Summary
Motivations intensify over hours or days, promoting goals that are achieved in minutes or hours, causing satiety that persists for hours or days. Here we develop Drosophila courtship as a system to study these long-timescale motivational dynamics. We identify two neuronal populations engaged in a recurrent excitation loop, the output of which elevates a dopamine signal that increases the propensity to court. Electrical activity within the recurrent loop accrues with abstinence and, through the activity-dependent transcription factor CREB2, drives the production of activity-suppressing potassium channels. Loop activity is decremented by each mating to reduce subsequent courtship drive, and the inhibitory loop environment established by CREB2 during high motivation slows the reaccumulation of activity for days. Computational modeling reproduces these behavioral and physiological dynamics, generating predictions that we validate experimentally and illustrating a causal link between the motivation that drives behavior and the satiety that endures after goal achievement.
eTOC BLURB
Zhang et al. show that the male fly’s mating drive is stored in a recurrent excitation loop. Excitation of loop neurons drives the transcription factor CREB to create an inhibitory environment that prevents rapid reaccumulation after loop activity is reduced by mating. Prior motivation is therefore a direct cause of enduring satiety.
Introduction
Though subject to hormonal regulation, motivational states are stored, adjusted, and used for decision making within the central nervous system. Our understanding of these processes is fragmentary even in well-studied models, especially regarding the diversity of timescales involved [1,2]. In the control of mammalian feeding behavior, slow-acting hormonal signals are immediately overridden and a rapid decrease in hunger-promoting AgRP neuronal activity is triggered by the sensory detection of food [3–5]. The effects of prior AgRP neuronal activity are surprisingly long-lasting: animals continue to show signs of increased hunger at least 30 minutes after the cessation of optogenetic stimulation [6]. Similar slow, fast, and anticipatory dynamics are seen in the neuronal control of thirst [7,8]. Here we present a functional molecular- and circuit-level description of motivational changes in response to abstinence and fulfillment, over timescales of minutes and days, centering on neurons that produce Neuropeptide F (NPF): the Drosophila homolog of NPY that is released by AgRP neurons to promote sustained hunger [9].
The reproductive behaviors of Drosophila offer several advantages for the study of motivation. Primary among these is the ability to identify and characterize defined circuit elements, guided by the transcription factors Fruitless and Doublesex that label sexually dimorphic neurons [10–12]. In the circuitry underlying male courtship behavior, neuronal populations have been described that receive sensory input from the courtship target [13,14] and that transform this information into parallel excitatory and inhibitory signals in the central brain [15,16]. These signals are received by command neurons, called P1 [15–21], that drive courtship behavior when sufficiently stimulated. Motivational control over P1 activity is supplied by a local dopamine tone that is tuned to reflect the male’s recent mating history and reproductive potency, through mechanisms that were previously unknown [22]. This motivating dopaminergic signal is received directly by P1 and biases the relative impact of the sensory-derived excitatory and inhibitory signals, adjusting the probability of courtship for each encounter with a potential target [23]. Here we identify and characterize three new circuit elements that sustain the motivating dopaminergic signal in the absence of a female, reduce it in response to copulation, and then orchestrate its gradual recovery over several days.
Copulation Reporting Neurons detect matings and induce satiety
In this study we use three behavioral assays that have been developed to assess various aspects of male mating drive: (i) standard, one-male-one-female, 5-minute courtship assays that report the fraction of time spent courting, which approaches a ceiling value of 1 when a naive male is placed with a virgin female (modified from Siegel and Hall, 1979); (ii) tap-induced courtship probability assays [23], which are similar to standard assays but report the probability that each contact with a female will trigger a courtship bout, providing greater sensitivity and removing the ceiling effect of standard courtship assays to allow assessment of hypersexuality; and (iii) satiety assays [22] in which a male is placed with ~15 females and his mating behaviors are scored over 4.5 hours, either continuously (videotaped in a flat chamber) or at 30 minute intervals (by eye in a standard food vial).
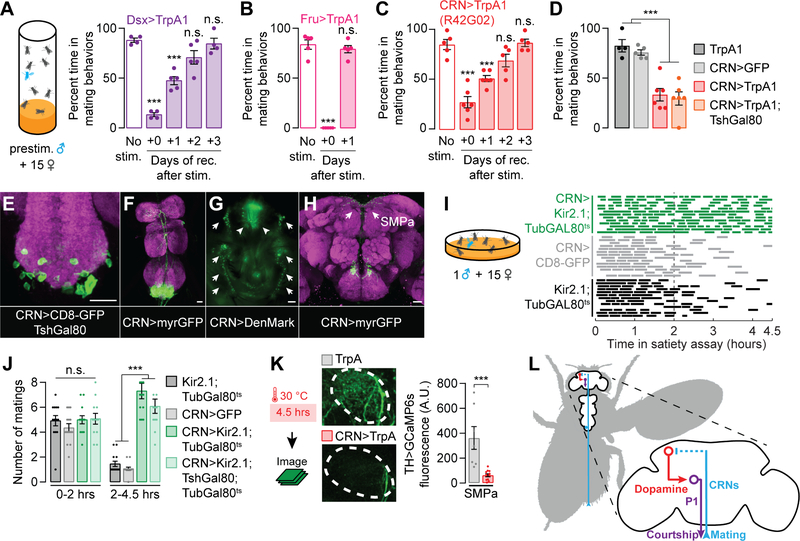
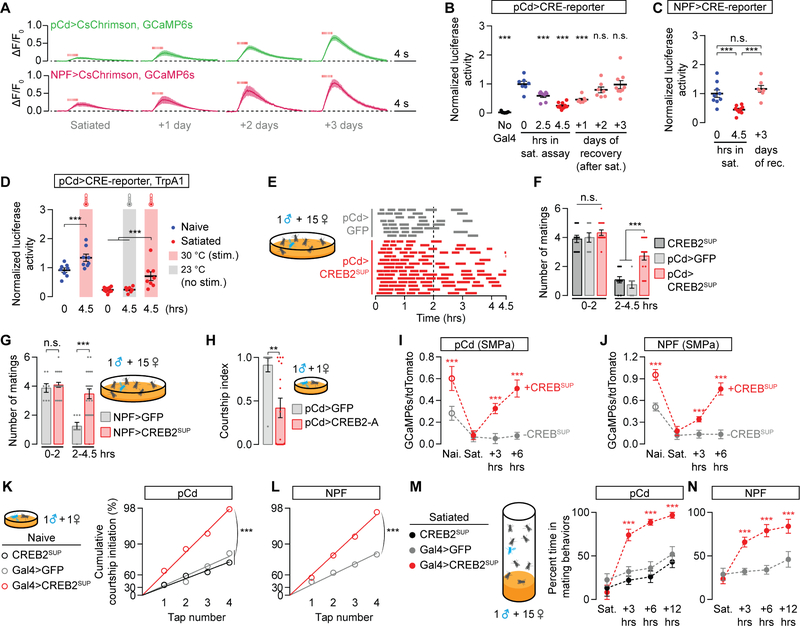
In the satiety assay, a naive male’s sexual activity declines with each mating, reaching maximum satiety after ~3–6 copulations [22]. To identify the neurons that trigger copulation-induced decrements in mating drive, we attempted to artificially induce satiety in naïve males through thermogenetic stimulation [25] of sexually dimorphic neurons, labeled by the transcription factors Fruitless (Fru) or Doublesex (Dsx) [10–12]. After pre-stimulating either the ~500 Dsx neurons or the ~2500 Fru neurons for 4.5 hrs (the length of a satiety assay), males showed a dramatic reduction in mating behaviors (Figure 1A and 1B). While the Fru>TrpA1 males recovered their mating behaviors relatively soon after the cessation of stimulation, Dsx>TrpA1 males returned to sexual behavior only gradually, over several days, similar to the time-course of natural satiety (Figure 1A and 1B) [22]. To identify the specific Dsx neurons responsible for inducing artificial satiety, we screened a set of Gal4 lines that subdivide the Dsx population [26]. We isolated a set of ~50 neurons labeled by R42G02-Gal4 that recapitulate the enduring satiety effect after stimulation (Figure 1C and S1A). Locomotion was not affected by the activation of these neurons (Figure S1B), arguing against a non-specific behavioral impairment. The induction of artificial satiety using R42G02-Gal4 was prevented by blocking TrpA1 expression in the Dsx+ neurons (Figure S1C), which only overlap in the abdominal ganglion of the ventral nervous system (VNS) [26]. The relevant neurons are likely to be cholinergic as an RNAi transgene targeted against the acetylcholine transporter prevented artificial satiety (Figures S1D–S1G). Artificial satiety was unaffected when about half of the abdominal R42G02 neurons were subtracted using Teashirt-Gal80 (TshGal80; Clyne and Miesenböck, 2008) (Figure 1D), which blocks Gal4 activity in many, but not all, neurons of the VNS. This allowed us to provisionally ascribe the satiety-inducing phenotype to a population of ~25 neurons with cell bodies localized to the abdominal ganglion (Figures 1E). These neurons project dendrites to the genitalia and branching axon tracts to the top of the brain (Figures 1F–1H and S1H–S1L), a configuration that suggests the reporting of copulation status to mating drive circuitry. We therefore refer to this subset of R42G02 neurons as the Copulation Reporting Neurons (CRNs), though we suspect that only a subpopulation mediates the satiety-promoting effect. When the CRNs were silenced or were unable to release acetylcholine, baseline courtship drive was unaffected (Figure S1M), but the males became largely insatiable for the duration of a 4.5-hour satiety assay (1 male housed with ~15 virgin females) (Figures 1I–1J and S1N). These results are consistent with an inability to report copulations to the brain. Though in these experiments the CRNs were activated or silenced for long periods of time, toward the end of this study we will show that their activity is only transiently necessary during mating to incrementally induce satiety (Figure 7).
Figure 1. Copulation Reporting Neurons (CRNs) induce a persisting decrease in mating drive.
(A–C) Thermogenetic prestimulation (4.5 hrs) of all Doublesex (Dsx) neurons (A), all Fruitless (Fru) neurons (B), or the CRNs labeled by R42G02-Gal4 (C) induces an immediate loss of mating behaviors, but only the Dsx- and CRN-stimulated flies take days to recover from the stimulation (***p<0.001 compared to the no-stim group†, one-way ANOVA, A: n = 4–5 groups of 5–7 males, B: n = 5 groups, C: n = 5–6 groups). Throughout this figure, sexual activity was scored in the satiety assay (cartoon) to allow comparison with naturalistic satiety [22]. See Figure S1A for data using other Dsx-related Gal4 lines. (D) CRN-induced artificial satiety is not blocked by TshGal80, a Gal4 inhibitor expressed in many VNS neurons, allowing us to more closely approximate the relevant population within the VNS (one-way ANOVA, n = 4–6 groups of 5–7 males). (E–H) The CRNs have cell bodies in the abdominal ganglion (E-F)‡ and project dendrites (arrows) around the male’s external genitalia (arrowheads) (G), and axons to the top of the brain (H); neuropil in magenta. (I–J) Conditionally silencing the CRNs throughout the assay prevents satiety, which is most apparent in the second half of the assay (2–4.5 hours). This effect is not blocked by TshGal80 (one-way ANOVA, n = 12–14 males). Matings of individual flies are represented by dashes within single rows in I. (K) Thermogenetic stimulation of the CRNs decreases calcium activity (measured with GCaMP6s) in dopaminergic projections to the SMPa (t-test, n = 7–11 male brains). See Figure S1P for fluorescence data from an unrelated brain region. (L) A circuit model for how CRNs relay mating information to induce satiety. †*p<0.05, **p<0.01, ***p<0.001 for all figures. Error bars represent 1 S.E.M. in all figures. ‡Unless otherwise stated, all scale bars represent 20 µm. See Table S1 for the numbers of animals and experimental conditions used in each experiment. The image stacks for the max projections in this and other figures are uploaded to Mendeley Data.
Figure 7. Recurrent loop dynamics make the reporting of goal achievement robust to duration.
