Abstract
Neutrophils are an important component of the innate immune system and provide a front line of defense against bacterial infection. Although most bacteria are killed readily by neutrophils, some bacterial pathogens have the capacity to circumvent destruction by these host leukocytes. The ability of bacterial pathogens to avoid killing by neutrophils often involves multiple attributes or characteristics, including the production of virulence molecules. These molecules are diverse in composition and function, and collectively have the potential to alter or inhibit neutrophil recruitment, phagocytosis, bactericidal activity, and/or apoptosis. Here, we review the ability of bacteria to target these processes.
Keywords: Bacterial pathogenesis, Virulence, Phagocytosis, Neutrophil, Host defense, Innate immunity
Introduction
The human innate immune system is comprised of many components that function in concert to defend against microorganisms. These components include physical barriers, antimicrobial peptides, freely secreted antimicrobial proteins present in the blood, mucous secretions, and interstitial fluid, and leukocytes. Together, these innate immune system elements protect against infection from many types of microorganisms, including bacteria.
The skin is an important physical barrier to microorganisms, and skin cells (or other cell types within or around the skin) produce many antimicrobial molecules that contribute to the host defense against bacteria [1]. Most skin antimicrobial peptides, such as β-defensins and cathelicidins, are comprised of approximately 15–20 amino acid residues, and have a net positive charge. These peptides interact readily with the negatively charged surface of bacteria, and, in turn, inhibit growth and/or are bactericidal [1, 2]. Individuals with defects in the regulation of these peptides are more susceptible to skin infections [2]. Microorganisms, whether commensal or pathogenic, can gain access to normally sterile host tissues and the bloodstream by way of an entry portal. Such portals include breaches in the skin barrier, e.g., abrasions, lacerations, and burns, and also access through mucous membranes. Importantly, invading bacteria trigger the release of proinflammatory molecules that amplify the inflammatory response and, in turn, the innate immune response.
The complement system, antibody, collectins, ficolins, and pentraxins are important noncellular antimicrobial factors that contribute significantly to the host defense against invading microbes [3, 4]. For example, individuals with complement deficiencies are more susceptible to recurrent bacterial infections than people with a fully functional complement system [4, 5]. The complement membrane attack complex (C5b-C9) forms a cytolytic pore in the bacterial cell envelope membrane, and thus has direct bactericidal activity [4]. Other complement fragments, including C3b and iC3b, bind the surface of bacteria and, in the context of antibodies, promote efficient phagocytosis by macrophages, monocytes, and neutrophils [4]. Thus, it should be no surprise that some bacterial pathogens have evolved molecules that inhibit components of the complement cascade and/or sequester or degrade antibodies.
Proinflammatory molecules such as chemokines and bacterial molecules recruit leukocytes to sites of infection. Phagocytic leukocytes, i.e., polymorphonuclear leukocytes (PMNs or neutrophils) and mononuclear phagocytes (monocytes, macrophages, and dendritic cells), are critical for defense against invading microorganisms. These cells ingest and kill bacteria, or provide a link between innate and adaptive immunity that is important for long-term protection against reinfection. Among phagocytic leukocytes, PMNs are the most numerous and have arguably the greatest bactericidal capacity. PMNs kill bacteria by using oxygen-dependent and oxygen independent processes [6]. Oxygen-dependent bactericidal activity involves the production of superoxide by an enzyme complex known as NADPH oxidase [6]. Superoxide is converted to other reactive oxygen species (ROS) that are effective at killing bacteria. PMNs from individuals with genetic defects in NADPH oxidase are susceptible to severe and recurrent bacterial and fungal infections [6]. In addition to the production of ROS, PMNs possess an array of antimicrobial peptides and proteins that have bactericidal activity. These molecules are contained within cytoplasmic granules and are typically delivered to bacteria-containing vacuoles (phagosomes) following phagocytosis. PMN ROS and granule proteins work together to kill ingested microbes.
Although PMNs are highly effective at killing most bacteria, some pathogens have evolved means to evade killing by these leukocytes. A comprehensive review of the immune evasion molecules produced by all types of microbes is beyond the scope of this article. Instead, we highlight selected molecules used by bacteria to circumvent killing by neutrophils.
Bacterial Strategies to Evade Neutrophil Function
Neutrophil Recruitment
Pathogen recognition and the subsequent recruitment of neutrophils to sites of infection are key elements of the host defense against bacterial disease. Neutrophil recruitment is a multistep process that includes extravasation of bloodstream neutrophils to distal sites of infection and/or injury, mobilization of neutrophils from bone marrow reserves, and increased hematopoiesis as needed. Invading pathogens and their signature pathogen-associated molecular patterns (PAMPs) are recognized by host pattern recognition receptors (PRRs), which include Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) proteins. Receptor ligation triggers the production of a variety of proinflammatory host cytokines and chemokines, such as interleukin (IL)-8, IL-1α, and IL-β, CXCL1 (GROα), CXCL2 (MIP2α), CXCL5 (ENA78), tumor necrosis factor (TNF), granulocyte-colony stimulating factor (G-CSF), or granulocyte-macrophage colony stimulating factor (GM-CSF). These molecules serve as chemoattractants and promote neutrophil recruitment to infected tissues.
Neutrophil migration is also driven by pathogen-derived molecules such as N-formylated peptides (e.g., formyl-methionyl-leucyl-phenylalanine, fMLF) that are byproducts of bacterial protein synthesis. Besides a handful of resident immune cells, the majority of neutrophils have to be recruited from blood, and this involves a process known as extravasation. Neutrophil extravasation is divided into 4 major phases: rolling, adhesion, crawling, and transmigration. Each step is tightly regulated by cross-talk between lymphocytes and endothelium, and involves the interaction of numerous integrins and adhesion molecules. Neutrophil exposure to host- and pathogen-derived proinflammatory molecules as well as interactions with activated endothelia primes these phagocytes for enhanced function. Historically, priming was defined as the ability of primary agonists such as lipopolysaccharide to enhance ROS production by a secondary stimulus. During this process, neutrophils undergo mobilization of secretory vesicles, partial mobilization of specific granules, and partial assembly of NADPH oxidase which enhances neutrophil function in response to a second stimulus [7].
