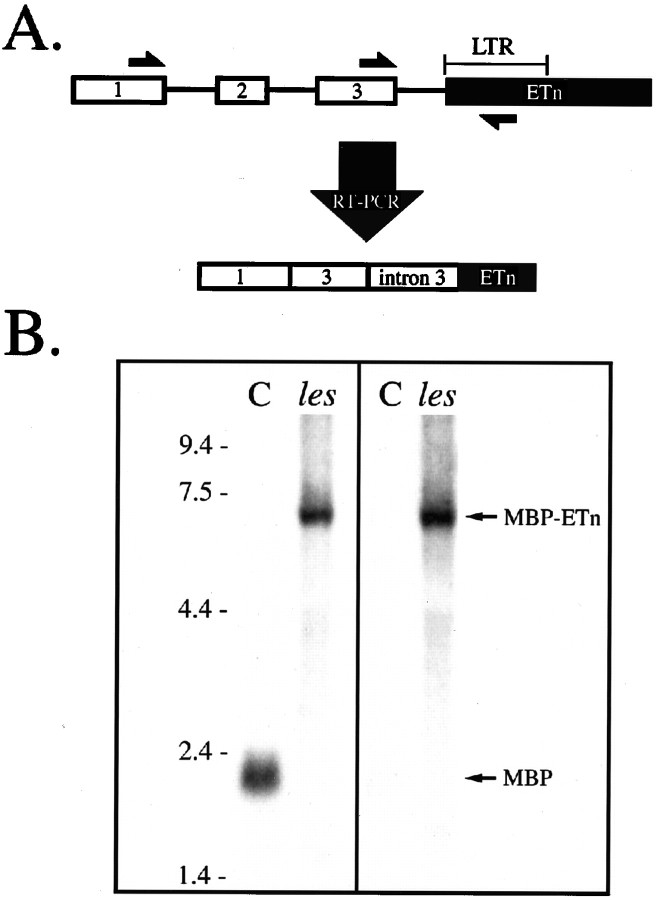
Molecular characterization of murine mutants arising from the insertion of an ETn element into various genes has demonstrated that gene inactivation was frequently a result of aberrant splicing events that generated chimeric transcripts in which coding sequences from the disrupted host gene were fused to sequences in the retrotransposon. To determine whether similar abnormalities had occurred in the processing of les MBP transcripts, we analyzed RNA extracted from mutant brains, spinal cords, and sciatic nerves by RT-PCR using primers complementary to sequences in Mbp exons 1 or 3 and the LTR of the ETn element (Fig.6A). A single PCR product was generated in each of these reactions that was not observed when control RNA was used (data not shown). Sequencing of these PCR products revealed that they consisted of MBP exons fused to the 0.4 kb of MBP intron 3 that lies just upstream of the insert and a portion of the retrotransposon LTR. When these PCR products were subsequently used to probe Northern blots of RNA extracted from the CNS and PNS ofles and controls, a single prominent transcript of ∼7 kb was observed in mutants, whereas the 2.2 kb MBP mRNA was detected in controls (Fig. 6B, left). Although the expression of the chimeric MBP–ETn transcript was abundant in the CNS and PNS of the les rat, it was not detected in RNA extracted from non-nervous system tissue (e.g., liver), indicating that transcription remained under the regulation of the MBP promoter (data not shown). This observation is consistent with results from previous studies that demonstrated that sequences within the LTR promote abundant transcription of the ETn element only during very early stages of embryonic development (Soniga et al., 1987).
Fig. 6.
Chimeric MBP transcripts containing ETn sequences are abundantly expressed in the CNS and PNS of the lesrat. A, Schematic of PCR strategy used to identify chimeric mRNA detected in les rats. Approximate locations of primers in MBP exons 1 and 3 and the LTR of the ETn element used in PCR are indicated by half arrows. Cloning and sequencing of the PCR products revealed that they consisted of Mbp exons 1 and/or 3 fused to portions ofMbp intron 3 and ETn sequences. B, Autoradiograms of Northern blots in which RNA (20 μg) extracted from brains of 4-week-old mutant (les) and control (C) rats was hybridized with32P-labeled probes specific either for one of the chimericles PCR products (left;Mbp exon 3–intron 3–ETn) or a 362 bpEcoRI fragment of the ETn element (right).
