Proteasomal-regulated chromatin remodeler mediates cocaine relapse during abstinence.
Abstract
Neuroadaptations in the nucleus accumbens (NAc) underlie cue-induced cocaine craving that intensifies (“incubates”) during abstinence and is believed to contribute to persistent relapse vulnerability. Changes in gene expression often govern perpetual behavioral abnormalities, but epigenetic plasticity during prolonged abstinence from drug exposure is poorly understood. We examined how E3 ubiquitin ligase TRIM3 dysregulates chromatin remodeler INO80 to mediate cocaine craving during prolonged abstinence. We found that INO80 expression increased in the NAc on abstinence day 30 (AD30) but not on AD1 following cocaine self-administration. Furthermore, TRIM3, which mediates degradation of INO80, was reduced on AD30, along with TRIM3-INO80 interaction. Viral-mediated gene transfer of INO80 or TRIM3 governed cocaine craving during prolonged abstinence. Lastly, chromatin immunoprecipitation followed by massively parallel DNA sequencing identified INO80-mediated transcriptional regulation of predicted pathways associated with cocaine plasticity. Together, these results demonstrate a novel ubiquitin-proteasomal-epigenetic mechanism by which TRIM3-INO80 mediates cocaine craving during prolonged abstinence.
INTRODUCTION
It is difficult for individuals with cocaine use disorder to achieve abstinence. However, the greatest challenge is avoiding relapse that is often precipitated by drug-associated cues even after long durations of abstinence. Both in humans with cocaine use disorder and in preclinical rodent relapse models, cue-evoked cocaine craving intensifies, or “incubates,” over a period of abstinence (1, 2). This progressive enhancement of cue saliency during drug abstinence is believed to increase propensity or susceptibility to relapse.
Incubated cocaine craving is mediated by maladaptive cellular plasticity in several mesolimbic brain regions, including the nucleus accumbens (NAc), a structure where many pathways involved in reward and motivation converge (3). Much of the work on incubated cocaine craving has focused on adaptations in synaptic transmission in the NAc (4). Enduring behavioral adaptations are often mediated by changes in gene expression, suggesting that epigenetic mechanisms play a presumptive role in persistent relapse vulnerability. Previous studies have reported abstinent-dependent changes in gene expression (5, 6) and DNA methylation (7) in the NAc, and intra-NAc injections of a DNA methyltransferase inhibitor abolished cue-induced cocaine seeking on AD30 (7). Beyond DNA methylation, epigenetic mechanisms that underlie incubated cocaine craving are unknown.
Gene expression is regulated, in part, by adenosine triphosphate (ATP)–dependent chromatin remodelers that govern histone composition of nucleosomes. Chromatin remodelers can be simple, involving as few as one protein, or can be more complicated multisubunit complexes. The inositol auxotroph 80 (INO80) complex is a multimeric chromatin remodeler from the SWI2/SNF2 (switch/sucrose nonfermentable) superfamily composed of 15 subunits including the adenosine triphosphatase (ATPase) subunit INO80 (8). INO80 regulates gene transcription by histone exchange (9, 10), nucleosome sliding (8), and nucleosome spacing (11, 12). However, the role of INO80-mediated gene expression in neuroadaptations following drug exposure is currently unknown.
There are numerous processes that regulate epigenetic mechanisms. The ubiquitin-proteasome system (UPS) is a multifaceted network of ubiquitin, enzymes, ligases, and proteasome complexes that is involved in proteolytic and nonproteolytic processes that affect genomic events. The UPS is involved in histone modification, DNA methylation, and proteolysis of proteins that alter gene expression (13–15). Through the proteolytic pathway, E3 ubiquitin ligases (E3) mediate polyubiquitination of substrates to be recognized by the proteasome complex for degradation (16), a process that is being actively pursued for its therapeutic potential (17). The UPS is involved in cocaine-associated plasticity in the NAc (18–20), but its role in mediating epigenetic plasticity and cocaine craving during prolonged abstinence has not been examined.
In the present study, we determined that INO80 protein expression in the NAc was increased during prolonged but not acute abstinence [abstinence day 30 (AD30) but not AD1]. Furthermore, viral-mediated manipulation of INO80 bidirectionally mediated the expression of cocaine craving on AD30. We then examined E3 ubiquitin ligase (E3) tripartite motif-containing protein 3 (TRIM3), of which INO80 is a substrate (21). We found that TRIM3 protein expression and interaction with INO80 are decreased on AD30, indicating that the ubiquitin-proteasomal regulation of INO80 is impaired during prolonged abstinence from cocaine. Furthermore, viral-mediated manipulation of TRIM3 also altered cocaine craving. We then performed chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) on AD30 to examine INO80 genomic enrichment patterns and found that INO80 binds to genes associated with cocaine-induced plasticity. These studies provide the first evidence that ubiquitin-proteasomal–regulated epigenetic plasticity mediates relapse vulnerability during prolonged abstinence.