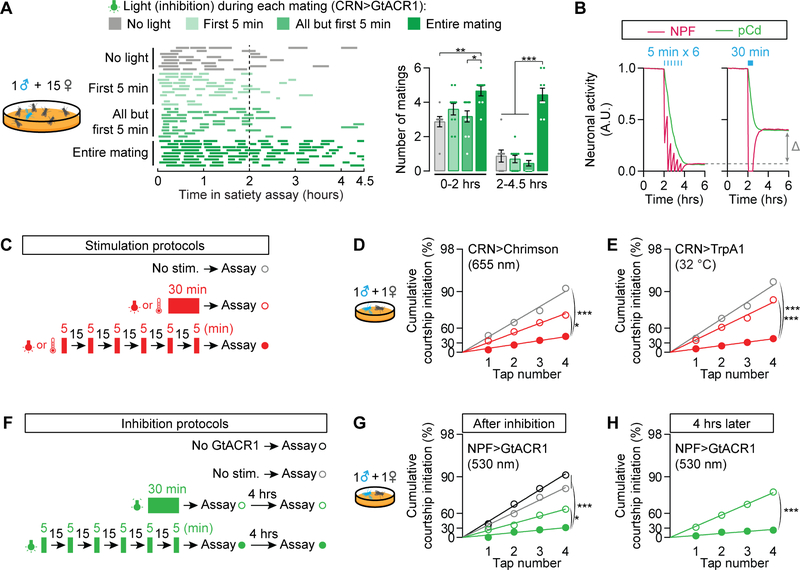
(A) The CRNs must be silenced throughout the entirety of copulation, but not between copulations, to prevent satiety (one-way ANOVA, n = 8–11 males). See Figure S6B for verification of hyperpolarization with GtACR1. (B) In the computational model, spaced CRN stimulations more effectively induce satiety than does a single pulse of equivalent duration. (C) Stimulation strategy for D–E. Flies experience either no stimulation, a single 30-min block of CRN stimulation, or 6 bouts of 5-min CRN stimulations. (D-E) Spaced optogenetic (D) or thermogenetic (E) stimulation of the CRNs induces stronger satiety (closed red circles) than collapsed stimulation of the same duration (open red circles) (bootstrap, D: n = 33–37 males, E: n = 29–44). (F) Silencing strategy for G–H. Flies experience either no silencing, a single 30-min block of NPF silencing, or 6 bouts of 5-min NPF silencing. See Figure S6C for verification of neuronal silencing with GtACR1. (G) Spaced optogenetic silencing of NPF neurons induces stronger satiety (closed green circles) than collapsed silencing of the same duration (open green circles) (bootstrap, n = 32–35 males). (H) Satiety induced by spaced NPF silencing persists for at least 4 hours (bootstrap, n = 28–33 males). See also Table S1.
Dopaminergic activity in the anterior of the Superior Medial Protocerebrum (SMPa) is a functional neuronal correlate of mating drive, modulating the response of P1 courtship command neurons to stimulation by female perceptual input [22,23]. This dopamine signal is maintained ex vivo and progressively decreases after repeated matings (Figure S1O) [22]. CRN stimulation caused a persisting decrement in dopaminergic activity in the SMPa (Figures 1K and S1P, see Figure 1H for SMPa location) and did not revert the loss of mating drive in 3–4 day old DopR2 mutant males (Figure S1Q) which cannot process the motivating dopamine signal [22]. These results suggest that the CRNs induce satiety by translating the detection of copulation into an enduring decrement in the dopamine tone received by P1 courtship command neurons (Figure 1L).
pCd and NPF neurons promote sustained increases in mating drive
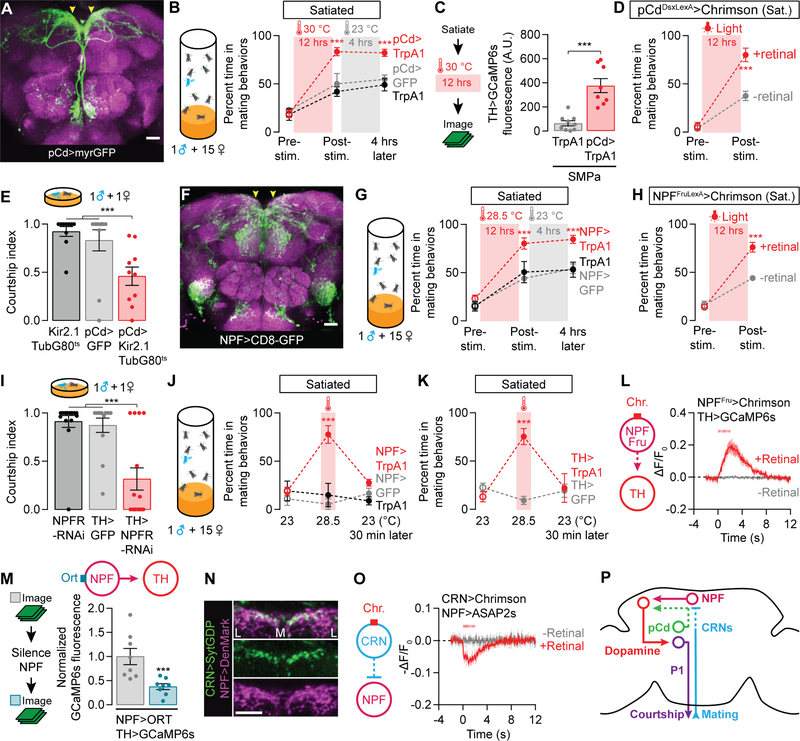
Though CRN axons project to the SMPa, they do not appear close enough to directly contact the courtship-motivating dopamine neurons (our observations). We therefore searched for neurons that might convert CRN excitation into a decrement in dopaminergic activity. We again screened the collection of Gal4 lines that subsets the Dsx neurons [26] looking for lines with the opposite phenotype of the CRNs: hastening recovery from satiety when stimulated (Figures S2A). Stimulating the neurons labeled by R41A01-Gal4 (Figure 2A) for twelve hours resulted in a complete and lasting reversion of both sexual behavior (Figure 2B and S2B) and dopaminergic activity in the SMPa (Figures 2C and S2C–S2D), without influencing general locomotor activity (Figure S2E). This recovery of mating drive is much faster than seen in control animals, which is only slightly sped up by the elevated temperature. The accelerated recovery is also seen when the stimulation is restricted to Dsx+ neurons within this line and is not blocked by TshGal80. These intersectional manipulations circumscribe a population of ~7 neurons in the brain that has been previously named pCd (Figures 2D and S2F) [26]. Silencing the pCd neurons lowered mating drive (Figure 2E) but, surprisingly, their acute stimulation had no effect on courtship behavior (Figure S2G). Moreover, the immediate electrical activity of the pCd neurons was unaffected by acute CRN stimulation (Figure S2H). These results suggest that courtship motivation is only influenced by pCd activity over long timescales, and reciprocally, that pCd activity is responsive to reproductive goals only well after they have been achieved. We therefore searched for neurons that are more immediately responsive to the CRN activation, and whose activity directly controls the motivating dopamine tone.
Figure 2. pCd and NPF neurons recharge mating drive after satiety.
(A–B) Prolonged thermogenetic stimulation of pCd neurons (A) accelerates the recovery from satiety, an effect that lasts long after the stimulation ceases (B) (two-way ANOVA, n = 4–8 groups of 5–7 males). Arrowheads point to the SMPa. See Figure S2A for data using other Dsx-related Gal4 lines. (C) Thermogenetic stimulation of pCd neurons accelerates the recovery of calcium levels in dopaminergic projections at the SMPa (t-test, n = 10–11 male brains). See Figure S2C for sample images and Figure S2D for fluorescence data from an unrelated brain region. (D) Prolonged optogenetic stimulation of Dsx+ pCd neurons speeds the recovery of mating drive (flies that have not been fed the obligate chromophore retinal serve as a control group; two-way ANOVA, n = 5 groups 5–7 males each). (E) Conditional, long-term silencing of pCd neurons in adult males decreases courtship (one-way ANOVA, n = 10–11 males). (F–H) Prolonged thermogenetic stimulation of all NPF neurons (G) or the Fru+ subset (H) that projects to the SMPa (arrowheads in F, also see Figure S2K), accelerates recovery from satiety (two-way ANOVA, G: n = 5–6 groups of 5–7 males, H: n = 5 groups). (I) Knockdown of NPFR in dopaminergic neurons decreases courtship (one-way ANOVA, n = 13–16). (J–K) Short-term (20 min) thermogenetic stimulation of NPF (J) or dopaminergic (K) neurons reverts satiety, but this effect is time-locked to the stimulation (two-way ANOVA, J: 5–8 groups of 5–7 males, K: n = 5–6 groups). (L) Optogenetic stimulation (8 ms pulses, 15.5 Hz for 2 s) of the Fru+ NPF neurons excites dopamine neurons projecting to the SMPa (n = 5–7 male brains). (M) Chemogenetic silencing of NPF neurons reduces the baseline calcium activity in dopaminergic projections to the SMPa (one-way ANOVA with data in Figures S3A–S3B, n = 8). See Figures S3A–S3B for controls and pCd-silencing experiments. (N) The ascending axons of the CRNs terminate near NPF dendrites in the SMPa. Letters L and M delineate the medial-lateral axis. (O) Optogenetic stimulation (8 ms pulses, 15.5 Hz for 2 s) of the CRNs causes hyperpolarization of the NPF projections at the SMPa (n = 5–8 male brains). (P) The CRNs decrease mating drive through inhibition of NPF neurons. See also Tables S1 and S2.
In mammals, Neuropeptide Y (NPY) is released from AgRP neurons and increases the motivation to eat both during and after AgRP stimulation [9,28,29]. Given the phenomenological similarities between mammalian hunger and fly mating drive, and that in the fly NPF signals directly to dopaminergic neurons in the motivational control of memory formation [30], we examined a possible role for NPF in courtship motivation. Overnight stimulation of NPF neurons (Figure 2F) led to a persisting recovery of mating drive without affecting locomotion (Figure 2G and S2I–S2J), similar to the effects seen with pCd stimulation (Figure 2B). This drive-promoting effect is mediated by the ~4 Fru+ NPF neurons per hemisphere (Figures 2H and S2K–S2M) which output their activity directly to the motivating dopaminergic neurons through the NPF Receptor (NPFR) (Figures 2I and S2N–S2O). This finding is supported by the close proximity of NPF axons to dopaminergic fibers in the SMPa (Figure S2P) as well as recent sequencing results which detected the NPFR in sexually dimorphic dopamine neurons (Table S2, see STAR Methods) [31,32]. Unlike the pCd neurons, and consistent with a direct connection to dopaminergic neurons, acute stimulation of the NPF neurons mimicked the drive-promoting effect of dopaminergic stimulation (Figures 2J–2K and S2Q). As expected, acute stimulation of all NPF neurons, or just the Fru+ subset, elevated dopaminergic activity in the SMPa (Figures 2L and S2R–S2T) and its behavioral effects required the DopR2 receptor (Figure S2U). The calcium increase in the dopamine neurons after NPF stimulation began as rapidly as could be captured by our frame rate (15.5 fps; Figure S2T, see STAR Methods). When NPF neurons were acutely silenced using the histamine-gated chloride channel Ort [33], the calcium activity in the dopaminergic projections at the SMPa was reduced (Figures 2M). This reduction was stronger than that observed following pCd silencing (Figure S3A and S3B), again indicative of a direct NPF-to-dopamine connection.
The ascending CRN projection terminates in close proximity to NPF dendrites (Figure 2N), suggesting transmission of the signal that decreases mating drive in response to copulation. Though acetylcholine (the likely CRN neurotransmitter) is generally thought of as excitatory in the fly, it can also be inhibitory [34]. Consistent with this idea, the NPF neurons were rapidly hyperpolarized by CRN stimulation (Figures 2O and S3C–S3D), an effect not seen in pCd neurons (Figure S2H). While conclusive proof of direct connectivity will likely require reconstruction from electron microscopy, at a minimum these results demonstrate a flow of information from the initiation of copulation, through the CRNs, causing inhibition of the NPF neurons and a resultant decrease in the activity of the dopaminergic neurons that determine the propensity to enter the courtship state (Figure 2P).
Recurrent excitation between pCd and NPF neurons accumulates and stores mating drive
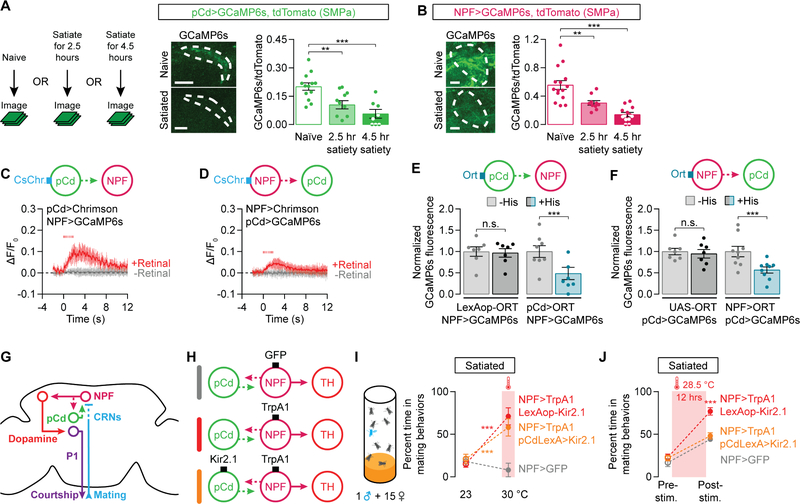
Perhaps the most striking feature of pCd and NPF neurons is their enduring impact on motivation after the cessation of stimulation, an effect also seen with AgRP stimulation [6,35], and that requires NPY release [9]. In the pCd and NPF neurons, these persisting effects require more than 6 hours of stimulation (Figure S3E) and are not seen following stimulation of the dopaminergic or P1 neurons (Figures S3F–S3G). Since recurrent activity maintains network states for motor planning and working memory [36–38], and sustains bouts of courtship in flies [23] and aggression in mice [39], we hypothesized that pCd and NPF neurons might form a recurrent excitatory loop that accumulates and sustains mating drive. Like the dopaminergic neurons, pCd and NPF projections to the SMPa have high levels of baseline calcium in naïve, highly-motivated males, and their activities gradually decline as males become satiated (Figures 3A–3B). Reciprocal manipulation and monitoring of pCd and NPF activities was strongly indicative of a recurrent circuit logic: stimulating one population rapidly (within ~100 ms) increased calcium activity in the other (Figures 3C–3D and S3H–S3K, see STAR Methods for estimating delay times) and silencing one population decreased the other’s activity (Figures 3E and 3F). A recurrent circuit is also anatomically suggested by the intermingled axons and dendrites of the two populations in the SMPa (Figure S3L). However, we again emphasize that we cannot say with certainty that the pCd and NPF neurons are directly connected; in fact, our results below lead us to predict that there are other neuronal populations within this recurrent circuitry. Stimulation of the dopaminergic neurons did not alter the activity of either pCd or NPF neurons (Figures S3M–S3N), consistent with the inability of dopamine to promote an increase in motivation that outlasts the stimulation (Figure S3F). Together, these results suggest that a recurrent excitation loop involving pCd and NPF neurons sustains the activity that i) is inhibited by the CRNs during copulation, and ii) instructs the activity of dopaminergic neurons (Figure 3G). Neurons that respond to the output of the loop (dopaminergic and P1) can influence mating behavior in real time, but changes in their activities have no enduring effect on the system.