The size of the chimeric mRNA coupled with our inability to detect it with the MBP probe indicated that a substantial portion of the ETn element was present in this transcript. To confirm this hypothesis, we further analyzed RNA by Northern blots with a probe derived from a 0.36 kb EcoRI fragment located ∼1.7 kb downstream of the ETn LTR primer used in the initial blots (Fig. 6B,right). A single band of ∼7 kb was again observed inles, whereas no corresponding band was evident in controls. Similar results were observed when a second probe derived from a 0.36 kb PstI restriction fragment located ∼4.2 kb downstream of the ETn LTR primer was used (data not shown), confirming that ETn sequences constitute the majority of the aberrantly spliced MBP transcript.
DISCUSSION
The diverse group of spontaneous mutants known as the myelin mutants has proved to be invaluable in providing new insights into roles CNS myelin proteins play in oligodendrocyte development and myelination. Here, we identify a novel defect in a new myelin mutant that causes a more severe dysmyelinating phenotype (Kwiecien et al., 1998) than that observed in any of the other myelin mutants. Myelin deficits in the les rat result from the insertion of an endogenous retrotransposable element into a noncoding region of theMbp gene that disrupts normal splicing, leading to the synthesis of nonfunctional transcripts. Although studies directly linking the insertion with myelin deficits in les are forthcoming, evidence gathered thus far strongly supports the notion that the lack of myelin observed throughout the CNS of this mutant is caused by the retroviral-mediated inactivation of the MBP gene. First, the autosomal recessive manner in which the dysmyelinating phenotype is inherited and its complete penetrance in homozygous animals (les/les) indicate that a single defective gene is responsible. Second, the severity of the myelin deficit is consistent with the absence of an essential structural component of the myelin sheath. Third, comparable reductions in MBP levels have been shown in other mutants (shi andshimld) to produce a CNS-dysmyelinating phenotype similar to that observed in les, whereas myelination of the PNS remains relatively unaffected.
The identification of the genetic defect in the les rat is unique not only because of its effect on myelination but also because it represents the first evidence that the ETn family of retrotransposons exists in the rat genome. In addition, because these elements lack the ability to encode proteins required for transposition, its presence in the MBP gene supports previous studies demonstrating that these retrotransposons are capable of movement into new sites when supplied with the appropriate trans-acting factors. The ETn elements were first identified in mice (Brulet et al., 1983) in which several hundred copies are estimated to be dispersed throughout the murine genome. The identification of such a retroviral-like element in rat is not unexpected given that >10% of the mammalian genome is thought to contain a variety of proviruses that are related to infectious retroviruses (Keshet et al., 1998). The apparent sequence divergence between the rat and murine ETn elements is also not surprising given the obscure origin of retroviral-like elements that are hypothesized to be derived either from a retrovirus whose internal sequences degenerated or from the recombination between two retroviruses or solitary LTRs (Soniga et al., 1987).
Recent interest in retroviruses and related retrovirus-like elements has arisen because of their highly recombinant nature and potential to act as insertional mutagens. In this regard, the ability of ETn elements to alter drastically the expression of genes in which they have inserted has been well documented in several spontaneous murine mutants (Herrmann et al., 1990; Steinmeyer et al., 1991; Adachi et al., 1993; Mitreiter et al., 1994; Moon and Friedman, 1997). In many of these mutants, the ETn element has inserted into a noncoding region similar to what has occurred in the MBP gene of the les rat. Gene inactivation in these murine mutants has been shown to result from either aberrant splicing or truncation of mRNA mediated by cryptic splice acceptor sites or the single polyadenylation signal located within the LTR of the ETn element. The detection of improperly spliced MBP mRNA along with chimeric transcripts containing portions of the MBP gene fused to retrotransposon sequences confirms that the rat ETn element uses a similar mechanism to disrupt MBP expression inles. In addition, the fact that these chimeric transcripts are present at such abundant levels further indicates that the retrotransposon does not directly interfere with normal gene transcription but rather alters the processing of nascent MBP mRNA. Our ability to detect very low levels of appropriately spliced MBP mRNAs inles is consistent with previous in vitroexperiments demonstrating that although ETn elements frequently suppress the expression of genes in which they insert via missplicing, normal mRNA processing can occur, resulting in low but detectable levels of properly spliced transcripts (Adachi et al., 1993).