RESULTS
Dysregulation of INO80 in the NAc during prolonged abstinence
We first trained adult male Sprague-Dawley rats to self-administer cocaine or saline under short-access (1 hour/day × 10 days) or extended-access (6 hours/day × 10 days) regimens (Fig. 1, A to C), which produce distinct patterns of drug intake (Fig. 1C) and cellular and behavioral plasticity during abstinence (22). On AD30, when incubated cocaine craving is maximally expressed following extended-access self-administration (fig. S1, A to C) (2), NAc tissues were collected (fig. S2A) and prepared for subcellular fractions (fig. S2B). We found that expression of the INO80 protein is increased on AD30 in the NAc nuclear (P1) fraction of rats following extended-access, but not short-access, self-administration (Fig. 1D). To examine whether these changes were dependent on a period of prolonged abstinence, we collected and fractionated NAc tissue on AD1 following extended-access self-administration (Fig. 1, A and B) but found no change in INO80 expression (Fig. 1D). These results demonstrate that INO80 is altered on AD30, but not on AD1, specifically after regimens with longer daily sessions of cocaine self-administration that are thought to model compulsive drug seeking in individuals with cocaine use disorder.
Fig. 1. INO80 in the NAc is dysregulated during prolonged abstinence following extended-access cocaine self-administration and mediates cocaine craving.
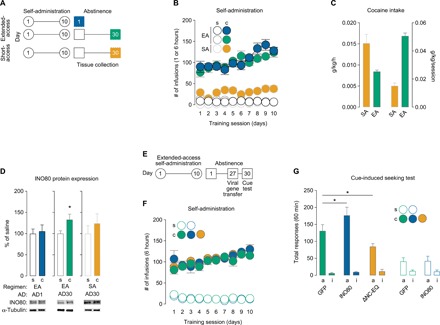
(A) Schematic of experimental timeline for short- and extended-access self-administration and tissue collection during abstinence. (B) Self-administration training behavior for extended-access [AD1: two-way repeated-measures analysis of variance (ANOVA), treatment × session: F9,190 = 4.724, P < 0.0001, n = 10 to 11 rats per group; AD30: two-way repeated-measures ANOVA, treatment × session: F9,170 = 2.734, P = 0.005, n = 9 to 10 rats per group] and short-access (two-way repeated-measures ANOVA, treatment: F1,90 = 198.8, P < 0.0001, n = 5 to 6 rats per group) paradigms. (C) Cocaine intake for short- and extended-access self-administration (g/kg per hour: t test, t14 = 4.059, P = 0.001; g/kg per session: t test, t14 = 10.87, P < 0.0001). (D) INO80 protein expression in NAc P1 fractions (AD1 after extended-access: t test, t10 = 0.296, P = 0.773; AD30 after extended-access: t test, t14 = 2.213, P = 0.044; AD30 after short-access: t test, t9 = 0.792, P = 0.449; n = 5 to 8 rats per group). (E) Schematic of experimental timeline for extended-access self-administration, viral-mediated gene transfer, and cue-induced seeking test. (F) Extended-access self-administration training behavior before viral-mediated gene transfer for cocaine- (two-way repeated-measures ANOVA, treatment: F2,26 = 0.274, P = 0.763, n = 9 to 10 rats per group) and saline-treated rats (F1,9 = 0.139, P = 0.718, n = 4 to 7 rats per group). (G) Total active and inactive responses during a 60-min cue-induced seeking test on AD30 after extended-access self-administration for cocaine-treated rats [one-way ANOVA, lever × treatment F2,26 = 7.361; Fisher protected least significant difference (PLSD) test for total active lever: GFP versus INO80, P = 0.018; GFP versus ΔNC-EQ, P = 0.018; n = 9 to 10 rats per group] and saline-treated rats (F1,9 = 0.025; Fisher PLSD test for total active lever: GFP versus INO80, P = 0.92; n = 4 to 7 rats per group). Data are means ± SEM. *P < 0.05. s, saline; c, cocaine; EA, extended-access self-administration; SA, short-access self-administration; P1, nuclear fraction; ΔNC-EQ, catalytically inactive mutant INO80; a, active lever; i, inactive lever.
