Abstract
The accumulation of DNA in the cytosol serves as a key immunostimulatory signal associated with infections, cancer and genomic damage1,2. Cytosolic DNA triggers immune responses by activating the cGAS/STING pathway3. The binding of DNA to the cytosolic enzyme cGAMP synthase (cGAS), activates its enzymatic activity, leading to the synthesis of a second messenger, cyclic[G(2’,5’)pA(3’,5’)] (2’3’-cGAMP)4-7. 2’3’-cGAMP, a cyclic dinucleotide (CDN), activates the protein ‘stimulator of interferon genes’ (STING)8, which in turn activates the transcription factors IRF3 and NF-κB promoting the transcription of genes encoding type I interferons and other cytokines and mediators that stimulate a broader immune response. Exogenous 2’3’-cGAMP produced by malignant cells9 and other CDNs, including CDNs produced by bacteria10-12 and synthetic CDNs used in cancer immunotherapy13,14, must traverse the cell membrane to activate STING in target cells. How these charged CDNs pass through the lipid bilayer is unknown. Here we used a genome-wide CRISPR interference screen to identify the reduced folate carrier SLC19A1, a folate-organic phosphate antiporter, as the major transporter for CDNs. CDN uptake and functional responses are inhibited by depleting SLC19A1 from human cells and enhanced by overexpressing SLC19A1. In both human cell lines and primary cells ex vivo, CDN uptake is inhibited by folates, as well as two medications approved for treatment of inflammatory diseases, sulfasalazine and the antifolate methotrexate. The identification of SLC19A1 as the major transporter of CDNs into cells has implications for the immunotherapeutic treatment of cancer13, host responsiveness to CDN-producing pathogenic microorganisms11, and potentially in certain inflammatory diseases.
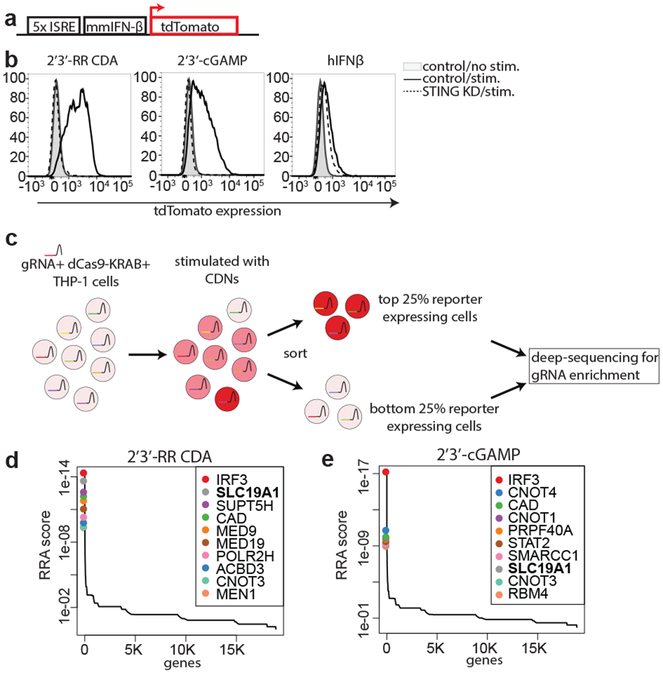
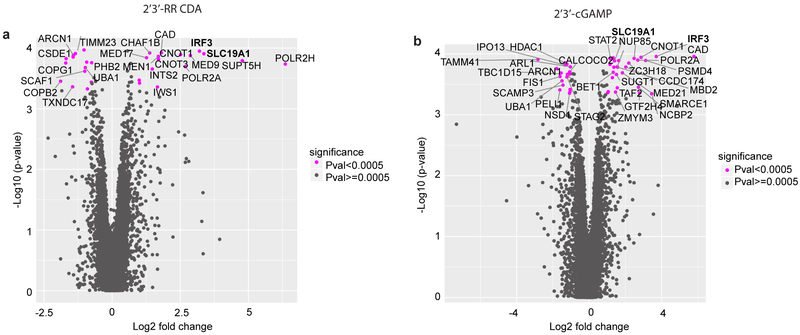
To systematically identify genes involved in intracellular transport of CDNs, we performed a genome-wide CRISPR interference (CRISPRi) forward genetic screen in the monocytic THP-1 cell line. To visualize STING activation, THP-1 cells were transduced with a CDN-inducible reporter construct (Fig. 1a, b). In line with previous results, the synthetic CDN 2’3’ RR-S2 cyclic di-AMP (2’3’-RR CDA, Extended Data Fig. 1a) induced a more potent response than 2’3’-cGAMP13 and the response to both CDNs was several fold higher than the response to human interferon-β (hIFN-β). The response to CDNs was completely dependent on STING expression (Fig. 1b), implying that the reporter primarily reported cell-intrinsic STING activity. For the screen, THP-1 cells expressing dCas9-BFP-KRAB and transduced with a genome-wide gRNA library were stimulated separately with 2’3’-RR CDA or 2’3’-cGAMP and the highest and lowest quartiles of reporter-expressing cells were sorted by flow cytometry, before deep sequencing to identify gRNAs enriched in each population (Fig. 1c and 1d, Extended Data Fig. 1b and 2, Tables S1 and S2). The two screens yielded many common hits, but there were differences, such as numerous hits in the 2’3’-cGAMP screen including STAT2, IRF9, IFNAR1, and IFNAR2 (Table S2). Hence, the 2’3’-RR CDA screen may have been mostly dependent on intrinsic STING signaling, whereas the 2’3’-cGAMP screen may have been partly dependent on autocrine/paracrine IFN-β signaling.
Figure 1.
Genome-wide CRISPRi screen for host factors necessary for cyclic dinucleotide (CDN) stimulation. a, schematic overview of tdTomato-reporter. tdTomato expression is driven by interferon-stimulatory response elements (ISRE) followed by a mouse minimal interferon beta (mmIFN-β) promoter. b, Control THP-1 cells and STING-depleted THP-1 cells were incubated with 2’3’-RR CDA (1.67 μg/ml), 2’3’-cGAMP (10 μg/ml) or hIFN-β (100 ng/ml). After 20h, tdTomato reporter expression was analyzed by flow cytometry. Data are representative of three independent experiments with similar results. c, Schematic overview of the genome-wide CRISPRi screen. A genome-wide library of CRISPRi guide RNA (gRNA)-expressing THP-1 cells was stimulated with CDNs. 20h after stimulation, cells were sorted into a tdTomato-low group (lowest 25% of cells) and a tdTomato-high group (highest 25% of cells). DNA from the sorted cells was deep sequenced to reveal gRNA enrichment in the two groups. d-e, Distribution of the robust rank aggregation (RRA) score in the comparison of hits enriched in the reporter-low versus reporter-high groups of THP-1 cells stimulated with (d) 2’3’-RR CDA or (e) 2’3’-cGAMP. Each panel represents combined results of two independent screens.
In both screens, the top hits in the hypo-responsive population (i.e. the genes most important for robust responses to CDNs) included the transcription factor IRF3, which acts directly downstream of STING. A gRNA for STING was also enriched in hyporesponsive cells from both screens, though other STING gRNAs were not, presumably because they were ineffective at interfering with STING expression (Table S1 and S2).
SLC19A1 was one of the most significant hits in both screens. SLC19A1 is a folate-organic phosphate antiporter that transports folates, structurally similar antifolates and a variety of organic phosphates encompassing, among others, thiamine derivatives and nucleotides15,16. Folate import is coupled to organic phosphate export and extensive inhibition and exchange phenomena have been demonstrated17-19.
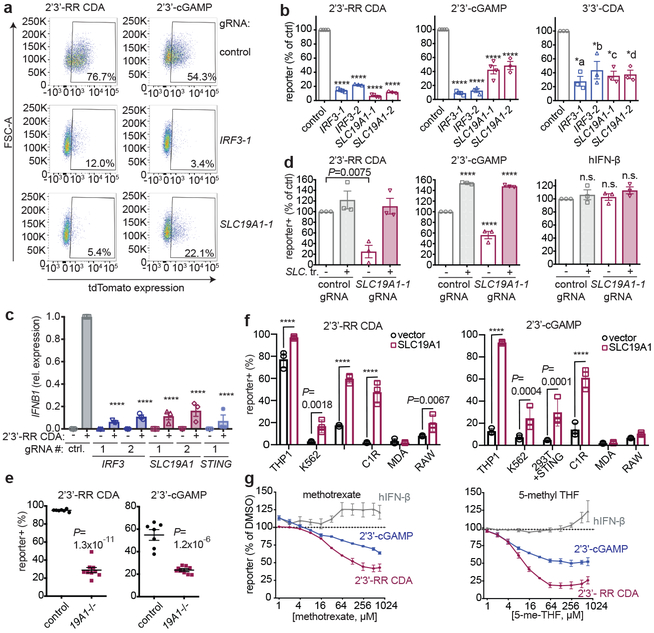
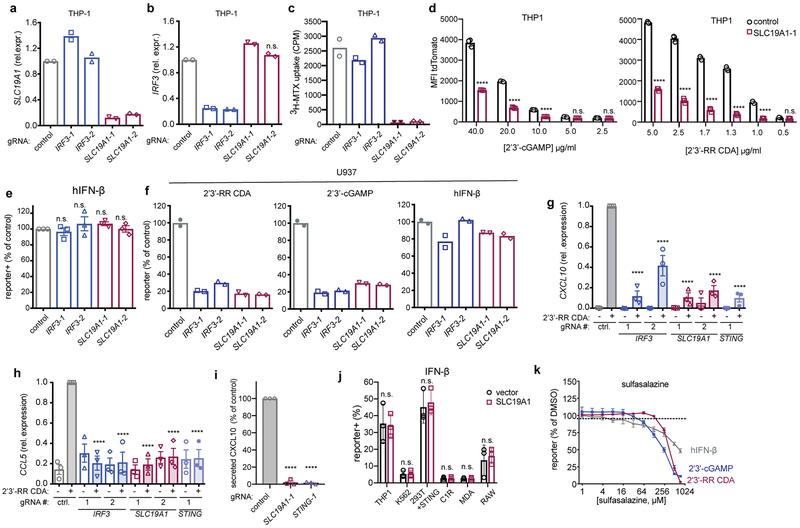
To validate the role of SLC19A1 in CDN stimulation, the top two enriched SLC19A1-targeting gRNAs from the 2’3’-RR CDA screen were used to stably deplete SLC19A1 in THP-1 cells expressing dCas9-KRAB (Extended Data Fig. 3a). SLC19A1-depleted cells grew normally but uptake of the SLC19A1 substrate methotrexate was nearly abolished20 (Extended Data Fig. 3c). SLC19A1-depleted THP-1 or U937 cells, like IRF3-depleted cells (Extended Data Fig. 3b), were substantially defective in reporter responses induced by 2’3’-cGAMP, 2’3’-RR CDA, and 3’3’-CDA (a bacterial CDN that stimulates STING), but responded normally to hIFN-β stimulation (Figs. 2a, 2b, Extended Data Figs. 3d-f). SLC19A1-depletion, like depletion of IRF3 and STING, also inhibited CDN-induced expression of direct downstream target genes in the STING pathway, including IFNB1 and the chemokines CCL5 and CXCL1021,22 (Fig. 2c, Extended Data Fig. 3g-i). Transduction of SLC19A1 in depleted THP-1 cells rescued CDN responsiveness (Fig. 2d). SLC19A1 disruption using the conventional CRISPR/Cas9 system similarly decreased responsiveness to CDNs in THP-1 cells (Fig. 2e).
Figure 2.
