Abstract
Interleukin (IL)-16 is a proinflammatory cytokine that has attracted widespread attention because of its ability to block HIV replication. We describe the identification and characterization of a large neuronal IL-16 precursor, NIL-16. The N-terminal half of NIL-16 constitutes a novel PDZ domain protein sequence, whereas the C terminus is identical with splenocyte-derived mouse pro-IL-16. IL-16 has been characterized only in the immune system, and the identification of NIL-16 marks a previously unsuspected connection between the immune and the nervous systems. NIL-16 is a cytosolic protein that is detected only in neurons of the cerebellum and the hippocampus. The N-terminal portion of NIL-16 interacts selectively with a variety of neuronal ion channels, which is similar to the function of many other PDZ domain proteins that serve as intracellular scaffolding proteins. Among the NIL-16-interacting proteins is the class C α1 subunit of a mouse brain calcium channel (mbC α1). The C terminus of NIL-16 can be processed by caspase-3, resulting in the release of secreted IL-16. Furthermore, in cultured cerebellar granule neurons undergoing apoptosis, NIL-16 proteolysis parallels caspase-3 activation. Cerebellar granule neurons express the IL-16 receptor CD4. Exposure of these cells to IL-16 induces expression of the immediate-early gene,c-fos, via a signaling pathway that involves tyrosine phosphorylation. This suggests that IL-16 provides an autocrine function in the brain. Therefore, we hypothesize that NIL-16 is a dual function protein in the nervous system that serves as a secreted signaling molecule as well as a scaffolding protein.
Keywords: IL-16, PDZ, caspase-3, cerebellar granule neurons, hippocampus, c-fos
Interleukin (IL)-16 (also known as lymphocyte chemoattractant factor; Cruikshank et al., 1994) is a T-cell-derived cytokine with pleiotropic, pro-inflammatory effects in the immune system (for review, see Cruikshank et al., 1998). IL-16 has gained considerable attention recently because of its ability to inhibit the replication of the human immunodeficiency virus (HIV) in infected T-cells by interfering with HIV mRNA expression (Baier et al., 1995; Zhou et al., 1997). IL-16 is a 121-amino-acid peptide secreted by T-lymphocytes. It is derived by proteolytical cleavage of an 80 kDa precursor molecule, pro-IL-16 (Baier et al., 1997). In vitro, the processing of pro-IL-16 to yield IL-16 is catalyzed by caspase-3 (Zhang et al., 1998). This cleavage reaction is reminiscent of the processing of pro-IL-1 by caspase-1 (Black et al., 1988; Kostura et al., 1989).
IL-16 exerts its effects via binding to a cell surface receptor, CD4 (Cruikshank et al., 1994; Center et al., 1996). CD4 was described originally as a protein expressed primarily in T-cells, where it is involved in antigen recognition in the context of class II major histocompatibility molecules (for review, see Parnes, 1989). Subsequently, CD4 expression was described in neurons, glia, and microglia throughout the brain (Funke et al., 1987; Perry and Gordon, 1987; Peudenier et al., 1991; Omri et al., 1994). Pyramidal neurons of the hippocampus as well as granule neurons of the dentate gyrus, the cerebellum, and the olfactory bulb display particularly high levels of CD4 expression (Omri et al., 1994). However, to date the CD4 ligand IL-16 has been detected only in the immune system (Chupp et al., 1998;Keane et al., 1998). Here we report the identification and characterization of a neuronal variant of IL-16, NIL-16.
Although the C-terminal region of NIL-16 is identical to splenocyte-derived mouse pro-IL-16 (Keane et al., 1998), the N-terminal portion of the molecule constitutes a novel protein sequence. NIL-16 is a cytosolic multi-PDZ domain protein that is expressed postnatally and throughout adulthood. Its mRNA is localized to neurons of the cerebellum and the hippocampus. The N terminus of NIL-16 physically interacted with the cytoplasmic domains of a variety of neuronal ion channels. Moreover, NIL-16 could be coimmunoprecipitated with the NMDA receptor subunit 2A (NR2A) from transfected cells. Therefore, we speculate that NIL-16 may be involved in the targeting and clustering of neurotransmitter receptors, which would be similar to the function of several other PDZ domain proteins (for review, seePonting et al., 1997; Sheng and Wyszynski, 1997). In contrast, the C terminus of NIL-16 could be proteolysed by caspase-3, which resulted in the release of secreted mature IL-16. In cultured cerebellar granule neurons (CGN) induced to undergo apoptosis, NIL-16 proteolysis was associated with caspase-3 activation. Moreover, IL-16 treatment of cultured CGN resulted in the induction of the immediate-early gene,c-fos, indicating that IL-16 is capable of initiating a signaling cascade in neurons. Therefore, NIL-16 may have dual functions in the nervous system, serving as a signaling molecule that is secreted after caspase-3 induction and as a cytosolic scaffolding protein that anchors ion channels in the membrane.
MATERIALS AND METHODS
Molecular cloning and sequence analysis of NIL-16. An artificial PDZ domain binding peptide containing the C-terminal sequence V-S-D-L was used as bait in a yeast two-hybrid screen of a mouse cerebellum cDNA library (Kurschner and Morgan, 1995), as described elsewhere (Kurschner et al., 1998). A partial cDNA of 925 bp in length was isolated in the screen. Full-length cDNA was obtained by DNA hybridization as described (Kurschner and Morgan, 1995) from a mouse cerebellum cDNA library in λZAP II (Kurschner and Morgan, 1995). The original cDNA isolate served as a probe. Analysis of DNA sequencing data was performed with the GCG Sequence Analysis Software Package (Version 9.1-UNIX) of the Genetics Computer Group (Madison, WI). Advanced BLAST searches with NIL-16 protein sequences were done via the National Center for Biotechnology Information BLAST server atwww.ncbi.nlm.nih.gov/BLAST.
Analysis of NIL-16 mRNA tissue distribution. Northern blot analysis was performed with 4 μg of total RNA from different mouse tissues. A cDNA fragment corresponding to NIL-16 codons 1058–1195 was random 32P-labeled, using the Oligolabeling Kit (Amersham Pharmacia Biotech, Piscataway, NJ) and served as a probe. In situ hybridization analysis was performed as described (Simmons et al., 1989) with 12-μm-thick sagittal cryosections of mouse brain. A33P-labeled riboprobe corresponding to NIL-16 codons 1058–1195 was used. Hybridization was at 60°C. After hybridization the sections were dehydrated and mounted with Permount (Fisher Scientific, Pittsburgh, PA) (for photomicrograph in Fig.2D). Alternatively, dehydration and mounting were preceded by counterstaining with 0.1% toluidine blue in H2O (for photomicrographs in Fig. 3).
Fig. 2.
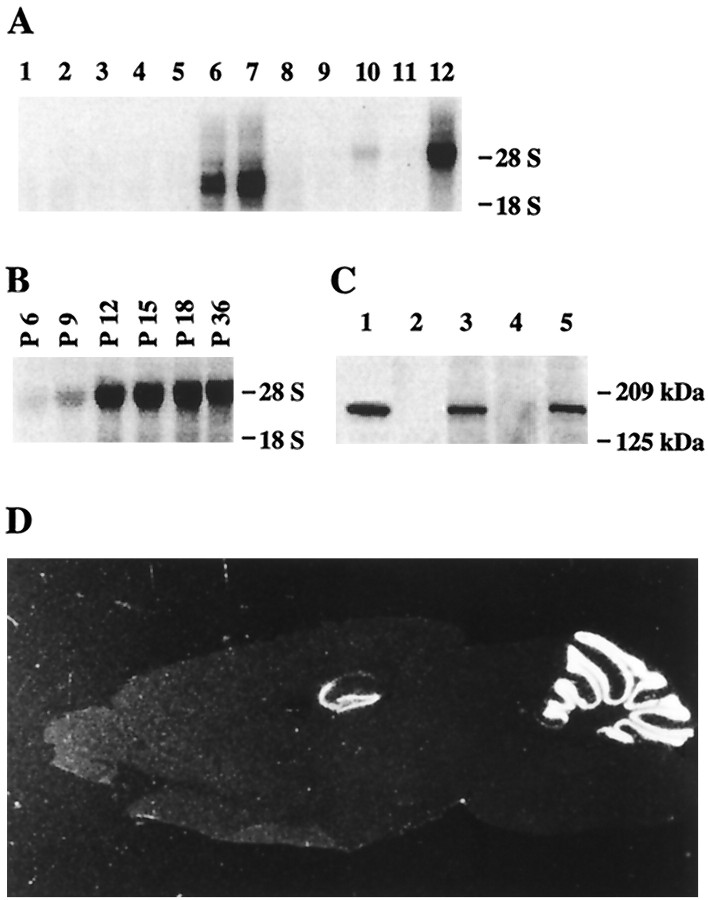
Tissue distribution of NIL-16. A, Northern blot analysis of mouse tissue. RNA samples were electrophoresed in an agarose gel, blotted onto a nylon membrane, and hybridized with a cDNA probe for NIL-16 codons 1058–1195. Lane 1, Skeletal muscle; lane 2, stomach; lane 3, liver; lane 4, heart; lane 5, testis; lane 6, spleen; lane 7, thymus;lane 8, lung; lane 9, kidney; lane 10, total brain; lane 11, brain without cerebellum; lane 12, cerebellum. The position of 28S and 18S ribosomal RNA bands are indicated on the right.B, Northern blot analysis of mouse cerebellum at different ages. Northern analysis was as described in A. The different ages are indicated above the respective lanes. C, Western analysis of NIL-16 in different brain regions. Lysates were prepared from P18 mouse brain. Brain lysate proteins were loaded at 100 μg per lane. An antibody specific for IL-16 was used for detection. Lane 1, Extract (3 μl) of NIL-16-transfected COS-7 cells (for size control); lane 2, eye; lane 3, cerebellum; lane 4, cortex; lane 5, hippocampus. Positions of molecular weight markers are indicated on the right.D, In situ hybridization analysis of a sagittal cryosection of adult mouse whole brain with a cRNA antisense-probe for NIL-16 codons 1058–1195. The analysis detected NIL-16 mRNA exclusively in the hippocampus and in the cerebellum.