INO80 in the NAc mediates cocaine craving during prolonged abstinence
Next, we asked if INO80 expression in NAc had a role in cocaine relapse behavior. To examine the behavioral role of INO80 in cocaine craving during prolonged abstinence (Fig. 1D), rats underwent self-administration training and were then pseudorandomly separated into groups based on training performance (Fig. 1E). Using viral-mediated gene transfer, we infused herpes simplex virus (HSV)–INO80 (fig. S3A) (23), HSV-INO80 ΔNC-EQ (mutant INO80 that lacks DNA-dependent ATPase activity) (fig. S3A) (23), or HSV-GFP (green fluorescent protein) directly into the NAc (fig. S3B) on AD27 to virally manipulate INO80 protein expression (fig. S3C). Rats were then returned to operant chambers for a craving test (20) on AD30 when viruses were maximally expressed (24). Viral-mediated expression of INO80 enhanced total active responses, whereas HSV-INO80 ΔNC-EQ reduced the number of total active responses compared to HSV-GFP controls in cocaine-treated rats (Fig. 1F). Viral-mediated expression of INO80 did not affect responses in saline-treated rats (Fig. 1F), and importantly, 30 days after food self-administration (fig. S4, A and B), neither HSV-INO80 nor HSV-INO80 ΔNC-EQ affected cue-induced food seeking (fig. S4C) or locomotor activity (fig. S4D).
Proteasomal degradation of INO80 in the NAc is decreased during prolonged abstinence
Next, we sought to determine the cellular mechanism by which INO80 is regulated during prolonged abstinence. The UPS targets proteins for proteolysis through the conjugation of ubiquitin to substrates, which is coordinated by E3s (16). INO80 was previously identified as a substrate of TRIM3, an E3 that has been implicated in hippocampal synaptic plasticity and memory (21). Following extended-access cocaine self-administration, we found a decrease in TRIM3 protein expression in NAc P1 fractions on AD30 (Fig. 2A). In contrast, we found no change in TRIM3 in NAc P1 on AD30 following short-access self-administration or on AD1 following extended-access self-administration (Fig. 2A). We found that TRIM3 protein expression in NAc crude synaptosomal (P2) fractions was significantly decreased on AD1 but unchanged on AD30 following extended-access cocaine self-administration (fig. S6A). Correspondingly, TRIM3 synaptic substrate γ-actin (21), but not Shank or GKAP (25), was increased in P2 fractions on AD1 but unchanged on AD30 in rats that self-administered cocaine compared to saline controls (fig. S6B). These results suggest that subcellular expression of TRIM3 is decreased in P2 fractions on AD1 but decreased in P1 fractions on AD30 following cocaine exposure (see Discussion).
Fig. 2. TRIM3 regulates the expression of INO80 during prolonged abstinence and mediates cocaine craving.
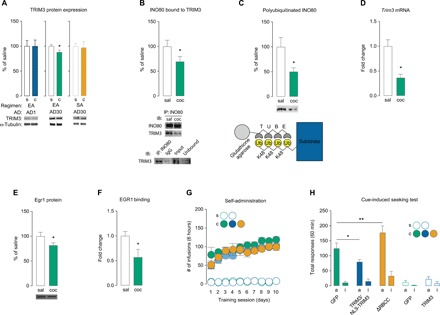
(A) TRIM3 protein expression in NAc P1 fractions (extended-access AD1: t test, t16 = 0.009, P = 0.992; extended-access AD30: t test, t13 = 2.479, P = 0.028; short-access AD30: t test, t10 = 0.177, P = 0.863; n = 5 to 9 rats per group). (B) Co-immunoprecipitation of INO80 with TRIM3 on AD30 (t test, t13 = 1.878, P = 0.042; n = 7 to 8 rats per group). (C) Polyubiquitinated levels of INO80 on AD30 measured by K48-TUBEs (t test, t18 = 2.451, P = 0.012; n = 10 rats per group) and schematic of anti–K48-TUBEs. (D) Trim3 mRNA expression in NAc on AD30 (t test, t8 = 4.245, P = 0.003; n = 5 rats per group). (E) EGR1 protein expression in NAc P1 fractions on AD30 (t test, t15 = 1.915, P < 0.05; n = 8 to 9 rats per group). (F) EGR1 binding along the TRIM3 promoter on AD30 (t test, t14 = 2.365, P < 0.05; n = 3 samples per group, 7 rats per sample). (G) Self-administration training behavior before viral-mediated gene transfer (two-way repeated-measures ANOVA, treatment group: F1,70 = 0.849, P = 0.849, n = 6 to 11 rats per group; saline treatment: F1,70 = 0.036, P = 0.849, n = 5 rats per group). (H) Total active and inactive responses during a 60-min cue-induced seeking test on AD30 for rats that self-administered cocaine (one-way ANOVA, lever × treatment F2,44 = 5.588; Fisher PLSD test for total active lever: GFP versus TRIM3/NLS-TRIM3, P = 0.034; GFP versus ΔRBCC, P = 0.003; n = 6 to 11 rats per group) and saline (F1,16 = 0.168; Fisher PLSD test for total active lever: GFP versus TRIM3, P = 0.227; n = 4 to 7 rats per group). Data are means ± SEM. *P < 0.05 and **P < 0.01. K48, lysine-48; NLS, nuclear localizing signal; ΔRBCC, catalytically inactive mutant TRIM3; IP, immunoprecipitation; IB, immunoblotting; IgG, immunoglobulin G.