SLC19A1 is required for CDN-induced reporter expression. a, dCas9-KRAB-expressing THP-1 cells transduced with non-targeting gRNA (control), IRF3-1 gRNA or SLC19A1-1 gRNA were exposed to 2’3’-RR CDA (1.67 μg/ml) or 2’3’-cGAMP (15 μg/ml). 20h later, tdTomato expression was analyzed by flow cytometry. Representative dot plots of three independent experiments are shown. b, THP-1 cells expressing the indicated CRISPRi gRNAs or non-targeting gRNA (control), were stimulated with 2’3’-RR CDA (1.67 μg/ml), 2’3’-cGAMP (10 μg/ml), or 3’3’-CDA (20 μg/ml). After 18-22h, tdTomato expression was quantified as in (a). c, Induction of IFNB mRNA in control (non-targeting gRNA) THP-1 cells or THP-1 cells expressing the indicated CRISPRi gRNAs after 5h stimulation with 5 μg/ml 2’3’-RR CDA. d, Control THP-1 cells and SLC19A1–1 gRNA expressing THP-1 cells transduced with SLC19A1 (SLC. tr.) were stimulated with 2’3’-RR CDA (1.67 μg/ml), 2’3’-cGAMP (15 μg/ml), or hIFN-β (100 ng/ml) and analyzed as in (a). e, Control THP-1 cells (n=7 clonal lines) and SLC19A1−/− cells (19A1−/−; n=9 clonal lines) were exposed to 2’3’-RR CDA (2.22 μg/ml), 2’3’-cGAMP (10 μg/ml), and tdTomato reporter expression was analyzed as in (a). Mean ± SEM are shown. f, Various cell lines expressing a control vector or an SLC19A1 expression vector were stimulated and analyzed as in (b). g, THP-1 cells were incubated with increasing concentrations of the competitive inhibitors methotrexate, 5-methyl tetrahydrofolate (5-me-THF) or DMSO as vehicle control, before stimulating with 2’3’-RR CDA (1.25 μg/ml), 2’3’-cGAMP (15 μg/ml) or hIFN-β (100 ng/ml). Cells were analyzed as in (a). For each stimulant, the data were normalized to the DMSO controls. In panels b-d and f-g, mean ± SEM of n=3 biological replicates are shown. Statistical analyses were performed using one-way ANOVA followed by Dunnett’s post-test for the comparison to stimulated control cells (b-d), unpaired two-tailed Student’s t tests for (e), or two-way ANOVA followed by uncorrected Fisher’s LSD tests (f). *a P = 0.0002; *b P = 0.0013; *c P = 0.0005; *d P =0.0006; ****P ≤ 0.0001; n.s. not significant.
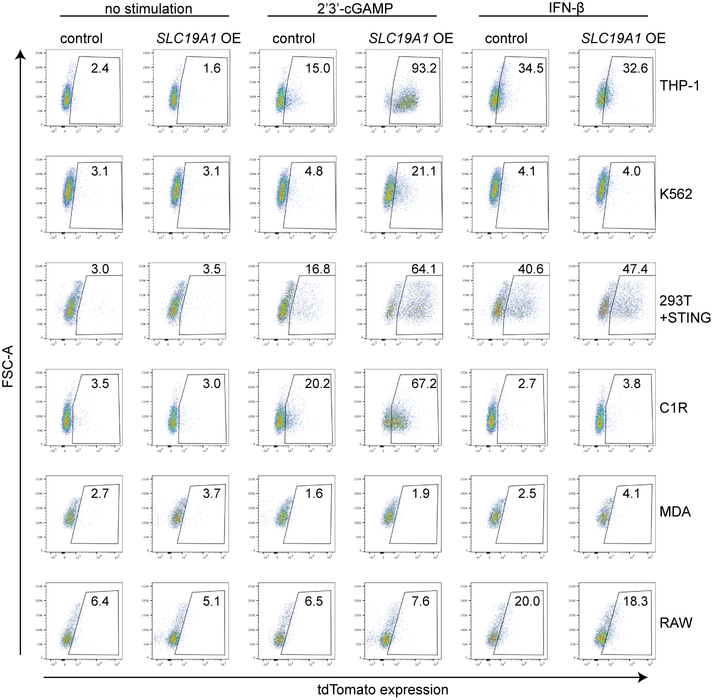
SLC19A1 overexpression robustly increased CDN responsiveness in WT THP-1 cells and in cell lines that normally responded poorly or not at all to CDN stimulation, including C1R, K562 and 293T (pre-transduced with STING) (Fig. 2f and Extended Data Fig. 3j and 4). Together, our data show reduced CDN responses in SLC19A1-deficient cells and amplified responses in SLC19A1 overexpressing cells. Inhibitor experiments showed that the known SLC19A1 substrates methotrexate and 5-methyl-tetrahydrofolic acid (5-me-THF)15 blocked stimulation of THP-1 cells by 2’3’-cGAMP or 2’3’-RR CDA, at concentrations only modestly higher than those that inhibit uptake of folate derivatives23, but did not inhibit reporter responses to hIFN-β (Fig. 2g). At high concentrations, sulfasalazine, a non-competitive SLC19A1 inhibitor23, blocked responses to CDNs and to hIFN-β stimulation, suggesting a broader effect on reporter activation (Extended Data Fig. 3k).
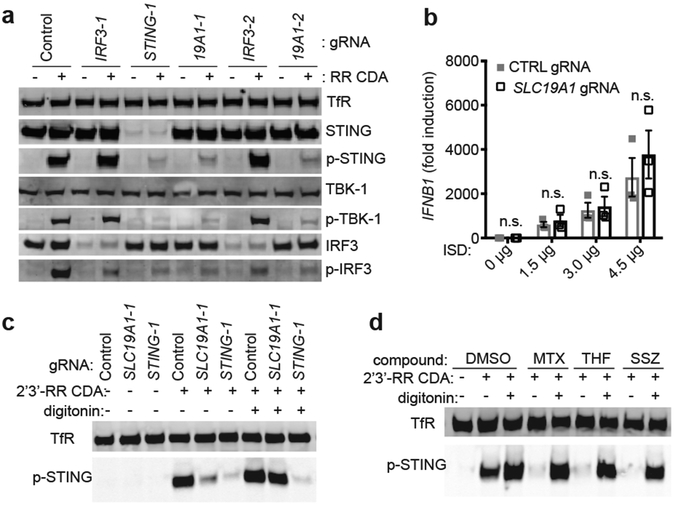
To directly assess the effect of SLC19A1 on STING pathway activation (Extended Data Fig. 5a), we employed immunoblotting to evaluate phosphorylation of STING, IRF3 and TBK1 induced by a 2 hr exposure to 2’3’-RR CDA in control (non-targeting gRNA) versus CRISPRi-depleted cells (Fig. 3a). As expected, STING-depletion inhibited phosphorylation of both TBK1 and IRF3, whereas IRF3-depletion did not inhibit phosphorylation of TBK1 or STING. SLC19A1-depletion, in contrast, resulted in major defects in phosphorylation of STING, TBK1 and IRF3, indicating that SLC19A1 acts upstream of STING.
Figure 3.
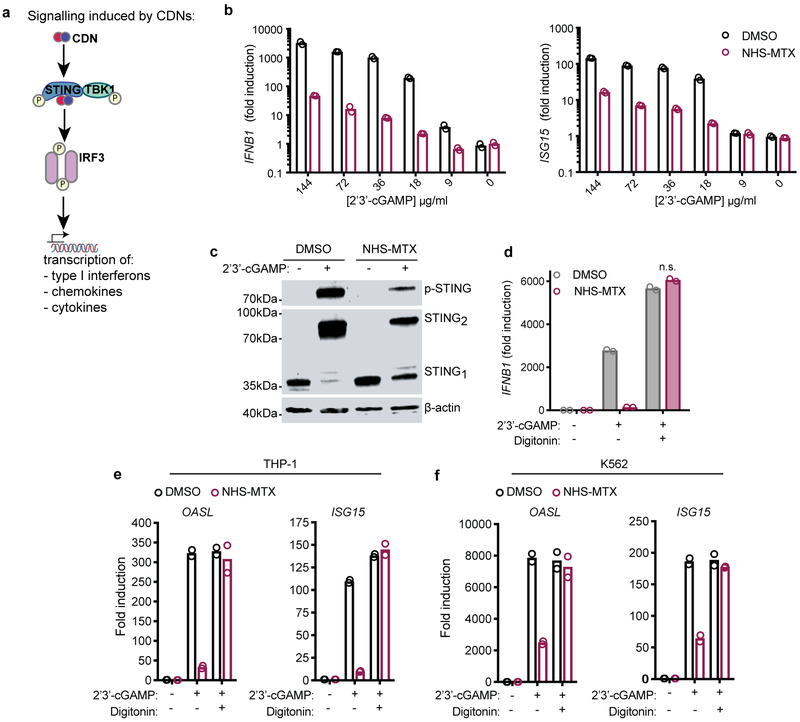
SLC19A1 is critical for STING-dependent responses to exogenous CDNs but not when CDNs are provided intracellularly. a, Immunoblot analysis of (phospho-) protein expression in control THP-1 cells or THP-1 cells expressing the indicated CRISPRi gRNAs. Cells were stimulated for 2h with 10 μg/ml 2’3’-RR CDA (RR CDA) or left unstimulated. TransferrinR.: Transferrin receptor; p-TBK1: TKB1 phosphorylated at position Ser172; p-IRF3: IRF3 phosphorylated at position Ser296; p-STING: STING phosphorylated at position Ser366. b, Control THP-1 cells or SLC19A1-depleted THP-1 cells were transfected with increasing amounts of interferon-stimulatory DNA (ISD) for 3h and the induction of IFNB1 mRNA was measured by RT-qPCR. c, cells were stimulated as in (a) in the absence or presence of digitonin (5 μg/ml). d, control THP-1 cells were stimulated as in (c) with the addition of the indicated SLC19A1 inhibitors (all at 750 μM): methotrexate, 5-methyl tetrahydrofolate (5-me-THF), or sulfasalazine or DMSO as vehicle control. Panels a, c, and d are representative of n=3 biological replicates; for gel source data, see Supplementary Figure 1. In panel b, data are means ± SEM of n=3 biological replicates and statistical analyses were performed using a two-way ANOVA followed by Sidak’s post-test (n.s. not significant).
Protein levels of STING, TBK1, and IRF3 were unaltered in SLC19A1-depleted cells. Notably, SLC19A1-depleted cells responded normally when transfected with DNA (Fig. 3b). Furthermore, when transport was bypassed by permeabilizing SLC19A1-depleted cells with digitonin, STING was phosphorylated completely normally in response to CDNs (Fig. 3c). Similarly, the inhibitory effects on CDN-induced STING phosphorylation and gene expression of SLC19A1 blockers, including methotrexate, 5-me-THF, and the covalent SLC19A1 inhibitor N-hydroxysuccinimide (NHS)-methotrexate (see Methods), were bypassed when the cells were permeabilized with digitonin (Fig. 3d, Extended Data Fig. 5b-e). Thus, STING functioned normally in SLC19A1-depleted or inhibited cells when CDNs or DNA were introduced directly into the cytosol, consistent with a role for SLC19A1 in CDN transport.