Fig. 3.
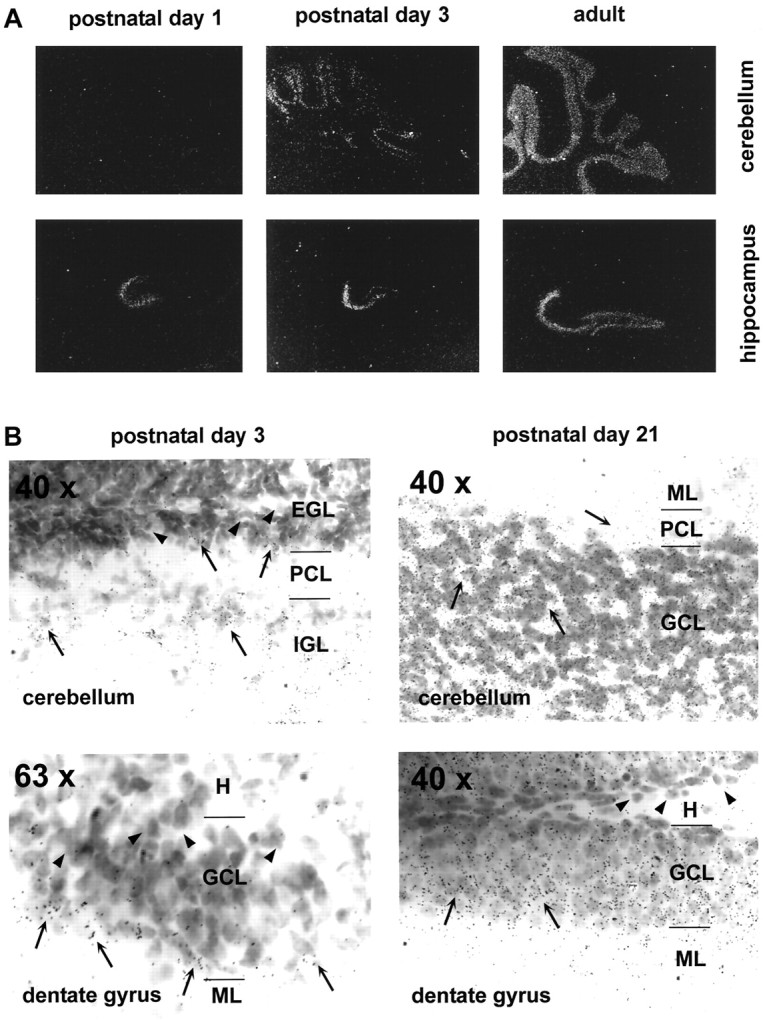
Expression pattern of NIL-16 during postnatal development of cerebellum and hippocampus. In situ hybridization analysis was performed as described in Figure 2D, with sagittal cryosections of P1, P3, P21, and adult brain, respectively. A, Dark-field photomicrographs of P1, P3, and adult (as indicatedabove the panels) cerebellum and hippocampus (labeled at the right of the panels) taken with a 10× magnification objective. B, Bright-field photomicrographs of P3 and P21 (indicated above the panels) cerebellum and dentate gyrus (labeled at the bottom left corner in the panels). Magnification is indicated at the top left corner in the panels. Arrowheads point to NIL-16-negative cells;arrows mark NIL-16-positive cells. NIL-16 expression was found predominantly in postmitotic neurons, whereas NIL-16-negative cells were found mainly in regions containing undifferentiated and proliferative cells. EGL, External granular layer;GCL, granule cell layer; H, hilus;IGL, internal granular layer; ML, molecular layer; PCL, Purkinje cell layer.
Preparation and culture of mouse cerebellar granule neurons.Primary cultures of CGN were prepared from postnatal day 7 (P7) cerebellum. Cerebellar explants were incubated for 5 min in 1 ml of solution I [HBSS, 0.3% bovine serum albumin, and (in mm) 14 glucose, 15 HEPES, 4.2 NaHCO3, and 1.5 MgSO4·7 H2O; all reagents were obtained from Sigma, St. Louis, MO] containing 1% trypsin (Sigma). Subsequently, the explants were washed with 4 ml of solution I, supplemented with 0.04% DNaseI (Sigma, catalog number D5025), 0.25% trypsin inhibitor (Sigma, catalog number T9003), and 1.5 mm MgSO4·7 H2O (Sigma) and then were transferred to 1 ml of the same solution. Cells were dissociated by trituration and seeded at a density of 2 × 105CGN/cm2 on 60 mm culture dishes (catalog number 25010, Corning Costar, Cambridge, MA) coated with poly-l-lysine (Sigma). CGN were maintained in Neurobasal medium that was supplemented with B27, penicillin/streptomycin, and 0.5 mm glutamine (all from Life Technologies, Gaithersburg, MD) and that contained 25 mm KCl.
To induce apoptosis, we replaced the medium with Neurobasal medium, supplemented as above, but containing only 5 mmKCl. For the inhibition of caspase-3 activation the medium was supplemented with 30 μmZ-Val-Ala-Asp(OMe)-CH2F (Z-VAD; Enzyme Systems Products, Livermore, CA).
Cultures prepared for the exposure of CGN to exogenous IL-16 were seeded in 60 mm dishes with medium containing 5 mm KCl for 2 d. Subsequently, the medium was replaced with fresh medium containing 0.1 μm recombinant mouse IL-16 (PharMingen, San Diego, CA). In experiments involving inhibitors of signal transduction pathways, the culture medium was removed 2 d after seeding and replaced with 1.7 ml of fresh medium or medium containing one of the following inhibitors: (1) 100 μm7-nitroindazole plus 100 μmNG-nitro-l-arginine methyl ester dihydrochloride (both from Research Biochemicals, Natick, MA); (2) 30 μm herbimycin A (Biomol, Plymouth Meeting, PA); (3) 100 μm PD58059 (Calbiochem, San Diego, CA); (4) 20 μm SB202190 (Calbiochem). Cultures were incubated for 3 hr before the addition of 400 μl of medium containing the same inhibitors and 3.6 μg of IL-16 (0.1 μm final IL-16 concentration). Incubation was continued for 1 hr. Subsequently, protein extracts were prepared as described below.
Immunofluorescence staining of mouse cerebellar granule neurons.Immunofluorescence staining was performed on CGN grown on coverslips for 7 d in medium containing 25 mmK+. Cells were fixed with 4% paraformaldehyde in 0.1 m phosphate buffer for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with 1 mg/ml bovine serum albumin for 60 min. They were incubated with a mouse monoclonal antibody against IL-16 (clone 14.1; PharMingen) at 1:100 dilution and a rabbit polyclonal antibody against NR2C (catalog number A-6475; Molecular Probes, Eugene, OR) at 1:500 dilution for 60 min. Antibody binding was visualized by an anti-mouse antibody conjugated with Alexia 546 and an anti-rabbit antibody conjugated with Alexia 488 (both from Molecular Probes). Samples without the addition of each primary antibody were used as negative controls for the nonspecific binding and the cross-activation of fluoroprobes.
Protein preparations and Western blot analysis. For the characterization of NIL-16 protein in various brain regions, tissue lysates from P18 mice were prepared from eye, cerebellum, cortex, and hippocampus, respectively. Fresh frozen tissues were homogenized in cold tissue lysis buffer [containing (in mm) 50 Tris-HCl, pH 7.5, 250 NaCl, and 1 EDTA plus 0.1% NP-40 and 20% glycerol], using a Dounce homogenizer with a loose pestle. Lysates were cleared by centrifugation in a 5415C microcentrifuge (Eppendorf, Hamburg, Germany) at 13,000 rpm for 15 min. Potein samples (100 μg) were separated by SDS-PAGE on a 7.5% gel. For size comparison, 3 μl of an extract prepared from NIL-16-transfected COS-7 cells (see below) was included in the analysis. Proteins subsequently were electroblotted onto an Immobilon-P membrane (Millipore, Bedford, MA), and NIL-16 immunoreactivity was detected by Western analysis as described [Bonifacino et al. (1998), chapter 6.2], with the anti-IL-16 antibody 14.1 (PharMingen) serving as the primary antibody. The ECL Western blotting analysis system (Amersham Pharmacia Biotech) was used for the visualization of immunoreactive protein bands.
For protein analysis in CGN undergoing apoptosis, CGN were prepared as described above and cultured for 8 d in medium containing 25 mm K+. Apoptosis was induced by switching to a culture medium containing 5 mmK+ in the presence or absence of Z-VAD (see above). At various times after the medium switch, cells (two 60 mm dishes of CGN per time point) were detached by scraping and were lysed for 10 min on ice in 150 μl of cell lysis buffer [containing (in mm) 50 HEPES, pH 7.5, 150 NaCl, and 1 EDTA plus 0.5% sodium deoxycholate supplemented with Complete protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN)]. Lysates were cleared as described above. The 20 μg lysate proteins were separated by SDS-PAGE on a 7.5% gel and subjected to Western analysis with the anti-IL-16 antibody, 14.1, as described above. In parallel, 20 μg protein samples were separated by SDS-PAGE on a 4–20% gradient gel and analyzed by Western blot, using a rabbit polyclonal anti-caspase-3 antibody (PharMingen, catalog number 67341A), which recognizes the inactive nonprocessed form of caspase-3 (p32) as well as the 17 kDa subunit of active caspase-3 (p17).