Figure 3. Recurrent excitation between pCd and NPF neurons recovers and sustains mating drive.
(A–B) Satiety decreases calcium activity in the projections of pCd (A) and NPF (B) neurons to the SMPa (dashed lines, drawn using the tdTomato channel) (one-way ANOVA, A: n = 6–8 male brains, B: n = 13–15). (C–D) Optogenetic stimulation (8 ms pulses, 15.5 Hz for 2 s) of either pCd (C) or NPF (D) neurons evokes calcium transients in the other population (C: n = 5–6 male brains, D: n = 6). See Figures S3J and S3K for similar experiments that use chemogenetic stimulation. (E–F) Chemogenetic silencing of either pCd (E) or NPF (F) neurons with the histamine-gated chloride channel Ort [33] decreases calcium activity in the other population (normalized to average levels before Histamine application; one-way ANOVA, E: n = 8 male brains each, F: n = 9 each). (G) A recurrent loop between pCd and NPF neurons holds, adjusts, and outputs mating drive information. (H) Experimental designs for I and J. (I) Acute thermogenetic stimulation of NPF neurons reverts satiety (red), even when pCd neurons are silenced (orange) (two-way ANOVA, n = 5 groups of 5–7 males each). (J) Silencing pCd neurons (orange) blocks the recharging effect of prolonged NPF stimulation (red) (two-way ANOVA, n = 5–6 groups of 5–7 males). See also Tables S1 and S2.
The behavioral and physiological results described above are in close agreement, with one exception: acute pCd stimulation elevates NPF activity (Figure 3C) but cannot immediately induce courtship from satiated males (Figure S2G). Consistent with these behavioral results, acute pCd stimulation does not significantly elevate dopaminergic activity in satiated males (Figure S3O), whereas satiety had little effect on the ability of NPF neurons to elevate dopaminergic activity (Figure S2R). These results may be explained if pCd neurons activate only a subset of NPF neurons, or by their indirect effect on dopaminergic neurons. Regardless, it is clear that the pCd neurons are critical for the long-term accumulation and maintenance of mating drive: stimulating the NPF neurons while silencing the pCd neurons (Figure 3H) acutely reverted satiety (Figure 3I), but the enduring post-stimulation effect was not seen (Figure 3J). Inversely, silencing NPF neurons abolishes the recharging effects of pCd stimulation (Figure S3P). These results show that it is the recurrent circuitry, rather than either pCd or NPF neurons alone, that holds and adjusts the motivational activity over hours and days.
CREB2 activity in recurrent circuitry slows the re-accumulation of mating drive
The sub-second ramping of electrical activity within recurrent circuitry seen in other systems (e.g., Inagaki et al [40]) is orders of magnitude faster than the stimulation duration required to permanently revert satiety (Figures 2B, 2G, and S3E). This suggests that activity ramping, on its own, is unlikely to account for the extremely slow recovery from satiety. By monitoring the calcium response to optogenetic stimulation of pCd or NPF neurons we found that the intrinsic excitability of loop neurons is dramatically reduced by satiety and gradually recovers over three days (Figure 4A) This suggests that molecular changes intrinsic to the loop instruct the satiety state, as in circadian rhythms and memory [41,42]. The activity-dependent transcription factor CREB functions in both circadian rhythms and memory [43–46], and its fly homolog, CREB2, was detected in pCd and NPF neurons (Table S2, see STAR Methods) [31,32]. Using a luciferase reporter of CREB2’s transcriptional activity [46,47] in pCd neurons, we found a striking correlation between mating drive and CREB2 activity, which was high in naive males, gradually declined during the satiety assay, and slowly recovered over three days (Figure 4B). In naïve animals, CREB2 activity was decreased to satiety-like levels by expression of a truncated, dominant-negative version of CREB2 (CREB2DN) (Figure S4A) [48], attesting the specificity of the reporter. Similar CREB2 activity dynamics were seen in NPF neurons, but not in other courtship-regulating neurons (Figures 4C and S4B–S4C), and CREB2 activity was elevated by experimental stimulation of loop neurons (Figure 4D). To test the impact of CREB2 on mating drive, we disrupted its function in loop neurons using three independent methods: (i) the transcription-suppressing B isoform of CREB2 (CREB2SUP) (Figures 4E and 4F) [46], (ii) CREB2DN (Figure S4D), and (iii) a CREB2-RNAi construct (Figure S4E). Expressing any of these three transgenes in the pCd neurons slowed the onset of satiety induced by repeated matings. A similar effect was seen in NPF neurons (Figure 4G), but not in the non-loop courtship-drive circuitry we examined (Figures S4F–S4G). Inversely, when the transcription-promoting isoform A of CREB2 was overexpressed in pCd neurons, mating drive was suppressed (Figure 4H).
Figure 4. CREB2 activation during high motivation prolongs subsequent satiety.
(A) Optogenetic stimulation (8 ms pulses, 15.5 Hz for 2 s) of pCd or NPF neurons evoked increasingly large calcium transients as males recovered from satiety (n = 6–7 brains). (B) CREB2 activity in pCd neurons tracks mating drive (one-way ANOVA, n = 7–11 groups of 3 brains; activity is normalized to the average signal from naïve males). See Figure S4A for validation of the CREB reporter. Table S4 for raw luciferase data. (C) CREB2 activity in NPF neurons also tracks mating drive (one-way ANOVA, n = 7–9 groups of 3 male brains). (D) Thermogenetic stimulation of pCd neurons increases CREB2 activity in both naïve and satiated animals (one-way ANOVA, n = 7–10 groups of 3 male brains). See Figure S4B for no-TrpA1 controls (E–G) Expressing the transcription-suppressing isoform of CREB2 (CREB2SUP) in pCd (E-F) and NPF neurons (G) slows the onset of satiety (one-way ANOVA, E: n = 8–15 males, F: n = 8–15, G: n = 6–21). (H) Overexpressing the transcription-promoting isoform A of CREB2 in pCd neurons decreases courtship in naïve males (one-way ANOVA, n = 10–16 males). (I–J) Expression of a transcription-suppressing version of CREB2, CREB2SUP, increases the baseline SMPa calcium activity of both pCd (I) and NPF neurons (J) and accelerates its recovery after satiety (one-way ANOVA, I: n = 9–13 males, J: n = 10–13). (K–L) CREB2SUP expression in either pCd (K) or NPF (L) neurons increases baseline mating drive as measured by tap-induced-courtship probabilities. We use this assay to remove the ceiling effect for naïve males in standard courtship assays. We do not plot error bars for tap-induced courtship assays due to distortions caused by the log-scaled y-axis (bootstrap, see STAR Methods, K: n = 33–48 males, L: n = 42–44). (M–N) CREB2SUP expression in either pCd (M) or NPF (N) neurons accelerates recovery from satiety (two-way ANOVA, M: n = 4–5 groups of 5–7 males, N: n = 5 groups each). See also Table S1.
The above results show that CREB2 has a satiety-promoting role in pCd and NPF neurons—but also that CREB2 is activated by electrical excitation in the recurrent loop. Paradoxically then, CREB2 would seem to promote satiety when the male has high mating drive. To explain this apparent contradiction, we hypothesized two functions for CREB2 as a feedback inhibitor, analogous to its role in the mammalian nucleus accumbens [45,49]: (i) to prevent runaway excitation in the loop and keep mating drive within an appropriate range; and (ii) to restrain post-mating reaccumulation of activity in the pCd/NPF loop and slow the recovery of drive. Consistent with function (i), suppressing CREB2 activity in the pCd or NPF neurons of naïve males increased their baseline calcium activity and mating drive (Figures 4I–4L). Though these animals could eventually be sated, their behavioral recovery from satiety (Figures 4M–4N) and physiological recovery of loop activity (Figures 4I–4J), was much faster than seen in control males, consistent with function (ii). These findings support the idea that excitation-driven CREB2 activity restrains peak mating drive and tunes the recovery from satiety to match the time required for the replenishment of reproductive fluids after repeated matings [22].
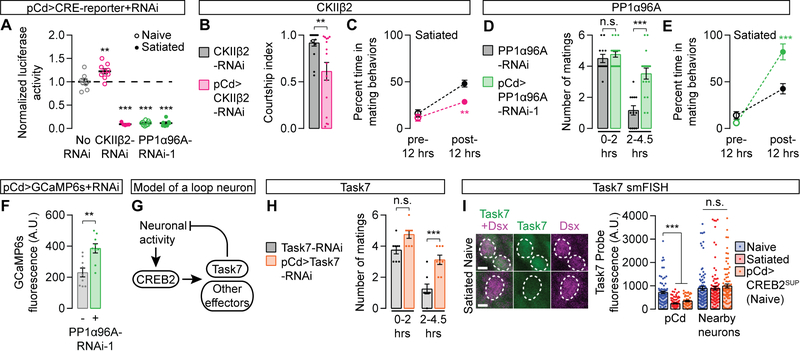
In mammals, phosphorylation at Serine-133 is critical for CREB activity [50]. In the fly brain, this site appears to be constitutively phosphorylated, and a different set of inhibitory phosphorylation sites have been suggested to regulate CREB2’s transcriptional activity [51]. In an RNAi screen of 30 candidate kinases and phosphatases that had previously been implicated in CREB regulation (see STAR Methods), we identified two opposing modifiers of CREB2 activity in mating drive control: the kinase CK2βII and the phosphatase PP1α96A (Figures S5A and Table S2). RNAi knockdown of CK2βII in pCd neurons increased baseline CREB2 activity, decreased mating drive, and slowed recovery from satiety (Figures 5A–5C), consistent with its CREB2-suppressing effect in biochemical studies [51]. Inversely, knocking down PP1α96A with two independent RNAi lines nearly eliminated detectable CREB2 activity in naïve males (Figures 5A and S5B), slowing the onset of, and hastening recovery from, satiety (Figures 5D–5E and S5C–S5D). PP1α96A knockdown in NPF neurons also increased mating drive (Figure S5E), suggesting similar CREB2-regulating machinery in both populations of loop neurons. PP1α96A does not activate CREB2 through an indirect effect on neuronal activity, as knocking down PP1α96A increased baseline calcium activity in pCd neurons (Figure 5F), a predicted consequence (not cause) of reduced CREB2 activation. This separation of CREB2 activity (low) from loop activity (high) in PP1α96A knock-downs suggests that electrical activity may work through phosphorylation to influence CREB2 activity in these neurons (Figure 5G). The identification of multiple regulators of CREB2 activity within the loop underscores the ability of cells to tune CREB activity to meet the temporal needs of circuits underlying a range of slowly-evolving brain functions (Figure 5G).
Figure 5. Transcriptional control of motivation by CREB2.
(A) RNAi knockdown of CKIIβ2 modestly increases, while PP1α96A-RNAi strongly decreases, CREB2 activity in pCd neurons of naïve males (statistical notations show comparisons with the no-RNAi group, one-way ANOVA, n = 5–9 groups of 3 male brains). (B–C) RNAi knockdown of CKIIβ2 in pCd neurons decreases courtship (B) and slows the recovery from satiety (C) (B: t-test, n = 16 males each; C: two-way ANOVA, n = 5 groups of 5–7 males). (D–E) Knocking down PP1α96A in pCd neurons slows the onset of satiety (D) and accelerates the recovery from satiety (D: one-way ANOVA, n = 12–17 males; E: two-way ANOVA, n = 5 groups of 5–7 males). (F) Knocking down PP1α96A in pCd neurons increases baseline calcium activity (t-test, n = 9 males each). (G) Model for a CREB2-imposed inhibitory environment in the loop during high motivation. (H) Knocking down Task7 in pCd neurons slows the onset of satiety (one-way ANOVA, n = 8 males each). (I) Satiety decreases Task7 transcript levels in pCd neurons (identified by location in the brain and Dsx smFISH probes, magenta), but not in neighboring Dsx− neurons. A similar decrease in Task7 transcript level is seen in the pCd neurons of naive males that express CREB2SUP (one-way ANOVA, n = 50–134 neurons from 9–17 brains). Scale bar represents 2 µm. See also Figure S5 and Tables S1–S4.