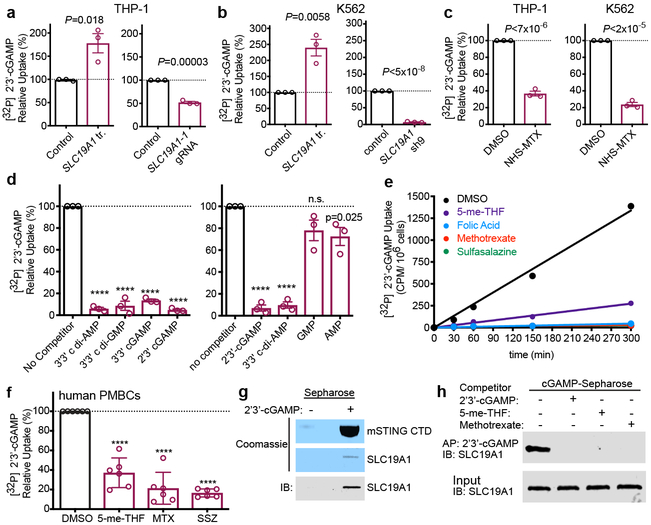
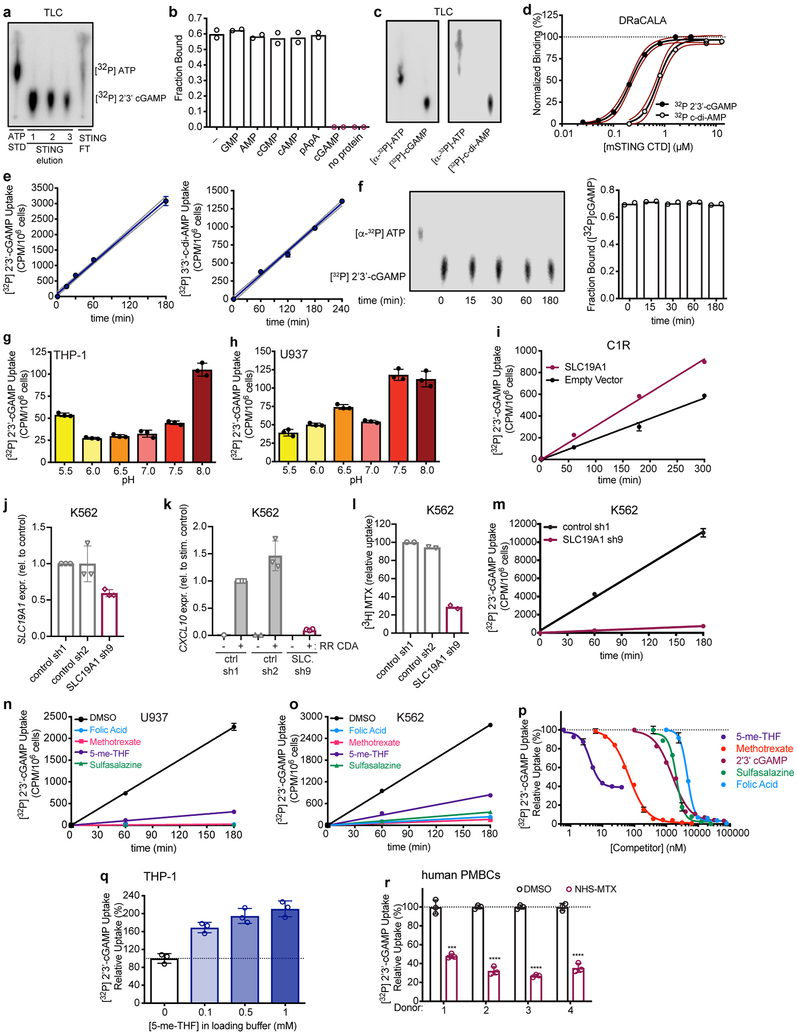
To directly test the impact of SLC19A1 on transport, we monitored cellular uptake of [32P]-labeled CDNs (Extended Data Fig. 6a-h). SLC19A1 overexpression resulted in a two to three-fold enhancement of [32P] 2’3’-cGAMP uptake by THP-1, K562 and C1R cells (Fig. 4a and b, Extended Data Fig. 6i). Conversely, SLC19A1-depletion reduced the uptake of [32P] 2’3’-cGAMP in THP-1 cells and K562 cells (Fig. 4a and b, Extended Data Fig. 6j-m). Furthermore, we observed much reduced 2’3’-cGAMP influx in THP-1 and K562 cells treated with the irreversible, covalent SLC19A1 inhibitor NHS-methotrexate (Fig. 4c). 2’3’-cGAMP uptake by THP-1 cells was also inhibited by excess unlabeled 2’3’-cGAMP as well as by the bacterial CDNs 3’3’-cGAMP, 3’3’-CDA and 3’3’ c-di-GMP. Thus, CDN interactions with the transporter are not highly specific for the 2’3’ linkage or the specific nucleotides (Fig. 4d). The nucleoside monophosphates AMP and GMP, the major ENPP-1 hydrolysis products of 2’3’-cGAMP, slightly inhibited [32P] 2’3’-cGAMP uptake (Fig. 4d)24 consistent with previous observations that AMP, other nucleotides, and organic phosphates in general, inhibit SLC19A1-mediated transport 17,18. Our findings indicated that SLC19A1 broadly interacts with CDNs irrespective of the phosphodiester linkages or the base content but that CDN breakdown products have a limited effect on CDN uptake.
Figure 4.
SLC19A1 transports CDNs. a, Normalized [32P] 2’3’-cGAMP uptake after one hour by THP-1 monocytes transduced with empty vector (control) or SLC19A1 expression vector (left panel), or transduced with a non-targeting control CRISPRi gRNA or SLC19A1 CRISPRi gRNA (right panel). b, Normalized [32P] 2’3’-cGAMP uptake after one hour by K562 cells transduced with empty vector (control) or SLC19A1 expression vector (left panel), or transduced with a non-targeting control shRNA (control) or SLC19A1 shRNA (right panel). c, Normalized [32P] 2’3’-cGAMP uptake after one hour in DMSO or NHS-methotrexate (MTX) (5 μM) treated THP-1 (left panel) and K562 (right panel) cells. d, Normalized [32P] 2’3’-cGAMP uptake after one hour by THP-1 monocytes in the presence and absence of 100 μM competing, unlabeled cyclic dinucleotides (left panel) or 200 μM competing, unlabeled nucleotides (right panel). e, Time course of [32P] 2’3’-cGAMP uptake in THP-1 monocytes in the presence and absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. f, Normalized [32P] 2’3’-cGAMP uptake after three hours in human PBMCs from six healthy donors in the presence and absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. g, Coomassie staining and western blot analysis of pulldowns by 2’3’-cGAMP (+) or control (−) Sepharose of mSTING-CTD or hSLC19A1. h, Western blot analysis of hSLC19A1 affinity purification (AP) with 2’3’-cGAMP Sepharose in the absence (−) or presence (+) of free, unbound 2’3’-cGAMP, 5-me-THF, or methotrexate (250 μM). In panels a-d, data are means ± SEM of n=3 biological replicates. In panel e, data are means ± SD of n=3 technical replicates and data are representative of three independent experiments. In panel f, data are means ± SD of n=6 healthy donors conducted over two independent experiments. Panels g and h are representative of two independent experiments; for gel source data, see Supplementary Figure 1. Statistical analyses were performed using unpaired, two-tailed Student’s t-tests (a-d), or a one-way ANOVA followed by a Tukey’s post-test (f). ****P ≤ 0.0001.
Sustained inhibition of 2’3’-cGAMP uptake occurred when folic acid, 5-me-THF, methotrexate, and sulfasalazine were added to the culture medium of various cell lines (Fig. 4e, Extended Data Fig. 6n-o). In terms of IC50, sulfasalazine (IC50= 2.1 μM) and folic acid (IC50= 4.8 μM) were in the same range as 2’3’-cGAMP (IC50= 1.89 μM) (Extended Data Fig. 6p), similar to the dosage of 2’3’-cGAMP required for STING activation and interferon responses (10–20 μM) (Extended Data Fig. 3d). 5-me-THF (IC50 = 4.1 nM) and methotrexate (IC50 = 54.8 nM) were much more potent inhibitors, in line with their much higher affinity for SLC19A1 binding in comparison to folic acid (Extended Data Fig. 6p)17. Interestingly, we did not observe complete inhibition of [32P] 2’3’-cGAMP uptake by 5-me-THF, as compared to the other (anti-) folates. In line with an antiporter mechanism of uptake, preloading the cells with 5-me-THF, which trans-stimulates SLC19A1 import25, augmented 2’3’-cGAMP influx (Extended Data Figure 6q). These observations establish that CDN import is altered by known substrates and inhibitors of the SLC19A1 transporter in a wide range of human cell lines.
We next monitored 2’3’-cGAMP uptake in primary healthy adult human peripheral blood mononuclear cells. Treatment of PBMCs with NHS-methotrexate, or excess, unlabeled 5-me-THF, methotrexate, and sulfasalazine all strongly inhibited [32P] 2’3’-cGAMP uptake (Fig. 4f and Extended Data Fig. 6r). These data generalize our findings to normal human blood cells.
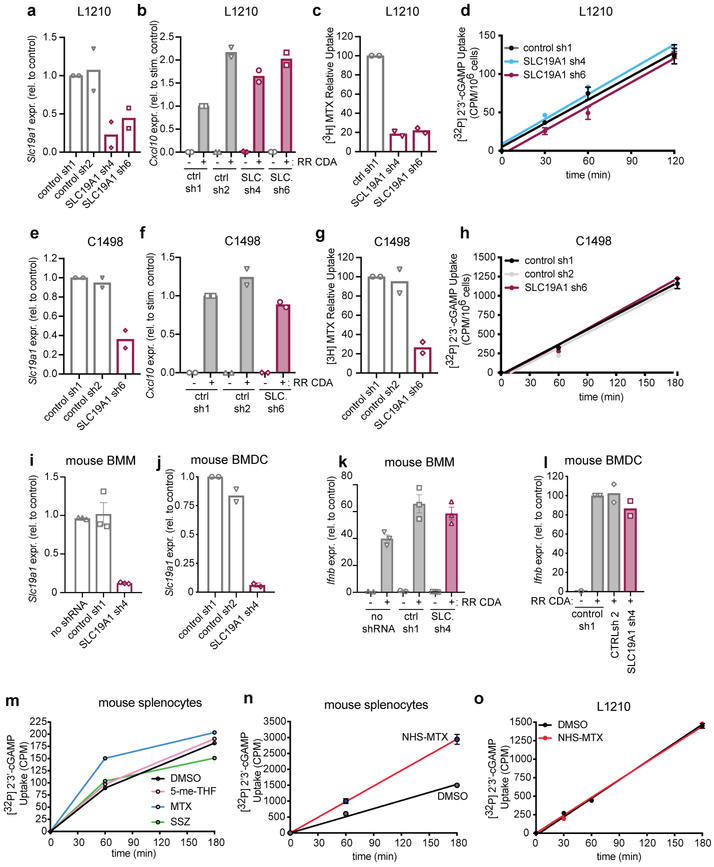
In contrast to these results in human cells, neither CDN uptake nor CDN-induced Cxcl10 expression was inhibited by depleting Slc19a1 expression in the murine C1498 or L1210 cell lines, the latter of which has been extensively studied in the context of SLC19A1-mediated transport (Extended Data Fig. 7)17,18,26. Slc19a1-depletion in mouse bone marrow-derived macrophages or dendritic cells also did not block Ifn gene expression induced by CDNs (Extended Data Fig. 7i-l). Furthermore, anti-folates that inhibit SLC19A1, including methotrexate, did not inhibit 2’3’-cGAMP uptake by murine splenocytes (Extended Data Fig. 7m-o). Collectively, these results suggested that SLC19A1 expression and function is essential for uptake of the metazoan CDN 2’3’-cGAMP by human cells, including cell lines and primary cells ex vivo, but not by the mouse cells studied. Therefore, there is likely another potent CDN transporter expressed by mouse cells.
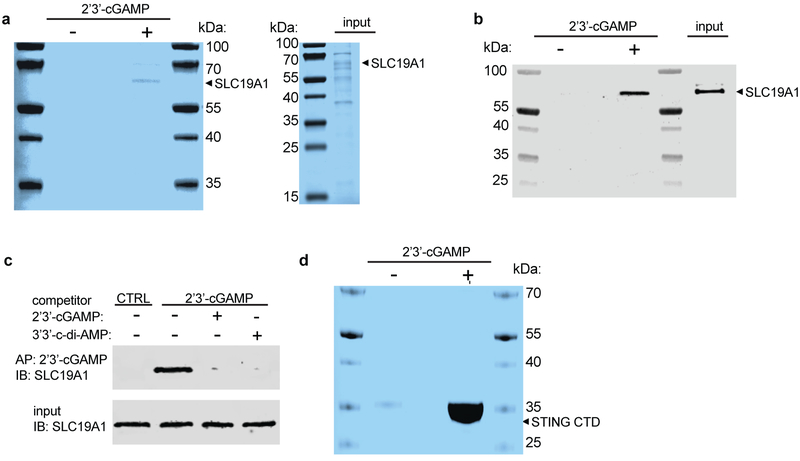
SLC19A1-mediated import of CDNs would require a direct interaction with the CDN. Consistent with this hypothesis, His-tagged SLC19A1 was precipitated by 2’3’-cGAMP immobilized on Sepharose beads (Fig. 4g Extended Data Fig. 8a-b). This interaction was competitively disrupted by free, unbound, 2’3’-cGAMP and 3’3’-CDA (Fig. 4h and Extended Data Fig. 8c) as well as by 5-me-THF and methotrexate (Fig. 4h). These data suggest that CDNs interact with SLC19A1, consistent with the proposed role of SLC19A1 as a CDN transporter. Taken together, our results demonstrate that SLC19A1 is a CDN transporter in human cells, required for exogenous CDN-mediated type I interferon activation.
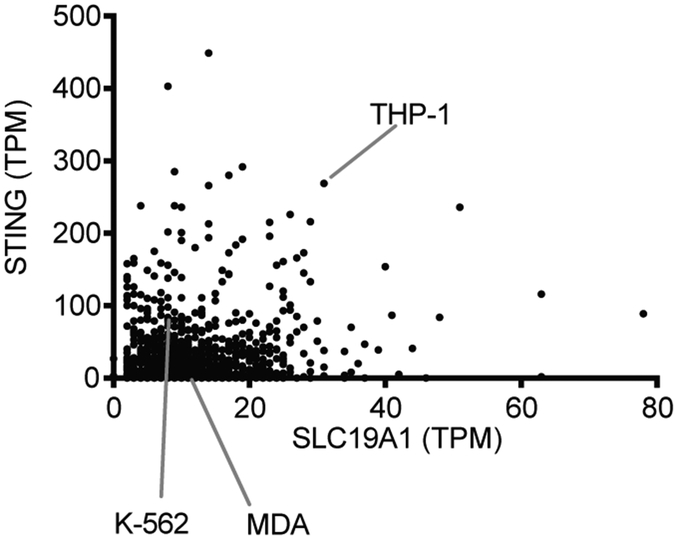
The response to CDNs was relatively weak in most cell lines we tested and was increased by overexpression of SLC19A1 or permeabilization of cells (Figs. 2f and 3c). Indeed, among a large set of cell lines, THP-1 cells are among the highest in expressing both SLC19A1 and STING (Extended Data Fig. 9). It is likely that SLC19A1 expression and STING expression are each important predictors of the responsiveness to CDN stimulation by cell lines and tumors.