The ultimate use of the les rat and certainly all of the myelin mutants lies in their ability to help us better understand myelination and the proteins involved in this process. Although there is an extensive array of mutations of the Plp gene (Duncan, 1995), only one previous Mbp mutant, the shimouse, has been reported. In the case of the PLP mutants, the wide variety of missense mutations have a variable effect on myelination of the CNS, and these differing phenotypes have been instructive in examining the role of PLP/DM-20 in myelin formation. In a similar manner, the les rat has a novel mutation that produces a distinctly different phenotype from that of the shi mouse (Kwiecien et al., 1998), and this should provide additional insights into the role MBP plays in oligodendrocyte function. The most notable of these differences is the ability of oligodendrocytes in each of these mutants to generate a myelin sheath. In les, although oligodendrocytes are initially able to synthesize some myelin, they appear to lose this function quickly and become unable to initiate even primitive axonal ensheathment. By contrast, the increased numbers of myelinated axons observed in older shi mice (Inoue et al., 1983) indicate that oligodendrocytes from this mutant retain their ability to produce myelin and ensheath axons.
The inability of oligodendrocytes to myelinate axons in lesmay be related to dysfunctional forms of MBP generated from chimeric MBP transcripts. A similar phenotype is not observed in shibecause of the large deletion in the Mbp gene that essentially prevents any MBP mRNA from being synthesized. One possible explanation for how novel MBP isoforms generated in lescould disrupt myelination can be derived from in vitrostudies indicating that MBP may be involved in mediating extracellular signals that influence the stability of microtubular structures in membrane sheets (Dyer et al., 1994). Because the oligodendrocyte cytoskeleton has been shown to be important in the formation of membranous sheets in vitro (Dyer, 1993), the presence of novel MBP isoforms could modulate microtubule stability in mutant oligodendrocytes, altering their ability to ensheath and myelinate axons.
Another notable difference between les and shi is the pleiotropic effects each of the MBP mutations has on the expression of other myelin genes. In shi, levels of MBP mRNA are reduced to <5% of normal, whereas levels of PLP mRNA are diminished to 30–55% of normal (Sorg et al., 1987). The fact that we observed near normal expression of major myelin genes (Plp andCnp) in les despite a severe lack of myelin and dramatically reduced levels of MBP indicates that the molecular signal coordinating the expression of Mbp with other myelin genes remains intact in this mutant. The concept of coordinated myelin gene expression was first proposed based on the precise temporal manner in which the various myelin proteins are expressed in oligodendrocytes before the formation of the myelin sheath (Campagnoni, 1988; Lemke, 1992). The identification of various Plp and Mbpmutants that exhibited pleiotropic changes in myelin gene expression further supported the hypothesis of coordinated gene expression. More recent studies showing that putative regulatory elements in promoter regions of the Plp, Mbp, and Cnp genes contain similar DNA sequences provide additional molecular evidence of coordinated regulation (Asipu and Blair, 1994). Thus, in spite of its severe dysmyelination, the les rat may actually provide a model system to identify factors involved in coordinating myelin gene expression.