To identify the targets of CREB2 activity in loop neurons, we examined all nine inhibitory ion channels that contain putative CREB binding sites (cAMP Response Elements, or CREs) in their cis-regulatory DNA (see STAR Methods). We found a strong drive-promoting phenotype when knocking down the potassium leak channel subunit Task7 in pCd neurons (Figure 5H). Drosophila Task7 functions in heteromeric two-pore potassium channels [52], and its transcripts have been detected in pCd neurons (Table S2) [31,32]. To ensure that changes in motivation caused by Task7 do not reflect a trivial consequence of potassium channel manipulation, but a mechanism actually employed by loop neurons, we fluorescently labeled Task7 transcripts in situ [53]. A strong correlation between Task7 probe aggregation and CREB2 activity was seen in pCd neurons: high in naïve males, low in satiated males, and low in naïve males with experimentally reduced CREB2 activity (Figure 5I). RNAi knockdown of Task7 in the pCd neurons caused a similar decrease in Task7 probe aggregation (Figure S5F). These results identify Task7 as a key effector through which CREB2, during periods of high motivation, imposes an inhibitory environment that will sustain satiety after goal achievement. This suggests the idea that the protein lifetimes of potassium channels, which have generally been measured in the tens of hours [54,55], may be critical determinants of the time over which motivation recovers following satiety.
Modeling the rapid sating and gradual recovery of mating drive
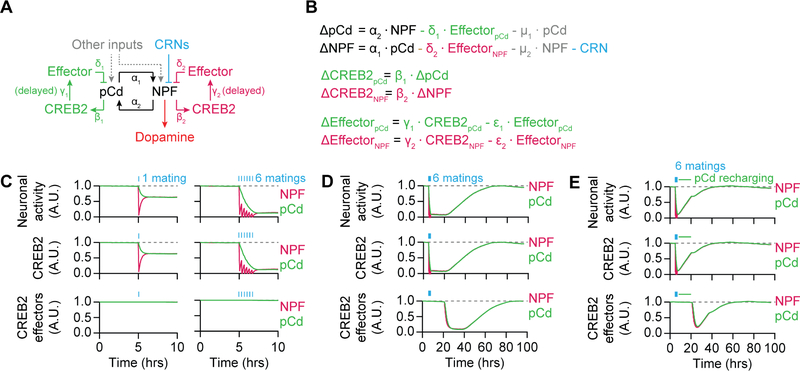
To ask whether the circuit and molecular mechanisms we have identified can function together to reproduce the behavioral and physiological dynamics of this system, we generated a simple, linear, computational model (Figures 6A and 6B; see STAR Methods). In the model, the NPF and pCd populations excite one another and receive additional excitatory inputs. Two experimental observations support the inclusion of these additional inputs: i) silencing either pCd or NPF neurons only modestly reduces the other’s activity (Figures 3E and 3F); and ii) activation of the CRNs rapidly hyperpolarizes NPF neurons, but no immediate effect is seen on pCd neurons (Figures 2O, S2H, S3C, and S3D). CREB2’s transcriptional activity in pCd and NPF neurons is driven by electrical activity—but, since CREB2’s targets (e.g. Task7) must be transcribed, translated, processed, and transported, the inhibitory impact of CREB2 activation is delayed relative to changes in loop activity (Figures 6A and 6B). Critically, since CREB2’s transcriptional targets likely endure after loop activity falls, we introduced a delay in their decline after mating. The resulting model equilibrates at a steady state (high mating drive) in which both pCd and NPF neurons have stable electrical, CREB2 transcriptional, and CREB2 effector, activities (Figure S6A). Copulation disrupts this equilibrium by triggering an immediate decrement in NPF neuronal activity which, in turn, causes a slower decline in pCd activity (Figure 6C). Over time, CREB2’s inhibitory effectors are gradually removed, allowing loop activity to recover and re-equilibrate (Figure 6D). Sustained stimulation of loop neurons overcomes the lingering CREB2 effectors (Figure 6E), in accord with the recharging experiments in Figures 2B and 2G, and similar to the enduring effects of NPY signaling from AgRP neurons on mammalian feeding behavior [6,9].
Figure 6. Modeling the circuit and molecular dynamics of mating drive.
(A–B) Representation (A) and implementation (B) of the model (see STAR Methods). (C) Left: A single mating immediately decreases electrical and CREB2 activity in NPF neurons, with a consequent but delayed reduction in pCd activity. CREB2 effectors are modeled as remaining high for several hours after CREB2 activity decreases. Right: 6 sequential matings cause a dramatic reduction in loop activity. See Figure S6A for model equilibration (D) After repeated matings (blue bars), both pCd and NPF neurons show decreased electrical and CREB2 activities, which then recover over 3 days. The presumed slow turnover in CREB2 effectors such as Task7 delays their removal relative to the decline in CREB2 transcriptional activity, postponing the recovery of electrical activity in the loop. (E) Stimulating pCd neurons (green line in legend) after 6 matings (blue bars) accelerates the recovery from satiety. See also Table S1.
Translating goal achievement into standardized decrements in motivation
The transfer of sperm occurs at 5–7 minutes into copulation, after which the male may truncate the ~23-minute mating in response to threats ([56,57] and our observations). The number of recent matings is therefore a more accurate estimator of remaining reproductive capacity than is the summed duration of matings. Consistent with these reproductive dynamics, we previously found that 5-minute matings induce the same decrement in mating drive as full-length matings [22]. To more closely examine the relationship between the CRNs that detect matings and the resulting satiety, we optogenetically silenced the CRNs during the first 5 minutes of each mating, or during all but the first 5 minutes, using GtACR1 (Figure S6B) [58]. Neither of these protocols produced the insatiable phenotype seen when the CRNs were silenced throughout matings (Figures 7A), indicating that satiety is not a reflection of the timing or duration of CRN activation, but the number of discrete CRN activations. Since the light was turned off between matings, these results also show that CRN activity is only required during copulation to induce sustained satiety.
We examined our computational model for insight into this copulation counting phenomenon. In the model, CRN-mediated suppression of NPF activity during mating initially creates a disparity between NPF and pCd neurons that slowly averages out to an intermediate level in both populations (Figure 7B). The degree of satiety resulting from spaced periods of inhibition, allowing time for re-equilibration in between pulses, is larger than that seen if the same total duration of inhibition is delivered continually and then allowed to re-equilibrate (Figure 7B). To test if a similar phenomenon occurs in actual flies, we optogenetically stimulated the CRNs of naïve males with either six 5-min light pulses, spaced by 20 minutes, or with one 30-min pulse (Figure 7C). As predicted by the model, the spaced stimulation had a much stronger satiating effect (Figure 7D). Similar effects were seen with pulses of thermogenetic CRN stimulation (Figure 7E), or optogenetic inhibition of the NPF neurons (Figures 7F–7H and S6C). Though we cannot rule out a contribution of adaptation effects for long-lasting manipulations, these results provide a plausible explanation for the ability of motivational circuitry to record and respond to a discrete event (mating), with standardized effects on future behavior. The molecular and circuit dynamics described here can buffer against noisy sensory inputs and unimportant details, resulting in the binary categorization of events as either achieving a goal—or not.
Discussion
“…when the gathering desire is sated, then for a while comes a little respite in their furious passion. Then the same madness returns, the old frenzy is back upon them…”. Though Lucretius (99–55 BCE) refers to human mating drive in De rerum natura, these lines could well describe the dynamics of motivations promoting diverse behaviors in even more diverse species. One insight relevant to our work is the distinction he draws between satiety and respite: we find that neuronal circuitry reporting goal achievement induces the initial satiety, while CREB2 effectors maintain the enduring respite. Both mechanisms reduce electrical activity held in a recurrent loop, but, as the Roman poet and philosopher may have intuited, their different timescales are produced by separate underlying mechanisms.
The best understood long-timescale brain functions are memory and circadian rhythms, both of which make use of the activity-dependent transcription factor CREB (CREB2 in flies) [43,45]. We show that CREB2 is activated by electrical activity within a drive-promoting recurrent loop containing pCd and NPF neurons. CREB is also activity-regulated in AgRP neurons [59], adding to the long list of similarities with the NPF neurons described here. In the pCd/NPF loop, CREB2 generates an inhibitory environment, at least in part by increasing the transcription of the leak potassium channel Task7. This negative feedback limits and stabilizes peak motivation. The involvement of transcriptional mechanisms introduces a delay in the onset of CREB2’s impact, but the lingering products of CREB2 activation may be more important for understanding motivational dynamics. Potassium channels, and their inhibitory conductances, can endure for days after being transcribed and translated [54]; Kir2.1, for example, has a half-life of 18–35 hours [55]. Protein lifetime and stability are central to both memory and circadian timing; we suggest that the regulated and diverse turnover rates of effector molecules may be used to generate enduring effects of tailored duration for a wide range of long-lasting behavioral and cognitive processes.
Though we believe we have uncovered much of the core molecular and circuit logic behind the maintenance, sating, and recovery of mating drive in male flies, gaps and limitations in our understanding remain. The CRNs hyperpolarize the NPF neurons within tens of milliseconds, indicating a direct connection, but this effect is not immediately relayed to the pCd neurons, pointing to the existence of unidentified loop neurons. We do not yet know how the CRNs become activated during mating, nor how the NPF receptor adjusts the output of the dopaminergic neurons. Within the neuronal populations that we treat as functional units, it is likely that there are subpopulations with specific and unexplored roles. Within the loop, our understanding of how electrical activity is transduced to stimulate CREB2 is incomplete, and there likely exist other CREB2 effectors that work alongside Task7 to slow and limit the accumulation of electrical activity. We tuned our model to match the time-course of mating drive, but many of the important parameters (e.g. Task7 protein half-life) remain to be directly measured. While we are certain that investigation of these details will reveal additional and unexpected mechanisms, the outline we provide here can produce the behavioral and physiological dynamics when modeled computationally, arguing for a large degree of accuracy in its essentials.
In both memory and circadian timing, external events drive CREB activation; in this motivational system, internally generated loop dynamics determine CREB2’s activity level. This difference relates to a potentially important insight: the respite after initial satiety is a direct consequence of the animal’s motivational state before goal achievement. This may be a useful concept for thinking about the relationship between the motivational and satiety states controlling many behaviors. It may, for example, help explain why we derive satisfaction from achieving long-sought goals or why goal achievement in the absence of motivation (e.g. snacking) does not produce appreciable satiety [60].
LEAD CONTACT AND MATERIALS AVAILABILITY
Contact M.A.C. (Michael.Crickmore@childrens.harvard.edu) for fly strains and other resources in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Stocks
Fly husbandry was performed as previously described [22]. Flies were maintained on conventional cornmeal-agar medium under 12-hr light/12-hr dark cycles at 25 °C and ambient humidity. Due to the focus of this study, all tested flies were adult males. Unless otherwise stated, males were collected on the day of eclosion and group-housed away from females for 3–4 days before testing. Virgin females were generated using a hs-hid transgene integrated on the Y-chromosome [61,62]. The following fly strains were backcrossed into a standard w1118 background (Bloomington 5905) for 5 generations to normalize baseline courtship behavior: NPF-Gal4, NPF-RNAi, CREB2-RNAi, VAChT-RNAi, and UAS-Kir2.1. All behavioral experiments were carried out within the first 10 hours of each light day-phase. Detailed genotypes of all strains used in the paper are listed in Table S1 available online.
METHOD DETAILS
Generating R42G02-p65AD Flies
The R42G02 fragment (a gift from the Rubin Lab) [26] was subcloned into the pBPp65ADZpUw backbone vector using the Gateway cloning system. The resulting construct was inserted into the attP40 locus on Chromosome II (Rainbow Transgenic Flies). The transformants were verified with PCR.
Courtship Assays
Courtship assays were carried out in the same setting as previously described [22,61]. A male fly (3–4 days old) and a w1118 virgin female fly were videotaped in cylindrical courtship chambers (10 mm diameter and 3 mm height) at 23 °C and ambie nt humidity. The same setup was used for the analyses of tap-induced courtship initiations (but analyzed differently; see below).
For thermogenetic experiments using TrpA1, half of the males were assayed at 23 °C, and the other half at a higher temperature (28.5 °C or 30 ° C). For optogenetic experiments using CsChrimson or GtACR1 [58,63], newly eclosed male flies were transferred to aluminum-foil-wrapped vials containing rehydrated potato flakes (Carolina Biological Supply, 173200) and 100 µL of all-trans-retinal stock solution (50 mM in ethanol, Sigma-Aldrich, R2500) for 3 days before experiments. In these experiments, half of the males were assayed under ambient light conditions, and the other half under 655-nm red light (~100 µW/mm2). For conditional silencing experiments involving TubGal80ts (e.g., Figure 2E), male flies were moved to 30 °C after eclosion and kept there (isolated from females) until the assay, which took place at 23 °C.
For the thermogenetic and optogenetic experiments in Figures 7C–7H, flies were treated with red light (655 nm, ~100 µW/mm2), green light (530 nm, ~100 µW/mm2), or heat (32 °C) until the assay, which took place at 23 °C and under ambient light. These treatments were either delivered in bulk (30 min) or pulses (5 min with 15 min between pulses). The assays took place immediately after the treatment, except for the one shown in Figure7H, which was carried out 4 hours later to demonstrate the persistence of the induced satiety.