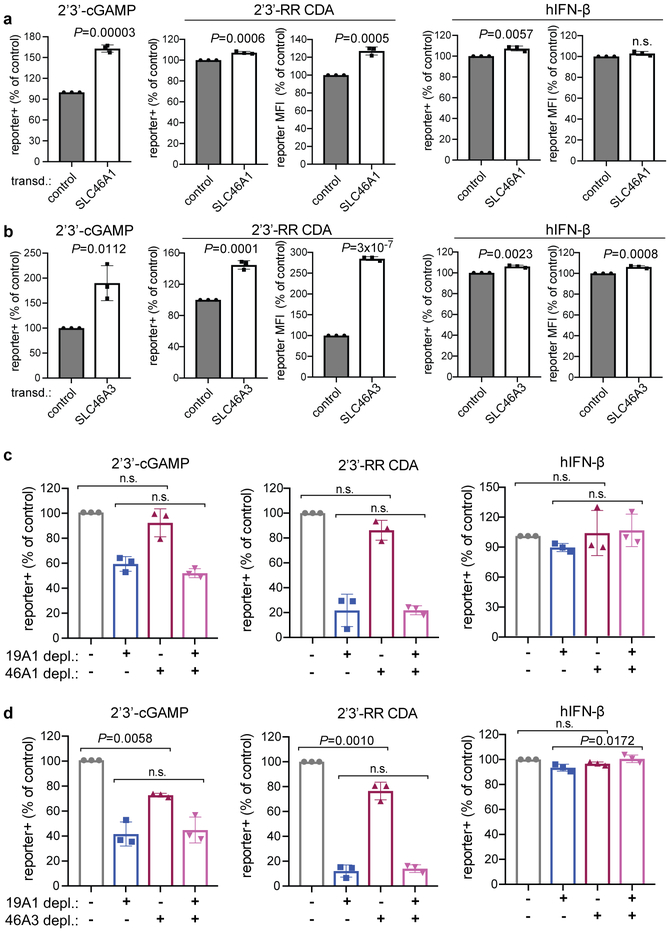
Methotrexate, folic acid and sulfasalazine almost completely blocked CDN uptake and/or stimulation, whereas CDN stimulation was not completely inhibited in SLC19A1-null cells. This implied that another transporter sensitive to these drugs may play a role in CDN uptake. Although it was not a hit in our screen, overexpression of SLC46A1, which encodes the only other known folate transporter15, did increase responses to CDNs (Extended Data Fig. 10a). However, depletion of SLC46A1 (approximately 90% reduction in mRNA) had little or no effect on CDN stimulation, even when combined with SLC19A1 depletion (Extended Data Fig. 10c). SLC46A3, another transporter, was a hit in our screen. Overexpression of SLC46A3 increased the response to CDNs (Extended Data Fig. 10b) and depletion of SLC46A3 (90% effective) had a minor effect on reporter induction by both CDNs (Extended Data Fig. 10d). However, depleting both SLC19A1 and SLC46A3 together did not reduce responses more than depletion of SLC19A1 alone (Extended Data Fig. 10d), suggesting that neither SLC46A3 nor SLC46A1 are responsible for most of the residual CDN transport in SLC19A1-depleted cells.
Our findings extend the spectrum of organic phosphates that utilize SLC19A1 to 2’3’-cGAMP, 2’3’-RR CDA and probably other CDNs, by the direct measurement of their transport and the impact of extra- and intra-cellular folates on their uptake in human cells by this route. In this context, it could play an important role in the anti-tumor and adjuvant effects of CDNs administered to human patients. It may also be important in cell-to-cell transport of CDNs in immune responses or immune pathology. For example, the SLC19A1 inhibitors methotrexate and sulfasalazine are first line treatments in rheumatoid arthritis, and are often used to treat inflammatory bowel diseases (IBD), including ulcerative colitis and Crohn’s disease27-29. Although no direct evidence for this is available in humans, studies in mouse models of IBD raise the possibility that host cells import CDNs produced by intestinal bacteria, activating STING in a cGAS-independent fashion30. It remains to be determined whether SLC19A1 plays a role in such processes in humans. In conclusion, we have identified SLC19A1 as a CDN transporter in humans with potential relevance in the context of cancer immunotherapy and immunopathology.
Methods
Cell culture
All cell lines were cultured at 37°C in humidified atmosphere containing 5% CO2 with media supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 0.2 mg/ml glutamine, 10 μg/ml gentamycin sulfate, 20 mM Hepes and 10% FCS. THP-1, C1R, and K562 cells were cultured in RPMI medium, and 293T, 293T transfected with hSTING (293T+hSTING), MDA-MBA-453 (MDA), and RAW macrophages were cultured in DMEM medium. Human embryonic kidney 293F cells (HEK293F), derived from a human fetus, were grown in FreeStyle 293 Media supplemented with GlutaMax (GIBCO) at 37°C in the presence of 5% CO2 in a shaking incubator.THP-1, K562, 293T cells, and RAW macrophages were present in the lab at the time this study began. MDA cells were obtained from the Berkeley Cell Culture Facility. C1R cells were a generous gift from Veronika Spies (Fred Hutchinson Cancer Center, Seattle WA). HEK293F cells were a generous gift from David Veesler (University of Washington, Seattle WA). 293T+hSTING cells were generated at Aduro Biotech Inc. L1210 cells were obtained from ATCC (CCL-219) and cultured in DMEM medium including 10% horse serum (Gibco, Cat.# 26–050-088). L1210 cells were authenticated by ATCC. MDA-MBA-453 were authenticated by the Berkeley Cell Culture Facility using karyotyping and/or PCR, other cell lines were not authenticated. All cell lines were negative for mycoplasma contamination.
Antibodies and reagents
The following antibodies were obtained from Cell Signaling Technology: rabbit-anti-human TBK1 mAb (clone D1B4, used 1:500 for immunoblot [IB]), rabbit-anti-human phospho-TBK1 mAb (clone D52C2, 1:1000 for IB), rabbit-anti-human STING mAb (clone D2P2F, 1:2000 for IB), rabbit-anti-human phospho-STING mAb (clone D7C3S, used 1:1000 for IB), rabbit-anti-human phospho-IRF3 mAb (clone 4D4G, 1:1000 for IB). Antibodies obtained from LI-COR Biosciences: goat-anti-mouse IgG IRDye 680RD conjugated (cat. #: 926–68070, used 1:5000), donkey-anti-rabbit IgG IRDye 800CW conjugated (cat. #: 926–32213), donkey-anti-rabbit IgG IRDye 680RD (cat. #: 926–68073). Other antibodies: rabbit-anti-human IRF3 mAb (Abcam, cat. #: EP2419Y, used 1:2000 for IB), mouse-anti-human transferrin receptor mAb (Thermo Fischer Scientific, clone H68.4, used 1:1000 for IB), rabbit-ant-human SLC19A1 pAb (BosterBio, cat. #: PB9504, used 0.4 μg/ml for IB), APC-conjugated mouse-anti-human CD55 mAb (BioLegend, clone JS11, used 1:50 for flow cytometry), mouse-anti-human CD59 mAb (BioLegend clone p282, used 1:250 for flow cytometry), APC-conjugated goat-anti-mouse IgG (BioLegend, cat. #: 405308, used 1:100 for flow cytometry). Reagents used: 5-methyl tetrahydrofolic acid (Cayman Chemical Company, cat. #: 16159), methotrexate (Cayman Chemical Company, cat. #: 13960), folic acid (Cayman Chemical Company, cat. #: 20515), sulfasalazine (Sigma-Aldrich, cat. #: S0883), polybrene (EMD Millipore, cat. #: TR1003G), 3’3’-cyclic-di-AMP (3’3’ CDA) (Invivogen, cat. #: tlrl-nacda), 2’3’-RR c-di-AMP (2’3’-RR-S2 CDA) and 2’3’- cyclic-di-GMP-AMP (2’3’-cGAMP) (generous gifts from Aduro Bioscience Inc.), human interferon-β (PeproTech, cat. #: 300–02B), mouse interferon-β1 (BioLegend, cat. #: 581302). Antibiotic selection: puromycin (Sigma-Aldrich, cat. #: P8833), blasticidin (Invivogen, cat. #: ant-bl-1, used at 10 μg/ml), zeocin (Invivogen, cat. #: ant-zn-1, used at 200 μg/ml).
Plasmids
A gBLOCK gene fragment (Integrated DNA Technologies, Inc.) encoding the tdTomato reporter gene driven by the interferon stimulatory response elements (ISREs) and the minimal mouse interferon-β promoter was cloned into a dual promoter lentiviral plasmid using Gibson assembly. This lentiviral plasmid co-expressed the Zeocin resistance gene and GFP via a T2A ribosomal skipping sequence controlled by the human EF1A promoter, and was generated as described previously31.
For rescue and overexpression of the folate-organic phosphate antiporter SLC19A115,26,32-35, the proton-coupled folate transporter SLC46A115, or the maytansine transporter SLC46A336, a gBLOCK gene fragment encoding SLC19A1 (gene ID 6573, transcript 1), SLC46A1 (gene ID 113235) or SLC46A3 (gene ID 283537) was cloned by Gibson assembly into a dual promoter lentiviral plasmid co-expressing the Blasticidin resistance gene and the fluorescent gene mAmetrine.
For CRISPR interference (CRISPRi)-mediated depletions, cells were transduced with a lentiviral dCas9-HA-BFP-KRAB-NLS expression vector (Addgene plasmid #102244).
For screen validation using individual gRNAs, gRNAs (Table S3) were cloned into the same expression plasmid used for the gRNA library (“pCRISPRia-v2”, Addgene plasmid #84832, a gift from Jonathan Weissman). The lentiviral gRNA plasmid co-expressed a puromycin resistance gene and blue fluorescence protein (BFP) via a T2A ribosomal skipping sequence controlled by the human EF1A promoter. The CRISPRi gRNAs introduced into this vector by Gibson assembly were expressed from a murine U6 promoter. For expression of multiple gRNAs, additional gRNAs were introduced in a separate vector that co-expressed the blasticidin resistance gene and mAmetrine via a T2A ribosomal skipping sequence under the control of a human EF1A promoter.
Conventional CRISPR gRNAs (see Table S3) were cloned into a selectable lentiviral CRISPR/Cas9 vector. This lentiviral vector included a human codon-optimized S. pyogenes Cas9 co-expressing the puromycin resistance gene via a T2A ribosome skipping sequence under the control of a minimal human EF1A promoter31,37.
Lentiviral production and transduction
Lentivirus was produced by transfecting lentiviral plasmids and 2nd generation packaging/polymerase plasmids into 293T cells using TransIT-LT1 Reagent (Mirus Bio LLC). Virus-containing supernatants were harvested 72h later, centrifuged to remove cell debris, and filtered using a 0.45 μm PES filter. Filtered virus supernatant was used to transduce target cells by spin-infection (800 x g for 90min at 33°C) in the presence of 8 μg/ml polybrene. After spin-infection, virus and polybrene containing medium was diluted 1:1 with fresh medium. 72 hours after transduction, cells were sorted based on fluorescence expression using a BD FACSAria cell sorter, or selected with relevant selection reagent for at least 7 days.
2’3’-RR CDA and 2’3’-cGAMP screens
THP-1 cells co-expressing the tdTomato reporter, GFP, and dCas9-BFP were single cell sorted to select for a THP-1 cell clone with efficient dCas9-BFP-knockdown capacity. Clones were transduced with lentiviral vectors encoding gRNAs targeting GFP, CD55 or CD59. After 1 week on puromycin (2 μg/ml) selection, CD55, CD59 and GFP expression were quantified using the BD LSR Fortessa flow cytometer. A clone that showed the highest reduction in all three marker genes was selected for the screens.