The identity of potential coordination factors has recently become the object of much speculation. One possible candidate under current investigation is MBP. Detection of low levels of MBP in oligodendrocytes before the appearance of myelin sheaths (Sternberger et al., 1978; Dubois-Dalcq et al., 1986; Monge et al., 1986) provided the first evidence that MBP isoforms are not functionally equivalent and thus might perform functions other than the well characterized role in myelin compaction. Studies demonstrating that relative levels of different MBP isoforms change during development (Carson et al., 1983;Mathisen et al., 1993) and are differentially localized within subcellular domains of the oligodendrocyte further support the hypothesis that certain isoforms could serve as an initiator and/or regulator of myelin gene expression. More recent studies demonstrating that MBP isoforms can be detected in the oligodendrocyte cytoplasm and nucleus both before axonal ensheathment and after myelination provide additional evidence in support of MBP isoforms having regulatory functions (Hardy et al., 1996). Although alternative functions have not been assigned to any MBP isoforms, emerging evidence indicates that exon 2 may provide a nuclear localization signal that enables MBP to be actively targeted to the nucleus (Alliquant et al., 1991; Pedraza, 1997). Our ability to identify low but detectable levels of normal MBP transcripts in les argues for the possibility that adequate levels of MBP are being translated and transported into the nucleus where they can initiate myelination and maintain the continued expression of myelin genes in this mutant. Alternatively, chimeric MBP transcripts identified in les could retain this function because they contain proximal Mbp exons. Although we have identified chimeric transcripts containing Mbp exons 1 and 3, studies are currently underway to determine whether transcripts are present that contain exon 2 that would facilitate transport of a putative regulatory protein into the nucleus. Although evidence linking MBP with the coordination of myelin gene expression remains indirect, the novel phenotype observed in lesfurther implicates that MBP is somehow involved in this process.
The question remains of course whether chimeric mRNAs detected inles are actually translated into proteins. The monoclonal antibody used in this study to identify MBP was specific for residues encoded by exon 5, and because this exon does not appear to be present in chimeric transcripts, we would not expect it to recognize the corresponding proteins. Further studies are underway using antibodies that recognize residues in MBP exons 1–3 (Hardy et al., 1996) to help resolve this question. Our inability to detect MBP immunohistochemically raises the additional question of whether MBP mRNA may actually serve as the signal that regulates myelin gene transcription. Consistent with this hypothesis, certain MBP transcripts have been detected in the murine embryonic CNS that were shown not to be translated into peptides (Mathisen et al., 1993). Recent studies identifying transcription factors in murine brain extracts that can complex with RNA and bind to cis elements in promoters of myelin genes (Haque et al., 1995) could provide an appropriate mediator by which MBP mRNA could directly influence the expression of myelin genes.
Although both les and shi have provided conclusive evidence of MBP’s role in compaction of the myelin sheath in the CNS, the identification in les of chimeric MBP transcripts capable of encoding exons previously associated with nuclear localization suggests that this mutant should be useful in further exploring the role MBP plays in oligodendrocyte function and myelinogenesis.
Footnotes
This study was supported by the National Multiple Sclerosis Society (NMSS) Grant RG2927-A7. J.M.K. was the recipient of an NMSS postdoctoral fellowship. We are grateful to Dr. Ian Griffiths for critical review of this paper. We acknowledge the excellent technical assistance of Randy Hoffman and Cynthia Cutshall.
The sequences of both wt Mbp intron 3 and that containing the ETn element in les have been deposited with EMBL/GenBank data libraries under accession numbers AF075578 andAF076337, respectively.
Correspondence should be addressed to Dr. Ian D. Duncan, University of Wisconsin, School of Veterinary Medicine, 2015 Linden Drive West, Madison, WI 53706.
Dr. O’Connor’s present address: The Women’s Health Research Institute, Wyeth-Ayerst Research, 145 King of Prussia Road, Radnor, PA 19087.
Dr. Kwiecien’s present address: Central Animal Facility, Faculty of Health Sciences, McMaster University, 1200 Main Street West, Hamilton, Ontario L8N 3Z5, Canada.