Satiety Assay and Reversal
Satiety assays were used to test if a neuronal population (e.g., dopaminergic neurons) has an acute motivation-promoting role and were carried out as previously described, with minor modifications [22]. Individual male flies were placed with ~15 virgin females in a food vial at 23 °C and ambient humidity for 4.5 hours. Mating behav iors (courtship and copulation) were scored manually at time points: 0.5, 1, 4 and 4.5 hours. 4.5-hour satiety assays were used to satiate males for courtship and imaging experiments.
To thermogenetically revert satiety, the temperature was raised after the last time point, and mating behaviors were scored 20 minutes after the incubator reached the appropriate temperature. We typically start at 30 °C and lower the temperature if the flies show seizure-like behaviors (e.g., dopaminergic stimulation, NPF stimulation in our early experiments using a particular incubator). In some experiments (e.g., Figure 2J), we took an additional time point 30 min after lowering the temperature back to 23 °C.
For Figure 7A, we optogenetically silenced CRNs with green light either only during matings or during specific parts of matings. This was done by performing the experiments under dim ambient light and transferring vials with copulating pairs into a green-lit environment (530 nm, ~100 µW/mm2) at the appropriate time (e.g., first 5 minutes of matings, all but the first 5 minutes, etc.). Green LEDs were oriented perpendicular to the vials so the food in the vials would not block the light. However, due to the curvature of the vials (and the resulting optical properties), it was difficult to maintain spatially uniform light intensities during the experiment. Perhaps for this reason, we did not observe the immediate termination of mating that we saw by silencing the 42G02-Gal4 neurons with strong, uniform green light. This immediate termination is not due to the CRNs within this Gal4 line as it is prevented by TshGal80 (our observations).
In some experiments, we used a flattened variant of the satiety assay for flies that take longer to satiate (e.g., Figure 1I). The main difference is that, instead of standard food vials, these assays were performed in cylinders (~2.75 cm diameter and ~4 mm height) with floors made of fly food (but see Boutros et al [61] for details). These assays allow video filming and scoring but are more difficult to carry out in bulk than regular satiety assays. In our experience, flies mate slightly more often in the smaller space, presumably due to more tapping behaviors and chances to initiate courtship [23]. For conditional silencing experiments involving TubGal80ts (e.g., Figure 1I), male flies were moved to 30 °C a fter eclosion and kept there (isolated from females) until just before the assay, which took place at 23 °C.
Artificial Satiety Assays
To search for satiety-inducing neurons, we performed thermogenetic experiments using males expressing TrpA1 in subsets of sexually dimorphic neurons at 27 °C (Fru-Gal4) or 30 °C (all other Gal4 lines) for 4.5 hours. This satiety assay was carried out at 23 °C. Satiety was then measured with an abbreviated satiety assay using only 2 time-points: 0.5 and 1 hour. We used the satiety assay in these experiments for comparison to naturalistic satiety. A within-group (5–7 males per group) average was generated for each time point and then averaged together.
Recovery Assays
Recovery assays were used to test if a neuronal population (e.g., pCd neurons) controls the recovery of motivation. Satiated males were isolated from females for 12–13 hours before their drive recovery was re-assessed using an abbreviated assay that consists of only 2 time-points: 0.5 and 1 hour. The recovery took place at either ambient temperature (e.g., Figure 4M) or with thermogenetic stimulation (e.g., Figure 2B). The second, abbreviated assay was always carried out with new females at 23 °C, either immediately a fter the temperature had re-equilibrated (which usually takes ~15 minutes) or after a 4-hour delay to demonstrate the persistence of drive recovery. In Figures 1A and 1B, we use recovery to validate the authenticity of the artificially induced satiety (as opposed to transient incapacitation). In these experiments, the following recovery time points were used instead (all with new females): immediately after satiety, 24, 48 and 72 hours later.
To measure of the recovery of hypersexual males (e.g., Figure 4M), which take longer to satiate, we ensured the initial satiety of these males by extending the satiating step by an additional 4–6 hours.
Locomotion Assays
Locomotion assays were carried out as described previously [22]. Briefly, male flies (some of which have been pre-stimulated) were individually aspirated into the courtship chambers (see above) at 23 °C or 30 °C and allowed to explore the chambers for 1 min before their locomotion was recorded for 10 min.
Antibody Staining and Confocal Microscopy
Antibody staining was performed as described previously [22]. Briefly, fly brains were dissected in Schneider’s medium and immediately fixed in 4% paraformaldehyde dissolved in PBS with 0.3% Triton X-100 (PBST) for 20 minutes. After 3× 20-minute washes with PBST, the brains were incubated with the primary antibodies diluted in PBST (1:1000 chicken anti-GFP, Aves Labs, GFP-1020 and 1:7 mouse anti-nc82 (to label neuropil (magenta)), Developmental Studies Hybridoma Bank) at 4 °C for 48 hours. After another round of 3× 20-minute washes with PBST, the brains were incubated with the secondary antibody diluted in PBST (1:400 donkey anti-chicken, Jackson ImmunoResearch 703–545-155 and 1:400 donkey anti-mouse, Jackson ImmunoResearch, 715–165-150) at 4 °C for 48 hours. Then, after a final round of 3× 20-minute washes, the brains were mounted on a glass slide using standard procedures. Confocal sections were acquired using an Olympus Fluoview 1000 microscope at 3-µm intervals.
Baseline Calcium Imaging of Dissected Brains
Fly brains expressing GCaMP6s [64] and, whenever possible (e.g., when it would not confound other optical techniques), tdTomato, were dissected from 3–6-day-old males in HL3.1 solution [65] and mounted anterior-side down onto the base of a glass-bottom Petri dish (MatTek, P35G-1.5–20-c) housing 3 mL HL3.1 saline, as previously described [22]. Confocal sections (4–7 sections at 3-µm intervals) were taken from the anterior of the superior medial protocerebrum (SMPa). Whenever possible, we also co-expressed and imaged tdTomato in the same cells, to normalize the GCaMP6s signal. Whenever possible, we also imaged from unrelated brain regions (the mushroom body) to show that the changes are specific to the SMPa. For chemogenetic experiments involving ORT, after the first round of imaging, 20 µL of 7.5 mM histamine was carefully pipetted along the inside edge of the dish into the saline, to reach a final concentration of 50 µM, and a second round of imaging was performed 5 minutes after. For no-Gal4 controls, the same procedures were applied to male heads that did not express ORT (although containing the transgene). For thermogenetic experiments involving TrpA1, the flies were either stimulated at 30 °C for 4.5 hours (e.g. , Figures 1K), or satiated and then stimulated for 12 hours (e.g., Figure 2C) before experiments, which took place at 23 °C.
Wide-field Activity Imaging with P2X2 Stimulation
An upright Leica DM5500 B scope was used for these calcium imaging experiments. We used GCaMP6s [64] to report trans-synaptic excitation and ASAP1 [66] for inhibition. Brains of 3-to-6 day old males were dissected and mounted posterior-side down on a coated Petri dish (Thermo Scientific 150318) containing 3 mL of HL3.1 solution [65], as described before [22]. This preparation preserves dopaminergic, NPF and pCd neurons, but severs the ascending axons of CRNs. GCaMP6s or ASAP1 fluorescence of dopaminergic, NPF and pCd projections at SMPa were identified with baseline fluorescence and imaged for 150 frames at 1 frame/second. At Frame #40, 20 µM 150 mM ATP solution (pH pre-adjusted, Sigma-Aldrich, A2383) was carefully pipetted along the inside edge of the Petri dish to reach a final concentration of 1 mM. For controls, the same procedures were applied to male heads that did not express P2X2 (though they contained the UAS/LexAop transgene).
Two-photon calcium imaging with CsChrimson stimulation
Two-photon laser-scanning microscopy was performed using a resonant-scanning microscope (NeuroLabWare, 15.5 frames/second and 675 frames per experiment) that is described in [67]. All experiments were performed with a 16× 0.8 numerical aperture objective (Nikon) at 8x digital zoom. Shortly after eclosion, flies were transferred to vials containing rehydrated potato flakes (Carolina Biological Supply, 173200) and 100 µL of all-trans-retinal stock solution (Sigma, R2500; 35 mM in ethanol) for at least 3 days prior to experiments. Fly brains were dissected and mounted in a Petri dish containing 4 mL HL3.1 solution as described above. GCaMP6s was excited at 910 nm and 8–17 mW at the sample. The power was kept minimal to each genotype in order to avoid two-photon activation of CsChrimson.
For each experiment, basal GCAMP6s fluorescence was collected for 10 seconds prior to optogenetic stimulations. A 655-nm LED is used to stimulate CsChrimson from below the Petri dish at about 1 mw/mm2. A train of 30 8-ms light pulses was delivered at the beginning of 30 consecutive frames (e.g., Frames 156–185) over the span of ~2 seconds. Right before each light pulse, a shutter circuit is activated to protect the photomultiplier tube (PMT) for the duration of each light pulse as well as an additional 2-ms buffer time after to account for the relatively slow turn-off times of LEDs. As a result, the first 10 ms (about the top ~20% rows) in each of the 30 stimulation frames was blank. To image samples during the stimulations, we therefore placed the regions of interest at the bottom of the field of view.
Two-photon voltage imaging with CsChrimson stimulation or GtACR1 inhibition
Two-photon voltage imaging experiments were conducted in a similar fashion as the calcium imaging experiments above. For imaging of CRN cell bodies (Figure S6B), the ventral nervous system was dissected out instead of the brain. ASAP2s was excited at 910 nm (8 mW) using the same 2-photon microscope. The stimulation of CsChrimson was done exactly as described above. For experiments using GtACR1 silencing, we used 10 5-ms pulses of 465-nm light instead (~20 µW/mm2). The pulses were delivered at the beginning of 10 consecutive frames, spanning ~0.65 seconds, and the shutter circuit protects the PMT for the duration of the pulses plus a 5-ms buffer time. We shortened the pulse widths and allocated more buffer time in these experiments to protect the green-light PMT, as the dichroic mirror in front of the PMT allows more blue light (e.g., 465 nm) through than red.
Measuring CREB2 Activity
To measure CREB2 activity we used a FLP-dependent luciferase construct driven by the cAMP-response element (CRE) promoter [47], with UAS-FLP driven in the neurons under examination. The luciferase assays were performed using a commercial kit (Promega, E1910). Brains of 3–6-day-old males were dissected in HL3.1 solution and transferred, in groups of 3, into 50 µL dissociation solution for 5 minutes at −20 °C. Then, a 20 µL supernatant of the dissociation solution was added to 50 µL substrate solution, and the luciferase activity was measured in a photoluminometer (Turner Designs TD-20/20) over a 5-second window.
For experiments involving satiety, males were satiated as described above before dissection. For experiments involving TrpA1, males were either thermogenetically stimulated at 30 °C for 4.5 hours, or satiated and then thermogenetically stimulated, before dissection.
Fluorescent in situ hybridization
Brains dissected from overnight-satiated and naïve wild-type Canton-S males were used for these experiments. Single-molecule fluorescent in situ hybridization (smFISH) was performed following the published protocol [53], with the exceptions that we omitted 1) the overnight incubation in 100% ethanol and 2) the bleaching steps using sodium borohydride. Fluorescence-tagged Task7 (Quasar 570) and Dsx (Quasar 670) probes were designed and manufactured using a commercial source (LGC Biosearch Technologies, see Table S3 for probe sequences). Confocal sections were acquired using an Olympus Fluoview 1000 microscope at 3-µm intervals.
Screening for Molecular Inputs and Outputs of CREB2
To search for molecular modulators of CREB2 activity in pCd neurons, we conducted a targeted screen through genes that have been previously shown to do so in other systems [43,44,51,68,69]: PKA subunits (PKA-R1, PKA-R2, PKA-C1, PKA-C2, PKA-C3), Ras family (Ras-64B, Ras-85D), PKC family (PKC-53E, PKC-98E, aPKC), MAPK family (bsk, rl, p38A, p38B, p38C), RSK family (S6k, S6kII), Akt (Akt1), CaMKII, Casein Kinase family (CK1α, CKIIα, CKIIβ, CKIIβ2), Calcineurin family (CanA1, CanB, CanB2), PP1 family (PP1–13C, PP1–87B, PP1α−96A).
To begin searching for transcriptional targets of CREB2 that suppress neuronal activity, we first carried out a genomic search for either palindromic (TGACGTCA) or partial CRE sites (AGACGTCA, GGACGTCA, CGACGTCA, TAACGTCA, TTACGTCA, TCACGTCA, TGTCGTCA, TGGCGTCA, TGCCGTCA) that are located within 1 kB upstream of transcription initiation sites (FlyBase data release 6.05) [44]. Then we tested all putative inhibitory channels from this list by knocking them down in pCd neurons: Sandman, CG34396, ClC-a, ClC-b, ClC-c, CG5404, CG6938, Irk2, and Task7. Out of all the channels tested, only knockdown of Task7 resulted in hypersexual males, and we therefore focused on this channel. The MATLAB code used for the genomic search is available online.