For the detection of Fos and Jun expression in CGN exposed to exogenous IL-16, the CGN culture medium was changed and replaced with a medium containing no IL-16 or 0.1 μm recombinant mouse IL-16. Cells were lysed at various times after the medium change. The 50 μg lysate proteins were separated by SDS-PAGE on an 8% gel. Fos was detected with the rabbit polyclonal antibody, Fos2.2 (Miao and Curran, 1994) (gift from Dr. Tom Curran, St. Jude Children’s Research Hospital, Memphis, TN). Jun was detected with the rabbit polyclonal antibody anti-c-Jun/AP-1 (Ab-1; Calbiochem). An NIH-3T3 cell extract (New England Biolabs, Beverly, MA) served as a control for Jun. The control for phosphorylated Jun was a UV-irradiated NIH-3T3 cell extract (New England Biolabs).
Yeast two-hybrid analysis. Yeast two-hybrid analysis (Fields and Song, 1989) was performed in the Saccharomyces cerevisiae strain, S260 (gift from Dr. Steve Dalton, University of Adelaide, South Australia, Australia), which contains a LacZreporter gene integrated into its genome (Kurschner and Morgan, 1996). A fusion of the LexA DNA binding domain with NIL-16 codons 1–525 was created in the vector Y.LexA (Kurschner and Morgan, 1996) (gift from Dr. Steve Dalton) and served as bait in the two-hybrid analysis. Fusions of the Herpes simplex virus protein VP16 transcriptional activation domain with the cytoplasmic domains of various NMDA receptor subunits and inward rectifier potassium channels (Kir) were created by RT-PCR as described previously (Kurschner et al., 1998). cDNA sequences of voltage-gated potassium channels (Kv) as well as the α1 subunit of a mouse brain class C calcium channel (mbC α1) were amplified from mouse brain by RT-PCR, using primers designed from published DNA sequences (for the respective GenBank accession numbers, see Table 1). PCR products were inserted into the yeast expression vector pSD.10a (Dalton and Treisman, 1992) (gift from Dr. Steve Dalton) in frame with the VP16 transcriptional activator domain. The VP16 fusion constructs served as prey in the yeast two-hybrid analysis. Two-hybrid analysis of pairs of cloned cDNAs was performed as described (Kurschner and Morgan, 1996).
Table 1.
Interaction of NIL-16 with transmembrane ion channels in the yeast two-hybrid system
| Ion channel | GenBank accession number | Residues included in prey construct | C-terminal sequence | NIL-16 binding |
|---|---|---|---|---|
| NR1–3a | U08265 | 738–922 | PSVSTVV | − |
| NR2A | D10217 | 1300–1464 | PSIESDV | + |
| NR2B | U11419 | 1289–1482 | SSIESDV | + |
| NR2C | U08259 | 975–1250 | SSLESEV | + |
| NR2D | U08260 | 1096–1323 | SSLESEV | + |
| Kir1.1a | X72341 | 230–391 | ETDDTQM | − |
| Kir2.1 | X73052 | 252–428 | LRRESEI | + |
| Kir2.3 | U11075 | 280–445 | YRRESAI | + |
| Kir4.1 | U27558 | 1–379 | SVRISNV | + |
| Kir4.2 | Y10745 | 245–375 | LLQQSNV | + |
| Kir6.2 | U73626 | 254–390 | ISPDSLS | − |
| Kv1.1 | Y00305 | 409–495 | SKLLTDV | − |
| Kv1.2 | M30440 | 411–499 | TKMLTDV | − |
| Kv1.3 | M30441 | 434–528 | KKIFTDV | − |
| Kv1.4 | X16002 | 563–655 | KAVETDV | − |
| Kv1.5 | L22218 | 506–602 | TSRETDL | − |
| Kv1.6 | M27159 | 460–530 | KRMLTEV | − |
| Kv4.1 | M64226 | 409–651 | TVKISSL | + |
| Kv4.2 | S64320 | 407–630 | IVRVSAL | + |
| Kv4.3 | U42975 | 404–636 | VVKVSVL | + |
| mbC α1 | L01776 | 1847–2139 | RSYVSNL | + |
The C-terminal cytoplasmic domains of various NMDA receptor subunits (NR), inward rectifier potassium channels (Kir), voltage-gated potassium channels (Kv), or the α1 subunit of the voltage-sensitive class C calcium channel (mbC α1) were coexpressed with NIL-16 amino acids 1–525 (serving as bait) in yeast and assayed for interaction in the two-hybrid system. The ion channel constructs were cloned by RT-PCR with primers designed from published sequences (see GenBank accession numbers). The amino acid residues included in the ion channel constructs are indicated. The C-terminal amino acid sequences are shown for all constructs. +, Blue yeast colony color in the yeast two-hybrid assay, indicating protein interaction with NIL-16; −, white yeast colony color after 6 hr observation time, indicating the lack of NIL-16 binding.
GST-pull-down assays. A cDNA encoding mbC α1 amino acids 1847–2139 was inserted into the vector pT7βplink (Dalton and Treisman, 1992) (gift from Dr. Steve Dalton) for in vitro transcription/translation. In vitrotranscription/translation constructs of NR2D amino acids 839–1323 as well as full-length Kir4.1 (amino acids 1–379), Kir4.2 (amino acids 1–375) and a mutated Kir4.1, which contained a serine to glycine mutation at position 377 and a valine to alanine exchange at residue 379 (Kir4.1mutC), were described elsewhere (Kurschner et al., 1998). Proteins were translated in vitro, using the TNTCoupled Reticulocyte Lysate System (Promega, Madison, WI). Reactions were done in the presence of [35S]methionine (Amersham Pharmacia Biotech) in a 50 μl volume, using 2 μg of plasmid DNA as a template. NIL-16 codons 1–297 were cloned into the vector pGEX-4T-1 (Amersham Pharmacia Biotech) and expressed inEscherichia coli as a glutathione S-transferase (GST) fusion protein. “Empty” pGEX-4T-1 vector encoded the GST protein alone and was expressed as a control. GST and GST-NIL-16 were purified from bacterial cell lysates as described in Ausubel et al. [(1995), section 20.2]. GST-pull-down assays were performed according to standard protocols described in Ausubel et al. [(1995), section 20.2], using 23 μl of in vitro-translated [35S]methionine-labeled proteins and equal amounts (in 25 μl) of GST or GST-NIL-16, respectively, bound to glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech). Precipitated protein complexes were separated by SDS-PAGE on a 12% polyacrylamide gel. The in vitro translation reactions (0.3 μl) were loaded as input controls. The gel was dried and autoradiographed.
Caspase-3 cleavage assay. Full-length NIL-16 cDNA, as well as an EcoRV/EcoRI restriction fragment corresponding to codons 705–1322 of NIL-16 (codons 7–624 of splenocyte-derived pro-IL-16), was inserted into the vector pT7βplink (Dalton and Treisman, 1992) for in vitro transcription. Capped in vitro transcripts were generated as described (Bonifacino et al., 1998) in the absence of radionucleotides. Of each transcript 0.4 μg was used separately as a template in 20 μl ofin vitro translation reactions with the Rabbit Reticulocyte Lysate System (Promega, Madison, WI) in the presence of [35S]methionine (Amersham Pharmacia Biotech). Translation products were split into two 10 μl aliquots each, and 15 μl of caspase-3 cleavage buffer [containing (in mm) 20 HEPES, pH 7.4, 100 NaCl, 20 DTT, and 0.5% NP-40] was added to each aliquot. One aliquot per transcript received 0.5 μl (100 ng) purified, active recombinant human caspase-3 (PharMingen). All four aliquots were incubated at 37°C for 2 hr. Subsequently, the translation and cleavage products were separated by SDS-PAGE on a 4–20% gradient gel. Protein bands were visualized by autoradiography.
Transfection of COS-7 cells; preparation of COS-7 extracts and conditioned medium. Full-length NIL-16 cDNA as well as codons 705–1322 of NIL-16 (codons 7–624 of splenocyte-derived pro-IL-16) was cloned into the mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, CA). COS-7 cells were transiently transfected with one of these constructs or with pcDNA3 vector, respectively, using LipofectAMINE (Life Technologies). Transfected cells were maintained overnight in DMEM (Bio Whittaker, Walkersville, MD) supplemented with 2 mm glutamine (Life Technologies) and 20% fetal calf serum (Harlan, Indianapolis, IN). Subsequently, the cells were washed with serum-free medium and incubated with serum-free medium for 48 hr. Thereafter, the conditioned medium was removed and concentrated 100-fold by centrifugation in Centricon-10 concentrators (Amicon, Beverly, MA). The transfected COS-7 cells were lysed for 10 min on ice in 1 ml of cell lysis buffer, and the lysates were cleared as described above. Then 10 μl of lysate and concentrated medium, respectively, were separated by SDS-PAGE on a 4–20% gradient gel and subjected to Western blot analysis with the anti-IL-16 antibody 14.1, as described above.
Coimmunoprecipitation (Co-IP) assay. COS-7 cells were transiently transfected as described above with full-length NIL-16 (in pcDNA3; Invitrogen) in the presence or absence of full-length NR2A (in pTracer-CMV; Invitrogen) (a gift from Dr. Jim Boulter, UCLA, Los Angeles, CA). Cells were lysed as described (Lin et al., 1998) in Co-IP buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% sodium deoxycholate supplemented with Complete protease inhibitor cocktail; Boehringer Mannheim). Immunoprecipitation was performed as described (Lin et al., 1998), using 0.6 μg of rabbit anti-NR2A antibodies (catalog number A-6473; Molecular Probes) and protein A–Sepharose CL-4B beads (Sigma). Precipitated proteins were analyzed for the presence of NIL-16 by Western analysis with the antibody 14.1 as described above.