Two separate cultures of THP-1 cells were transduced with the human genome-wide CRISPRi v2 library38. Each culture was divided in two and screened separately after treatment with 2’3’-RR CDA or 2’3’-cGAMP, followed by selection and analysis. Hence, each screen was performed twice, with independent library transductions. For each transduction, the THP-1 clone was expanded to 3.2 × 108 cells and transduced with the human genome-wide CRISPRi v2 library38, which contains approximately 100,000 gRNAs targeting around 20,000 genes. Sufficient cells were transduced and propagated to maintain at least 5 × 107 transduced (BFP+) cells, representing 500x coverage of the gRNA library. The transduction efficiency was around 20% to minimize the chance of multiple lentiviral integrations per cell. Two days after transduction, cells were cultured in the presence of puromycin for two days and one additional day without puromycin. 4 × 108 cells were seeded to a density of 106 cells/ml and stimulated with 2’3’-RR CDA (2 μg/ml) or 2’3’-cGAMP (15 μg /ml). 20h later, cells were harvested, washed in PBS, and sorted based on BFP expression (presence of gRNAs), GFP expression (presence of reporter) and tdTomato expression using the BD Influx cell sorter and BD FACSaria Fusion cell sorter. The cells were sorted into two populations based on tdTomato expression: the highest 25% of tdTomato expressing cells (hyper-responsive population) and lowest 25% of tdTomato expressing cells (hypo-responsive population). During sorting, all cells were kept at 4°C. After sorting, cells were counted: the sorted populations contained 1.5 – 2 × 107 cells, and the unsorted control contained 1 – 1.5 × 108 cells. Cells were washed in PBS, and cell pellets were stored at −80°C until further processing.
gDNA isolation and sequencing
Genomic DNA was isolated from sorted cells using NucleoSpin Blood kits (Macherey-Nagel). PCR was used to amplify gRNA cassettes with Illumina sequencing adapters and indexes as described previously39. Genomic DNA samples were first digested for 18 hours with SbfI-HF (NEB) to liberate a ~500 bp fragment containing the gRNA cassette. The gRNA cassette was isolated by gel electrophoresis as described previously39 using NucleoSpin Gel and PCR Clean-up kits (Macherey-Nagel), and the DNA was then used for PCR. Custom PCR primers are listed in Supplementary Table 5. Indexed samples were pooled and sequenced on an Illumina HiSeq-2500 for the 2’3’RR CDA screen and an Illumna HiSeq-4000 for the 2’3’-cGAMP screen using a 1:1 mix of two custom sequencing primers (Supplementary Table 5). Sequencing libraries were pooled proportional to the number of sorted cells in each sample. The target sequencing depth was at least 2,000 reads/gRNA in the library for unsorted “background” samples, and at least 10 reads/cell in sorted samples.
Screen data analysis
CRISPRi samples were analyzed using the Python-based ScreenProcessing pipeline (https://github.com/mhorlbeck/ScreenProcessing). Normalization using a set of negative control genes and calculations of phenotypes and Mann-Whitney p-values were performed as described previously38,40. Briefly, Illumina 50bp single end sequencing reads for pooled sublibraries one to four and five to seven were trimmed to 29bp and guides were quantified by counting exact matches to the CRISPRi v2 human library guides. Phenotypes were calculated as the log2 fold change in enrichment of an sgRNA in the high and low samples versus background as well as high versus low, normalized by median subtracting non-targeting sgRNAs40,41. Phenotypes from sgRNAs targeting the same gene were collapsed into a single sensitivity phenotype for each gene using the average of the top three scoring sgRNAs (by phenotype absolute value). For genes with multiple independent transcription start sites (TSSs) targeted by the sgRNA libraries, phenotypes and p-values were calculated independently for each TSS and then collapsed to a single score by selecting the TSS with the lowest Mann-Whitney p-value. Counts from the ScreenProcessing pipeline were then used as input to the MAGeCK program to obtain FDR scores for filtering (see Table S2). ). We integrated multiple gRNAs per gene comparing the hyporesponsive and hyperresponsive populations calculated as robust rank aggregation scores (RRA) as depicted in Fig. 1d and e. Similar results were obtained when each sorted population was compared to unsorted cells (Table S2).
Genes were also ranked by individual gRNAs with the greatest enrichment/depletion between the hypo-responsive and hyper-responsive libraries. gRNA read counts were normalized to library sequencing depth by converting to read counts per million total reads. For each gRNA, the ratio between the read counts for the hypo-responsive and hyper-responsive libraries was determined and averaged between replicates. For hypo-responsive gene rankings, each gene was ranked by the single corresponding gRNA with the highest hypo-to-hyper ratio (see Table S1, ‘highest ratio hypo/hyper’ column). For hyper-responsive gene rankings, each gene was ranked by the single corresponding gRNA with the lowest hypo-to-hyper ratio (see Table S1, ‘lowest ratio hypo/hyper’ column). Gene-level phenotypes are available as Supplemental Materials (Table S1 and S2).
CDN and IFN-β stimulation assays
The week prior to stimulation experiments, cells were cultured at the same density. The day before stimulation, cells were seeded to 0.5 × 106 cells/ml. Cells were stimulated with CDNs or IFN- β in 48W plates using 50,000 cells/well in 300 μl medium. After 18–24h, cells were transferred to a 96W plate and tdTomato expression was measured by flow cytometry using a high throughput plate reader on a BD LSR Fortessa. For stimulations in the presence of sulfasalazine, 5-methyl tetrahydrofolate, or methotrexate, cells were stimulated in 48W plates using 20,000 cells/well in 300 μl medium. Cells were incubated with compounds or DMSO as vehicle prior to stimulations with CDNs or IFN-β. 18–24h after stimulation, tdTomato reporter expression was quantified by flow cytometry using a high throughput plate reader on a BD LSR Fortessa.
Production of SLC19A1 knockout cell lines
As an alternative approach to corroborate the role of SLC19A1 in CDN responses, SLC19A1 was disrupted in THP-1 cells using the conventional CRISPR/Cas9 system. THP-1 cells expressing the tdTomato reporter were transduced with a CRISPR/Cas9 lentiviral plasmid encoding a control gRNA or a gRNA targeting SLC19A1 at a region critical for transport42 (see Table S3). Transduced cells were selected using puromycin for 2 days and single cell sorted using a BD FACSAria cell sorter. Control cells and SLC19A1-targeted cells were selected that had comparable forward and side scatter by flow cytometry analysis. Genomic DNA was isolated from clones using the Qiamp DNA minikit (Qiagen), and the genomic region surrounding the SLC19A1 gRNA target site was amplified by PCR using primers 5’-TTCTCCACGCTCAACTACATCTC-3’ and 5’-CAGCATCCGCGCCAGCACTGAGT-3’. PCR product was cloned into 5-alpha competent bacteria (New England Biolabs, cat. #C2987) using a TOPO TA cloning kit (Thermo Fischer Scientific, cat. # 450641) according to manufacturer’s instructions. After blue/white screening, a minimum of 10 colonies were sequenced per THP-1 clone, and sequences were analyzed using SeqMan (Lasergene DNASTAR). Nine independent THP-1 clones with out-of-frame mutations at the SLC19A1 gRNA target site were selected for further experiments. These clones were all significantly less sensitive to CDN stimulation when compared to seven control clones that received a non-targeting gRNA (Fig. 2e).
shRNA knockdown
For shRNA knockdown experiments, shRNA sequences were cloned into the pLKO.1 lentiviral expression vector. Cells were transduced with lentivirus and selected using 2 μg/ml puromycin for at least 5 days. As controls, shRNAs targeting GFP (shRNA1; TRCN0000231753) and luciferase (shRNA2 TRCN0000231737) were used. Mouse SLC19A1 targeting shRNA sequences: GCAGGTGACTAACGAGATCAT (shRNA 4), and CCGTATCTACTTCATATACTT (shRNA 6); human SLC19A1 targeting shRNA (shRNA 9): CGACGGTGTTCAGAATGTGAA. All shRNA knockdowns were confirmed by real-time qPCR. Depletion of SLC19A1 function was confirmed by showing decreased uptake of [3H]-methotrexate (see below and Extended Data Fig. 6l, 7c,g)
RT-qPCR
Cells were harvested and washed in ice-cold PBS. Cells were transferred to RNase-free microcentrifuge tubes and RNA was isolated using the RNeasy mini kit (Qiagen, cat. #: 74104) including a DNase I step (Qiagen, cat. #: 79254). RNA concentration was measured by NanoDrop (Thermo Fischer), and 1 μg of RNA was used as input for cDNA synthesis using the iScript cDNA synthesis kit (Bio-rad, cat. #: 1708890). cDNA was diluted to 20 ng/μl and 2.5 μl/reaction was used as input for the qPCR reaction. qPCR reactions were set up using SSOFast EvaGreen Supermix (Bio-Rad, cat. #: 1725200) according to the manufacturer’s recommendations, using 500 nM of each primer and following cycling conditions on a Bio-Rad C1000 Thermal Cycler: 2 min at 98°C, 40 repeats of 2 sec at 98°C and 5 sec at 55°C. Primers used to amplify the HPRT1, YHWAZ, CCL5, CXCL10, STING, IRF3, SLC19A1, SLC46A1, and SLC46A3-specific PCR products are listed in Table S4. The housekeeping genes HPRT1 and YHWAZ served as endogenous control for human cDNA samples and Gapdh and Ubc served as endogenous control for mouse cDNA samples.
For quantification of human IFNB1, Oasl, or Isg15 mRNA, RNA was extracted with the Nucleospin RNA Isolation Kit (Machery-Nagel) and reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). TaqMan real-time qPCR assays were used for quantification of human Ifnb1 (Hs01077958_s1), human Oasl (Hs00984387_m1), and human Isg15 (Hs01921425_s1). Actb (Hs01060665_g1) served as an endogenous control.
Synthesis of [32P] 2’3’-cGAMP and [32P] 3’3’-CDA
Radiolabeled 2’3’-cGAMP was enzymatically synthesized by incubating 0.33 μM α-[32P] ATP (Perkin-Elmer) with 250 μM unlabeled GTP, 1 μg of interferon stimulatory DNA 100mer (kindly provided by Daniel Stetson), and 1 μM of recombinant His-tagged 2’3’-cGAMP Synthase (cGAS) in binding buffer [40 mM Tris pH 7.5, 100 mM NaCl, 20 mM MgCl2] at 37°C overnight. The reaction was confirmed to have gone to completion by Thin Layer Chromatography (TLC) analysis (Extended Data Fig. 6a, b). Briefly, the 2’3’-cGAMP synthesis reaction was separated on Polygram CEL300 PEI TLC plates (Machery-Nagel) in buffer containing 1:1.5 (vol/vol) saturated (NH4)2SO4 and 1.5 M NaH2PO4 pH 3.6. The TLC plates were then air dried and exposed to a PhosphorImager screen for visualization using a Typhoon scanner (GE Healthcare Life Sciences). Next, the sample was incubated with HisPur Ni-NTA resin (Thermo Scientific) for 30 min in order to remove recombinant cGAS. The resultant slurry was transferred to a minispin column (Thermo Scientific) to elute crude [32P] 2’3’-cGAMP. Recombinant mSTING-CTD protein was used for further purification of synthesized [32P] 2’3’-cGAMP. 100 μM mSTING-CTD was bound to HisPur Ni-NTA resin and incubated with the remaining crude 2’3’-cGAMP synthesis reaction mixture for 30 min on ice. Following removal of the supernatant, the Ni-NTA resin was washed three times with cold binding buffer. The resin was then incubated with 100 μl of binding buffer for 10 min at 95 °C, and transferred to a minispin column to elute [32P] 2’3’-cGAMP. The resulting STING-purified [32P] 2’3’-cGAMP was evaluated by TLC analysis and determined to be ∼99% pure (Extended Data Fig. 6c).
Radiolabeled 3’3’-CDA was synthesized as described previously 43. Briefly, 1 μM α-[32P] ATP (Perkin-Elmer) was incubated with 1 μM of recombinant DisA in binding buffer at 37°C overnight. The reaction mixture was boiled for 5 min at 95°C and DisA was removed by centrifugation. Recombinant His-tagged RECON was then used to further purify the 3’3’-CDA reaction mixture. 100 μM His-tagged RECON was bound to HisPur Ni-NTA resin for 30 min on ice. The resin was washed three times with cold binding buffer and then incubated with 100 μl of binding buffer for 5 min at 95°C. The slurry was then transferred to a minispin column to elute [32P] c di-AMP. The purity of the radiolabeled 3’3’-CDA was assessed by TLC and determined to be ~98%.
Nucleotide-binding assays
The ability of radiolabeled 2’3’-cGAMP and 3’3’-CDA to bind recombinant STING was evaluated by DRaCALA (differential radial capillary action of ligand assay) analysis, as previously described44 (Extended Data Fig. 6d). Briefly, varying concentrations of recombinant STING were incubated with ~1 nM of radiolabeled cyclic dinucleotide in binding buffer for 10 min at room temperature. The reaction mixtures were blotted on nitrocellulose membranes and air dried for 15 min. The membranes were then exposed to a PhosphorImager screen and visualized using a Typhoon scanner.