Pharmacology
ATP was prepared in a 150 mM solution in HL3.1. Histamine was prepared as a 7.5 mM solution in HL3.1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of Courtship Behavior
The courtship index was manually scored as previously described [22,61]. It is defined as the fraction of time during which the male fly is engaged in mating behaviors (courtship and copulation) within 5 minutes of courtship initiation. A bout of courtship was scored as initiated when the male oriented toward the female, began tracking her, and unilaterally extended his wing to sing to her. A bout was scored as terminated if the male stopped tracking the female or turned away from her. Once courtship was terminated, it was almost never reinitiated within 10 seconds. Whenever possible, terminations were verified by confirming that the male did not resume singing when the female subsequently passed in front of him.
For experiments that measure tap-induced courtship probabilities, the experimenter who scored the videos was blind to the genotypes and experimental conditions. Taps are defined as a male foreleg touching any part of the female body. In our assays, courtship is always initiated by a tap, always within a few seconds, and most often within 500 ms. The details of this analysis can be found in Zhang et al [23]. Briefly, we calculated the cumulative courtship probabilities (y) after each tap (x). We plotted –Ln(1 – y) against x fitted a straight line that is forced through the point of origin ([0, 0]). The Y-axis ticks are labeled with the same log transformation. This fitted line represents the expected cumulative courtship-initiation probability if the tap-to-courtship transitions are modeled as a perfect coin (with varying odds). A steeper line represents higher courtship probability (i.e., high odds of the coin). From the slope of the line (s), we can calculate the per-tap courtship probability as 1 – e–s.
We used bootstrapping to perform hypothesis tests on courtship probabilities, with the underlying null hypothesis that these two datasets were generated with the same unknown courtship probability. Before bootstrapping, we first pooled two datasets together. Then, we resampled two bootstrapped datasets from the pooled data with the same sample size as in the experiments. Then, using the same linearization and linear regression procedures, we calculated new, bootstrapped courtship probabilities. We calculated their difference and repeated the resampling and re-calculations 100,000 times. The p-value was then calculated as the fraction of iterations that generate a difference in courtship probabilities at least as large as the experimental one. We used Bonferroni corrections to adjust the p-values for multiple comparisons. The MATLAB code to perform these calculations is available online.
Analysis of Satiety and Recovery Assays
For standard 4.5-hr satiety assays, individual males were paired with ~15 virgin females and mating behaviors (courtship and copulation) were scored manually at time points: 0.5, 1, 4 and 4.5 hours. 4.5-hour satiety assays were used to satiate males for courtship and imaging experiments. A within-group (5–7 males per group) average was generated for each time point. The first two (generally 0.5 and 1 hour) and last two (generally 4 and 4.5 hours) time points were averaged to generate Initial and End percentages, respectively.
Recovery assays were scored the same way as satiety assays, except that we report the average percentage of mating behaviors from 1) the last two time points in the satiating phase and 2) the first (and only) two time points immediately after recovery. Typically, control genotypes, which go through the same temperature shifts (if necessary), recover overnight to only ~50% mating behaviors. In experiments where we tested the persistence of recovered drive (e.g., Figure 2B), we also reported the average percentage of mating behaviors at a time point 4 hours after the conclusion of full 4.5-hr satiety assays.
Analysis of locomotion assays
Video segmentation of the male flies was done using FIJI with the MTrack2 plugin. The total distance traveled by each male over the 10-min assay is recorded and calibrated to cm using the dimensions of the chambers.
Quantifying Imaging Data
For quantification of baseline calcium level, after one person (S.X.Z.) took the images, another person (M.F., C.B., L.M., B.G., A.C., or E.G., see acknowledgments) selected regions of interest (ROIs) of ipsilateral SMPa, mushroom body medial lobe, and background (near the antennal lobe, where dopamine projections are sparse) using the tdTomato channel (e.g., Figures 3A) or the GCaMP6s channel (e.g., Figure 1K). ROI sizes were kept roughly the same between samples. Average pixel fluorescence in each ROI was calculated with a custom-written MATLAB script. Whenever possible, normalized fluorescence was calculated as (GCaMP6s_SMPa – GCaMP6s_background) / (tdTomato_SMPa – tdTomato_background). If normalization was not possible (i.e., no tdTomato), we reported the raw GCaMP6s fluorescence instead (GCaMP6s_SMPa – GCaMP6s_background). The same formulas were used for the mushroom body medial lobe as well. For experiments involving ORT, the same analysis was performed again on the post-histamine images. Post-histamine fluorescent intensities were normalized to their respective pre-histamine values. The MATLAB code to perform these step is available online.
For wide-field imaging using P2X2 stimulation, fluorescence data were analyzed by selecting a region of interest from a pre-stimulation frame of the lateral protocerebrum. The regions of interest were kept roughly the same size between experiments.
For smFISH quantification, we used a custom, semi-automated MATLAB script to segment images and measure fluorescence intensities. After selecting a general ROI that contained the pCd cell bodies (i.e., superior medial protocerebrum) using the Dsx channel, the software applies a series of common image-processing steps to both channels to segment out cells that express Task7 and/or Dsx: 1) Gaussian filtering to smooth noise, 2) using the “imopen” function to correct for background unevenness, 3) thresholding by intensity to identify puncta, and 4) thresholding by area to remove isolated pixels. Task7-positive puncta were then grouped by whether they are also co-labeled by Dsx. Finally, the shapes, sizes, and locations of the puncta were used to mask the original, unprocessed images to measure Task7 probe intensities (shown in Figure 5I). Given that these Dsx+ puncta have similar cluster size and location as pCd neurons, we refer to them as pCd in the paper. We were unable to do the same experiments for Fru+ NPF neurons because both Fru and NPF label cells that are largely unclustered and mixed with the non-intersectional populations by either marker alone. Background fluorescence was estimated by measuring the intensities in an unrelated brain region (the antennal lobe) and subtracted from the data. The MATLAB code to perform these calculations is available online.
For 2-photon imaging experiments, fluorescence data were analyzed in MATLAB using a semi-automated program modified from a published independent component analysis algorithm (Mukamel et al., 2009). The program is available online. Data are presented as ∆F/F0 for calcium imaging experiments and −∆F/F0 for voltage imaging experiments.
To estimate when the imaged neurons respond to CsChrimson stimulation (either directly or via an upstream neuron), we plotted the frame-by-frame fluorescence changes (e.g., Figure S2T). In these experiments, each trace was normalized to the max ∆F/F0 to enable cross-trial comparison and shuffling. Since it takes ~64 ms to scan a frame and each stimulation is delivered at the beginning of a frame, the response we recorded at the bottom of the first and second stimulation-frame cannot be later than 64ms – 8 ms (pulse width) = 56 ms and 64 ms x 2 – 8 ms = 120 ms, respectively, after the stimulation. To estimate the pre-stimulation noise levels, we calculated the 95% confidence interval of fluorescence fluctuations by bootstrapping the fluorescence data during the last second before stimulations. Specifically, we randomly sampled, with replacement, one value per trace from the 1-sec fluorescence data to calculate their average, and repeated this procedure 100,000 times to estimate the confidence interval (e.g., orange shading in Figure S2T). The response delay was calculated as the first time point at which the average trace (dark red) rises beyond the noise boundary (e.g., ≤56 ms if this occurs in the first frame of stimulation). We note that, occasionally the average trace transiently dips back into noise boundary (Figure S3H), presumably due to noise in the signal.
In the voltage-imaging experiments in which we imaged the same neurons that we also silenced with GtACR1, the GtACR1 construct also expresses voltage-insensitive EYFP in the same open reading frame [58]. Since EYFP is spectrally inseparable from ASAP2s in our microscopy setup, the reported –∆F/F0 values are decreased in amplitude, though this did not interfere with our conclusions. The same is true for the CsChrimson-mVenus construct that we used for Figure 2O.
Analyses of CREB2 Activity
Because CREB activity differs significantly between driver lines, we normalized the values to the mean of the driver-matched, naïve flies with no manipulation of CREB activity. The values before normalization are reported in Table S4, which also specifies the normalization scheme.
Because Tanenhaus et al. reported leakiness when using this reporter with a hs-FLP transgene, we performed experiments suppressing CREB activity in the same cells that express the reporter (Figure S4A) and saw little residual activity. This is consistent with Tanenhaus’ conclusion that the hs-FLP, not the reporter, was the source of the leakiness.
Modeling of Mating Drive Circuitry
In Figure 6, we illustrate the feasibility of our mating-drive model mathematically. Given the illustrative goal, we choose the simplest implementation for each step to minimize the degrees of freedom in the model. The model does not imply direct connections of the circuitry (e.g., whether pCd neurons synapse directly on NPF or not). The model is written as a set of 6 linear ordinary differential equations describing changes in NPF activity (∆NPF), pCd activity (∆pCd), CREB2 activity in NPF (∆CREB2NPF), CREB2 activity in pCd (∆CREB2pCd), CREB2 effectors in NPF (∆EffectorNPF), and CREB2 effectors in pCd (∆EffectorpCd). The bulk of the equations are written exactly as in Figure 6B, with the addition of Poisson noise terms added to both NPF and pCd activity (so their activity could build up as in Figure S6A). The MATLAB code for analysis is available online.
Changes in neuronal activity are written as a sum of synaptic input (e.g., +α2·NPF) and inhibition by CREB2 effectors (e.g., −δ1·EffectorpCd). For inhibition here and in the rest of the model, we have also used non-linear terms (e.g., writing inhibition of pCd by CREB2 Effector as −δ1·pCd·EffectorpCd instead of −δ1·EffectorpCd), with similar results. We settled on this version only because of its simplicity.
Changes in CREB2 activity are written as exactly tracking the corresponding neuronal activity (e.g., ∆CREB2pCd = β1·dpCd). This fits our observations of their relationship (Figures 3A–3B, and 4B–4D). This relation however may not hold true on a faster timescale (milliseconds), which our model does not use. We have also modeled changes in CREB2 activity as a function of neuronal activity (e.g., ∆CREB2pCd = β1·pCd – ζ1·CREB2pCd) with similar outcomes.
Changes in CREB2 effectors are written as a linear sum of CREB2 activity (e.g., +γ1·CREB2pCd) and Effector turnover (-ε1·EffectorpCd). However, since CREB2 is a transcription factor, it is unlikely to have an instantaneous impact on its effectors (e.g., Task7), so we built in a delay between CREB2 activity and changes in Effector levels. For the purpose of illustration, we arbitrarily chose 16 hours as the delay (i.e., CREB2 level now determines ∆Effector 16 hours later). The exact length of the delay does not affect the main conclusions of the model.
The satiating effect of matings is implemented by adding a negative constant to ∆NPF when the fly mates. Stimulation of pCd neurons is implemented by adding a positive constant to the ∆pCd term during the stimulation window (Figure 6E). We have also used other methods for both satiation (e.g., −η2·NPF) and stimulation (e.g., setting pCd activity at a fixed level during the stimulation window), again with similar results.
Finally, since activity drops in one population (e.g., NPF) do not instantaneously affect the activity of the other population (e.g., pCd) (Figures 2O, S2H and S3D), we suspect these neurons have mechanisms to sustain, at least to some extent, their current activity level. Among possible mechanistic explanations, this could be because both pCd and NPF neurons receive other inputs from outside the loop (suggested in Figures 6A), because they have persistent activity, or because they form recurrent sub-loops within each population. We implemented the sustaining effect by adding a proportional decay to both ∆pCd (-µ·pCd) and ∆NPF (-µ·NPF) (Figure 6B). This could be interpreted as either 1) persistent activity in both populations that decays exponentially over time, or 2) both neurons forming recurrent pCd-loops or NPF-loops (with or without additional populations), whose activity decays exponentially over time. We have also implemented this sustaining effect by other means, such as using a third population that receives excitation from both NPF and pCd neurons and also feedback on both, with similar results.
Reanalysis of single-cell sequencing data
In Table S2, we show a reanalysis of results from two published single-cell sequencing datasets in order to confirm the expression of certain genes in our neuronal populations (e.g., NPFR in dopamine neurons)[31,32]. The Croset et al. dataset was accessed as their Figure 1 source data 1 (https://doi.org/10.7554/eLife.34550.007). The Davie et al dataset was accessed via the NIH Gene Expression Omnibus (GSE107451). We used the 57K-neuron dataset. We performed our analyses on the raw, as opposed to normalized, expression data. Neurons were defined as cells that express both elav and nSyb. Dopaminergic neurons were defined as cells that are positive for elav (neuronal marker), nSyb (neuronal marker), and ple (Tyrosine Hydroxylase). NPF neurons are defined as cells that are positive for elav, nSyb, and NPF. Since there are no known exclusive markers for pCd neurons, we instead identified putative pCd neurons that are elav-, nSyb-, dsx-positive but fru-negative. This method has the caveat that it excludes 1–2 fru-positive pCd neurons and includes pC2, as well as some pC1 (but not P1) neurons. Using these criteria, we generally found similar results from the two datasets.
Additional Statistical Tests
One-way ANOVA, two-way ANOVA and two-tailed student’s t-test were performed using Prism 7. All tests are unpaired. All One-way ANOVA tests use post-hoc Tukey corrections. All Two-way ANOVA tests use post-hoc Bonferroni corrections. Error bars represent 1 S.E.M. for all figures.