RESULTS
Molecular cloning of NIL-16
A cDNA library made by using RNA extracted from adult mouse cerebellum was screened for proteins that bind to an artificial PDZ domain target peptide with the C-terminal sequence V-S-D-L, as described previously (Kurschner et al., 1998). One plasmid isolated in the screen contained a cDNA of 925 bp in length that encoded part of a novel protein. This cDNA served as a probe to isolate a full-length NIL-16 cDNA from a mouse cerebellum cDNA library. Several overlapping cDNAs were aligned (data not shown), revealing that the initial cDNA contained the first 297 codons of a protein of 1322 amino acids. BLAST searches indicated that the full-length protein contained four PDZ domains but no other known protein motifs (Fig.1). Amino acids 1–698 represented novel sequences, whereas amino acids 699–1322 were 99.4% identical (four mismatches; data not shown) to the published sequence of splenocyte-derived mouse pro-IL-16 (Keane et al., 1998) (GenBank accession number AF006001; Fig. 1). Therefore, the protein was termed “NIL-16” for “Neuronal IL-16.”
Fig. 1.
Primary sequence of NIL-16. PDZ domains areunderlined; GLGF motifs (Cho et al., 1992) areboxed; amino acid numbers are indicated atleft and right. The horizontal arrow marks the start of splenocyte-derived pro-IL-16 (Keane et al., 1998). The vertical arrow indicates the site of proteolytic cleavage of pro-IL-16 by caspase-3 (Zhang et al., 1998).
Tissue distribution of NIL-16
Northern blot analysis of mouse tissue was performed with a cDNA probe that corresponded to NIL-16 codons 1058–1195, which are contained within the published pro-IL-16 sequence (Fig. 1). This analysis detected the previously described (Keane et al., 1998) pro-IL-16 mRNA doublet of 2.5 and 3.5 kb in spleen and thymus RNA (Fig.2A). In addition, novel mRNA species of ∼5 kb were detected in total brain and cerebellum RNA (Fig. 2A). The size correlated well with the size of the full-length NIL-16 cDNA that we had cloned, which was 4994 bp in length (data not shown). Northern analysis of cerebellum RNA samples obtained from mice of different ages demonstrated that NIL-16 transcript levels increased steadily during postnatal cerebellar development. Expression was maximal at P12 and stayed constant thereafter (Fig. 2B).
In situ hybridization analysis of adult mouse brain, which used a cRNA probe that corresponded to NIL-16 codons 1058–1195, revealed striking expression of NIL-16 in the cerebellum and in the hippocampus. In the latter structure, NIL-16 was expressed strongly in the dentate gyrus as well as in the CA3 and CA4 regions of the hippocampus (Fig. 2D). Lower expression levels were found in CA1 (Fig. 2D). The identical expression pattern was detected with a cRNA probe corresponding to NIL-16 codons 1–130 (data not shown).
Western analysis of different brain regions that used a monoclonal antibody specific for IL-16 identified a protein of ∼180 kDa in cerebellar and hippocampal extracts (Fig. 2C). Lysates from COS-7 cells, transfected with full-length NIL-16 cDNA, contained an immunoreactive protein band of approximately the same size, suggesting that the cloned cDNA encoded the full-length protein (Fig.2C).
NIL-16 is expressed in postmitotic neurons
In situ hybridization analysis revealed that NIL-16 expression in the cerebellum started at P3 in the differentiated granule cells of the internal granular layer (IGL), whereas it was virtually absent from the mitotic and premigratory granule cells of the external granular layer (EGL) (Fig.3A,B). In the more mature cerebellum at P21, high levels of NIL-16 transcript were detected in the granule cell layer (GCL), whereas lower levels were observed in the Purkinje cell layer (PCL) (Fig. 3B). In the hippocampus, NIL-16 expression was detected first in the CA3 and CA4 regions at P1 and then in the suprapyramidal blade of the dentate gyrus at P3 (Fig.3A). Here, transcripts were restricted mainly to the most mature granule cells, which are located at the outer regions of the fascia (Fig. 3B). At P21, most granule neurons of the dentate gyrus were NIL-16-positive, and NIL-16-negative cells were found mainly in the region adjacent to the hilus (Fig. 3B), which contains undifferentiated and proliferating cells (for review, see Gaarskjaer, 1986; Stanfield and Cowan, 1988). Thus, within both the cerebellum and the hippocampus, NIL-16 transcripts were found predominantly in regions containing the most mature cell populations.
NIL-16 interacts with ion channels
Many PDZ domain proteins function as cytoskeletal elements that anchor transmembrane proteins to specialized submembranous locations. The C-terminal consensus sequence x-S/T-x-V/I, which is contained in a large number of neuronal ion channels, identifies proteins as possible PDZ ligands (Songyang et al., 1997). Therefore, we investigated the possibility that NIL-16 binds to glutamate receptor subunits, potassium channels, or the class C α1 subunit of a mouse brain calcium channel (mbC α1) in the yeast two-hybrid system (Table 1). In this assay, NIL-16 selectively associated with NMDA receptor 2 subunits, Kir2.0 and Kir4.0 family members, Kv4 channels, and mbC α1. In contrast, no binding to NMDA receptor 1–3a, Kir1.1a, Kir6.2, or Kv1 family members was observed. All of the ion channels that interacted with NIL-16 possessed the C-terminal consensus sequence x-S-x-V/I/L.
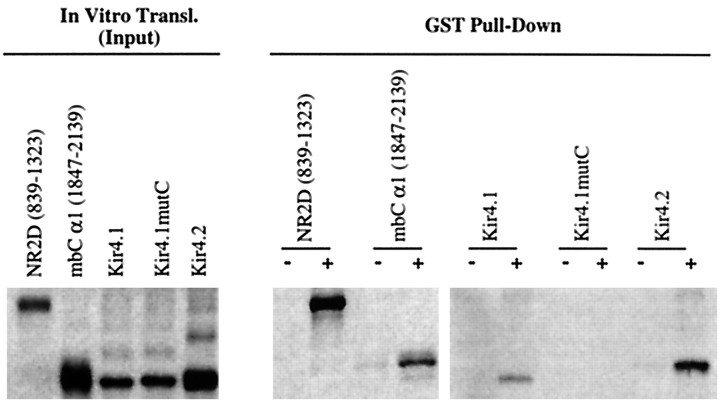
To confirm that NIL-16 can interact physically with ion channels, we performed pull-down experiments with a glutathione S-transferase (GST)-fused NIL-16 fragment containing the first PDZ domain (amino acids 1–297) and a selection of in vitro-translated [35S]methionine-labeled NIL-16 ligands. A mutated Kir4.1 construct, Kir4.1mutC, in which the C-terminal amino acid sequence S-N-V was changed to G-N-A (Kurschner et al., 1998), was included in the analysis. GST-NIL-16, but not GST alone, precipitated the C-terminal fragment of NR2D as well as full-length Kir4.1 (Fig.4). Similarly, although small amounts of Kir4.2 and the mbC α1 fragment were pulled down by GST alone, this precipitation was enhanced significantly by GST-NIL-16, indicating an association of NIL-16 with Kir4.2 and mbC α1. In contrast, the C-terminal mutant Kir4.1mutC did not associate with NIL-16 in this assay, suggesting that the interaction of NIL-16 with ion channels is dependent on the presence of the C-terminal consensus motif found in all of the identified NIL-16 ligands.
Fig. 4.
GST-pull-down analysis of NIL-16 with NIL-16-binding ion channels. cDNAs encoding NIL-16-interacting ion channels as well as Kir4.1mutC were translated in vitroin the presence of [35S]methionine. The cDNA templates used in the in vitro translation reactions are indicated above the lanes. The respective codons included in the NR2D and mbC α1 constructs are shown inparentheses. Kir constructs encoded the full-length proteins. NIL-16 (amino acids 1–297) was expressed and purified fromE. coli as a glutathione S-transferase (GST) fusion protein; unfused GST protein was expressed as a control.Left, Shown is 0.3 μl of the in vitrotranslation reactions (input controls). Right, GST-pull-down assays were performed with in vitro-translated proteins, as indicated abovethe lanes, and equal amounts of GST (−) or GST-NIL-16 (+), respectively, as bait. Precipitated proteins were analyzed by SDS-polyacrylamide electrophoresis and subsequent autoradiography.
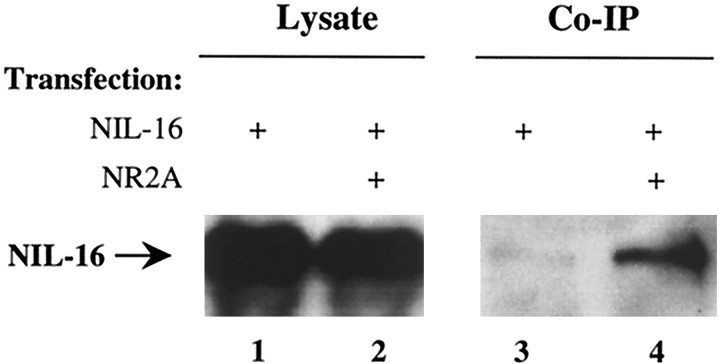
To demonstrate that NIL-16 is able to associate with its ligands inside a cell, we performed coimmunoprecipitation studies on transfected COS-7 cells. Cells were transfected with NIL-16 or cotransfected with NIL-16 and NR2A. Immunoprecipitation was done on cell lysates by using anti-NR2A antibodies. Precipitates were assayed for the presence of NIL-16 by immunoblotting. Although comparable amounts of NIL-16 were found in the lysates from both single- and cotransfected cells (Fig.5, left panel), only trace amounts were detectable in immunoprecipitates from cells that had been transfected with NIL-16 alone (Fig. 5, lane 3). This unspecific precipitation of NIL-16 was amplified greatly in preparations from cells that had been cotransfected with NR2A (Fig. 5,lane 4), demonstrating that overexpressed NIL-16 bound to NR2A in transfected COS-7 cells. We were unable to find conditions under which we could coimmunoprecipitate the two proteins from synaptic membrane preparations or CGN cultures (data not shown). This may be attributable to a relatively low abundance of the proteins or to a poor sensitivity of our reagents.