REFERENCES
- 1.Adachi M, Watanabe-Fukunaga R, Nagata S. Aberrant transcription caused by the insertion of an early transposable element in an intron of the Fas antigen gene of lpr mice. Proc Natl Acad Sci USA. 1993;90:1756–1760. doi: 10.1073/pnas.90.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alliquant B, Staugaitis SM, D’Urso D, Colman DR. The ectopic expression of myelin basic protein isoforms in Shiverer oligodendrocytes: implications for myelinogenesis. J Cell Biol. 1991;113:393–403. doi: 10.1083/jcb.113.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asipu A, Blair GE. Regulation of myelin basic protein-encoding gene transcription in rat oligodendrocytes. Gene. 1994;150:227–234. doi: 10.1016/0378-1119(94)90431-6. [DOI] [PubMed] [Google Scholar]
- 4.Barbarese E, Barry C. Expression and localization of myelin basic protein in oligodendrocytes and transfected fibroblasts. J Neurochem. 1988;51:1737–1745. doi: 10.1111/j.1471-4159.1988.tb01153.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernier L, Alvarez F, Norgard E, Raible D, Mentaberry A, Schembri J, Sabatini D, Colman D. Molecular cloning of a 2′,3′-cyclic nucleotide 3′-phosphodiesterase: mRNAs with different 5′ ends encode the same set of proteins in nervous and lymphoid tissues. J Neurosci. 1987;7:2703–2710. doi: 10.1523/JNEUROSCI.07-09-02703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biddle F, March E, Miller JR. Research news. Mouse News Lett. 1973;48:24. [Google Scholar]
- 7.Bird TD, Farrell DF, Sumi SM. Brain lipid composition of the shiverer mouse (genetic defect in myelin development). J Neurochem. 1978;31:387–391. doi: 10.1111/j.1471-4159.1978.tb12479.x. [DOI] [PubMed] [Google Scholar]
- 8.Boison D, Büssow H, D’Urso D, Müller H-W, Stoffel W. Adhesive properties of proteolipid protein are responsible for the compaction of CNS myelin sheaths. J Neurosci. 1995;15:5502–5513. doi: 10.1523/JNEUROSCI.15-08-05502.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brophy PJ, Boccaccio GL, Colman DR. The distribution of myelin basic protein mRNAs within myelinating oligodendrocytes. Trends Neurosci. 1993;16:515–521. doi: 10.1016/0166-2236(93)90196-s. [DOI] [PubMed] [Google Scholar]
- 10.Brulet P, Kaghad M, Xu Y, Croissant O, Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci USA. 1983;80:5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. J Neurochem. 1988;51:1–14. doi: 10.1111/j.1471-4159.1988.tb04827.x. [DOI] [PubMed] [Google Scholar]
- 12.Campagnoni AT, Macklin WB. Cellular and molecular aspects of myelin protein gene expression. Mol Neurobiol. 1988;2:41–90. doi: 10.1007/BF02935632. [DOI] [PubMed] [Google Scholar]
- 13.Carson JH, Nielson ML, Barbarese E. Developmental regulation of myelin basic protein expression in mouse brain. Dev Biol. 1983;96:485–492. doi: 10.1016/0012-1606(83)90185-9. [DOI] [PubMed] [Google Scholar]
- 14.Chernoff GF. Schiverer: an autosomal recessive mutant mouse with myelin deficiency. J Hered. 1981;72:128. doi: 10.1093/oxfordjournals.jhered.a109442. [DOI] [PubMed] [Google Scholar]
- 15.Danielson PE, Forss-Petter S, Brow MA, Calavetta L, Douglass J, Milner RJ, Sutcliffe JG. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 16.De Ferra F, Engh H, Hudson L, Kamholz J, Puckett C, Molineaux S, Lazzarini RA. Alternative splicing accounts for the four forms of myelin basic protein. Cell. 1985;43:721–727. doi: 10.1016/0092-8674(85)90245-4. [DOI] [PubMed] [Google Scholar]
- 17.Delaney KH, Kwiecien J, Wegiel J, Wisniewski HM, Percy DH, Fletch AL. Familial dysmyelination in a Long Evans rat mutant. Lab Anim Sci. 1995;45:547–553. [PubMed] [Google Scholar]