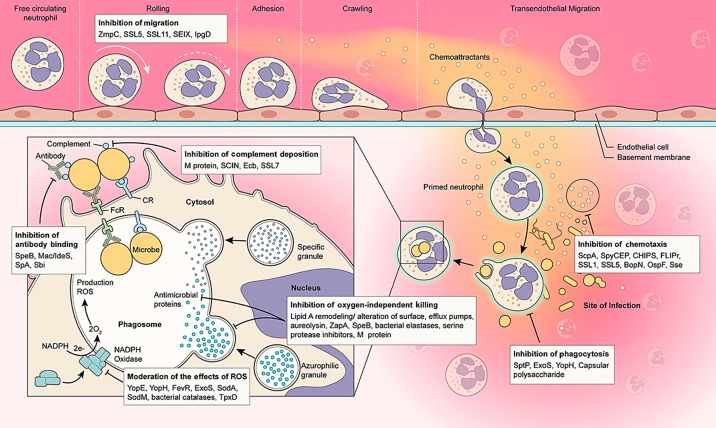
Pathogen success is often highly dependent on the ability to avoid recognition and killing by the host immune system. To that end, a number of bacterial pathogens produce molecules that have the ability to dampen neutrophil recruitment. Some of these molecules target chemoattractants. For example, Streptococcus pyogenes produces secreted peptidases known as ScpA and ScpC/SpyCEP that degrade C5a and IL-8, respectively [8, 9], and the streptococcal secreted esterase Sse, which inactivates platelet-activating factor [10, 11]. Inhibition of neutrophil recruitment is enhanced in hypervirulent S. pyogenes strains by mutation/deletion in CovRS, a 2-component gene regulatory system that controls the expression of multiple virulence factors, including SpyCEP and SsE [11, 12]. Streptococcus pneumoniae employs a slightly different mechanism to block neutrophil recruitment. This pathogen produces pneumococcal zinc metalloproteinase C (ZmpC), which targets the initial rolling step of neutrophil extravasation by cleavage of the N-terminal domain of P-selectin glycoprotein 1 (PSGL-1) [13].
Staphylococcus aureus, one of the most prominent gram-positive pathogens, possesses an arsenal of virulence factors that have the ability to counteract the initial steps of the innate immune response. For example, neutrophil activation and chemotaxis can be inhibited by the chemotaxis inhibitory protein of S. aureus (CHIPS), which binds to the C5a receptor and formyl peptide receptor (FPR), thereby blocking ligand interaction [14]. FPR and its homolog formyl peptide receptor-like 1 are additionally blocked by the S. aureus-secreted proteins known as FPRL1 inhibitory protein (FLIPr) and FLIPr-like protein [15, 16]. Staphylococcal superantigen-like protein (SSL3) inhibits TLR2, an important PRR for the recognition of S. aureus. Moreover, SSL5 and SSL11 and staphylococcal enterotoxin-like toxin X (SElX) inhibit neutrophil extravasation by blocking the interaction of PSGL-1 and P-selectin on the endothelium [17]. Recently, SSL1 and SSL5 were shown to inhibit neutrophil matrix metalloproteinases (MMP8 and MMP9), which subsequently inhibit MMP-dependent IL-8 cleavage to prevent potentiation and inhibit neutrophil migration [18].
Gram-negative pathogens often employ a flagellar and translocation-associated type III secretion system (T3SS) to evade the host immune system, including neutrophil recruitment. The T3SS is a plasmid-encoded and contact-dependent translocation mechanism that allows bacteria to inject effector molecules into host cell cytoplasm [19]. Pathogenic bacteria use this route to introduce virulence factors into host cells and as an adaptation to favor pathogen intracellular survival. Several bacterial factors contribute to the evasion of the immune system by impairing NF-κB signaling, and thereby altering the production of interleukins. For example, Bordetella effector BopN has the ability to stimulate the production of anti-inflammatory IL-10, which in turn inhibits neutrophil recruitment [20]. The OspF phosphatase of Shigella represses the transcription of multiple genes involved in the immune response, including IL-8, a potent neutrophil chemoattractant [21]. Moreover, the Shigella virulence factor IpgD induces phosphatidylinositol 5-phosphate (PI5P) production. The high levels of PI5P trigger ICAM-1 internalization and degradation in infected epithelial cells, and significantly affect neutrophil trafficking during infection [22].
Phagocytosis
The ability of neutrophils to ingest and subsequently kill invading microbes is essential for the maintenance of host health. Neutrophils remove bacterial and fungal pathogens through a process known as phagocytosis. Recognition of invading microbial pathogens is mediated by receptors present on the neutrophil surface, such as PRRs (e.g., TLRs) and opsonic receptors, which recognize host proteins that are deposited on the microbial surface. The ligation of PRRs initiates a complex series of molecular signals that modulate effector functions such as enhanced phagocytosis, killing, and the regulation of inflammation via cytokine production. Phagocytosis is most efficient in the presence of opsonins such as specific immunoglobulin (Ig)G and complement factors that directly mediate uptake (opsonophagocytosis). IgG or IgM bound to the microbial surface is recognized by C1q which activates the classical complement pathway. In addition, complement can be deposited on the microbial surface following activation of the alternative or mannose-binding lectin pathways. PMNs express distinct receptors for IgG (FcγRI, FcγRII, and FcγRIII) and opsonic complement molecules C3b and iC3b (CR1, CR3, and CR4). Efficient particle-binding is enhanced by simultaneous or sequential engagement of receptors on the phagocyte surface and precedes the internalization of pathogens. Actin polymerization is a requisite for phagocytosis and, in conjunction with progressive FcR binding, it provides the cytoskeletal framework to advance the plasma membrane of neutrophils over the particle and sequester them in phagosomes prior to killing.