Fig. 5.
Coimmunoprecipitation of NIL-16 and NR2A from transfected COS-7 cells. COS-7 cells were transfected with NIL-16 (lanes 1, 3) or cotransfected with NIL-16 and NR2A (lanes 2, 4). Aliquots of cell lysates were analyzed for NIL-16 expression by Western blot (left panel) or subjected to immunoprecipitation with anti-NR2A. Precipitates were subjected to Western analysis with anti-NIL-16 to detect coimmunoprecipitated NIL-16 (right panel). The position of NIL-16 in the gel is indicated on the left.
The subcellular localization of NIL-16 overlaps with that of NR2C
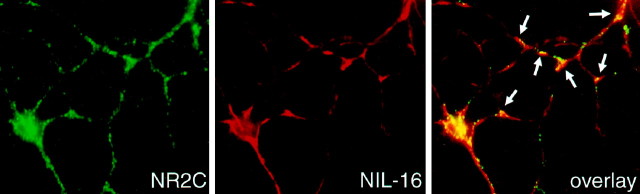
The subcellular localization of NIL-16 in cultured CGN was compared with that of one of its ligands, NR2C, using immunofluorescence (Fig. 6). Immunoreactivity to NR2C was found in punctate clusters throughout the cell bodies and the neurites. NIL-16 immunoreactivity also was detected in clusters as well as larger patches in both the cell bodies and the neurites. It overlapped partially with the distribution of NR2C. This colocalization was concentrated particularly at neuritic branch points.
Fig. 6.
Partial colocalization of NIL-16 and NR2C in cultured CGN. Cerebellar granule cells were double-stained with the mouse anti-IL-16 and the rabbit anti-NR2C antibodies. NR2C (green) displayed punctate staining pattern in the cell soma and dendrites. NIL-16 (red) was highly enriched at branching points of dendrites as well as soma. Particularly at these locations, NR2C and NIL-16 were colocalized, as indicated byarrows.
NIL-16 is a caspase-3 substrate
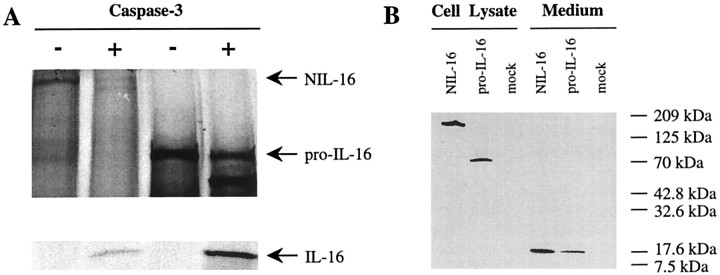
During the activation of CD4+T-cells, IL-16 release from its precursor correlated with caspase-3 activation (Wu et al., 1999). Moreover, it was established recently that splenocyte-derived pro-IL-16 is an in vitro substrate for caspase-3 (Zhang et al., 1998). To test if NIL-16 could be processed similarly by caspase-3, we incubated in vitro-translated radiolabeled full-length NIL-16 and pro-IL-16 in the presence or absence of active recombinant caspase-3. After separation by SDS-PAGE and autoradiography, NIL-16 cleavage products were detected in caspase-3-treated samples (Fig.7A). Moreover, a protein fragment of ∼18 kDa was found in both the NIL-16- and the pro-IL-16-containing samples. This indicated that, like pro-IL-16, NIL-16 was processed by caspase-3 to generate mature IL-16. The finding that the apparent sizes of the cleavage products was identical in caspase-3-treated samples of both NIL-16 and pro-IL-16 suggested that IL-16 was cleaved off the C termini of NIL-16 and pro-IL-16 at analogous sites.
Fig. 7.
Processing of NIL-16 by caspase-3.A, In vitro caspase-3 cleavage assay. Radiolabeled NIL-16 and pro-IL-16 were generated by in vitro translation and incubated in the presence or absence of recombinant, active caspase-3. Reaction products were separated by SDS-PAGE and analyzed by autoradiography. The positions of unprocessed NIL-16 and pro-IL-16 as well as that of the cleavage product IL-16 are indicated on the right. B, Processing of NIL-16 and pro-IL-16 in transfected COS-7 cells. COS-7 cells have an intrinsic caspase-3-like activity. Therefore, the processing of NIL-16 and pro-IL-16 was analyzed in these cells. Lysates of NIL-16-, pro-IL-16-, and mock-transfected cells as well as conditioned medium from the cultures were subjected to Western analysis. The primary antibody used in the experiment recognized the IL-16 epitope. Full-length proteins were detected in the respective cell lysates, whereas processed, mature IL-16 was found secreted into the medium of both NIL-16- and pro-IL-16-transfected cells. The positions of molecular weight markers in the gel are indicated on theright.
COS-7 cells have an intrinsic caspase-3-like activity, and they have been shown to secrete mature IL-16 after transfection with pro-IL-16 (Zhang et al., 1998). Therefore, we tested whether the cleavage of NIL-16 by caspase-3 in transfected COS-7 cells gave rise to a similarly secreted form of IL-16. COS-7 cells transfected with NIL-16 and pro-IL-16 as well as conditioned medium were subjected to Western analysis by using a monoclonal antibody specific for the IL-16 epitope. Unprocessed NIL-16 and pro-IL-16, respectively, were found only in the corresponding cell lysates (Fig. 7B). Processed, mature IL-16 was detected in the conditioned medium from both NIL-16- and pro-IL-16-transfected cells (Fig. 7B), demonstrating that processing of NIL-16, like that of pro-IL-16, resulted in the release of a secreted cleavage product.
NIL-16 processing parallels caspase-3 activation in primary cerebellar granule cell cultures undergoing apoptosis
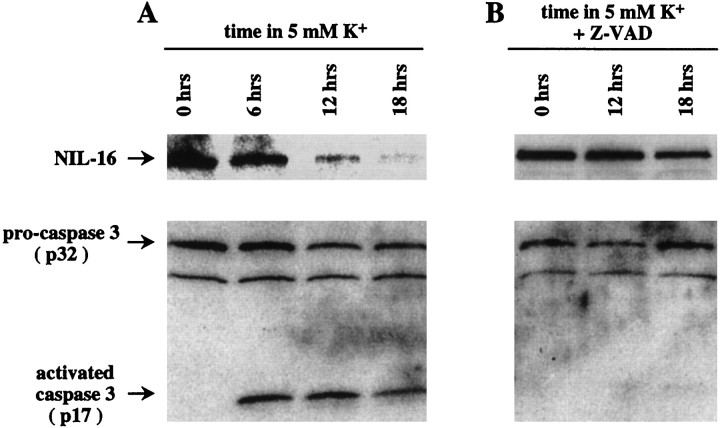
Postmitotic CGN can be cultured in vitro in the presence of 25 mm K+(depolarizing conditions), but they undergo a cell death program with morphological features of apoptosis when they are maintained in nondepolarizing conditions (5 mmK+) (Yan et al., 1994). One feature of this paradigm of CGN apoptosis is the activation of caspase-3 (Armstrong et al., 1997; Ni et al., 1997; Marks et al., 1998), which occurs via cleavage of the 32 kDa proenzyme into the 12 and 17 kDa subunits of the active enzyme (Fernandes-Alnemri et al., 1994;Nicholson et al., 1995; Tewari et al., 1995). Therefore, we assayed the processing of NIL-16 in apoptotic CGN. CGN cultures were prepared from P7 mouse cerebellum and kept under depolarizing conditions for 8 d. Subsequently, the medium was switched to nondepolarizing conditions, and the cells were lysed for protein preparation at various time points (Fig. 8). The lysates were examined by Western analysis for the presence of NIL-16 and caspase-3 proteins, respectively. The anti-IL-16 antibody 14.1 that was used in this assay recognizes the IL-16 epitope, which is cleaved off the C terminus of NIL-16 by caspase-3. The anti-caspase-3 antiserum recognizes both the inactive pro-form of caspase-3 (p32) and the p17 subunit of processed, active caspase-3. As shown in Figure 8A, inactive pro-caspase-3 was present at all of the time points that were analyzed, whereas the p17 subunit of active caspase-3 was found only after the induction of apoptosis. During the time course that was examined, the levels of full-length NIL-16 was diminished progressively (Fig.8A). In contrast, when the caspase-3 inhibitor Z-VAD was included in the culture medium, both caspase-3 activation and NIL-16 processing were reduced dramatically (Fig.8B). This suggested that NIL-16 was processed by caspase-3 during apoptosis of CGN.
Fig. 8.
Processing of NIL-16 and caspase-3 activation in cultured cerebellar granule neurons (CGN) after the induction of apoptosis. A, Apoptosis of CGN was induced by a medium switch to nondepolarizing conditions. Cell lysates were prepared at various times thereafter (indicated above the lanes) and subjected to Western analysis. The antibody used for the detection of NIL-16 recognized the IL-16 epitope, which is cleaved off the C terminus by caspase-3. The anti-caspase-3 antibody recognized both the inactive proenzyme (p32) and the p17 subunit of the active caspase. Caspase-3 activation was detected at all time points analyzed after the medium switch. In parallel, immunoreactivity against full-length NIL-16 was diminished progressively. B, The caspase inhibitor Z-VAD was included in the medium at the time of the switch to nondepolarizing conditions. Western analysis of cell lysates was performed as described in A. Under these conditions the caspase-3 activation and NIL-16 processing were greatly reduced. The positions of NIL-16, pro-caspase-3, and active caspase-3 (p17) in the gel are indicated on the left.