- 18.Doolittle DP, Schweikart KM. Myelin deficient, a new neurological mutant in the mouse. J Hered. 1977;68:331–332. doi: 10.1093/oxfordjournals.jhered.a108850. [DOI] [PubMed] [Google Scholar]
- 19.Dubois-Dalcq M, Behar TN, Hudson L, Lazzarini RA. Emergence of three myelin proteins in oligodendrocytes cultured without neurons. J Cell Biol. 1986;102:384–392. doi: 10.1083/jcb.102.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan ID. Inherited disorders of myelination of the central nervous system. In: Ransom BR, Kettenmann HR, editors. Neuroglial cells. Oxford UP; New York: 1995. pp. 990–1009. [Google Scholar]
- 21.Duncan ID, Lunn KF, Holmgren B, Urba-Holmgren R, Brignolo-Holmes L. The taiep rat: a myelin mutant with an associated oligodendrocyte microtubular defect. J Neurocytol. 1992;21:870–884. doi: 10.1007/BF01191684. [DOI] [PubMed] [Google Scholar]
- 22.Dupouey P, Jacque C, Bourre JM, Cesselin F, Privat A, Baumann N. Immunochemical studies of myelin basic protein in shiverer mouse devoid of major dense line of myelin. Neurosci Lett. 1979;12:113–118. doi: 10.1016/0304-3940(79)91490-3. [DOI] [PubMed] [Google Scholar]
- 23.Dyer CA. Novel oligodendrocyte transmembrane signaling systems. Mol Neurobiol. 1993;7:1–22. doi: 10.1007/BF02780606. [DOI] [PubMed] [Google Scholar]
- 24.Dyer CA, Philibotte TM, Wolf MK, Billings-Gagliardi S. Myelin basic protein mediates extracellular signals that regulate microtubule stability in oligodendrocyte membrane sheets. J Neurosci Res. 1994;39:97–107. doi: 10.1002/jnr.490390112. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;32:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths IR. Myelin mutants: model systems for the study of normal and abnormal myelination. Bioessays. 1996;18:789–797. doi: 10.1002/bies.950181005. [DOI] [PubMed] [Google Scholar]
- 27.Haque NS, Buchberg AM, Khalili K. Isolation and characterization of MRF-1, a brain-derived DNA-binding protein with a capacity to regulate expression of myelin basic protein gene. J Biol Chem. 1995;269:31149–31156. [PubMed] [Google Scholar]
- 28.Hardy RJ. Molecular defects in the dysmyelinating mutant quaking. J Neurosci Res. 1998;51:417–422. doi: 10.1002/(SICI)1097-4547(19980215)51:4<417::AID-JNR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 29.Hardy RJ, Lazzarini RA, Colman DR, Friedrich VL., Jr Cytoplasmic and nuclear localization of myelin basic proteins reveals heterogeneity among oligodendrocytes. J Neurosci Res. 1996;46:246–257. doi: 10.1002/(SICI)1097-4547(19961015)46:2<246::AID-JNR13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 31.Holmgren B, Urba-Holmgren R, Riboni L, Vega-SaenzdeMiera EC. Sprague Dawley rat mutant with tremor, ataxia, tonic immobility episodes, epilepsy and paralysis. Lab Anim Sci. 1989;39:226–228. [PubMed] [Google Scholar]
- 32.Holz A, Schwab ME. Developmental expression of the myelin gene MOBP in the rat nervous system. J Neurocytol. 1997;26:467–477. doi: 10.1023/a:1018529323734. [DOI] [PubMed] [Google Scholar]
- 33.Hudson LD. Molecular biology of myelin proteins in the central and peripheral nervous systems. The Neurosciences. 1990;2:483–496. [Google Scholar]
- 34.Hudson LD, Berndt JA, Puckett C, Kozak CA, Lazzarini RA. Aberrant splicing of proteolipid protein mRNA in the dysmyelinating jimpy mutant mouse. Proc Natl Acad Sci USA. 1987;84:1454–1458. doi: 10.1073/pnas.84.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue Y, Inoue K, Terashima T, Mikoshiba K, Tsukada Y. Developmental changes of oligodendroglia in the posterior funiculus of “shiverer” mutant mouse spinal cord, with special reference to myelin formation. Anat Embryol (Berl) 1983;168:159–171. doi: 10.1007/BF00315814. [DOI] [PubMed] [Google Scholar]