To more closely examine the relationship of INO80 with TRIM3 on AD30, we conducted co-immunoprecipitation (Co-IP) in NAc tissue, which revealed decreased interaction of INO80 with TRIM3 in rats that self-administered cocaine compared to saline controls on AD30 (Fig. 2B). Furthermore, we used tandem ubiquitin binding entities (TUBEs) (26) to quantify polyubiquitinated (lysine [K]-48–specific; Fig. 2C) INO80 bound for proteasomal degradation and found a significant decrease in polyubiquitinated IN080 on AD30 in rats that self-administered cocaine compared to saline controls (Fig. 2C). These findings support the idea that TRIM3-mediated proteasomal degradation of INO80 is reduced during prolonged abstinence.
Next, we sought to examine how TRIM3 may be regulated. In addition to a decrease in TRIM3 protein expression, we used quantitative real-time polymerase chain reaction (qPCR) and found that Trim3 mRNA is reduced in cocaine-treated rats compared to saline controls (Fig. 2D and table S1), suggesting that TRIM3 expression may be mediated at the transcriptional level. Immediate early gene early growth response 1 (EGR1) is predicted to regulate transcription of Trim3 based on identified binding sites for EGR1 within the Trim3 promoter region. EGR1 is required for long-lasting, cocaine-induced behavioral alterations (27), and a previous study reported that Egr1 mRNA is reduced in the NAc during prolonged abstinence when incubation is observed following discrete-trial self-administration (5). In accordance, we found a reduction in EGR1 protein in NAc P1 fractions on AD30 in rats that self-administered cocaine compared to saline controls (Fig. 2E). Using quantitative ChIP, we found that EGR1 binding at an EGR1 site along the Trim3 promoter (table S2) was decreased on AD30 in rats that self-administered cocaine compared to saline controls (Fig. 2F), indicating that EGR1 may regulate Trim3 transcription during prolonged abstinence.
E3 TRIM3 in the NAc mediates cocaine craving during prolonged abstinence
We then aimed to determine the behavioral role of TRIM3 (Fig. 1D). Rats were trained to self-administer cocaine or saline (Fig. 2G) and then received intra-NAc injections (fig. S3B) of viral-mediated gene transfer of HSV-TRIM3 (fig. S5, A to C), HSV–nuclear localizing signal (NLS)–TRIM3 (because of the differences in subcellular TRIM3 expression during abstinence; fig. S5, A, D, and E), or HSV-ΔRBCC-TRIM3 (mutant TRIM3 that lacks the RING/B-box/coiled-coil domain that contains ubiquitin ligase activity) (fig. S5, A and F) (21). On AD30, we conducted a cue-induced craving test. We found that HSV-TRIM3 or HSV-NLS-TRIM3 similarly attenuated cocaine craving (fig. S7), so the groups were combined (Fig. 2G). In contrast, HSV-ΔRBCC-TRIM3 enhanced cocaine craving (Fig. 2G). Viral-mediated manipulation of TRIM3 did not affect cue-induced seeking in saline-treated rats (Fig. 2G), cue-induced food seeking (fig. S4C), or locomotor activity (fig. S4D).
Genomic INO80 enrichment patterns in the NAc during prolonged withdrawal
INO80 is a critical transcription mediator, so we next examined the genomic enrichment patterns of INO80 in the NAc during prolonged abstinence. We performed ChIP-seq on AD30 following extended-access self-administration. Analysis of genomic distributions of INO80 enrichment events in rats that self-administered cocaine revealed that the largest percentage of events unique to cocaine exposure occurred within the gene bodies, followed by promoter regions (Fig. 3A and data S1). Subsequent gene enrichment analysis (KEGG 2019) identified pathways associated with neurotrophin signaling, alcoholism, dopaminergic synapse function, amphetamine addiction, and long-term potentiation (Fig. 3B and data S1). Genome browser tracks were created for a number of cocaine-mediated INO80 downstream target genes that demonstrated (i) significant differential enrichment for INO80 comparing cocaine versus saline groups and (ii) significant peaks in both saline- and cocaine-treated animals (see Materials and Methods). These genes included transcription factor Ying-Yang-1 (Yy1), histone demethylase Jumonji domain-containing protein 6 (Jmjd6), and pre-mRNA splicing factor DExD-box helicase 39B (Ddx39b; see Discussion and Fig. 3C). INO80 enrichment was increased for Yy1, Jmjd6, and Ddx39b (Fig. 3C), which we then validated via qPCR (Fig. 3D and table S1). Viral-mediated manipulation of INO80 expression altered these genes (Fig. 3D).
Fig. 3. INO80 genomic enrichment during prolonged abstinence.