Inasmuch as the process of phagocytosis is predicated by PMN recognition of microbial pathogens, it is not surprising that pathogens have evolved strategies to limit or prevent binding and uptake. One of the primary mechanisms to prevent recognition is through the masking of surface epitopes, thereby preventing the binding of antibodies and the deposition of complement on the bacterial surface. The ability of bacterial pathogens to prevent/evade complement deposition and subsequent activation has 3 potential consequences for pathogen survival: (1) it serves as a mechanism to limit direct complement mediated lysis/killing of the microbe; (2) (and perhaps more pertinent for interactions with PMNs) it prevents direct recognition and opsonophagocytosis of the pathogen and consequent exposure to intracellular neutrophil microbicidal agents; and (3) it interferes with downstream complement signaling cascades (e.g., an inflammatory response). One of the most common strategies for bacterial pathogens to mask surface antigens is by simply expressing an enveloping polysaccharide capsule [23]. There are many examples of encapsulated bacteria that have been described as inhibiting neutrophil phagocytosis including Streptococcus spp., Neisseria meningitidis, Klebsiella spp., Escherichia coli, Pseudomonas aeruginosa, and Haemophilus influenzae. Bacterial pathogens also mask epitopes through structural modifications that prevent recognition by PRRs. For example, several gram-negative bacterial pathogens such as Yersinia spp. and Salmonella typhimurium modify lipid A structure to inhibit recognition by TLR4. Bacteria can also interfere with complement regulatory proteins as an evasion strategy to limit opsonization. For example, the sequestration of complement regulatory factor H by N. meningitidis impairs complement activation by the alternative pathway which favors bacterial survival [24]. Furthermore, the surface M protein of S. pyogenes impairs the binding of opsonic fragment C3b to the cell surface by inhibiting complement regulatory proteins, such as C4b-binding protein, factor H, and factor H-like protein [25]. S. pyogenes also secretes Mac/IdeS, a host-receptor mimetic of the leukocyte β2-integrin Mac-1 that has 2 distinct immune evasion properties that function in concert to inhibit opsonophagocytosis [26, 27]. Mac/IdeS interacts with CD16 and Mac-1 at the neutrophil plasma membrane to block the binding of IgG to CD16, and streptococcal Mac is a cysteine protease that degrades IgG. S. aureus produces a number of complement inhibitors that interfere with opsonophagocytosis, including staphylococcal complement inhibitor (SCIN), extracellular complement-binding protein (Ecb), and staphylococcal superantigen-like protein (SSL7). Bacterial pathogens can also interfere with antibody opsonization through protease degradation of immunoglobulin by factors such as SpeB and the aforementioned IdeS, albeit interference by proteolytic activity requires high concentrations of proteins in vivo [28]. In addition, some bacteria can produce immunoglobulin-binding proteins (e.g., SpA and Sbi of S. aureus) that are capable of sequestering antibodies [29] and thus, potentially, inhibiting opsonophagocytosis.
As indicated above, evasion of neutrophil recognition is a common and successful strategy employed by many bacterial pathogens to survive in the host. Alternatively, some pathogens have developed methods to actively inhibit phagocytosis following neutrophil recognition. Many gram-negative pathogens use a T3SS to deliver effector proteins into the host cell to promote the establishment of infection. Among the type III effector molecules, the GTPase activating proteins (GAPs) such as Yersinia YopE are capable of targeting small RhoA family G proteins, leading to the impairment of the actin cytoskeleton, and thus actively inhibiting phagocytosis. Salmonella enterica SptP and P. aeruginosa ExoS are similar GAPs that target the actin cytoskeleton. Yersinia expresses an ad ditional type III effector protein tyrosine phosphatase (PTPase), YopH, that blocks immediate early Ca2+ signaling in neutrophils and impairs phagocytosis [30], and, together with YopE, has recently been shown to interfere with neutrophil degranulation [31].
Bactericidal Activity
Neutrophils use oxygen-dependent and oxygen independent processes to kill ingested microorganisms. The phagocytosis of bacterial pathogens leads to the formation of potent antimicrobial ROS, such as superoxide radicals, hydrogen peroxide, hypochlorous acid, hydroxyl radicals, and chloramines. In addition, cytoplasmic granules fuse with bacteria-containing phagosomes and enrich the vacuole lumen with antimicrobial peptides and proteases. Thus, the potent antimicrobial activity of the neutrophil is a collaborative effort between highly proteolytic and degradative enzymes, cationic molecules, and ROS.
In activated neutrophils, a membrane-bound NADPH-dependent oxidase generates high levels of superoxide. In unstimulated neutrophils, subunits of the NADPH oxidase complex are separated in cytosol (p40phox, p47phox, p67phox, and Rac2) and membrane compartments (flavocytochrome b558, Rap1A). During phagocytosis, the cytosolic components translocate to the plasma and/or phagosome membrane and associate with flavocytochrome b558, a transmembrane heterodimer comprised of gp91phox and p22phox, to form the active oxidase. The oxidase transfers electrons from cytosolic NADPH to intraphagosomal molecular oxygen to produce superoxide. Superoxide anion is short-lived and dismutates rapidly to hydrogen peroxide and forms other secondary products, such as hypochlorous acid, hydroxyl radical, and singlet oxygen, which are effective microbicidal compounds. ROS are cytotoxic and cause damage to proteins, membrane lipids, and nucleic acids. The NADPH oxidase is essential for the host defense against bacterial and fungal pathogens, as inherited defects in components of this enzyme system predispose individuals to severe and/or fatal infections.