Cerebellar granule neurons induce c-fos in response to IL-16
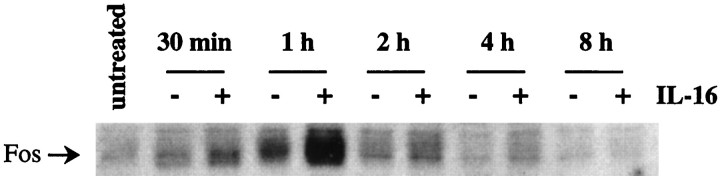
Granule cells of the cerebellum are among the neurons that have been shown to express the IL-16 receptor CD4 (Omri et al., 1994). Therefore, we investigated whether cultured CGN had the potential to respond to exogenous IL-16. To this end, the culture medium was replaced with medium containing no IL-16 or 0.1 μmrecombinant mouse IL-16, respectively, and CGN were incubated for various lengths of time (Fig. 9). A CGN culture that had not been subjected to medium change served as a control (untreated). Low levels of Fos were detected in lysates from untreated cells. Medium change alone induced a slight increase in Fos protein, which was apparent between 30 min and 2 hr after a medium change. However, stimulation with IL-16 resulted in a dramatic upregulation of Fos expression, which peaked at 1 hr after medium change. This effect was transient, because Fos levels in CGN treated with IL-16 for 8 hr were similar to those found in untreated cells (Fig. 9).
Fig. 9.
Induction of c-fos in cerebellar granule neurons (CGN) by incubation with IL-16. CGN culture medium was changed and replaced with medium containing no IL-16 (−) or 0.1 μm recombinant mouse IL-16 (+). A culture not subjected to medium change (untreated) served as a control for basal levels of Fos in CGN cultures before the medium change. Cells were lysed at various times after the start of exposure to IL-16, as indicated above the lanes. Levels of Fos protein expression were determined by Western analysis. The position of Fos in the gel is indicated on the left.
Induction of c-fos in cerebellar granule neurons involves tyrosine phosphorylation
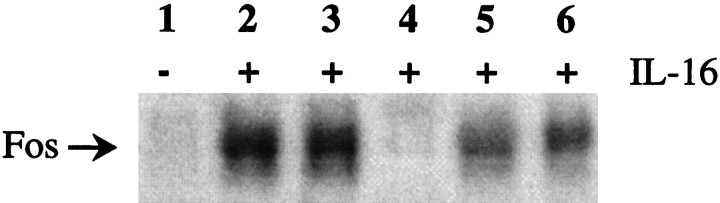
To reveal the signaling pathways involved in mediating Fos upregulation, we performed IL-16 treatment of CGN in the presence and absence of various signaling pathway inhibitors (Fig.10). Cells were pretreated with the inhibitors for 3 hr before IL-16 was added to a final concentration of 0.1 μm. After 1 hr of continued incubation the Fos levels in cell lysates were determined by Western analysis. Although basal levels of Fos were low in the absence of IL-16 (Fig. 10, lane 1), IL-16 treatment resulted in a marked upregulation of Fos expression (lane 2). This effect was abolished completely by the tyrosine kinase inhibitor herbimycin A (lane 4), indicating that tyrosine phosphorylation is required for IL-16 signaling in this system. In contrast, neither the inhibition of nitric oxide synthase by treatment with a combination of 7-nitroindazole and NG-nitro-l-arginine methyl ester dihydrochloride (lane 3) nor the inhibition of mitogen-activated protein kinase (MAPK) kinase (MEK) by PD98059 (lane 5) nor the inhibition of p38-MAPK by SB202190 (lane 6) interfered significantly with Fos induction.
Fig. 10.
Inhibition of IL-16-mediated Fos upregulation by the tyrosine kinase inhibitor herbimycin A. CGN cultures were incubated for 3 hr with or without signaling pathway inhibitors, followed by exposure to 0.1 μm IL-16 (lanes 2–6) for 1 hr in the presence or absence of inhibitors. Cell lysates were assayed for Fos expression by Western analysis.Lane 1, Pretreatment with medium only, no IL-16 treatment; lane 2, pretreatment with medium only;lane 3, pretreatment with 7-nitroindazole (nNOS inhibitor) andNG-nitro-l-arginine methyl ester dihydrochloride (NOS inhibitor); lane 4, pretreatment with herbimycin A (tyrosine kinase inhibitor); lane 5, pretreatment with PD58059 (MEK inhibitor); lane 6, pretreatment with SB202190 (p38 MAPK inhibitor). The position of Fos in the gel is indicated on theleft.
IL-16 treatment does not induce Jun phosphorylation in cerebellar granule neurons
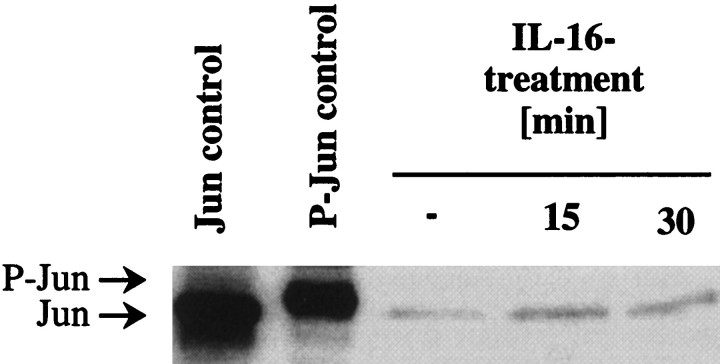
Studies in activated macrophages showed that IL-16 induced rapid and transient phosphorylation of Jun in these cells, which peaked 15 min after the start of treatment (Krautwald, 1998). Therefore, we investigated whether IL-16 had a similar effect on cultured CGN. CGN were treated with 0.1 μm IL-16 for 15 and 30 min, respectively. A CGN culture that had not been exposed to IL-16 served as a control for Jun expression in CGN. Cell extracts were subjected to Western analysis with an anti-Jun antibody (Fig.11). An NIH-3T3 cell extract was a control for Jun protein (lane 1), and a UV-irradiated National Institutes of Health-3T3 cell extract was a control for phosphorylated Jun (P-Jun, lane 2). Figure 11 shows that IL-16 treatment for 15 or 30 min did not lead to Jun phosphorylation in CGN. Similarly, longer IL-16 treatments for up to 8 hr did not affect the phosphorylation state of Jun (data not shown).
Fig. 11.
IL-16 treatment does not induce Jun phosphorylation in cultured CGN. CGN were left untreated (−) or treated with 0.1 μm IL-16 for 15 and 30 min, respectively. Cell extracts were subjected to Western analysis with an antibody that recognizes both Jun and phosphorylated Jun (P-Jun). An NIH-3T3 cell extract was used as a control for Jun protein, and a UV-irradiated NIH-3T3 cell extract was used as a control for P-Jun. The positions of Jun and P-Jun in the gel are indicated on the left.
DISCUSSION
It has been recognized for some time that the immune system and the CNS share a variety of signaling molecules that regulate cellular function. For example, cytokines such as IL-1 and IL-2 and their receptors, which originally were described in the immune system, also are expressed by neurons in the brain (Cunningham et al., 1992;Yan et al., 1992; Farrar et al., 1987). The identification of NIL-16 as a neuronal variant of the lymphokine IL-16 marks another molecular parallel between the immune and the nervous systems.
Mature IL-16 is the only known secreted member of the PDZ protein family. However, like in the case of IL-1, the mechanism of secretion remains elusive, because neither IL-1 nor IL-16 possesses a leader sequence that would target it for conventional release pathways. IL-16 exerts its function in the immune system via binding to its cell surface receptor, CD4, on T-lymphocytes, eosinophils, and monocytes (for review, see Center et al., 1996). Although neuronal CD4 expression was reported previously (Funke et al., 1987; Omri et al., 1994), its ligand IL-16 has not been described in the brain. Because the function of CD4-expressing immune cells is different from that of neurons and glia, CD4 receptor engagement by IL-16 is likely to have different functional consequences in the CNS. Notably, whereas IL-16 has been demonstrated to induce Jun phosphorylation in activated macrophages (Krautwald, 1998), it did not do so in cultured CGN. Nonetheless, CGN responded to IL-16 by upregulating expression of the transcription factor Fos, indicating that an IL-16 signaling pathway exists in these cells. In activated macrophages, p38 MAPK becomes rapidly phosphorylated after IL-16 stimulation (Krautwald, 1998). However, signaling via p38 MAPK does not seem to be a major pathway in the upregulation of Fos in CGN, because the inhibitor SB202190 did not interfere significantly with Fos expression. In contrast, the IL-16-signaling pathway in CGN is likely to involve tyrosine phosphorylation, because the tyrosine kinase inhibitor herbimycin A completely abolished Fos upregulation. In T-lymphocytes the binding of IL-16 to CD4 results in the activation of the src family tyrosine kinase p56lck (Ryan et al., 1995), which is bound tightly to the cytoplasmic tail of CD4 (Rudd et al., 1988;Veillette et al., 1988). Therefore, it is conceivable that, in CGN, the activation of a p56lck-like kinase is part of the signaling pathway that leads to Fos upregulation. However, further experiments have to be performed to clarify this point.