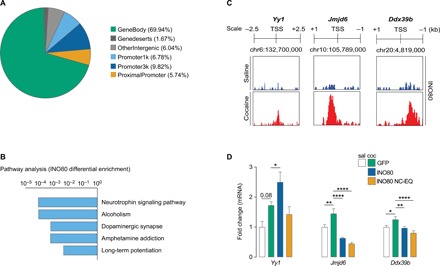
(A) Genomic distribution of INO80 differential enrichment events in NAc tissue from rats that self-administered cocaine. (B) KEGG 2019 pathway enrichment analysis of INO80 differential enrichment events in rats that self-administered cocaine. (C) Genome browser track sections for selected genes that displayed significant differential enrichment for INO80 comparing rats that self-administered cocaine versus saline. (D) qPCR analysis of Yy1 (one-way ANOVA, treatment: F3,28 = 5.962, P = 0.003; Fisher PLSD test: saline GFP versus cocaine GFP, P = 0.08; cocaine GFP versus cocaine INO80, P = 0.037), Jmjd6 (F3,28 = 24.31, P < 0.0001; saline GFP versus cocaine GFP, P = 0.004; cocaine GFP versus cocaine INO80, P < 0.0001; cocaine GFP versus cocaine ΔNC-EQ, P < 0.0001), and Ddx39b (F3,29 = 7.351, P = 0.0008; saline GFP versus cocaine GFP, P = 0.025; cocaine GFP versus cocaine INO80, P = 0.006; cocaine GFP versus cocaine ΔNC-EQ, P < 0.0001) in the NAc 24 hours after cue-induced seeking test on AD30 (n = 6 to 10 rats per group). Data are means ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001.
DISCUSSION
Here, we have demonstrated the essential role of a UPS-regulated chromatin remodeler in cocaine craving during prolonged abstinence. Following extended-access cocaine self-administration, INO80 expression was increased in the NAc on AD30 when incubated cocaine craving is expressed (2), which we found may be governed by a reduction in TRIM3-mediated proteasomal degradation. Viral-mediated gene expression of INO80 or TRIM3 bidirectionally regulated cocaine craving on AD30. Examination of genomic enrichment of INO80 in the NAc revealed INO80 binding along genes involved in cocaine-associated plasticity in cocaine-treated rats. Together, this study provides the first evidence of a concerted action of a proteasomal-mediated epigenetic mechanism in persistent relapse vulnerability.
Short- and extended-access cocaine self-administration procedures lead to different cellular and behavioral plasticity. Extended-access cocaine self-administration has been observed to produce escalation of cocaine intake, enhanced motivation for cocaine, increased cocaine seeking despite adverse consequences, and distinct neuroadaptations (4, 22), suggesting that extended-access procedures more closely model compulsive drug-seeking and drug-taking characteristics of substance use disorder. Here, we observed that rats self-administered more cocaine per hour in the short-access regimen, but rats took significantly more cocaine per session in the extended-access regimen. Thus, divergent neuroadaptations in short- and extended-access paradigms that occur during abstinence can be attributed to a greater duration and/or total cocaine intake. However, a previous study found that accumulation of calcium-permeable AMPA receptors in the NAc, which mediate expression of incubated cocaine craving (28), depends on drug session duration rather than the total amount of cocaine intake (or the total number of sessions) (29). Together, these results indicate that longer durations of cocaine exposure result in abstinence-associated differences in chromatin architecture in the NAc, via an INO80-dependent mechanism, that mediates incubated cocaine craving.
Cocaine use disorder involves potentially lifelong behavior abnormalities. The persistence of these behavioral changes suggests that long-lasting alterations in gene expression may contribute to the disease phenotype. Numerous studies have examined epigenetic mechanisms, including chromatin modifications, that regulate cocaine-induced neural and behavioral plasticity (30), but only a small number of studies have focused specifically on chromatin remodeling complexes and their associated subunits (31–34). These studies in the NAc found that BAZ1A and BAZ1B, accessory subunits of the Imitation Switch (ISWI) family of chromatin remodeling complexes, alter behavioral responses to cocaine (32, 34), and BRG1, a core subunit of ATP-dependent SWI/SNF complexes, mediates cocaine seeking on AD7 following short-access cocaine self-administration (33). ISWI and SWI/SNF, along with INO80 and other subfamilies, comprise the SWI2/SNF2 ATPase superfamily that are all related by the SWI2/SNF2 ATPase at their catalytic core. Despite the conserved motor domain, SWI2/SNF2 enzymes display structural versatility and execute a broad spectrum of remodeling reactions (35), but these diverse complexes have not been examined in cocaine-induced neuroadaptations during prolonged abstinence.