Supplementary Material
Table S4. Raw data from luciferase-based assays of CREB activity in this paper. Related to Figures 4 and 5.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP | Aves Labs | GFP-1020, AB_10000240 |
| Mouse anti bruchpilot (nc82) | Developmental Studies Hybridoma Bank | nc82, AB_2314866 |
| Alexa Fluor 488 Donkey anti-Chicken IgY | Jackson ImmunoResearch Labs | 703–545-155, AB_2340375 |
| Cy3 Donkey anti-mouse IgG (H+L) | Jackson ImmunoResearch Labs | 715–165-150, AB_2340813 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| All-trans-retinal | Sigma-Aldrich | R2500 |
| ATP disodium salt hydrate | Sigma-Aldrich | A2383 |
| Critical Commercial Assays | ||
| Luciferase reporter assay | Promega | E1910 |
| Deposited Data | ||
| Drosophila brain single-cell sequencing dataset | [31] | https://doi.org/10.7554/eLife.34550.007 |
| Drosophila brain single-cell sequencing dataset | [32] | GEO: GSE107451 |
| Images of neurons that control mating drive in male Drosophila | This paper | DOI: 10.17632/4c44xb5zyh.1 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w1118/Dp(2;Y), P{hs-hid}Y | Bloomington Drosophila Stock Center (BDSC) | BDSC: 24638 |
| D. m.: wild-type Canton S | [70] | FlyBase: FBsn0000274 |
| D. m.: dsx-Gal4: w1118;;TI{Gal4}dsxKI.Gal4 | [11] | FlyBase: FBti0168641 |
| D. m.: Fru-Gal4: w*;; TI{GAL4}fru[GAL4.P1.D]/TM3, Sb1 | BDSC | BDSC: 66696 |
| D. m.: w*; P{UAS-TrpA1(B).K}attP16 | BDSC | BDSC: 26263 |
| D. m.: CRN-Gal4: w1118; P{GMR42G02-Gal4}attP40 | [26] | N/A |
| D. m.: y1, w*; PinYt/CyO; P{UAS-mCD8::GFP.L}LL6 | BDSC | BDSC: 5130 |
| D. m.: Tsh-Gal80: w1118; P{GAL80}tshGAL80 | Gift from J.H. Simpson | FlyBase: FBti0114123 |
| D. m.: w1118; P{UAS-DenMark}2 | BDSC | BDSC: 33062 |
| D. m.: w1118; P{y[+t7.7] w[+mC]=8xLexAop2-IVS-Syt1::smGdP-HA}su(Hw)attP1 | BDSC | BDSC: 62213 |
| D. m.: w1118; L1/CyO; P{w[+mC]=UAS-DenMark}3, P{w[+mC]=UAS-syt.eGFP}3 | BDSC | BDSC: 33065 |
| D. m.: y1,w*; PBac{y[+mDint2] w[+mC]=13XLexAop2–6XmCherry-HA}VK00018/CyO; Dr1/TM6C, Sb1, Tb1 | BDSC | BDSC: 52272 |
| D. m.: w1118;; P{UAS-IVS-myr::GFP}attP2 | BDSC | BDSC: 32197 |
| D. m.: w1118; P{UAS-eGFP-Kir2.1}/CyO | [71] | FlyBase: FBtp0110871 |
| D. m.: w*;; P{tubP-GAL80ts}2/TM2 | BDSC | BDSC: 7017 |
| D. m.: TH-Gal4: w1118;; P{ple-GAL4.F}3 | [72] | FlyBase: FBti0072936 |
| D. m.: CRN-LexA: w1118; P{GMR42G02-LexA4}attP40 | Gift from Bruce Baker | BDSC: 61540 |
| D. m.: w1118;; P{20xUAS-IVS-GCaMP6s}VK00005 | BDSC | BDSC: 42749 |
| D. m.: w1118; P{LexAop-TrpA1} | [73] | BDSC: FBtp0084378 |
| D. m.: pCd-Gal4: w1118;; P{GMR41A01-GAL4}attP2 | BDSC | 39425 |
| D. m.: pCd-LexA: w1118; P{GMR41A01-LexA}attP40 | BDSC | 54787 |
| D. m.: w1118; P{UAS-ASAP1}attP40 | BDSC | BDSC: 65412 |
| D. m.: w*; PBac{y[+mDint2] w[+mC]=20XUAS-ASAP2s}VK00005 | BDSC | BDSC: 76247 |
| D. m.: w1118; P{y[+t7.7] w[+mC]=13XLexAop2-IVS-CsChrimson.mVenus}attP40 | BDSC | BDSC: 55138 |
| D. m.: w1118;; P{13xLexAop2-IVS-Syn21-CsChrimson-tdTomato}VK00005 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w1118;; P{lexAop-P2X2.Y} | [74] | FlyBase: FBtp0093385 |
| D. m.: w1118, y1;; P{NPF-Gal4.1}1 | BDSC | BDSC: 25682 |
| D. m.: w1118; P{20xUAS>myrTopHat2>Syn21-CsChrimson-tdTomato}su(Hw)attP5 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w*;; TI{FLP}fru[FLP]/TM3, Sb1 | BDSC | BDSC: 66870 |
| D. m.: w1118; P{y[+t7.7] w[+mC]=8XLexAop2-FLPL}attP2 | BDSC | BDSC: 55819 |
| D. m.: w*; wgSp−1/CyO; TI{lexA::VP16}fruP1.LexA/TM6B, Tb1 | BDSC | BDSC: 66698 |
| D. m.: w*;; TI{lexA::p65}dsxlexA::p65 | [75] | Flybase: FBti0201624 |
| D. m.: w*; P{y[+t7.7] w[+mC]=8XLexAop2-IVS-GAL80-WPRE}attP2 | BDSC | BDSC: 32213 |
| D. m.: NPF-RNAi: y1, v1;;P{TRiP.JF02555}attP2 | BDSC | BDSC: 27237 |
| D. m.: w1118; P{UAS-Dcr-2.D}2 | BDSC | BDSC: 24650 |
| D. m.: NPFR-RNAi: w1118; P{KK112704}VIE-260B | VDRC | VDRC: 107663 |
| D. m.: elav-Gal4: P{GawB}elavc155 | BDSC | BDSC: 458 |
| D. m.: w1118; P{13xLexAop2-IVS-Syn21-opGCaMP6s}su(Hw)attP5 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w1118;; P{UAS-Rnor\P2rx2.L} | [76] | FlyBase: FBtp0021869 |
| D. m.: w1118; TH-LexA | [77] | N/A |
| D. m.: +;; DopR2attP | [70] | N/A |
| D. m.: w1118;; P{13xLexAop2-IVS-Syn21-opGCaMP6s}su(Hw)attP1 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w1118;; P{20xUAS-IVS-Syn21-opGCaMP6s}su(Hw)attP1 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w*;; P{y[+t7.7] w[+mC]=10XUAS-IVS-myr::tdTomato}attP2 | BDSC | BDSC: 32221 |
| D. m.: w1118;P{UAS-FLP.U};P{CRE>mCherry>luciferase} | [47] | FlyBase: FBtp0083995, FBtp0065911 |
| D. m.: UAS-CREB2SUP: w1118; P{w[+mC]=dCREB-B-UAS}94 | BDSC | BDSC: 7220 |
| D. m.: UAS-CREB2-A: w*;; P{UAS-CrebB-17A-a.cor}T7.1 | BDSC | BDSC: 9232 |
| D. m.: w1118; P{20xUAS-IVS-Syn21-opGCaMP6s}su(Hw)attP5 | Gift from B.D. Pfeiffer and D.J. Anderson | N/A |
| D. m.: w1118; P{10XUAS-IVS-myr::tdTomato}attP40 | BDSC | BDSC: 32222 |
| D. m.: PP1α96A-RNAi-1: y1, v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS02154}attP40 | BDSC | BDSC: 40906 |
| D. m.: Task7-RNAi: w1118; P{GD3628}v8564 | VDRC | VDRC: 8564 |
| D. m.: w1118;; P{20xUAS-IVS-Syn21-CsChrimson-tdTomato}attP2 | [78] | N/A |
| D. m.: w1118;; P{UAS-IVS-GtACR1}attP2 | [58] | N/A |
| D. m.: w1118; P{UAS-IVS-GCaMP6s}attP40 | BDSC | BDSC: 42746 |
| D. m.: VAChT-RNAi: w1118; P{GD3054}v40918 | VDRC | VDRC: 40918 |
| D. m.: CRN-p65AD: w1118; P{GMR42G02-p65AD}attP40 | This paper | N/A |
| D. m.: w*; P{w[+mC]=UAS-2xEGFP}AH2; P{w[+mC]=ChAT-GAL4.DBD}J8A1 | BDSC | BDSC: 23869 |
| D. m.: w1118; P{UAS-ort.2HA} | [79] | FlyBase: FBtp0093981 |
| D. m.: w1118; P{LexAop-ort.HA} | [80] | FlyBase: FBtp0126450 |
| D. m.: CREB2-RNAi: y1, v1; P{TRiP.HMJ30249}attP40/CyO | BDSC | BDSC: 63681 |
| D. m.: CKIIβ2-RNAi: w1118; P{KK103402}VIE-260B | VDRC | VDRC: 102633 |
| D. m.: PP1α96A-RNAi-2: y1, sc*, v1; P{y[+t7.7] v[+t1.8]=TRiP.HMS02477}attP40 | BDSC | BDSC: 42641 |
| D. m.: P1-B-Gal4: w1118;; P{GMR71G01-GAL4}attP2 | BDSC | BDSC: 39599 |
| D. m.: UAS-CREBDN: w1118;; P{w[+mC]=Cbz}4/TM3, Sb1 Ser1 | BDSC | BDSC: 7222 |
| Oligonucleotides | ||
| FISH probes targeting Task7 (tagged with Quasar-570) | LGC Biosearch Technologies | See Table S3 for sequences |
| FISH probes targeting Dsx (tagged with Quasar-670) | LGC Biosearch Technologies | See Table S3 for sequences |
| Recombinant DNA | ||
| R42G02 DNA fragment | Gift from Gerry Rubin | N/A |
| Software and Algorithms | ||
| ROI-based fluorescence quantification package (MATLAB) | [22] | https://github.com/CrickmoreRoguljaLabs/Blind-image-analysis |
| Courtship-initiation analysis package (MATLAB) | [23] | https://github.com/CrickmoreRoguljaLabs/tapping-codes |
| ROI-based FISH analysis (MATLAB) | This paper | https://github.com/CrickmoreRoguljaLabs/ROI_FISH_analysis |
| Models for fly mating drive dynamics (MATLAB) | This paper | https://github.com/CrickmoreRoguljaLabs |
| Predict putative CREB-regulated genes (MATLAB) | This paper | https://github.com/CrickmoreRoguljaLabs/CREB_analysis |
| FIJI | [81] | https://fiji.sc/ |
| MTrack2 (plugin for FIJI) | Ron Vale lab | https://valelab4.ucsf.edu/~stuurman/IJplugins/MTrack2.html |
| Other | ||
| Phosphate Buffered Saline 10x | MediaTech | 46–013-CM |
| Triton X-100 | Sigma-Aldrich | X100 |
| Potato flakes | Carolina Biological Supply | 173200 |
| Nunc Petri dishes | Thermo Scientific | 150318 |
| Glass-bottom Petri dishes | MatTek | P35G-1.5–20-c |
| 655-nm LED | Luxeon Star LEDs | LXM3-PD01–0350 |
| 530-nm LED | Luxeon Star LEDs | LXML-PM01–0100 |
| Arduino MEGA | Arduino | A000067 |
| Raspberry PI Model B+ | Raspberry PI | Model B+ |
| 2-photon microscope (including the 465-nm LED) | Neurolabware | Standard |
HIGHLIGHTS.
Copulation Reporting Neurons in male Drosophila detect matings and induce satiety
A recurrent excitation loop promotes recovery from satiety over days
Loop activity uses CREB to transcribe the inhibitory channel subunit TASK7
Prior motivation generates the inhibitory environment that sustains satiety
Acknowledgments:
We thank Mark Andermann, Steve Flavell, and the members of the Rogulja and Crickmore labs for comments on the manuscript. Corey Harwell and Chuck Weitz provided equipment for wide-field microscopy and luciferase assays, respectively. Mark Andermann and Andrew Lutas provided equipment and technical support for two-photon imaging experiments. Michelle Frank, Christine Boutros, Lauren Miner, Ben Gorko, Ariadna Corredera, Ethan Glantz, and Blake Karavas helped with experiments. The unpublished 13xLexAop-IVS-Syn21-opGCaMP6s (II), 13xLexAop-IVS-Syn21-opGCaMP6s (III), 20xUAS-IVS-Syn21-opGCaMP6s (II), 20xUAS-IVS-Syn21-opGCaMP6s (III), 20xUAS>myrTopHat2>Syn21-CsChrimson-tdTomato (II), 13xLexAop2-IVS-Syn21-CsChrimson-tdTomato (III) lines were gifts from Barret Pfeiffer and David Anderson. The R42G02 DNA fragment and R42G02-LexA were gifts from Gerry Rubin and Bruce Baker, respectively. S.X.Z. is a Stuart H.Q. & Victoria Quan Fellow at Harvard Medical School. D.R. is a New York Stem Cell Foundation-Robertson Investigator. This work was supported by The New York Stem Cell Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no conflict of interest.