In addition to the similarity in tissue distribution of IL-1 and IL-16 (both being expressed in the immune system as well as the CNS), another striking parallel between the two peptides involves their release from a precursor via proteolytical cleavage involving a caspase. In the case of IL-1β, maturation is achieved by way of caspase-1 (Cerretti et al., 1992; Thornberry et al., 1992), whereas IL-16 is released from its precursor by caspase-3 cleavage in both the immune (Zhang et al., 1998;Wu et al., 1999) and the nervous systems (this study). The involvement of caspase-3 during apoptosis was recognized initially in the immune system (Fernandes-Alnemri et al., 1994). However, high expression levels of caspase-3 have been found during postnatal brain development (Ni et al., 1997), indicating that this protease may play a role in developmental cell death in the brain. Studies of caspase-3-deficient mice, which display profound deficiencies in brain development and decreased apoptosis in the brain (Kuida et al., 1996), support this notion. A low level of caspase-3 expression persists throughout adulthood (Ni et al., 1997), suggesting that caspase-3 has additional functions after the naturally occurring cell death program is completed. Because the caspase-3 expression pattern in adult brain overlaps strikingly with that of NIL-16 in the hippocampus and the cerebellum (Ni et al., 1997), we propose that, in the adult, caspase-3 functions in the processing of NIL-16 to mature secreted IL-16. It remains to be determined whether, after cleavage, the N-terminal portion of NIL-16 stays in the cytosol or whether it is subject to secretion or degradation. However, we conclude from the coimmunoprecipitation study with NR2A (see Fig. 5) as well as from Western analysis of lysates from transfected COS-7 cells (see Fig.7B) that at least a part of all cellular full-length NIL-16 molecules is localized within the cell.
In contrast to the findings with other cytokines and neurotrophins such as IL-10, IL-13, and ciliary neurotrophic factor, which are capable of delaying apoptosis of cultured CGN (de Luca et al., 1996), IL-16 had no effect on the time course of apoptosis in CGN, as judged by Hoechst 33258 staining (data not shown). This was similar to the observations made in HIV-infected T-cells (Baier et al., 1995) and in activated macrophages (Krautwald, 1998). Therefore, the processing of NIL-16 by caspase-3 in the adult brain may have functional consequences that are unrelated to the cell death program. Given the strikingly restricted expression pattern of NIL-16 in the regions of the brain that can undergo synaptic plasticity, such as long-term potentiation and long-term depression, it is reasonable to hypothesize that IL-16 may contribute to in these processes. This would be similar to the effects of IL-1β, which inhibited synaptic strength and long-term potentiation in the hippocampus (Bellinger et al., 1993; Katsuki et al., 1990; Coogan and O’Connor, 1997).
Although our data suggest that NIL-16 can be processed by caspase-3 to give rise to a secreted signaling molecule with autocrine effects on cerebellar granule neurons, our data also show that NIL-16 may function as a molecular anchor for transmembrane proteins. PDZ domains [also called “GLGF domains,” for the relatively conserved tetrapeptide Gly-Leu-Gly-Phe contained in their primary sequence (Cho et al., 1992)] have been described as protein interaction motifs that are found in an emerging class of structurally related proteins. These proteins typically function as intracellular cytoskeletal elements that cluster their ligands at specific subcellular locations (for review, see Ponting et al., 1997; Sheng and Wyszynski, 1997). This is achieved via an interaction between a PDZ domain and the C terminus of a respective ligand (Songyang et al., 1997). Indeed, NIL-16 was found in clusters throughout the cytosol of cultured CGN. Moreover, NIL-16 was able to associate with a battery of ion channels that possess the C-terminal consensus sequence x-S-x-V/I/L. Among the NIL-16-interacting proteins was the α1 subunit of a mouse brain class C calcium channel (mbC α1) (Ma et al., 1992). This constitutes the first report of an interaction between a class C calcium channel and a PDZ domain protein.
It is not surprising to observe that the tissue distribution and subcellular localization of NIL-16 overlap only partially with the NIL-16 binding proteins, because these ligands typically can associate with a variety of different PDZ domain proteins. For example, one prototype of the PDZ domain protein family, PSD-95/SAP90 (Cho et al., 1992; Kistner et al., 1993), binds several of the NIL-16 binding proteins, including NMDA receptor subunits (Kornau et al., 1995;Niethammer et al., 1996) and Kir channels (Cohen et al., 1996; Horio et al., 1997). PSD-95/SAP90 is confined mainly to dendrites, but it is virtually absent from cell bodies (Cho et al., 1992; Kornau et al., 1995). Consequently, its subcellular distribution overlaps only in dendrites with, for example, NMDA receptor subunits, which also are found in cell bodies. In contrast, NIL-16 partially colocalized with NR2C in both the dendrites and the cell bodies. Therefore, it is conceivable that interactions between different PDZ domain proteins with a given ion channel occur selectively at certain subcellular locations. Indeed, targeting of ion channels to a specific region in the cell membrane may be a unique property of an individual PDZ domain protein. Studies in mutant mice in which single PDZ domain proteins have been eliminated will be required to investigate this possibility.
Interestingly, all of the observed protein interactions involved the N-terminal neuron-specific portion of NIL-16, although we did not detect associations of ion channels with either of the two C-terminal PDZ domains (data not shown). In the case of the fourth PDZ domain, which constitutes the major part of mature IL-16, this is not surprising, because NMR structures of IL-16 revealed an unusual GLGF cleft with an occluded peptide binding site (Muhlhahn et al., 1998). Therefore, this portion of NIL-16 is unlikely to serve as a cytosolic anchor for transmembrane proteins in a manner that involves the typical association with a C-terminal x-S/T-x-V/I/L motif. However, it is not excluded that this domain engages in other types of intracellular protein interactions. This possibility, as well as the unveiling of ligands of the third PDZ domain in NIL-16, will be the focus of future studies.
In summary, we hypothesize that NIL-16 has dual functions in the CNS as a secreted signaling molecule and as a scaffolding protein. Moreover, it is conceivable that ligand–receptor interactions and intracellular signaling are coupled via NIL-16. Indeed, receptor occupation may regulate NIL-16 processing. Future experiments are aimed at addressing this question.
Footnotes
This work was supported in part by National Institutes of Health Cancer Center Support CORE Grant P30 CA21765 and by the American Lebanese Syrian Associated Charities. We thank Drs. Jim Morgan and Tom Curran for critical reading of this manuscript.
Correspondence should be addressed to Dr. Cornelia Kurschner, Department of Developmental Neurobiology, St. Jude Children’s Research Hospital, Memphis, TN 38105.
REFERENCES
- 1.Armstrong RC, Aja TJ, Hoang KD, Gaur S, Bai X, Alnemri ES, Litwack G, Karanewsky DS, Fritz LC, Tomaselli KJ. Activation of the CED3/ICE-related protease CPP32 in cerebellar granule neurons undergoing apoptosis but not necrosis. J Neurosci. 1997;17:553–562. doi: 10.1523/JNEUROSCI.17-02-00553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology, 3rd Ed. Greene and Wiley Interscience; New York: 1995. [Google Scholar]
- 3.Baier M, Werner A, Bannert N, Metzner K, Kurth R. HIV suppression by interleukin-16. Nature. 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 4.Baier M, Bannert N, Werner A, Lang K, Kurth R. Molecular cloning, sequence, expression, and processing of the interleukin 16 precursor. Proc Natl Acad Sci USA. 1997;94:5273–5277. doi: 10.1073/pnas.94.10.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellinger FP, Madamba S, Siggins GR. Interleukin 1β inhibits synaptic strength and long-term potentiation in the rat CA1 hippocampus. Brain Res. 1993;628:227–234. doi: 10.1016/0006-8993(93)90959-q. [DOI] [PubMed] [Google Scholar]
- 6.Black RA, Kronheim SR, Cantrell M, Deeley MC, March CJ, Prickett KS, Wignall J, Conlon PJ, Cosman D, Hopp TP, Mochizuki DY. Generation of biologically active interleukin-1β by proteolytic cleavage of the inactive precursor. J Biol Chem. 1988;263:9437–9442. [PubMed] [Google Scholar]
- 7.Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM. Current protocols in cell biology. In: Morgan KS, editor. Current protocols. Wiley; New York: 1998. pp. 11.2.11–11.2.12. [Google Scholar]
- 8.Center DM, Kornfeld H, Cruikshank WW. Interleukin 16 and its function as a CD4 ligand. Immunol Today. 1996;17:476–481. doi: 10.1016/0167-5699(96)10052-i. [DOI] [PubMed] [Google Scholar]
- 9.Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Van Ness K, Greenstreet TA, March CJ, Kronheim SR, Druck T, Cannizzaro LA, Huebner K, Black RA. Molecular cloning of the interleukin-1β converting enzyme. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 10.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 11.Chupp GL, Wright EA, Wu D, Vallen-Mashikian M, Cruikshank WW, Center DM, Kornfeld H, Berman JS. Tissue and T-cell distribution of precursor and mature IL-16. J Immunol. 1998;161:3114–3119. [PubMed] [Google Scholar]