NHS-methotrexate synthesis and affinity labeling
N-hydroxysuccinimide (NHS)-methotrexate was prepared as previously described by Henderson and Zeveley45. Briefly, methotrexate (2.2 mg) was acidified by the addition of HCl and dried under vacuum. Next, acidified methotrexate, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (7.8 mg), and NHS (5.8 mg) were dissolved in 1 mL of anhydrous, tissue-culture grade dimethylsulfoxide and incubated for 2 hours at 23°C. The activated reagent was used immediately by incubating cells with 5 μM NHS-methotrexate in RPMI 1640 medium (GIBCO) supplemented with 10 mM HEPES, 1 mM sodium pyruvate and 2 mM L-Glutamine (Thermo Fisher) for 30 minutes at 37°C. The cells were then recovered by centrifugation, washed twice in treatment medium, and re-suspended in pre-warmed RPMI 1640 medium (GIBCO) containing 10% (v/v) heat-inactivated FBS (HyClone) and supplemented with 10 mM HEPES, 1 mM sodium pyruvate and 2 mM L-Glutamine (Thermo Fisher) to a final cell density of 1 X 106 – 1 X 107 cells per ml.
Nucleotide-uptake assays
For transport assays, cells were collected by centrifugation and washed in Dulbecco’s Phosphate-Buffered Saline (DPBS) (Life Technologies). The cell pellets were re-suspended in pre-warmed RPMI 1640 medium (GIBCO) containing 10% heat-inactivated FBS (HyClone) and supplemented with 10 mM HEPES, 1 mM sodium pyruvate and 2 mM L-Glutamine (Thermo Fisher) to a final cell density of 1 × 107 cells per ml. Uptake of 1 nM [32P] 2’3’-cGAMP and 3’3’-CDA was assayed in cell suspensions at 37°C over the indicated time points. At the end of each time point, transport was quenched by the addition of cold DPBS. Cells were washed three times with cold DPBS, followed by lysis in 50 μl of cold deionized water. The cell lysates were then transferred to 5 ml of liquid scintillation cocktail (National Diagnostics) and the associated radioactivity was measured by liquid scintillation counting using a LS6500 Liquid Scintillation Counter (Beckman Coulter). For each sample, [32P] 2’3’-cGAMP (counts per minute) was normalized to cell count (106 cells/ sample). For competition experiments, cells were pre-incubated with indicated concentrations of “cold” unlabeled ligand or inhibitor for 15 minutes prior to the addition of 1 nM “hot” [32P] 2’3’-cGAMP. Cells were then collected at the indicated time points and processed as described above.
2’3’-cGAMP was incorporated into THP-1 cells at a linear rate over at least three hours of incubation (Extended Data Fig. 6e), during which time the [32P] 2’3’-cGAMP was not hydrolyzed or modified (Extended Data Fig 6f). 2’3’-cGAMP uptake was most efficient at a pH range of 7.5 to 8.0 in both THP-1 and U937 cells (Extended Data Fig. 6g, h), consistent with a neutral pH optimum for SLC19A115.
[3H]-methotrexate uptake assays
Tritium-labeled methotrexate (Vitrax Cat. # VT 145) had a specific activity of 40.6 Ci/mmole. [3H] methotrexate uptake in SLC19A1 knockdown and overexpressing cells was performed in RPMI complete medium. [3H] methotrexate competition assays were performed in MHS buffer (20 mM HEPES, 235 mM sucrose adjusted to pH7.4 using MgO). Cells were washed and resuspended in the appropriate buffer to 4×106 cells/ml. Cells (106/ condition) were exposed to 12.3 nM [3H]-methotrexate (1.25 μCi) at 37 °C and 5% CO2 for the duration indicated in the figure legends. Cells were subsequently washed 4 times in ice cold PBS to stop uptake and remove extracellular [3H]-methotrexate. Cells were lysed in 250 μl ice cold water and 200 μl was mixed with 4ml scintillation liquid. 50 μl was used for protein quantification using the Bicinchoninic acid (BCA) assay (Thermo Scientific Cat. # 23235). Radioactivity (counts per minute) was measured for 3 min/sample using by liquid scintillation counting using a LS6500 Liquid Scintillation Counter (Beckman Coulter). Counts per minute were normalized to total protein. For competition assays, cells were incubated for 15 min at 37°C and 5% CO2 with the indicated ligand in MHS buffer prior to addition of [3H] methotrexate.
ELISA
Cells were stimulated with 2’3’-RR CDA (2 μg/ml) for 20h. Supernatant was collected and centrifuged to remove cells. CXCL10 was quantified in the supernatant using the CXCL10 ELISA kit (BioLegend Cat. # 439904) according to the manufacturer’s recommendations.
Digitonin permeabilization
Cells were counted, washed and resuspended to 2×106 cells/ml. Cells (106/condition) were permeabilized using 5 μg/ml digitonin (Sigma Cat. # D-141; stock dissolved in H2O) and stimulated with 2’3’-RR CDA (10 μg/ml) and the indicated SLC19A1 (competitive) inhibitor for 2h at 37°C and 5% CO2. Cells were subsequently washed in ice cold PBS, lysed in RIPA buffer and used for Western blot protein analysis.
Protein expression and purification
Full-length human SLC19A1 cDNA with a C-terminal 8 X His-tag was subcloned into a dual promoter lentiviral vector (see above). Recombinant His-tagged SLC19A1 was expressed using a FreeStyle 293 Expression System. Briefly, 293F cells (1 × 106 cells per ml) grown in FreeStyle 293 Media supplemented with GlutaMax (GIBCO) were transfected with the SLC19A1 expression construct (1 μg plasmid DNA per ml of cells) using PEI transfection reagent. Transfected cells were grown for 72 hours in a shaking incubator at 37°C in 5% CO2. Three days after transfection, the cells were harvested by centrifugation and washed in DBPS. Cell pellets were then re-suspended in lysis buffer [25 mM Tris pH 8.0, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride] supplemented with HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) and lysed by sonication. The cell lysate was supplemented with 2% (w/v) n-dodecyl-β-D-maltoside (DDM) and rotated for 2h at 4°C. The cell lysates were centrifuged at 15,000 rpm for 1h at 4 °C to remove cell debris, and the detergent-soluble fraction was incubated with HisPur Ni-NTA resin for 1h at 4 °C. The resin was washed with 100 column volumes of wash buffer [25 mM Tris pH 6.0, 150 mM NaCl, 30 mM imidazole, 5% glycerol (v/v), and 0.05% DDM (w/v)], and bound proteins were eluted in elution buffer [25 mM Tris pH 6.0, 150 mM NaCl, 300 mM imidazole, 5% glycerol (v/v), and 0.05% DDM (w/v)]. The resulting proteins were analyzed by SDS-PAGE followed by Coomassie staining and immunoblotting to confirm expression and purification of His-tagged SLC19A1.
Recombinant cGAS, DisA, mSTING-CTD, and mRECON were expressed and purified as previously described43,44,46. Briefly, plasmids for cGAS, DisA, mSTING-CTD, and mRECON expression were transformed into Rosetta (DE3) pLysS chemically competent cells. Overnight cultures of the resulting transformed bacteria were inoculated into 1.5 L of LB broth at a 1:100 dilution. Bacterial cultures were grown at 37°C to OD600 0.5 followed by overnight induction at 18°C with 0.5 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). Cells were harvested and lysed in PBS supplemented with 1 mM PMSF and soluble protein was purified using nickel-affinity chromatography followed by gel filtration chromatography (S‐300, GE Healthcare, Piscataway, New Jersey, USA). After SDS-PAGE analysis, the purified proteins were concentrated in storage buffer [40 mM Tris pH 7.5, 100 mM NaCl, 20 mM MgCl2, 25% glycerol (v/v)] and stored at −80 °C.
Synthesis of 2’3’-cGAMP Sepharose
2’3’-cGAMP was enzymatically synthesized using recombinant cGAS as described previously7,44. Approximately, 100 mg of purified 2’3’-cGAMP was dissolved in PBS to 200 μM. The pH of the solution was adjusted to 7.5 with NaOH, and the resulting solution was added directly to washed epoxy-activated Sepharose and incubated at 56°C for 2 days. The Sepharose was washed and the absorbance spectrum of 50% slurry was measured to ensure nucleotide coupling. HPLC analysis of the remaining uncoupled nucleotide ensured no degradation of 2’3’-cGAMP occurred during the 2-day incubation. The remaining epoxy groups were blocked with ethanolamine following the instructions provided by GE. In parallel with this blocking step, fresh epoxy-activated Sepharose was also treated with ethanolamine to generate control resin.
2’3’-cGAMP pulldowns
Following nickel affinity purification, recombinant His-tagged SLC19A1 was incubated with 100 μl of ethanolamine- or 2’3’-cGAMP-conjugated Sepharose beads for 4h at 4°C with rotation, as described previously44,46. Beads were washed three times with wash buffer [25 mM Tris pH 6.0, 150 mM NaCl, 5% glycerol (v/v), and 0.05% DDM (w/v)], and bound proteins were eluted by boiling in SDS-PAGE sample loading buffer for 5 min at 95°C. The soluble fraction was then removed and analyzed by SDS-PAGE followed by Coomassie Blue staining and immunoblotting. As a positive control, recombinant His-tagged mSTING-CTD was incubated with ethanolamine- or 2’3’-cGAMP-conjugated Sepharose beads, as described above (Fig 4g and Extended Data Fig. 8d). Beads were washed three times with binding buffer, and then boiled in SDS-PAGE sample loading buffer for 5 min at 95°C. The soluble fraction was then analyzed by SDS-PAGE followed by Coomassie Blue staining.
Cell lysis and immunoblotting
For anti-SLC19A1 immunoblotting, cells were lysed and proteins were separated by SDS-PAGE as described above in the ‘2’3’-cGAMP pulldowns’ paragraph. SDS-PAGE-separated proteins were transferred onto nitrocellulose membranes (Bio-Rad) at 30V overnight at 4°C. Membranes were then air-dried for 1h and blocked in 5% Blotto, non-fat milk (NFM, Santa Cruz Biotechnology) in 1 X TBS. Membranes were probed in 5% Bovine Serum Albumin (Fisher) in 1 X TBS-T with anti-SLC19A1 Picoband antibody (Boster Bio).
For protein detection using all other antibodies, cells were counted, washed with PBS and lysed in RIPA buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS) including cOmplete ULTRA protease inhibitors (Sigma-Aldrich cat. #: 05892791001), phosphatase inhibitors (Biomake, cat. # B15001) and 50mM DTT. Cells lysates were mixed with 4x NuPage LDS sample buffer (Invitrogen cat. #: NP0007), pulse sonicated and incubated at 75°C for 5min. Lysates were loaded onto Bolt 4–12% Bis-Tris Plus SDS-PAGE gels (Invitrogen cat. #: NW04125BOX). SDS-PAGE separated proteins were transferred onto Immobilon-FL PVDF membranes (EMD Millipore) at 100V for 1h at 4°C. Membranes were blocked in 4% NFM, and probed in 1% NFM overnight at 4°C with primary antibody. Membranes were subsequently washed 3 times in 1x-TBS-T and probed with secondary antibody for 1h at RT protected from light. Membranes were washed 2 times in TBS-T, once in TBS, and blots were imaged using an Odyssey CLx System (LI-COR).
Mice
Male and female C57BL/6J mice (4–20 weeks old) were obtained from The Jackson Laboratory. All of the mice were maintained in specific pathogen free conditions by the Department of Comparative Medicine at the University of Washington School of Medicine or at the University of California Berkeley. All experimental procedures using mice complied with all relevant ethical regulations and were approved by the Institutional Animal Care and Use Committee of the University of Washington, or the University of California Berkeley, and were conducted in accordance with institutionally approved protocols and guidelines for animal care and use.
Isolation of Mouse Splenocytes and bone marrow-derived dendritic cells and macrophages
For isolation of murine splenocytes, spleens were removed from mice, strained through a 70 μm cell strainer, and homogenized into a single cell suspension using ice cold PBS supplemented with 3% FCS. Red blood cells were lysed by resuspending spleen cells in Red Blood Cell Lysing Buffer (Sigma) and incubating on ice for 10 min. Splenocytes were washed, resuspended in RPMI 1640 medium (GIBCO) supplemented with 10% (v/v) heat-inactivated FBS (HyClone), 10 mM HEPES, 1 mM sodium pyruvate, 2 mM L-Glutamine (Thermo Fisher), 100 U/ml penicillin, 100 μg/ml streptomycin, and used immediately for [32P] 2’3’-cGAMP uptake assays.