Inasmuch as ROS are a critical component of neutrophil killing of bacterial pathogens, it is not unexpected that there are diverse mechanisms to either inhibit ROS production or to detoxify these molecules. As discussed above, NADPH oxidase is a multicomponent complex that is unassembled and inactive in resting cells. The oxidase components are translocated to the plasma or phagosomal membrane upon activation, and the subunits are assembled into a functional complex. Several bacterial pathogens have exploited the complexity of the assembly process, to either inhibit ROS production or misdirect the oxidase complex away from the phagosome. Anaplasma phagocytophilum is one of the few obligate intracellular pathogens with a tropism for neutrophils; part of its success is attributed to its ability to evade NADPH oxidase-mediated killing. A. phagocytophilum does not elicit a pronounced increase of intracellular ROS upon neutrophil uptake, and it has been shown to inhibit the delivery of flavocytochrome b558 to the phagosomal/vacuolar membrane [32]. The intracellular pathogen Francisella tularensis also excludes flavocytochrome b558 from the neutrophil phagosome, resulting in pronounced inhibition of ROS production [33]. By contrast, neutrophil phagosomes containing Opa-negative N. gonorrhoeae accumulate flavocytochrome b558, but exhibit defects in the recruitment of p47phox and p67phox, thus resulting in a reduced production of ROS [34]. Helicobacter pylori employs a somewhat different strategy and, following phagocytosis, targets localization of the NADPH oxidase to the neutrophil outer membrane, thus directing ROS to the extracellular space [35]. Pathogenic Yersiniae utilize the T3SS to deploy effector molecules into the neutrophil cytoplasm to interfere with NADPH oxidase. As discussed above, YopE is a GTPase-activating protein and the molecule has recently been shown to inactivate Rac2, a key regulatory GTPase for activation of the NADPH oxidase [36]. The P. aeruginosa type III cytotoxic effector ExoS inhibits ROS production in human neutrophils, and, similar to Yersinia YopE, functions as a GTPase-activating protein. However, ExoS is a bifunctional protein that also contains a C-terminal ADP ribosyltransferase domain. ExoS has recently been shown to ADP-ribosylate RAS to inhibit PI3K signaling for the activation and assembly of the neutrophil NADPH oxidase [37]. Bacterial pathogens are capable of producing numerous enzymatic antioxidants to detoxify ROS and reduce damage caused by oxidative stress. For example, superoxide dismutases such as SodA and SodM produced by S. aureus are enzymes that catalyze the dismutation of superoxide to hydrogen peroxide. Hydrogen peroxide can be further decomposed to water and oxygen by catalases (e.g., S aureus KatA, and E. coli KatE and KatG). In addition, ROS can be broken down by enzymes of the thioredoxin system such as the S. pneumoniae thiol peroxidase TpxD [38], and those of the glutathione system such as the S. pyogenes glutathione peroxidase [39].
Oxygen independent microbicidal systems utilize antimicrobial peptides and enzymes to facilitate the killing and degradation of ingested microbes. In neutrophils, the antimicrobial peptides and proteins are stored primarily in azurophilic granules and specific granules in the cytoplasm. Neutrophil phagocytosis promotes mobilization of these granules, which subsequently fuse with phagosomes or the plasma membrane (exocytosis). Fusion of azurophilic granules with phagosomes enriches the vacuole lumen with a diversity of antimicrobial peptides, including α-defensins, cathepsins, proteinase-3, elastase, azurocidin, and lysozymes. Neutrophil α-defensins (HNP 1–4) are highly abundant in azurophilic granules and exhibit potent antimicrobial activity. The defensins are relatively small cationic polypeptides (3–5 kDa) that interact with negatively charged molecules at the pathogen surface and contribute to the permeabilization of bacterial membranes. In addition, PMN α-defensins and other antimicrobial peptides serve as an important bridge between the innate and adaptive immune systems, and are known to contribute to processes such as chemotaxis, wound repair, and stimulation of histamine release. Degranulation also enriches phagosomes with specific granule constituents such as flavocytochrome b558 (membrane) and lactoferrin (lumen), further contributing to antimicrobial potential. Lactoferrin sequesters iron needed for bacterial growth, and supplies iron required to generate neutrophil hydroxyl radicals. Lysozyme is universally recognized for the ability to degrade bacterial peptidoglycan, and is ubiquitous among the granule subtypes. Azurocidin, elastase, cathepsin G, and proteinase 3 are collectively known as serprocidins, a family of antimicrobial proteins with a structural homology with serine proteases. The proteins exhibit direct antimicrobial activity and, except for azurocidin (also known as CAP37), are serine proteases. Neutrophil elastase has been shown to cleave the outer membrane protein A (OmpA) of E. coli, thus disrupting membrane integrity and resulting in cell death [40]. Collectively, the oxygen-dependent and oxygen independent neutrophil microbicidal systems operate as an efficient system to prevent bacterial infection. Antimicrobial peptides (AMPs) are important in the host defense against microbial pathogens and a key component of the human innate immune response. There are 2 major AMP families in mammals: the cathelicidins (LL-37 is the sole member in humans), and the defensins (the α and β families). Prototypical AMPs have a net positive charge to facilitate interaction with the net negative charge of bacterial surfaces. These cationic AMPs target anionic lipids (e.g., phosphatidylglycerol and cardiolipins) and other anionic components (e.g., lipopolysaccharide and lipoteichoic acid) of the cell membrane. Once associated with the microbial surface, the amphipathic nature of the cationic AMPs enables insertion into the cell membrane, thereby disrupting membrane integrity and leading to osmotic lysis of the microbial cell. Inasmuch as AMPs are ubiquitous and are encountered frequently by bacterial pathogens, several different mechanisms have evolved for the evasion of AMP-mediated killing, such as alteration of the microbial surface charge, sequestration of AMPs by secreted and cell surface-associated molecules, energy-dependent membrane efflux pumps, and degradation by peptidases.