- 36.Kaatz KW, Bazzett TJ, Albin RL. A new, simple myelin stain. Brain Res Bull. 1992;29:697–698. doi: 10.1016/0361-9230(92)90141-j. [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki ES. Amplification of RNA. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic; San Diego: 1990. pp. 21–27. [Google Scholar]
- 38.Keshet E, Schiff R, Itin A. Mouse retrotransposons: a cellular reservoir of long terminal repeat (LTR) elements with diverse transcriptional specificities. Adv Cancer Res. 1998;56:215–251. doi: 10.1016/s0065-230x(08)60482-0. [DOI] [PubMed] [Google Scholar]
- 39.Kirschner D, Ganser AL. Compact myelin exists in the absence of basic protein in the shiverer mutant mouse. Nature. 1980;283:207–209. doi: 10.1038/283207a0. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S. Differential regulation of oligodendrocyte markers by glucocorticoids: post-transcriptional regulation of both proteolipid protein and transcriptional regulation of glycerol phosphate dehydrogenase. Proc Natl Acad Sci USA. 1989;86:6807–6811. doi: 10.1073/pnas.86.17.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwiecien JM, O’Connor LT, Goetz BD, Delaney KH, Fletch AL, Duncan ID. Morphological and morphometric studies of the dysmyelinating mutant, the Long Evans shaker rat. J Neurocytol. 1998;27:581–591. doi: 10.1023/a:1006922227791. [DOI] [PubMed] [Google Scholar]
- 42.Lemke G. Unwrapping the genes of myelin. Neuron. 1988;1:535–543. doi: 10.1016/0896-6273(88)90103-1. [DOI] [PubMed] [Google Scholar]
- 43.Lemke G. Myelin and myelination. In: Hall ZW, editor. An introduction to molecular neurobiology. Sinauer; Sunderland, MA: 1992. pp. 281–309. [Google Scholar]
- 44.Lunn KF, Fanarraga ML, Duncan ID. Myelin mutants: new models and new observations. Microsc Res Tech. 1995;32:183–203. doi: 10.1002/jemt.1070320303. [DOI] [PubMed] [Google Scholar]
- 45.Mathisen PM, Pease S, Garvey J, Hood L, Readhead C. Identification of an embryonic isoform of myelin basic protein that is expressed widely in the mouse embryo. Proc Natl Acad Sci USA. 1993;90:10125–10129. doi: 10.1073/pnas.90.21.10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mentaberry A, Adesnik M. Small basic proteins of myelin from central and peripheral nervous systems are encoded by the same gene. Proc Natl Acad Sci USA. 1986;83:1111–1114. doi: 10.1073/pnas.83.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitreiter K, Schmidt J, Luz A, Atkinson MJ, Hofler H, Erfle V, Strauss PG. Disruption of the murine p53 gene by insertion of an endogenous retrovirus-like element (ETn) in a cell line from radiation induced osteosarcoma. Virology. 1994;200:837–841. doi: 10.1006/viro.1994.1253. [DOI] [PubMed] [Google Scholar]
- 48.Molineaux SM, Engh H, de Ferra F, Hudson L, Lazzarini RA. Recombination within the myelin basic protein gene created the dysmyelinating shiverer mouse mutation. Proc Natl Acad Sci USA. 1986;83:7542–7546. doi: 10.1073/pnas.83.19.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monge M, Kadiiski D, Jacque CM, Zalc B. Oligodendroglial expression and deposition of four major myelin constituents in the myelin sheath during development. Dev Neurosci. 1986;8:222–235. doi: 10.1159/000112255. [DOI] [PubMed] [Google Scholar]
- 50.Moon BC, Friedman JM. The molecular basis of the obese mutation in ob2J mice. Genomics. 1997;42:152–156. doi: 10.1006/geno.1997.4701. [DOI] [PubMed] [Google Scholar]