Dynamic changes in gene expression have been reported in the NAc and other regions of the mesolimbic dopaminergic system during abstinence following cocaine self-administration (5, 6), but, unexpectedly, few studies have examined the role of epigenetic mechanisms in neuroadaptations and relapse behaviors during prolonged abstinence. One study reported abstinent-dependent alterations in NAc DNA methylation, of which pharmacological manipulation bidirectionally affected cocaine seeking on AD30 and persisted for 1 month (7). More recently, short hairpin RNA (shRNA)–mediated knockdown of DNA methyltransferase 3a (Dnmt3a2) in the NAc during acute abstinence attenuated the development of incubation (36). While these studies provide evidence of epigenetic mechanisms in incubated cocaine craving, the role of chromatin remodelers has not been examined. In the present study, we found that expression of INO80, the ATP subunit of the INO80 complex, was increased in the NAc on AD30 but not on AD1. Furthermore, viral-mediated expression of INO80 increased, while ΔNC-EQ decreased, cocaine seeking on AD30. These findings suggest that there are genes that are expressed or silenced by INO80 that mediate cocaine craving during prolonged abstinence following extended-access cocaine self-administration, and disruption of this gene expression pattern via ΔNC-EQ attenuates cocaine craving (see Discussion below on INO80 targets).
The UPS is involved in a variety of cocaine-associated adaptations in the NAc (18–20), and we previously demonstrated that E3 SMURF1 in the NAc mediates cocaine craving following short-access cocaine self-administration (20). Recently, INO80 was identified as a substrate of E3 TRIM3 (21), so we therefore investigated the relationship of TRIM3 and INO80 during prolonged abstinence. We found that TRIM3 protein was decreased on AD30 in cocaine-treated rats and that both the TRIM3 interaction with INO80 and polyubiquitinated INO80 were also decreased. Moreover, viral-mediated expression of TRIM3 or nuclear-localized TRIM3 decreased cocaine craving, while ΔRBCC, a catalytically inactive TRIM3 mutant that lacks ubiquitin ligase ability (21), increased cocaine craving. These results suggest that the abstinent-associated increase in INO80 expression may be due to a reduction in TRIM3-dependent proteolysis and that manipulation of this interaction regulates expression of cocaine craving. TRIM3 protein expression was decreased, and TRIM3 substrate γ-actin (21) was increased, on AD1 in the P1 fraction. γ-Actin is involved in cytoskeleton maintenance and stability (37), and increases in spine density in the NAc have been reported within 1 day following cocaine exposure (although spine density has not been specifically examined on AD1 after extended-access cocaine self-administration) (38). These results demonstrate that TRIM3 may mediate synaptic plasticity through γ-actin during early abstinence and epigenetic plasticity through INO80 during prolonged abstinence.
Alterations in the immediate early gene Egr1 during abstinence from various cocaine administration procedures have been well characterized (5, 39). EGR1 is predicted to regulate transcription of Trim3 based on identified EGR1 binding sites, and we found that Trim3 mRNA is decreased in cocaine-treated rats on AD30. Following a discrete-trial cocaine self-administration model that produces incubation of cocaine craving, a persistent decrease in NAc Egr1 gene expression in the NAc was reported after 1, 10, and 100 days of abstinence (5). In accordance, we found that EGR1 protein expression was decreased in the NAc on AD30, and moreover, EGR1 binding was decreased at an EGR1 site along the Trim3 promoter. These findings suggest that the reduction in TRIM3 expression during prolonged abstinence may be controlled by EGR1-mediated transcription of Trim3.
We then performed ChIP-seq and gene enrichment analysis for INO80 genomic binding patterns during prolonged abstinence, which identified numerous pathways associated with cocaine-induced plasticity including neurotrophin signaling (40) and long-term potentiation (41). We then attempted to identify legitimate cocaine-mediated INO80 target genes by using a very conservative approach in which we overlapped protein coding genes that displayed both significant differential (increased or decreased) enrichment in cocaine-treated rats compared to saline controls and significant peaks in both saline- and cocaine-treated animals in promoter regions. These strict criteria were used to attempt to reduce false positives. We validated three high-confidence INO80 targets (Yy1, Jmjd6, and Ddx39b) that each have unique functions. YY1 is a transcription factor that associates with INO80 to gain access to target promoters and initiate transcription (42), suggesting that an INO80-YY1 positive feedback loop may be activated during prolonged abstinence. JMJD6 is a histone demethylase that can demethylate histone H3 (43) and regulates nuclear factor κB (NF-κB) (44), both of which have been implicated in cocaine-associated plasticity (45, 46). DDX39B is an RNA helicase that promotes translation through regulation of preribosomal RNA levels (47). mRNA expression of these genes matched INO80 binding patterns, and viral manipulation of INO80 affected these targets. Together, these findings suggest that INO80 governs cocaine-induced neuroadaptations through multiple pathways.
Together, this study demonstrates a novel role for a chromatin remodeler, INO80, and E3 Trim3 in cocaine craving during prolonged abstinence (see schematic in fig. S8). Future studies will aim to better understand INO80 downstream genes that mediate cocaine craving during prolonged abstinence, which will provide further insight into the transcriptional plasticity that maintains persistent relapse vulnerability. Our findings also warrant a more comprehensive examination of the UPS and its regulation of addiction-related synaptic and transcriptional plasticity. This study lays the foundation for future work on these molecular mechanisms, which hold a great deal of therapeutic potential for the treatment of cocaine use disorder.