In general, bacterial surfaces are more negatively charged than those of the eukaryotic host, and thus the cationic AMPs exhibit a higher degree of attraction to the bacterial cell. One of the most commonly employed mechanisms of AMP resistance is through alteration of the surface charge with cationic molecules, thus promoting electrostatic repulsion. Gram-negative bacteria typically alter the surface charge by remodeling lipid A and the addition of phosphatidylethanolamine and 4-amino-4-deoxy-L-arabinose. Gram-positive bacteria often modify cell wall teichoic acids by D-alanylation to reduce the negative surface charge and enhance electrostatic repulsion and AMP resistance. Bacteria can also reduce the deleterious effects of AMPs by the use of efflux pumps. For example, the multiple transferable resistance transporter of N. gonorrhoeae provides resistance to LL-37 and other AMPs [41]. AMPs can also be rendered inactive by bacterial proteases. Numerous studies demonstrate that the cathelicidin LL-37 is degraded by metalloproteases such as S. aureus aureolysin, Proteus mirabilis ZapA, S. pyogenes SpeB, P. aeruginosa elastase, and Enterococcus faecalis gelatinase [42]. In addition to AMP resistance, pathogenic bacteria have also developed mechanisms to evade targeting by other neutrophil granule constituents, including serine proteases such as neutrophil elastase, cathepsin G, proteinase 3, and neutrophil serine protease-4. The primary mechanisms of serine protease resistance used by bacterial pathogens are the modification of serine protease substrates and the production of serine protease inhibitors [43].
Neutrophil extracellular traps (NETs) are web-like structures that consist of strands of decondensed chromatin decorated with the contents of neutrophil granules [44]. NETs can be released from either live or dying cells and, importantly, they have been reported to ensnare and kill various bacterial pathogens [45]. Although NETs have the potential to augment the innate host defense, most bacterial pathogens produce nucleases (e.g., Sda of S. pyogenes) that can destroy cell-free DNA that forms NETs [46].
Apoptosis
As discussed above, the high toxicity of ROS and enzymes from the neutrophil granules enables the immune system to successfully defend the host from a wide range of bacterial pathogens. However, these molecules have the potential to cause collateral damage to adjacent host tissue, if not tightly controlled. In steady-state conditions, aged or spent neutrophils are destined to undergo constitutive (spontaneous) apoptosis [47]. Neutrophils undergo multiple changes during spontaneous apoptosis, including chromatin condensation, DNA fragmentation, vacuolization of the cytoplasm, exposure of phosphatidyl serine on the surface of the cell, diminished proinflammatory and antimicrobial capabilities, and metabolic changes [48]. Intact apoptotic neutrophils are safely removed by macrophages via a nonphlogistic uptake process known as efferocytosis [48]. Two neutrophil apoptotic pathways have been described: (i) the intrinsic or stress-induced pathway associated with death-mediated by neutrophil mitochondria, and (ii) the extrinsic pathway activated by death receptor FAS (CD95) or TNF interactions. Spontaneous apoptosis relies heavily on the proapoptotic B cell lymphoma (Bcl-2) family of proteins and typically is associated with the intrinsic pathway. Upon apoptosis, Bcl-2-associated X (Bax) protein translocates from the cytosol to the mitochondria and interacts with the BH3-interacting domain death agonist (Bid). Bid induces conformational changes, oligomerization, and the anchoring of Bax that creates pores in the outer mitochondrial membrane. These pores allow the release of mitochondrial proapoptotic intermembrane space proteins that activate caspase-9 and displace the X-linked inhibitor of apoptosis (XIAP) [49]. In the extrinsic pathway, the activation of death receptors leads to the recruitment and activation of caspase-8 by the death-inducing signaling complex (DISC). Of note, caspase-8 can be activated independently from the extrinsic pathway, and can activate caspase-9 as a downstream effect of cleavage and the activation of Bid, thus demonstrating the interconnection between the intrinsic and extrinsic pathways. Moreover, both pathways converge at the effector caspase-3 level [47].
During bacterial infection, neutrophil ingestion of microbes prompts cells to undergo apoptosis in a process termed “phagocytosis-induced cell death” (PICD) [47]. PICD is highly dependent on ROS, Mac-1 (CD11b/CD18), and Fcγ receptor signaling [50, 51]. Nonlytic programmed cell death is highly advantageous to the host, since it allows the safe removal of effete neutrophils containing killed bacteria, and promotes the resolution of infection. In general, a select group of bacterial pathogens can either delay apoptosis to prolong host cell survival, or accelerate and/or redirect apoptosis causing cell lysis, and leading to prolonged inflammation and surrounding tissue destruction. There are very few intracellular pathogens that are neutrophil-tropic, attributed, primarily, to the short neutrophil lifespan. Nonetheless, the delay of neutrophil apoptosis is a crucial strategy for intracellular pathogen survival and is necessary to evade the host immune system. A. phagocytophilum, the etiological agent of human granulocytic anaplasmosis, and Chlamydia pneumoniae, a cause of acute respiratory disease, are examples of pathogens that can survive and replicate within neutrophils [52, 53]. The delay of neutrophil apoptosis by A. phagocytophilum is mediated in part by the increased phosphorylation of p38 mitogen-activated protein kinase (MAPK) and activation of phosphatidylinosytol 3-kinase (PI3K)/Akt [54, 55]. Apoptosis is delayed by affecting 2 main antiapoptotic proteins. Specifically, infected neutrophils maintain the expression of Mcl-1, a Bcl-2 family antiapoptosis protein that directly prevents Bax translocation, and the enhanced expression of cellular inhibitor of apoptosis (cIAP2), which regulates caspase activity by direct binding. Furthermore, infected neutrophils increase IL-8 production, which promotes neutrophil survival in an autocrine/paracrine manner [55]. F. tularensis is another intracellular bacterial pathogen that delays neutrophil apoptosis by affecting Bcl-2 family proteins. Namely, the pathogen impairs the processing and activation of caspase-8 and Bid, and significantly inhibits apoptosis by impairing translocation of Bax into the mitochondria [56, 57]. More recently, it has been suggested that F. tularensis-secreted bacterial lipoproteins play a role in apoptosis inhibition via a TLR-2 dependent route [58].