For generation of bone-marrow derived macrophages (BMM) and dendritic cells (BMDC), bones from the hind legs were removed and crushed to release the bone marrow. Bone marrow was washed in complete RPMI medium and filtered using a 70 μm cell strainer. Cells were incubated in Ammonium-Chloride-Potassium (ACK) lysis buffer for 2 minutes to remove red blood cells. Cells were subsequently resuspended in RPMI 1640 supplemented with 10% (v/v) heat-inactivated FBS (HyClone), 10 mM HEPES, 1 mM sodium pyruvate, 2 mM L-Glutamine (Thermo Fisher), 100 U mL-1 penicillin, 100 μg mL-1 streptomycin, and 5 ng/ml GM-CSF (Preprotech, Cat. # 315–03) to generate BMDCs or M-CSF conditioned medium to generate macrophages. Cells were transduced with shRNA-encoding lentiviruses on day 2 and 3 after bone-marrow isolation, and selected using 7 μg/ml puromycin for 3 days starting at day 5 after bone marrow isolation. Medium was refreshed every other day. Nine days after bone marrow isolation, non-adherent BMDCs or adherent BMM cells were harvested. Cells were stimulated with 5 μg/ml 2’3’-RR CDA for 5h prior to RNA extraction.
Isolation of human Peripheral Blood Mononuclear Cells (PBMCs)
Whole blood from healthy, human donors was collected in 10-ml EDTA blood tubes (Beckton Dickinson) from healthy adults with written informed consent. Bulk PBMCs were isolated using SepMate tubes (STEMCELL Technologies), according to the manufacturer’s instructions. The remaining cells were washed in PBS, resuspended in RPMI 1640 medium (GIBCO) supplemented with 10% (v/v) heat-inactivated FBS (HyClone), 10 mM HEPES, 1 mM sodium pyruvate, 2 mM L-Glutamine (Thermo Fisher), 100 U/ml penicillin, 100 μg/ml streptomycin, and used immediately for [32P] 2’3’-cGAMP uptake assays. The isolation of primary human cells complied with all relevant ethical regulations and was conducted under a protocol to K.B. Elkon which was approved by the University of Washington Institutional Review Board.
Extended Data
Extended Data Figure 1. Structures of cyclic dinucleotides (CDN) used in this study and gating strategy for the genome-wide CRISPRi screens.
a, Structures of the CDNs used in this study. b, Representative gating strategy for flow cytometry-based sorting of the CRISPRi library of reporter-expressing THP-1 cells stimulated with CDNs. Cells were gated based on their forward scatter (FSC) and side scatter (SSC) using gate P1. The P1 cells were subsequently selected for co-expression of blue fluorescent protein (BFP, fluorescent marker for the CRISPRi gRNAs) and GFP (marker for the expression of the reporter construct) (gate P2). Gate P3 excluded cell doublets present among P2 cells. Gate P4 selected for the lowest 25% of cells with respect to tdTomato expression and gate P5 selected for the highest 25%. c, Representative flow cytometry dot plots showing tdTomato expression in unstimulated cells or in cells stimulated for 20h with cells for 20h with CDN (2’3’-RR CDA). Data is representative of n=3 biological replicates.
Extended Data Figure 2. Results of genome-wide CRISPRi screens for host factors crucial for CDN stimulation.
Volcano plots of the gRNA-targeted genes enriched or depleted in the tdTomato reporter-low versus reporter-high groups after stimulation with 2’3’-RR CDA (a), or 2’3’-cGAMP (b). FC: fold change. Each panel represent the combined results of two independent screens. Calculations of phenotypes and Mann-Whitney p-values were performed as described in the Methods section.
Extended Data Figure 3. SLC19A1 is critical for CDN-induced gene expression.
a,b, mRNA expression levels of (a) SLC19A1 or (b) IRF3 in THP-1 cells expressing a CRISPRi vector and a control non-targeting gRNA or gRNAs targeting IRF3 or SLC19A1 (two gRNAs each). c, THP-1 cells described in panels a and b were exposed to [3H]-methotrexate (MTX). After 1h, radioactivity (counts per minute, CPM) in lysates of cell pellets was measured. CPMs were normalized to protein concentrations in the lysate. d, THP-1 cells expressing a control gRNA or SLC19A1 targeting gRNA were exposed to indicated concentrations of 2’3’-RR CDA or 2’3’-cGAMP. After 20h, the mean fluorescence intensity (MFI) of tdTomato was quantified by flow cytometry. e, THP-1 cells expressing the indicated CRISPRi gRNAs or non-targeting gRNA (control), were stimulated with hIFN-β (100 ng/ml). After 18-22h, tdTomato expression was quantified as in Fig. 2a. f, U937 cells expressing the indicated CRISPRi gRNAs or non-targeting gRNA (control), were stimulated with 2’3’-RR CDA (1.67 μg/ml), 2’3’-cGAMP (15 μg/ml), or hIFN-β (100 ng/ml). After 18–22h, tdTomato expression was quantified as in Fig. 2a. g, h, Induction of CXCL10 mRNA (g) or CCL5 mRNA (h) in control (non-targeting gRNA) THP-1 cells or THP-1 cells expressing the indicated CRISPRi gRNAs after 5h stimulation with 5 μg/ml 2’3’-RR CDA. i, CXCL10 protein expression in the supernatant of indicated gRNA-expressing THP-1 cells after exposure to 2 μg/ml 2’3’-RR CDA for 20h. j, Various cell lines expressing a control vector or an SLC19A1 expression vector were stimulated with hIFN-β (100 ng/ml). After 18-22h, tdTomato expression was quantified as in Fig. 2a. k, THP-1 cells were incubated with increasing concentrations of the non-competitive inhibitor sulfasalazine or DMSO as vehicle control, before stimulating with 2’3’-RR CDA (1.25 μg/ml), 2’3’-cGAMP (15 μg/ml) or hIFN-β (100 ng/ml). After 18-22h, tdTomato expression was quantified as in Fig. 2a. The data were normalized to the DMSO controls. In panels a-c and e-f, means of n=2 biological replicates are shown. In panel d and g-k, means ± SEM of n=3 biological replicates are shown. Statistical analyses were performed to compare each cell line to the control using a one-way ANOVA followed by Dunnetts’s post-test (panels d, and g-i), or a two-way ANOVA followed by uncorrected Fisher’s LSD tests (j). ****P ≤ 0.0001; n.s. not significant.
Extended Data Figure 4. SLC19A1 overexpression increases the response to CDNs in various cell lines.
Various cell lines expressing a control vector or an SLC19A1 expression vector (SLC19A1 OE) stimulated with 2’3’-cGAMP (10 μg/ml) (e) or hIFN-β (100 ng/ml) (or 100 ng/ml murine IFN-β in the case of RAW cells). After 20h, reporter expression was quantified by flow cytometry. Representative flow plots of n=3 biological replicates shown in Fig. 2f and Extended Data Fig. 3j.
Extended Data Figure 5. Covalent inhibition of SLC19A1 by NHS-methotrexate blocks STING activation.
a, Schematic overview of CDN-induced phosphorylation (P) of STING and downstream effectors TBK1 and IRF3. b, THP-1 monocytes pre-treated with DMSO or NHS-methotrexate (MTX) (5 μM) were treated with varying concentrations of 2’3’-cGAMP for 4h, and the amounts of IFNB1 or ISG15 transcripts were measured by RT-qPCR. c, Semi-native PAGE immunoblot analysis of STING dimerization and phosphorylation in DMSO and NHS-MTX (5 μM) pre-treated THP-1 monocytes stimulated with 100 μM 2’3’-cGAMP for 4h. For gel source data, see Supplementary Figure 1. d, DMSO and NHS-MTX (5 μM) treated THP-1 monocytes were treated with 100 μM 2’3’-cGAMP in the presence and absence of digitonin (5 μg/mL) for 4h, and the induction of IFNB1 mRNA was measured by RT-qPCR. e, f, DMSO (◯) and NHS-MTX (◯) (5 μM) pre-treated THP-1 monocytes (e) or K562 cells (f) were stimulated for 4h with 100 μM 2’3’-cGAMP, or not, in the presence or absence of digitonin (5 μg/mL), and the induction of OASL and ISG15 mRNA was measured by RT-qPCR. In panels b, d and e, data are means of n=2 technical replicates and data are representative of three independent experiments with similar results. In panel c, data are representative of three independent experiments with similar results.
Extended Data Figure 6. SLC19A1 is critical for CDN uptake in human cell lines and primary cells.
a, Thin layer chromatography (TLC) analysis of [32P] ATP standard (STD) and enzymatically synthesized [32P] 2’3’-cGAMP. 2’3’-cGAMP was purified on STING resin. Unbound nucleotides flowed through the resin (STING FT). Following four washes, the bound [32P] 2’3’-cGAMP was eluted over three fractions. b, DRaCALA binding analysis of [32P] 2’3’-cGAMP to STING C-terminal domain (CTD) in the presence of competing unlabeled nucleotides (200 μM). c, Thin layer chromatography (TLC) analysis of [32P]-ATP and enzymatically synthesized [32P]-2’3’-cGAMP and [32P]-c-di-AMP. d, Binding titration of [32P] 2’3’-cGAMP or [32P] c-di-AMP to mSTING C-Terminal Domain (CTD), determined with DRaCALA assays. Red dashed lines represent the 95% confidence interval for the non-linear regression. e, Time course of [32P] 2’3’-cGAMP (left panel) or [32P] 3’3’-CDA (right panel) uptake in THP-1 monocytes. f, TLC analysis (left panel) and STING-binding (DRaCALA) (right panel) of [32P] ATP standard, or [32P] 2’3’-cGAMP recovered from supernatants of THP-1 monocytes at the indicated time points. g, h, Effect of cell culture medium pH on [32P] 2’3’-cGAMP uptake in THP-1 (g) monocytes or U937 monocytes (h). i, Time course of [32P] 2’3’-cGAMP uptake by CIR cells transduced (tr.) with empty vector or SLC19A1 expression vector. j, mRNA expression levels of SLC19A1 (SLC.) in K562 cells expressing control shRNAs (sh1 and sh2) or an SLC19A1-targeting shRNA (sh9). k, mRNA expression levels of CXCL10 in K562 cells described in panel j, stimulated with 5 μg/ml 2’3’-RR CDA (CDN) for 5h. l, [3H]-Methotrexate uptake in K562 cells described in panel j, 1h after exposure to [3H]-Methotrexate. m, Time course of [32P] 2’3’-cGAMP uptake in K562 cells described in panel j. n, Time course of [32P] 2’3’-cGAMP uptake in U937 monocytes in the presence or absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. o. Time course of [32P] 2’3’-cGAMP uptake in K562 cells in the presence or absence of 500 μM competing, unlabeled (anti-) folates or sulfasalazine. p, Competition uptake assay of [32P] 2’3’-cGAMP uptake in THP-1 cells in the presence of varying concentrations of competing, unlabeled 5-me-THF (IC50 = 4.10 ± 0.16 nM), methotrexate (IC50 = 54.83 ± 5.08 nM), 2’3’-cGAMP (IC50 = 1.89 ± 0.11 μM), sulfasalazine ((IC50 = 2.06 ± 0.17 μM), and folic acid (IC50 = 4.79 ± 0.08 μM). q, Trans-stimulation of [32P] 2’3’-cGAMP influx in THP-1 cells by 5-me-THF. Cells were preloaded with indicated concentrations of 5-me-THF for 30 min. Cells were washed and incubated with [32P] 2’3’-cGAMP for one hour. r, Normalized [32P] 2’3’-cGAMP uptake after one hour in DMSO or NHS-methotrexate (MTX) (5 μM) treated human PBMCs from four healthy donors. In panel a and c, data are representative of three independent experiments with similar results. In panel b, data are means of n=2 technical replicates and are representative of three independent experiments. In panel d and f, data are means of n=2 technical replicates and are representative of two independent experiments. In panel e and m, data are means ± SD of n=3 technical replicates and are representative of three independent experiments. In panel g, h, i, and n-r, data are means ± n=3 technical replicates and are representative of two independent experiments. In panel j and k, data are means ± SEM of n=3 biologically independent experiments. In panel l, data are means of n=2 biologically independent experiments.