DATA AND SOFTWARE AVAILABILITY
MATLAB scripts and functions used to 1) quantify calcium transients, 2) analyze tap-induced courtship initiation, 3) analyze in situ hybridization data, 4) model motivational dynamics, and 5) predict the putative CREB-regulated genes can be found available online at https://github.com/CrickmoreRoguljaLabs.
REFERENCES
- 1.Berridge KC (2004). Motivation concepts in behavioral neuroscience. Physiol. Behav 81, 179–209. [DOI] [PubMed] [Google Scholar]
- 2.Woods SC, and Ramsay DS (2007). Homeostasis : Beyond Curt Richter. Appetite 49, 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Lin Y-CC, Kuo T-WW, and Knight ZA (2015). Sensory Detection of Food Rapidly Modulates Arcuate Feeding Circuits. Cell 160, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betley JN, Xu S, Cao ZFH, Gong R, Magnus CJ, Yu Y, and Sternson SM (2015). Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature 521, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandelblat-Cerf Y, Ramesh RN, Burgess CR, Patella P, Yang Z, Lowell BB, and Andermann ML (2015). Arcuate hypothalamic AgRP and putative POMC neurons show opposite changes in spiking across multiple timescales. Elife 4, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Lin Y-C, Zimmerman CA, Essner RA, and Knight ZA (2016). Hunger neurons drive feeding through a sustained, positive reinforcement signal. Elife 5, e18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman CA, Leib DE, and Knight ZA (2017). Neural circuits underlying thirst and fluid homeostasis. Nat. Rev. Neurosci 18, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmerman CA, Huey EL, Ahn JS, Beutler LR, Tan CL, Kosar S, Bai L, Chen Y, Corpuz TV, Madisen L, et al. (2019). A gut-to-brain signal of fluid osmolarity controls thirst satiation. Nature 568, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Essner RA, Kosar S, Miller OH, Lin Y-C, Mesgarzadeh S, and Knight ZA (2019). Sustained NPY signaling enables AgRP neurons to drive feeding. Elife 8, e46348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, and Baker BS (2005). Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400. [DOI] [PubMed] [Google Scholar]
- 11.Rideout EJ, Dornan AJ, Neville MC, Eadie S, and Goodwin SF (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci 13, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ, Tirián L, and Dickson BJ (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. [DOI] [PubMed] [Google Scholar]
- 13.Thistle R, Cameron P, Ghorayshi A, Dennison L, and Scott K (2012). Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, et al. (2013). Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clowney EJ, Iguchi S, Bussell JJ, Scheer E, and Ruta V (2015). Multimodal Chemosensory Circuits Controlling Male Courtship in Drosophila. Neuron 87, 1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallman BR, Kim H, and Scott K (2015). Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. Elife 4, e11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Philipsborn AC, Liu T, Yu JY, Masser C, Bidaye SS, and Dickson BJ (2011). Neuronal control of Drosophila courtship song. Neuron 69, 509–522. [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, Meissner GW, and Baker BS (2012). Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc. Natl. Acad. Sci. U. S. A 109, 10065–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohatsu S, and Yamamoto D (2015). Visually induced initiation of Drosophila innate courtship-like following pursuit is mediated by central excitatory state. Nat. Commun 6, 6457. [DOI] [PubMed] [Google Scholar]
- 20.Kohatsu S, Koganezawa M, and Yamamoto D (2011). Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron 69, 498–508. [DOI] [PubMed] [Google Scholar]
- 21.Kimura K, Hachiya T, Koganezawa M, Tazawa T, and Yamamoto D (2008). Fruitless and Doublesex Coordinate to Generate Male-Specific Neurons that Can Initiate Courtship. Neuron 59, 759–769. [DOI] [PubMed] [Google Scholar]
- 22.Zhang SX, Rogulja D, and Crickmore MA (2016). Dopaminergic Circuitry Underlying Mating Drive. Neuron 91, 168–181. [DOI] [PubMed] [Google Scholar]
- 23.Zhang SX, Miner LE, Boutros CL, Rogulja D, and Crickmore MA (2018). Motivation, perception, and chance converge to make a binary decision. Neuron 99, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegel RW, and Hall JC (1979). Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. U. S. A 76, 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou C, Pan Y, Robinett CCC, Meissner GWW, and Baker BSS (2014). Central Brain Neurons Expressing doublesex Regulate Female Receptivity in Drosophila. Neuron 83, 149–163. [DOI] [PubMed] [Google Scholar]
- 27.Clyne JD, and Miesenböck G (2008). Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell 133, 354–63. [DOI] [PubMed] [Google Scholar]
- 28.Atasoy D, Betley JN, Su HH, and Sternson SM (2012). Deconstruction of a neural circuit for hunger. Nature 488, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krashes MJ, Shah BP, Koda S, and Lowell BB (2013). Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab 18, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, and Waddell S (2009). A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139, 416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croset V, Treiber CD, and Waddell S (2018). Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 7, e34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davie K, Janssens J, Koldere D, Waegeneer M De, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, González-Blas CB, et al. (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu WW, and Wilson RI (2013). Transient and specific inactivation of drosophila neurons in vivo using a native ligand-gated ion channel. Curr. Biol 23, 1202–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren GR, Folke J, Hauser F, Li S, and Grimmelikhuijzen CJP (2015). The A- and B-type muscarinic acetylcholine receptors from Drosophila melanogaster couple to different second messenger pathways. Biochem. Biophys. Res. Commun 462, 358–364. [DOI] [PubMed] [Google Scholar]
- 35.Jikomes N, Ramesh RN, Mandelblat-Cerf Y, and Andermann ML (2016). Preemptive Stimulation of AgRP Neurons in Fed Mice Enables Conditioned Food Seeking under Threat. Curr. Biol 26, 2500–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang XJ (2008). Decision Making in Recurrent Neuronal Circuits. Neuron 60, 215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo ZV, Inagaki HK, Daie K, Druckmann S, Gerfen CR, and Svoboda K (2017). Maintenance of persistent activity in a frontal thalamocortical loop. Nature 545, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Lenek D, Dag U, Dickson B, and Keleman K (2017). Persistent activity in a recurrent circuit underlies courtship memory in Drosophila. Elife 7, e31425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stagkourakis S, Spigolon G, Williams P, Protzmann J, Fisone G, and Broberger C (2018). A neural network for intermale aggression to establish social hierarchy. Nat. Neurosci 21, 834–842. [DOI] [PubMed] [Google Scholar]
- 40.Inagaki HK, Fontolan L, Romani S, and Svoboda K (2019). Discrete attractor dynamics underlies persistent activity in the frontal cortex. Nature 566, 212–217. [DOI] [PubMed] [Google Scholar]
- 41.Song BJ, and Rogulja D (2017). SnapShot: Circadian Clock. Cell 171, 1468–1468.e1. [DOI] [PubMed] [Google Scholar]
- 42.Hall JC (2005). Systems Approaches to Biological Rhythms in Drosophila. In Methods in Enzymology, pp. 61–185. [DOI] [PubMed]
- 43.Lonze BE, and Ginty DD (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- 44.Mayr B, and Montminy M (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 45.Carlezon WA, Duman RS, and Nestler EJ (2005). The many faces of CREB. Trends Neurosci 28, 436–45. [DOI] [PubMed] [Google Scholar]
- 46.Yin JCP, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, and Quinn WG (1995). A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol. Cell. Biol 15, 5123–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanenhaus AK, Zhang J, and Yin JCP (2012). In vivo circadian oscillation of dCREB2 and NF-κB activity in the Drosophila nervous system. PLoS One 7, e45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eresh S, Riese J, Jackson DB, Bohmann D, and Bienz M (1997). A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J 16, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, and Malenka RC (2006). CREB modulates excitability of nucleus accumbens neurons. Nat. Neurosci 9, 475–7. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez GA, and Montminy MR (1989). Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680. [DOI] [PubMed] [Google Scholar]
- 51.Horiuchi J, Jiang W, Zhou H, Wu P, and Yin JCP (2004). Phosphorylation of Conserved Casein Kinase Sites Regulates cAMP-response Element-binding Protein DNA Binding in Drosophila. J. Biol. Chem 279, 12117–12125. [DOI] [PubMed] [Google Scholar]
- 52.Döring F, Scholz H, Kühnlein RP, Karschin A, and Wischmeyer E (2006). Novel Drosophila two-pore domain K+ channels: Rescue of channel function by heteromeric assembly. Eur. J. Neurosci 24, 2264–2274. [DOI] [PubMed] [Google Scholar]
- 53.Long X, Colonell J, Wong AM, Singer RH, and Lionnet T (2017). Quantitative mRNA imaging throughout the entire Drosophila brain. Nat. Methods 14, 703–706. [DOI] [PubMed] [Google Scholar]
- 54.Crane A, and Aguilar-Bryan L (2004). Assembly, Maturation, and Turnover of KATP Channel Subunits. J. Biol. Chem 279, 9080–9090. [DOI] [PubMed] [Google Scholar]
- 55.Okada M, Kano M, and Matsuda H (2013). The degradation of the inwardly rectifying potassium channel, Kir2.1, depends on the expression level: Examination with fluorescent proteins. Brain Res 1528, 8–19. [DOI] [PubMed] [Google Scholar]
- 56.Crickmore MA, and Vosshall LB (2013). Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell 155, 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilchrist AS, and Partridge L (2000). Why It is Difficult to Model Sperm Displacement in Drosophila melanogaster : The Relation between Sperm Transfer and Copulation Duration. Evolution (N. Y) 54, 534–542. [DOI] [PubMed] [Google Scholar]
- 58.Mohammad F, Stewart JC, Ott S, Chlebikova K, Chua JY, Koh T-W, Ho J, and Claridge-Chang A (2017). Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14, 271–274. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, and McKnight GS (2015). Hypothalamic PKA regulates leptin sensitivity and adiposity. Nat. Commun 6, 8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marmonier C, Chapelot D, Fantino M, and Louis-Sylvestre J (2002). Snacks consumed in a nonhungry state have poor satiating efficiency: Influence of snack composition on substrate utilization and hunger. Am. J. Clin. Nutr 76, 518–528. [DOI] [PubMed] [Google Scholar]
- 61.Boutros CL, Miner LE, Mazor O, and Zhang SX (2017). Measuring and Altering Mating Drive in Male Drosophila melanogaster. J. Vis. Exp, e55291. [DOI] [PMC free article] [PubMed]
- 62.Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- 63.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr R. a, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Y, Ueda A, and Wu C-F (2004). A Modified Minimal Hemolymph-Like Solution, HL3.1, for Physiological Recordings At the Neuromuscular Junctions of Normal and Mutant Drosophila Larvae. J. Neurogenet 18, 377–402. [DOI] [PubMed] [Google Scholar]
- 66.St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, and Lin MZ (2014). High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci 17, 884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess CR, Ramesh RN, Sugden AU, Levandowski KM, Minnig MA, Fenselau H, Lowell BB, and Andermann ML (2016). Hunger-Dependent Enhancement of Food Cue Responses in Mouse Postrhinal Cortex and Lateral Amygdala. Neuron 91, 1154–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sweatt JD (1997). Protooncogenes Subserve Memory Formation in the Adult CNS. Neuron 31, 671–674. [DOI] [PubMed] [Google Scholar]
- 69.Mayr BM, Canettieri G, and Montminy MR (2001). Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl. Acad. Sci. U. S. A 98, 10936–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keleman K, Vrontou E, Krüttner S, Yu JY, Kurtovic-Kozaric A, and Dickson BJ (2012). Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature 489, 145–149. [DOI] [PubMed] [Google Scholar]
- 71.Baines RA, Uhler JP, Thompson A, Sweeney ST, and Bate M (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci 21, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, and Birman S (2003). Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol 54, 618–27. [DOI] [PubMed] [Google Scholar]
- 73.Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, and Waddell S (2012). Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao Z, Macara AM, Lelito KR, Minosyan TY, and Shafer OT (2012). Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol 108, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou C, Franconville R, Vaughan AG, Robinett CC, Jayaraman V, and Baker BS (2015). Central neural circuitry mediating courtship song perception in male Drosophila. Elife 4, e08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lima SQ, and Miesenböck G (2005). Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121, 141–152. [DOI] [PubMed] [Google Scholar]
- 77.Berry JAA, Cervantes-Sandoval I, Chakraborty M, and Davis RLL (2015). Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell 161, 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoopfer ED, Jung Y, Inagaki HK, Rubin GM, and Anderson DJ (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. Elife 4, e11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karuppudurai T, Lin T-Y, Ting C-Y, Pursley R, Melnattur KV, Diao F, White BH, Macpherson LJ, Gallio M, Pohida T, et al. (2014). A Hard-wired Glutamatergic Circuit Pools and Relays UV Signals to Mediate Spectral Preference in Drosophila. Neuron 81, 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu WW, Mazor O, and Wilson RI (2015). Thermosensory processing in the Drosophila brain. Nature 519, 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Raw data from luciferase-based assays of CREB activity in this paper. Related to Figures 4 and 5.