Common bacterial pathogens such as E. coli, P. aeruginosa, and S. aureus induce neutrophil apoptosis following phagocytosis. In general, PICD is a host-driven process that promotes pathogen removal and the resolution of inflammation; however, some bacterial pathogens have evolved means by which to exploit this process to their benefit. For example, P. aeruginosa secretes a phenazine exotoxin, pyocyanin, that rapidly accelerates neutrophil apoptosis. This pathogen-driven premature apoptosis of neutrophils interferes with the ability of the immune system to eliminate invading bacteria. Pyocyanin-induced apoptosis is a ROS-dependent process that mediates dysfunction of the phagosome and leads to the release of cathepsin D, permeabilization of the mitochondrial membranes and, subsequently, reduced levels of Mcl-1 [59]. Moreover, pathogens such as S. aureus have the ability to redirect neutrophil fate towards a cytolytic path or cause direct cell lysis by secreted toxins. Some ingested S. aureus or S. pyogenes strains survive within human neutrophils and cause premature lysis that leads to the dissemination of viable bacteria, thus perpetuating the disease process. Lysis of PMNs infected with S. aureus resembles necroptosis [60]. This process is typically dependent on an interaction between receptor-interacting serine/threonine kinase (RIPK-1 and RIPK-3) and mixed-lineage kinase like protein (MLKL). Necroptosis induced by S. aureus is primarily dependent on RIPK3 activation, but is inhibited by necrostatin-1 (Nec-1), an antagonist of RIPK-1 [61]. In addition, neutrophils with ingested bacteria increase the expression of CD47 (a “don't eat me” signal) that interferes with macrophage efferocytosis and cytokine production [60]. Furthermore, S. aureus and S. pyogenes produce several leukotoxins (e.g., streptolysins O and S, and the staphylococcal leukotoxins HlgAB, LukDE, LukGH, and Panton-Valentine leukocidin [PVL]) that form pores in the neutrophil membrane and cause subsequent cytolysis [62, 63].
Concluding Comment
Many bacterial species, whether primary pathogens or commensal microbes, have the capacity to produce immune evasion molecules. The fundamental role played by these molecules is similar, to avoid detection and/or immune clearance by the host. A diverse array of bacterial immune evasion molecules exist, and they can potentially inhibit or moderate key host immune processes (Table 1; Fig. 1). In an era of increasing antibiotic resistance of bacteria, especially among commensal microbes that are opportunistic pathogens, alternative therapeutics and/or vaccines are needed. Targeting bacterial immune evasion molecules is one such approach. Efforts are ongoing to develop vaccines or immunotherapies that target these molecules for a number of human pathogens, but more work is needed in this area.
Table 1.
Selected bacteria and associated immune evasion molecules
| Process blocked or targeted | Bacteria (selected) | Selected molecules involved |
|---|---|---|
| Phagocytosis | Streptococcus pyogenes | M protein, Mac |
| Klebsiella pneumoniae | capsular polysaccharide | |
| Yersinia | YopE, YopH | |
| Recruitment | Staphylococcus aureus | CHIPS, FLIPr, SSL3, SElX |
| S. pyogenes | ScpA, ScpC, Sse, ZmpC | |
| Bordetella pertussis | BopN | |
| Shigella flexneri | OspF | |
| Antimicrobial peptide actions | S. aureus | aureolysin |
| S. pyogenes | SpeB | |
| Antibody function | S. aureus | SpA, Sbi |
| S. pyogenes | SpeB, IdeS | |
| NADPH oxidase activation and/or assembly | Francisella tularensis | FevR |
| Yersinia spp. | YopE, YopH | |
| Pseudomonas aeruginosa | ExoS | |
| Apoptosis/cell lysis | Anaplasma phagocytophilum | Ats-1 |
| F. tularensis | lipoproteins | |
| P. aeruginosa | pyocyanin | |
| S. pyogenes | streptolysin O/streptolysin S | |
| S. aureus | LukGH, PVL | |
| Lysosome or granule-phagosome | S. pyogenes | M protein |
| fusion | Yersinia pseudotuberculosis | YopE, YopH |
Fig. 1.
Bacteria produce molecules that target (inhibit) key neutrophil processes. The bacterial molecules shown are examples of those that inhibit or alter the indicated neutrophil function. See text for details.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgements
We thank Ryan Kissinger (NIAID) for assistance with Figure 1. The authors are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Takahashi T, Gallo RL. The critical and multifunctional roles of antimicrobial peptides in dermatology. Dermatol Clin. 2017;35:39–50. doi: 10.1016/j.det.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Braff MH, Gallo RL. Antimicrobial peptides: an essential component of the skin defensive barrier. Curr Top Microbiol Immunol. 2006;306:91–110. doi: 10.1007/3-540-29916-5_4. [DOI] [PubMed] [Google Scholar]
- 3.Doni A, Garlanda C, Mantovani A. Innate immunity, hemostasis and matrix remodeling: PTX3 as a link. Semin Immunol. 2016;28:570–577. doi: 10.1016/j.smim.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeley S, Kemper C, Le Friec G. The “ins and outs” of complement-driven immune responses. Immunol Rev. 2016;274:16–32. doi: 10.1111/imr.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Densen PR, Complement deficiencies . Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. In: Bennett JE, Dolin R, Blaser MJ, editors. Vol. 1. Philadelphia: Elsevier Saunders; 2015. pp. 93–115. [Google Scholar]
- 6.Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 7.McPhail LC, Clayton CC, Snyderman R. The NADPH oxidase of human polymorphonuclear leukocytes. Evidence for regulation by multiple signals. J Biol Chem. 1984;259:5768–5775. [PubMed] [Google Scholar]