Extended Data Figure 7. SLC19A1 expression or inhibition has no effect on CDN uptake and signaling in mouse cells.
a, mRNA expression levels of Slc19a1 in mouse L1210 cells expressing control shRNAs (sh1 and sh2) or Slc19a1-targeting shRNA (sh4 and sh6). b, mRNA expression levels of Cxcl10 in L1210 cells described in panel a stimulated with 5 μg/ml 2’3’-RR CDA (CDN) for 5h. c, [3H]-Methotrexate uptake in L1210 cells described in panel a 1h after exposure to [3H]-Methotrexate. d, Time course of [32P] 2’3’-cGAMP uptake in L1210 cells described in panel a. e, mRNA expression levels of Slc19a1 in mouse C1498 cells expressing control shRNAs (sh1 and sh2) or Slc19a1-targeting shRNA (sh6). f, mRNA expression levels of Cxcl10 in the C1498 cells described in panel e, stimulated with 5 μg/ml 2’3’-RR CDA (CDN) for 5h. g, [3H]-Methotrexate uptake in C1498 cells described in panel e 1h after exposure to [3H]-Methotrexate. h, Time course of [32P] 2’3’-cGAMP uptake in C1498 cells transduced with a non-targeting control shRNA (control) or Slc19a1 shRNA. i, j, mRNA expression levels of Slc19a1 in mouse bone marrow-derived macrophages (BMM) (i) or mouse bone marrow-derived dendritic cells (BMDCs) (j) transduced or not with control shRNAs (sh1 and 2) or an shRNA targeting Slc19a1. k, l, mRNA expression of the Cxcl10 in cells described in panels i and j stimulated with 5 μg/ml 2’3’-RR CDA (CDN) for 5h. m, Time course of [32P] 2’3’-cGAMP uptake in primary murine splenocytes in the presence and absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. n, Time course of [32P] 2’3’-cGAMP uptake in primary murine splenocytes pretreated or not with NHS-methotrexate (MTX) (5 μM). o, Time course of [32P] 2’3’-cGAMP uptake in L1210 cells pretreated or not with NHS-MTX (5 μM). In panels a-c, e-g, j and l, data are means of n=2 biologically independent experiments. In panel d, h, i, k, n and o, data are means ± SD of n=3 technical replicates and are representative of two independent experiments. In panel m, data are means of n=2 technical replicates and are representative of two independent experiments. In time course experiments (d, h, m, n, o), data are presented as counts per minute (cpm) normalized to cell count.
Extended Data Figure 8. 2’3’-cGAMP binds to SLC19A1.
a, Sodium dodecyl sulfate (SDS)-PAGE analysis followed by Coomassie blue staining of His-tagged human SLC19A1 (Ni-NTA affinity-purified) pull-downs with Sepharose beads coupled with 2’3’-cGAMP (+) or control Sepharose beads (−). Input is shown in the right panel. b, Western blots of the samples in a with anti-SLC19A1 antibody. c, Pull-downs of SLC19A1 competed with CDNs. His-tagged SLC19A1 was incubated with no CDN or with the indicated competing CDNs (250 μM) before pulldowns with 2’3’-cGAMP-Sepharose, followed by SDS-PAGE and Western blotting with an anti-SLC19A1 antibody. A pulldown with control Sepharose is shown for comparison. For gel source data, see Supplementary Figure 1. d, SDS-PAGE analysis followed by Coomassie blue staining of pull-downs of mSTING-C-Terminal Domain (CTD) with 2’3’-cGAMP (+) or control (−) Sepharose. In all panels, data are representative of two independent experiments with similar results.
Extended Data Figure 9. RNA-Seq data of STING and SLC19A1 mRNA expression in 934 human cancer cell lines available at the Cancer Cell Line Encyclopedia website.
Expression is presented as transcripts per kilobase million (TPM). Data is downloaded from the European Bioinformatics Institute Gene expression Atlas (URL: https://www.ebi.ac.uk/gxa/home). The data set included three of the cell lines we examined, as shown.
Extended Data Figure 10. The effect of SLC46A1 and SLC46A3 expression on CDN-induced reporter activation.
a-b, Enforced expression of SLC46A1 and SLC46A3 affects the responses of THP-1 cells to CDNs. Control THP-1 cells (transduced with empty expression vector) or SLC46A1-transduced THP-1 cells (a), or control THP-1 cells or SLC46A3-transduced cells (b) were stimulated with 2’3’-RR CDA (1.25 μg/ml), 2’3’-cGAMP (15 μg/ml) or hIFN-β (100 ng/ml). tdTomato reporter expression was measured by flow cytometry 18-22h after stimulation. c-d, SLC46A1 or SLC46A3 depletions had little or no effects on cellular responses to CDNs, and combining SLC46A1 or SLC46A3 depletions with SLC19A1 depletion, had no additional effect compared to SLC19A1-depletion by itself. THP-1 cells were transduced with non-targeting control CRISPRi gRNAs or SLC19A1-targeting CRISPRi gRNA in combination with a second control CRISPRi gRNA or SLC46A1-targeting CRISPRi gRNA in (c) or SLC46A3-targeting gRNA in (d). Cells were stimulated with 2’3’-RR CDA (1.67 μg/ml), 2’3’-cGAMP (10 μg/ml), or hIFN-β (100 ng/ml). tdTomato reporter expression was measured by flow cytometry 18-22h after stimulation. Combined data of three independent experiments. Statistical analysis was performed using unpaired two-tailed Student’s t tests (a-b), or one-way ANOVA followed by Tukey’s post-tests when comparing only the effects of depleting SLC46A1 (c) or SLC46A3 (d). Data are means ± SEM of n=3 independent replicates.
Supplementary Material
Acknowledgements
We thank Lily Zhang and Erik Seidel for lab and technical assistance, Hector Nolla and Alma Valeros for assistance with cell sorting, the UC Berkeley High Throughput Screening Facility for preparation of gRNA lentivirus, Adelle P. McFarland for assistance in the isolation of primary cells from mice, Shana L McDevitt for assistance with deep-sequencing, Jie An and Keith Elkon for assistance in the collection and isolation of primary peripheral blood leukocytes from healthy, human volunteers, and Raulet lab members, Russell Vance, Michel DuPage, Michiel van Gent, Jeremy Thorner and Andrea Van Elsas for helpful discussions. R.D.L. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. D.H.R. is supported by NIH grants R01-AI113041 and R01-CA093678. B.G.G. is supported by the IGI-AstraZeneca Postdoctoral Fellowship, J.J.W. is supported by the Pew Scholars Program in the Biomedical Sciences, the Lupus Research Alliance, and NIH grant 1R21AI137758–01. S.A.Z. is supported by grants from the University of Washington/Fred Hutchinson Cancer Research Center Viral Pathogenesis Training Program (AI083203), the University of Washington Medical Scientist Training Program (GM007266), as well as the Seattle ARCS foundation. J.E.C. is supported by the National Institute of Health New Innovator Awards (DP2 HL141006), the Li Ka Shing Foundation and the Heritage Medical Research Institute. This work employed the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10 OD018174, S10RR029668 and S10RR027303.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
Competing interests
D.H.R. is a co-founder of Dragonfly Therapeutics and served or serves on the scientific advisory boards of Dragonfly, Aduro Biotech, and Ignite Immunotherapy; he has a financial interest in all four companies and could benefit from commercialization of the results of this research. S.M.M. is, and G.E.K. was, an employee of Aduro Biotech.
Code availability
The CRISPRi screen sequences were analyzed using the Python-based ScreenProcessing pipeline. This custom code is available at: https://github.com/mhorlbeck/ScreenProcessing
Data availability
Raw sequencing data from the CRISPRi screens are available at NCBI GEO (GEO accession number GSE134371)
References
- 1.Ishii KJ et al. A toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7, 40–48 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Stetson DB & Medzhitov R Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Li T & Chen ZJ The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao P et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ablasser A et al. CGAS produces a 2′−5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diner EJ et al. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 3, 1355–1361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcus A et al. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 49, 754–763.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodward JJ, Lavarone AT & Portnoy DA C-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McWhirter SM et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206, 1899–1911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corrales L et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 11, 1018–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corrales L, McWhirter SM, Dubensky TW & Gajewski TF The host STING pathway at the interface of cancer and immunity. J. Clin. Invest. 126, 2404–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Z & Matherly LH Biology of the major facilitative folate transporters SLC19A1 and SLC46A1 Current Topics in Membranes 73, (Elsevier Inc., 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao R, Diop-Bove N, Visentin M & Goldman ID Mechanisms of Membrane Transport of Folates into Cells and Across Epithelia. Annual Review of Nutrition 31, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson GB & Zevely EM Structural requirements for anion substrates of the methotrexate transport system in L1210 cells. Arch. Biochem. Biophys. 221, 438–46 (1983). [DOI] [PubMed] [Google Scholar]
- 18.Goldman ID The characteristics of the membrane transport of amethopterin and the naturally occurring folates. Ann. N. Y. Acad. Sci. 186, 400–22 (1971). [DOI] [PubMed] [Google Scholar]
- 19.Yang CH, Sirotnak FM & Dembo M Interaction between anions and the reduced folate/methotrexate transport system in L1210 cell plasma membrane vesicles: directional symmetry and anion specificity for differential mobility of loaded and unloaded carrier. J. Membr. Biol. 79, 285–92 (1984). [DOI] [PubMed] [Google Scholar]
- 20.Goldman ID, Lichtenstein NS & Oliverio VT Carrier-mediated transport of the folic acid analogue, methotrexate, in the L1210 leukemia cell. J. Biol. Chem. 243, 5007–17 (1968). [PubMed] [Google Scholar]
- 21.Lin R, Heylbroeck C, Genin P, Pitha PM & Hiscott J Essential Role of Interferon Regulatory Factor 3 in Direct Activation of RANTES Chemokine Transcription. Mol. Cell. Biol. 19, 959–966 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brownell J et al. Direct, Interferon-Independent Activation of the CXCL10 Promoter by NF- B and Interferon Regulatory Factor 3 during Hepatitis C Virus Infection. J. Virol. 88, 1582–1590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen G et al. Sulfasalazine is a potent inhibitor of the reduced folate carrier: Implications for combination therapies with methotrexate in rheumatoid arthritis. Arthritis Rheum. 50, 2130–2139 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Kato K et al. Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nat. Commun. 9, 4424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman ID A model system for the study of heteroexchange diffusion: methotrexate-folate interactions in L1210 leukemia and Ehrlich ascites tumor cells. Biochim. Biophys. Acta 233, 624–34 (1971). [DOI] [PubMed] [Google Scholar]
- 26.Henderson GB, Grzelakowska-Sztabert B, Zevely EM & Huennekens FM Binding properties of the 5-methyltetrahydrofolate/methotrexate transport system in L1210 cells. Arch. Biochem. Biophys. 202, 144–9 (1980). [DOI] [PubMed] [Google Scholar]
- 27.Plosker GL & Croom KF Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs 65, 1825–49 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Kozuch PL & Hanauer SB Treatment of inflammatory bowel disease: A review of medical therapy. World J. Gastroenterol. 14, 354–377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajitha P, Biswas R, Sabitha M & Jayakumar R Methotrexate in the Treatment of Psoriasis and Rheumatoid Arthritis: Mechanistic Insights, Current Issues and Novel Delivery Approaches. Curr. Pharm. Des. 23, 3550–3566 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Ahn J, Son S, Oliveira SC & Barber GN STING-Dependent Signaling Underlies IL-10 Controlled Inflammatory Colitis. Cell Rep. 21, 3873–3884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Weijer ML et al. A high-coverage shRNA screen identifies TMEM129 as an E3 ligase involved in ER-associated protein degradation. Nat. Commun. 5, 3832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao R et al. Impact of the reduced folate carrier on the accumulation of active thiamin metabolites in murine leukemia cells. J. Biol. Chem. 276, 1114–8 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Zhao R, Gao F & Goldman ID Reduced folate carrier transports thiamine monophosphate: an alternative route for thiamine delivery into mammalian cells. Am. J. Physiol. Cell Physiol. 282, C1512–7 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Visentin M, Zhao R & Goldman ID Augmentation of reduced folate carrier-mediated folate/antifolate transport through an antiport mechanism with 5-aminoimidazole-4-carboxamide riboside monophosphate. Mol. Pharmacol. 82, 209–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson GB & Zevely EM Anion exchange mechanism for transport of methotrexate in L1210 cells. Biochem. Biophys. Res. Commun. 99, 163–9 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Hamblett KJ et al. SLC46A3 Is Required to Transport Catabolites of Noncleavable Antibody Maytansine Conjugates from the Lysosome to the Cytoplasm. Cancer Res. 75, 5329–40 (2015). [DOI] [PubMed] [Google Scholar]
- 37.van Diemen FR et al. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 12, e1005701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horlbeck MA et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, 1–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kampmann M, Bassik MC & Weissman JS Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat. Protoc. 9, 1825–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kampmann M, Bassik MC & Weissman JS Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc. Natl. Acad. Sci. 110, E2317–E2326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadlish H, Williams FMR & Flintoff WF Functional Role of Arginine 373 in Substrate Translocation by the Reduced Folate Carrier. J. Biol. Chem. 277, 42105–42112 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Huynh TN et al. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U. S. A. 112, E747–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sureka K et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158, 1389–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson GB & Zevely EM Affinity labeling of the 5-methyltetrahydrofolate/methotrexate transport protein of L1210 cells by treatment with an N-hydroxysuccinimide ester of [3H]methotrexate. J. Biol. Chem. 259, 4558–62 (1984). [PubMed] [Google Scholar]
- 46.McFarland AP et al. Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 46, 433–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.