MATERIALS AND METHODS
Subjects
Adult male Sprague-Dawley rats (250 to 275 g at the beginning of experiments; Envigo) were allowed to habituate to the colony room for 3 days upon arrival, which was held at a constant temperature and humidity level. Rats were singly housed following surgery on a reverse light/dark cycle and had ad libitum access to food and water. Behavioral testing took place during the dark phase of the 12-hour light/12-hour dark cycle. This study was conducted in accordance with the guidelines set up by the Institutional Animal Care and Use Committee of The State University of New York at Buffalo.
Drugs
Cocaine hydrochloride [a gift from the National Institute on Drug Abuse (NIDA)] was dissolved in sterile 0.9% saline. Cocaine solutions (4.5 mg/ml) were prepared on a weekly basis. Cocaine was delivered via a syringe pump, and injection volumes were adjusted according to body weight on a daily basis (48).
Jugular catheterization and patency testing
Rats were implanted with chronic indwelling jugular catheters, as has been previously described (49). Briefly, rats were anesthetized using ketamine and xylazine (60 and 5 mg/kg, intraperitoneally, respectively), the right external jugular vein was isolated, and the catheter was inserted. The other end of the catheter was fitted to a Vascular Access harness (Instech). Rats were allowed 3 to 5 days to recover following surgery, and catheters were flushed daily with 0.2-ml solution of enrofloxacin (4 mg/ml) in heparinized saline (50 IU/ml in 0.9% sterile saline) to preserve catheter patency. One day before acquisition training, each animal received an intravenous infusion of ketamine hydrochloride (0.5 mg/kg in 0.05 ml), and loss of muscle tone and righting reflex was confirmed to verify catheter patency. Only rats with patent catheters were used in behavioral and biochemical studies.
Acquisition of self-administration
Following recovery from jugular catheter surgery, rats were randomly assigned to train to self-administer cocaine (0.5 mg/kg per infusion) or saline as previously described (50). Briefly, rats were trained for 1 hour/day × 5 days in operant chambers (Med Associates) inside sound-attenuating cabinets that were equipped with two nose-poke holes. Responses in the active hole resulted in an infusion using a fixed ratio 1 (FR1) schedule of reinforcement, which was increased daily to an FR5 schedule. Infusions were accompanied by a 5-s illumination of the stimulus light above the active hole. The house light was extinguished for the duration of the infusion and 30-s timeout period. Responses in the inactive hole resulted in no programmed consequences. Following each session, catheters were flushed with heparinized saline and rats were returned to the colony room. The criterion for acquisition of cocaine self-administration was an average of eight infusions per day during the last three acquisition sessions.
Self-administration training
Following self-administration acquisition, rats were allowed to self-administer cocaine (0.5 mg/kg per infusion) or saline under an extended-access (6 hours/day) or short-access (1 hour/day) regimen using an FR5 schedule of reinforcement. Following training sessions, catheters were flushed and rats were returned to the colony room. Only rats that reached acquisition criterion and responded for cocaine (an average of >50 infusions per session) were used in successive phases of experiments.
Tissue collection and subcellular fractionation
Tissue was collected 1 or 30 days after self-administration training. Following rapid decapitation, brains were removed and sliced into 1-mm-thick sections using a brain matrix, and 2-mm tissue punches targeted at the NAc were collected, rapidly frozen on dry ice for immunoblotting and RNA isolation, or post-fixed for ChIP experiments.
Subcellular fractionation procedures were slightly modified from previous reports (19, 51). Briefly, samples were homogenized in Hepes-buffered sucrose [4 mM Hepes and 0.32 M sucrose (pH 7.4)] and centrifuged (1000 g, 15 min, 4 °C). The resulting pellet [nuclear fraction (P1)] was resuspended in radioimmunoprecipitation assay (RIPA) buffer [10 mM tris base, 150 mM NaCl, 1 mM EDTA, 1% SDS, 1% (v/v) Triton X-100, 24 mM Na deoxycholate, and X phosphatase inhibitor cocktail 2 (Sigma); X phosphatase inhibitor cocktail 3 (Sigma); and X proteasome inhibitor tablet (Roche) (pH 7.4)], and the supernatant (S1) was centrifuged (10,000 g, 30 min, 4 °C). The resulting pellet [crude synaptosomal fraction (P2)] was resuspended in RIPA buffer.