- 51.Newman SL, Kitamura K, Campagnoni AT. Identification of a cDNA coding for a fifth form of myelin basic protein in mouse. Proc Natl Acad Sci USA. 1987;84:886–890. doi: 10.1073/pnas.84.3.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okano H, Aruga J, Nakagawa T, Shiota C, Mikoshiba K. Myelin basic protein gene and the function of antisense RNA in its repression in myelin-deficient mutant mouse. J Neurochem. 1991;56:560–567. doi: 10.1111/j.1471-4159.1991.tb08186.x. [DOI] [PubMed] [Google Scholar]
- 53.Pedraza L. Nuclear transport of myelin basic protein. J Neurosci Res. 1997;50:258–264. doi: 10.1002/(SICI)1097-4547(19971015)50:2<258::AID-JNR14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 54.Peterson AC, Bray GM. Hypomyelination in the peripheral nervous system of shiverer mice and in shiverer-normal chimera. J Comp Neurol. 1984;227:348–356. doi: 10.1002/cne.902270305. [DOI] [PubMed] [Google Scholar]
- 55.Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3:191–197. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- 56.Privat A, Jacque C, Bourre JM, Dupouey P, Baumann N. Absence of the major dense line in myelin of the mutant mouse shiverer. Neurosci Lett. 1979;12:107–112. doi: 10.1016/0304-3940(79)91489-7. [DOI] [PubMed] [Google Scholar]
- 57.Roach A, Takahashi N. Chromosomal mapping of mouse myelin protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- 58.Roach A, Boylan K, Horvath S, Prusiner SB, Hood LE. Characterization of cloned cDNA representing rat myelin basic protein: absence of expression in brain of shiverer mutant mice. Cell. 1983;34:799–806. doi: 10.1016/0092-8674(83)90536-6. [DOI] [PubMed] [Google Scholar]
- 59.Rosenbluth J. Peripheral myelin in the mouse mutant shiverer. J Comp Neurol. 1980;193:729–739. doi: 10.1002/cne.901930310. [DOI] [PubMed] [Google Scholar]
- 60.Scherer SS. Molecular genetics of demyelination: new wrinkles on an old membrane. Neuron. 1997;18:13–16. doi: 10.1016/s0896-6273(01)80042-8. [DOI] [PubMed] [Google Scholar]
- 61.Shell B, Collins JT, Elenich LA, Szurek PF, Dunnick WA. Two subfamilies of murine retrotransposon ETn sequences. Gene. 1990;86:269–274. doi: 10.1016/0378-1119(90)90289-4. [DOI] [PubMed] [Google Scholar]
- 62.Soniga P, Wain-Hobson S, Bougueleret L, Tiollais P, Jacob F, Brulet P. Nucleotide sequence and evolution of ETn elements. Proc Natl Acad Sci USA. 1987;84:3768–3771. doi: 10.1073/pnas.84.11.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sorg BA, Smith MM, Campagnoni AT. Developmental expression of the myelin proteolipid protein and basic protein mRNAs in normal and dysmyelinating mutant mice. J Neurochem. 1987;49:1146–1153. doi: 10.1111/j.1471-4159.1987.tb10005.x. [DOI] [PubMed] [Google Scholar]
- 64.Steinmeyer K, Klocke R, Ortland C, Gronemeier M, Jockusch H, Grunder S, Jentsch TJ. Inactivation of muscle chloride channel by transposon insertion in myotonic mice. Nature. 1991;354:304–308. doi: 10.1038/354304a0. [DOI] [PubMed] [Google Scholar]
- 65.Sternberger NH, Itoyama Y, Kies W, Webster Hd. Immunocytochemical method to identify basic protein in myelin-forming oligodendrocytes of newborn rat CNS. J Neurocytol. 1978;7:251–263. doi: 10.1007/BF01217922. [DOI] [PubMed] [Google Scholar]
- 66.Trapp BD, Moench T, Pulley M, Barbosa E, Tennekoon G, Griffin JW. Spatial segregation of mRNA encoding myelin-specific proteins. Proc Natl Acad Sci USA. 1987;84:7773–7777. doi: 10.1073/pnas.84.21.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verdi JM, Campagnoni AT. Translational regulation by steroids. Identification of a steroid modulatory element in the 5′-untranslated region of the myelin basic protein messenger RNA. J Biol Chem. 1990;265:20314–20320. [PubMed] [Google Scholar]