- 8.Cleary PP, Prahbu U, Dale JB, Wexler DE, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60:5219–5223. doi: 10.1128/iai.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidalgo-Grass C, Mishalian I, Dan-Goor M, Belotserkovsky I, Eran Y, Nizet V, Peled A, Hanski E. A streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 2006;25:4628–4637. doi: 10.1038/sj.emboj.7601327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Yago T, Zhang N, Panicker SR, Wang Y, Yao L, Mehta-D'souza P, Xia L, Zhu C, McEver RP. L-selectin mechanochemistry restricts neutrophil priming in vivo. Nat Commun. 2017;8:15196. doi: 10.1038/ncomms15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng W, Minor D, Liu M, Lei B. Requirement and synergistic contribution of platelet-activating factor acetylhydrolase Sse and streptolysin S to inhibition of neutrophil recruitment and systemic infection by hypervirulent emm3 group A streptococcus in subcutaneous infection of mice. Infect Immun. 2017;85:e00530. doi: 10.1128/IAI.00530-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surewaard BG, Trzcinski K, Jacobino SR, Hansen IS, Vughs MM, Sanders EA, van der Ende A, van Strijp JA, de Haas CJ. Pneumococcal immune evasion: ZmpC inhibits neutrophil influx. Cell Microbiol. 2013;15:1753–1765. doi: 10.1111/cmi.12147. [DOI] [PubMed] [Google Scholar]
- 14.Haas PJ, de Haas CJ, Kleibeuker W, Poppelier MJ, van Kessel KP, Kruijtzer JA, Liskamp RM, van Strijp JA. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. J Immunol. 2004;173:5704–5711. doi: 10.4049/jimmunol.173.9.5704. [DOI] [PubMed] [Google Scholar]
- 15.Prat C, Bestebroer J, de Haas CJ, van Strijp JA, van Kessel KP. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol. 2006;177:8017–8026. doi: 10.4049/jimmunol.177.11.8017. [DOI] [PubMed] [Google Scholar]
- 16.Prat C, Haas PJ, Bestebroer J, de Haas CJ, van Strijp JA, van Kessel KP. A homolog of formyl peptide receptor-like 1 (FPRL1) inhibitor from Staphylococcus aureus (FPRL1 inhibitory protein) that inhibits FPRL1 and FPR. J Immunol. 2009;183:6569–6578. doi: 10.4049/jimmunol.0801523. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy AJ, Lindsay JA. Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect Genet Evol. 2013;19:7–14. doi: 10.1016/j.meegid.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Koymans KJ, Bisschop A, Vughs MM, van Kessel KP, de Haas CJ, van Strijp JA. Staphylococcal superantigen-like protein 1 and 5 (SSL1 and SSL5) limit neutrophil chemotaxis and migration through MMP inhibition. Int J Mol Sci. 2016;17:E1072. doi: 10.3390/ijms17071072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagamatsu K, Kuwae A, Konaka T, Nagai S, Yoshida S, Eguchi M, Watanabe M, Mimuro H, Koyasu S, Abe A. Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J Exp Med. 2009;206:3073–3088. doi: 10.1084/jem.20090494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 22.Boal F, Puhar A, Xuereb JM, Kunduzova O, Sansonetti PJ, Payrastre B, Tronchere H. PI5P triggers ICAM-1 degradation in Shigella infected cells, thus dampening immune cell recruitment. Cell Rep. 2016;14:750–759. doi: 10.1016/j.celrep.2015.12.079. [DOI] [PubMed] [Google Scholar]
- 23.Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MA. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovingh ES, van den Broek B, Jongerius I. Hijacking complement regulatory proteins for bacterial immune evasion. Front Microbiol. 2016;7:2004. doi: 10.3389/fmicb.2016.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 26.Lei B, DeLeo FR, Hoe NP, Graham MR, Mackie SM, Cole RL, Liu M, Hill HR, Low DE, Federle MJ, Scott JR, Musser JM. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat Med. 2001;7:1298–1305. doi: 10.1038/nm1201-1298. [DOI] [PubMed] [Google Scholar]
- 27.von Pawel-Rammingen U, Johansson BP, Bjorck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002;21:1607–1615. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okumura CY, Anderson EL, Dohrmann S, Tran DN, Olson J, von Pawel-Rammingen U, Nizet V. IgG protease Mac/IdeS is not essential for phagocyte resistance or mouse virulence of M1T1 group A streptococcus. MBio. 2013;4:e00499. doi: 10.1128/mBio.00499-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woof JM. Immunoglobulins and their receptors, and subversion of their protective roles by bacterial pathogens. Biochem Soc Trans. 2016;44:1651–1658. doi: 10.1042/BST20160246. [DOI] [PubMed] [Google Scholar]
- 30.Rolan HG, Durand EA, Mecsas J. Identifying Yersinia YopH-targeted signal transduction pathways that impair neutrophil responses during in vivo murine infection. Cell Host Microbe. 2013;14:306–317. doi: 10.1016/j.chom.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taheri N, Fahlgren A, Fallman M. Yersinia pseudotuberculosis blocks neutrophil degranulation. Infect Immun. 2016;84:3369–3378. doi: 10.1128/IAI.00760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlyon JA, Abdel-Latif D, Pypaert M, Lacy P, Fikrig E. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect Immun. 2004;72:4772–4783. doi: 10.1128/IAI.72.8.4772-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaffrey RL, Schwartz JT, Lindemann SR, Moreland JG, Buchan BW, Jones BD, Allen LA. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J Leukoc Biol. 2010;88:791–805. doi: 10.1189/jlb.1209811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnov A, Daily KP, Criss AK. Assembly of NADPH oxidase in human neutrophils is modulated by the opacity-associated protein expression state of Neisseria gonorrhoeae. Infect Immun. 2014;82:1036–1044. doi: 10.1128/IAI.00881-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen LA, Beecher BR, Lynch JT, Rohner OV, Wittine LM. Helicobacter pylori disrupts NADPH oxidase targeting in human neutrophils to induce extracellular superoxide release. J Immunol. 2005;174:3658–3667. doi: 10.4049/jimmunol.174.6.3658. [DOI] [PubMed] [Google Scholar]