- 12.Cohen NA, Brenman JE, Snyder SH, Bredt DS. Binding of the inward rectifier K+ channel Kir 2.3 to PSD-95 is regulated by protein kinase A phosphorylation. Neuron. 1996;17:759–767. doi: 10.1016/s0896-6273(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 13.Coogan A, O’Connor JJ. Inhibition of NMDA receptor-mediated synaptic transmission in the rat dentate gyrus in vitro by IL-1β. NeuroReport. 1997;8:2107–2110. doi: 10.1097/00001756-199707070-00004. [DOI] [PubMed] [Google Scholar]
- 14.Cruikshank WW, Center DM, Nisar N, Wu M, Natke B, Theodore AC, Kornfeld H. Molecular and functional analysis of a lymphocyte chemoattractant factor: association of biologic function with CD4 expression. Proc Natl Acad Sci USA. 1994;91:5109–5113. doi: 10.1073/pnas.91.11.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruikshank WW, Kornfeld H, Center DM. Signaling and functional properties of interleukin-16. Int Rev Immunol. 1998;16:523–540. doi: 10.3109/08830189809043007. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham ET, Jr, Wada E, Carter DB, Tracey DE, Battey JF, De Souza EB. In situ histochemical localization of type I interleukin-1 receptor messenger RNA in the central nervous system, pituitary, and adrenal gland of the mouse. J Neurosci. 1992;12:1101–1114. doi: 10.1523/JNEUROSCI.12-03-01101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 18.de Luca A, Weller M, Frei K, Fontana A. Maturation-dependent modulation of apoptosis in cultured cerebellar granule neurons by cytokines and neurotrophins. Eur J Neurosci. 1996;8:1994–2005. doi: 10.1111/j.1460-9568.1996.tb01343.x. [DOI] [PubMed] [Google Scholar]
- 19.Farrar WL, Kilian PL, Ruff MR, Hill JM, Pert CB. Visualization and characterization of interleukin 1 receptors in brain. J Immunol. 1987;139:459–463. [PubMed] [Google Scholar]
- 20.Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein CED-3 and mammalian interleukin-1β-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 21.Fields S, Song O-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 22.Funke I, Hahn A, Rieber EP, Weiss E, Riethmuller G. The cellular receptor (CD4) of the human immunodeficiency virus is expressed on neurons and glial cells in human brain. J Exp Med. 1987;165:1230–1235. doi: 10.1084/jem.165.4.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaarskjaer FB. The organization and development of the hippocampal mossy fiber system. Brain Res. 1986;396:335–357. doi: 10.1016/0165-0173(86)90004-4. [DOI] [PubMed] [Google Scholar]
- 24.Horio Y, Hibino H, Inanobe A, Yamada M, Ishii M, Tada Y, Satoh E, Hata Y, Takai Y, Kurachi Y. Clustering and enhanced activity of an inwardly rectifying potassium channel, Kir4.1, by an anchoring protein, PSD-95/SAP90. J Biol Chem. 1997;272:12885–12888. doi: 10.1074/jbc.272.20.12885. [DOI] [PubMed] [Google Scholar]
- 25.Katsuki H, Nakai S, Hirai Y, Akaji K, Kiso Y, Satoh M. Interleukin-1β inhibits long-term potentiation in the CA3 region of mouse hippocampal slices. Eur J Pharmacol. 1990;181:323–326. doi: 10.1016/0014-2999(90)90099-r. [DOI] [PubMed] [Google Scholar]
- 26.Keane J, Nicoll J, Kim S, Wu DM, Cruikshank WW, Brazer W, Natke B, Zhang Y, Center DM, Kornfeld H. Conservation of structure and function between human and murine IL-16. J Immunol. 1998;160:5945–5954. [PubMed] [Google Scholar]
- 27.Kistner U, Wenzel BM, Veh RW, Cases-Langhoff C, Garner AM, Appeltauer U, Voss B, Gundelfinger ED, Garner CC. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. J Biol Chem. 1993;268:4580–4583. [PubMed] [Google Scholar]
- 28.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 29.Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P, Hillman AG, Chartrain NA, Schmidt JA. Identification of a monocyte specific pre-interleukin 1β convertase activity. Proc Natl Acad Sci USA. 1989;86:5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krautwald S. IL-16 activates the SAPK signaling pathway in CD4+ macrophages. J Immunol. 1998;160:5874–5879. [PubMed] [Google Scholar]
- 31.Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 32.Kurschner C, Morgan JI. The maf proto-oncogene stimulates transcription from multiple sites in a promoter that directs Purkinje neuron-specific gene expression. Mol Cell Biol. 1995;15:246–254. doi: 10.1128/mcb.15.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurschner C, Morgan JI. Analysis of interaction sites in homo- and heteromeric complexes containing Bcl-2 family members and the cellular prion protein. Mol Brain Res. 1996;37:249–258. doi: 10.1016/0169-328x(95)00323-k. [DOI] [PubMed] [Google Scholar]
- 34.Kurschner C, Mermelstein PG, Holden WT, Surmeier DJ. CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol Cell Neurosci. 1998;11:161–172. doi: 10.1006/mcne.1998.0679. [DOI] [PubMed] [Google Scholar]
- 35.Lin JW, Wyszynski M, Madhavan R, Sealock R, Kim JU, Sheng M. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma WJ, Holz RW, Uhler MD. Expression of a cDNA for a neuronal calcium channel α1 subunit enhances secretion from adrenal chromaffin cells. J Biol Chem. 1992;267:22728–22732. [PubMed] [Google Scholar]
- 37.Marks N, Berg MJ, Guidotti A, Saito M. Activation of caspase-3 and apoptosis in cerebellar granule cells. J Neurosci Res. 1998;52:334–341. doi: 10.1002/(SICI)1097-4547(19980501)52:3<334::AID-JNR9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 38.Miao GG, Curran T. Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol Cell Biol. 1994;14:4295–4310. doi: 10.1128/mcb.14.6.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muhlhahn P, Zweckstetter M, Georgescu J, Ciosto C, Renner C, Lanzendorfer M, Lang K, Ambrosius D, Baier M, Kurth R, Holak TA. Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site. Nat Struct Biol. 1998;5:682–686. doi: 10.1038/1376. [DOI] [PubMed] [Google Scholar]
- 40.Ni B, Wu X, Du Y, Su Y, Hamilton-Byrd E, Rockey PK, Rosteck P, Jr, Poirier GG, Paul SM. Cloning and expression of a rat brain interleukin-1β-converting enzyme (ICE)-related protease (IRP) and its possible role in apoptosis of cultured cerebellar granule neurons. J Neurosci. 1997;17:1561–1569. doi: 10.1523/JNEUROSCI.17-05-01561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, Munday NA, Raju SM, Smulson ME, Yamin T-T, Yu VL, Miller DK. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis [see comments]. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 42.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omri B, Crisanti P, Alliot F, Marty MC, Rutin J, Levallois C, Privat A, Pessac B. CD4 expression in neurons of the central nervous system. Int Immunol. 1994;6:377–385. doi: 10.1093/intimm/6.3.377. [DOI] [PubMed] [Google Scholar]
- 44.Parnes JR. Molecular biology and function of CD4 and CD8. Adv Immunol. 1989;44:265–311. doi: 10.1016/s0065-2776(08)60644-6. [DOI] [PubMed] [Google Scholar]
- 45.Perry VH, Gordon S. Modulation of CD4 antigen on macrophages and microglia in rat brain. J Exp Med. 1987;166:1138–1143. doi: 10.1084/jem.166.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peudenier S, Hery C, Ng KH, Tardieu M. HIV receptors within the brain: a study of CD4 and MHC-II on human neurons, astrocytes, and microglial cells. Res Virol. 1991;142:145–149. doi: 10.1016/0923-2516(91)90051-4. [DOI] [PubMed] [Google Scholar]
- 47.Ponting CP, Phillips C, Davies KE, Blake DJ. PDZ domains: targeting signaling molecules to submembranous sites. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 48.Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF. The CD4 receptor is complexed in detergent lysates to a protein- tyrosine kinase (pp58) from human T-lymphocytes. Proc Natl Acad Sci USA. 1988;85:5190–5194. doi: 10.1073/pnas.85.14.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan TC, Cruikshank WW, Kornfeld H, Collins TL, Center DM. The CD4-associated tyrosine kinase p56lck is required for lymphocyte chemoattractant factor-induced T-lymphocyte migration. J Biol Chem. 1995;270:17081–17086. doi: 10.1074/jbc.270.29.17081. [DOI] [PubMed] [Google Scholar]
- 50.Sheng M, Wyszynski M. Ion channel targeting in neurons. BioEssays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 51.Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 52.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 53.Stanfield BB, Cowan WM. The development of the hippocampal region. Plenum; New York: 1988. [Google Scholar]
- 54.Tewari M, Quan LT, O’Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Yama/CPP32β, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 55.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano JR, Chin J, Ding GJ-F, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin R-T, Lee TD, Shively JE, MacCross M, Mumford RA, Schmidt JA, Tocci MJ. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 56.Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T-cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 57.Wu DM, Zhang Y, Parada NA, Kornfeld H, Nicoll J, Center DM, Cruikshank WW. Processing and release of IL-16 from CD4+ but not CD8+ T-cells is activation-dependent. J Immunol. 1999;162:1287–1293. [PubMed] [Google Scholar]
- 58.Yan GM, Ni B, Weller M, Wood KA, Paul SM. Depolarization or glutamate receptor activation blocks apoptotic cell death of cultured cerebellar granule neurons. Brain Res. 1994;656:43–51. doi: 10.1016/0006-8993(94)91364-1. [DOI] [PubMed] [Google Scholar]
- 59.Yan HQ, Banos MA, Herregodts P, Hooghe R, Hooghe-Peters EL. Expression of interleukin (IL)-1β, IL-6, and their respective receptors in the normal rat brain and after injury. Eur J Immunol. 1992;22:2963–2971. doi: 10.1002/eji.1830221131. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Center DM, Wu DMH, Cruikshank WW, Yuan J, Andrews DW, Kornfeld H. Processing and activation of pro-interleukin-16 by caspase-3. J Biol Chem. 1998;273:1144–1149. doi: 10.1074/jbc.273.2.1144. [DOI] [PubMed] [Google Scholar]
- 61.Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: inhibition of mRNA expression. Nat Med. 1997;3:659–664. doi: 10.1038/nm0697-659. [DOI] [PubMed] [Google Scholar]