- 36.Songsungthong W, Higgins MC, Rolan HG, Murphy JL, Mecsas J. ROS-inhibitory activity of YopE is required for full virulence of Yersinia in mice. Cell Microbiol. 2010;12:988–1001. doi: 10.1111/j.1462-5822.2010.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vareechon C, Zmina SE, Karmakar M, Pearlman E, Rietsch A. Pseudomonas aeruginosa effector ExoS inhibits ROS production in human neutrophils. Cell Host Microbe. 2017;21:611–618 e615. doi: 10.1016/j.chom.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajaj B, Yesilkaya H, Benisty R, David M, Andrew PW, Porat N. Thiol peroxidase is an important component of Streptococcus pneumoniae in oxygenated environments. Infect Immun. 2012;80:4333–4343. doi: 10.1128/IAI.00126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun. 2004;72:408–413. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belaaouaj A, Kim KS, Shapiro SD. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science. 2000;289:1185–1188. doi: 10.1126/science.289.5482.1185. [DOI] [PubMed] [Google Scholar]
- 41.Shafer WM, Qu X, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer ME, Shafer WM. On the in vivo significance of bacterial resistance to antimicrobial peptides. Biochim Biophys Acta. 2015;1848:3101–3111. doi: 10.1016/j.bbamem.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stapels DA, Geisbrecht BV, Rooijakkers SH. Neutrophil serine proteases in antibacterial defense. Curr Opin Microbiol. 2015;23:42–48. doi: 10.1016/j.mib.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 45.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013;122:2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 46.Storisteanu DM, Pocock JM, Cowburn AS, Juss JK, Nadesalingam A, Nizet V, Chilvers ER. Evasion of neutrophil extracellular traps by respiratory pathogens. Am J Respir Cell Mol Biol. 2017;56:423–431. doi: 10.1165/rcmb.2016-0193PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi SD, DeLeo FR. Role of neutrophils in innate immunity: a systems biology-level approach. Wiley Interdiscip Rev Syst Biol Med. 2009;1:309–333. doi: 10.1002/wsbm.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savill JS, Wyllie AH, Henson JE, Walport MJ, Henson PM, Haslett C. Macrophage phagocytosis of aging neutrophils in inflammation. Programmed cell death in the neutrophil leads to its recognition by macrophages. J Clin Invest. 1989;83:865–875. doi: 10.1172/JCI113970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maianski NA, Roos D, Kuijpers TW. Bid truncation, bid/bax targeting to the mitochondria, and caspase activation associated with neutrophil apoptosis are inhibited by granulocyte colony-stimulating factor. J Immunol. 2004;172:7024–7030. doi: 10.4049/jimmunol.172.11.7024. [DOI] [PubMed] [Google Scholar]
- 50.Watson RW, Redmond HP, Wang JH, Condron C, Bouchier-Hayes D. Neutrophils undergo apoptosis following ingestion of Escherichia coli. J Immunol. 1996;156:3986–3992. [PubMed] [Google Scholar]
- 51.Zhang B, Hirahashi J, Cullere X, Mayadas TN. Elucidation of molecular events leading to neutrophil apoptosis following phagocytosis: cross-talk between caspase 8, reactive oxygen species, and MAPK/ERK activation. J Biol Chem. 2003;278:28443–28454. doi: 10.1074/jbc.M210727200. [DOI] [PubMed] [Google Scholar]
- 52.Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–595. doi: 10.1128/jcm.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Zandbergen G, Gieffers J, Kothe H, Rupp J, Bollinger A, Aga E, Klinger M, Brade H, Dalhoff K, Maass M, Solbach W, Laskay T. Chlamydia pneumoniae multiply in neutrophil granulocytes and delay their spontaneous apoptosis. J Immunol. 2004;172:1768–1776. doi: 10.4049/jimmunol.172.3.1768. [DOI] [PubMed] [Google Scholar]
- 54.Choi KS, Park JT, Dumler JS. Anaplasma phagocytophilum delay of neutrophil apoptosis through the p38 mitogen-activated protein kinase signal pathway. Infect Immun. 2005;73:8209–8218. doi: 10.1128/IAI.73.12.8209-8218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkar A, Hellberg L, Bhattacharyya A, Behnen M, Wang K, Lord JM, Moller S, Kohler M, Solbach W, Laskay T. Infection with Anaplasma phagocytophilum activates the phosphatidylinositol 3-kinase/Akt and NF-κB survival pathways in neutrophil granulocytes. Infect Immun. 2012;80:1615–1623. doi: 10.1128/IAI.05219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz JT, Barker JH, Kaufman J, Fayram DC, McCracken JM, Allen LA. Francisella tularensis inhibits the intrinsic and extrinsic pathways to delay constitutive apoptosis and prolong human neutrophil lifespan. J Immunol. 2012;188:3351–3363. doi: 10.4049/jimmunol.1102863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCracken JM, Kinkead LC, McCaffrey RL, Allen LA. Francisella tularensis modulates a distinct subset of regulatory factors and sustains mitochondrial integrity to impair human neutrophil apoptosis. J Innate Immun. 2016;8:299–313. doi: 10.1159/000443882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinkead LC, Whitmore LC, McCracken JM, Fletcher JR, Ketelsen BB, Kaufman JW, Jones BD, Weiss DS, Barker JH, Allen LH. Bacterial lipoproteins and other factors released by Francisella tularensis modulate human neutrophil lifespan: effects of a TLR1 SNP on apoptosis inhibition. Cell Microbiol. 2018 doi: 10.1111/cmi.12795. doi: 10.1111/cmi.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prince LR, Bianchi SM, Vaughan KM, Bewley MA, Marriott HM, Walmsley SR, Taylor GW, Buttle DJ, Sabroe I, Dockrell DH, Whyte MK. Subversion of a lysosomal pathway regulating neutrophil apoptosis by a major bacterial toxin, pyocyanin. J Immunol. 2008;180:3502–3511. doi: 10.4049/jimmunol.180.5.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenlee-Wacker MC, Rigby KM, Kobayashi SD, Porter AR, DeLeo FR, Nauseef WM. Phagocytosis of Staphylococcus aureus by human neutrophils prevents macrophage efferocytosis and induces programmed necrosis. J Immunol. 2014;192:4709–4717. doi: 10.4049/jimmunol.1302692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greenlee-Wacker MC, Kremserova S, Nauseef WM. Lysis of human neutrophils by community-associated methicillin-resistant Staphylococcus aureus. Blood. 2017;129:3237–3244. doi: 10.1182/blood-2017-02-766253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of group A streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, Shabb DW, Diep BA, Chambers HF, Otto M, DeLeo FR. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One. 2011;6:e18617. doi: 10.1371/journal.pone.0018617. [DOI] [PMC free article] [PubMed] [Google Scholar]