Immunoblotting
Protein concentrations were determined, and a total of 10 μg of protein was loaded onto 10% tris/SDS-polyacrylamide gels for electrophoresis separation and then transferred to nitrocellulose membranes, blocked with Rockland Blocking Buffer (VWR International), and incubated overnight at 4 °C with primary antibodies diluted in Rockland Blocking Buffer: anti-INO80 (1:1000; Bethyl Laboratories Inc., A303-371A), anti-TRIM3 (1:100; Santa Cruz Biotechnology, sc-136363), anti-EGR1 (1:100; Santa Cruz Biotechnology, sc-515830), anti–γ-actin (1:1000; Abcam, ab123034), anti–pan-SHANK [1:500; University California (UC) Davis/National Institutes of Health (NIH) NeuroMab Facility, 73–089], anti-GKAP/pan-SAPAP (1:500; UC Davis/NIH NeuroMab Facility, 73–156), and anti–α-tubulin (1:10,000; Cell Signaling Technology, 3837). After washing with tris-buffered saline (TBS) containing 0.1% Tween 20 (TBS-T), membranes were incubated with IRDye secondary antibodies (1:5000; LI-COR) in Rockland Blocking Buffer for 1 hour at room temperature. The blots were imaged using the Odyssey Infrared Imaging System (LI-COR) and quantified by densitometry using ImageJ (NIH). α-Tubulin was used as a protein loading control.
Viral-mediated gene transfer
The following plasmids were used to make HSV vectors: pCMV-3XFLAG-hINO80 (23) (INO80) was a gift from J. Conaway (Addgene; plasmid no. 44149); pcDNA5RT/FLAG-INO80 ΔNC-EQ (amino acids 267 to 1261 E653Q; ΔNC-EQ), a catalytically inactive INO80 ATPase (23), was a gift from J. Conaway and R. Conaway (Addgene; plasmid no. 29437); TRIM3 (TRIM3; OriGene; RC211739); NLS-TRIM3 (TRIM3; OriGene; RC211739) with an H2B tag on the 5′ end to restrict NLS-TRIM3 to the nucleus; and pcDNA-myc-TRIM3-ΔRBCC (amino acids 283 to 744; ΔRBCC), a catalytically inactive TRIM3 lacking the RING/B-box/coiled-coil domain (21), was a gift from R. E. Van Kesteren [Vrije Universiteit Amsterdam]. HSV vectors were generated in p1005 transcription cassettes expressing GFP driven by a cytomegalovirus promoter to allow neuronal visualization. These HSV vectors exhibit maximal expression 3 to 5 days after injection (24). All viral tools were validated both in vitro and in vivo before use in behavioral experiments.
Cue-induced seeking test
Rats were returned to operant chambers on AD30 to measure cue-induced seeking. Tests were performed under extinction conditions (i.e., responding in the active hole delivered the infusion-paired light cue but no infusion) as previously described (20). The total number of responses in the active hole was the operational measure of seeking from which the affective state of craving was inferred.
Food self-administration and test
Food self-administration was conducted as previously described with minor modifications (52). Briefly, rats were singly housed on a light/dark cycle and were food-restricted to 10% of body weight. During the dark phase of the 12-hour light/12-hour dark cycle, rats underwent training for obtaining a food reward (45 mg of pellet; Bio-Serv) following lever press responses in two-lever operant chambers housed within sound-attenuating ventilated boxes (Coulbourn Instruments LLC). Each session consisted of daily 1-hour time period during which rats could press either the active or inactive lever under an FR1 schedule to earn a maximum of 50 food pellets. The response requirement was progressively increased to FR5 over a 10-day period. Following food training, rats were subjected to a forced abstinence period in home cages with ad libitum access to food. Twenty-seven days after training, rats were pseudorandomly assigned to receive bilateral intra-NAc injections of HSV-INO80, HSV-INO80 ΔNC-EQ, HSV-TRIM3, HSV-ΔRBCC-TRIM3, or HSV-GFP. Three days later (30 days after training), when viruses were maximally expressed, rats were then tested for cue-induced food seeking under extinction conditions.
Locomotor activity
Locomotor activity was conducted as previously described (20, 33). Briefly, transparent plastic cages fitted with infrared motion sensor system (AccuScan Instruments Inc.) were used to monitor distance traveled using VersaMax animal activity software (Omnitech Electronics Inc.). Locomotor activity was recorded during a 1-hour test during the overexpression of HSV-INO80, HSV-INO80 ΔNC-EQ, HSV-TRIM3, HSV-ΔRBCC-TRIM3, or HSV-GFP.
Co-immunoprecipitation
Co-IP was performed as previously described with slight modifications (28, 53). Briefly, NAc tissue punches from rats were homogenized in RIPA buffer. Antibody-bead complex was prepared by incubating anti-INO80 antibody (Bethyl Laboratories) to Protein G Sepharose 4 Fast Flow antibody purification resin (GE Healthcare) end-over-end at 4 °C for 4 hours. Whole-cell NAc protein (100 μg) was added to antibody-bead complex and incubated end-over-end overnight at 4 °C with rocking. Samples were then centrifuged, and the pellet obtained was washed three times with wash buffer (1× TBS and 0.1% Triton X-100). Following the final wash, the pelleted beads were suspended in sample buffer containing SDS and 2-mercaptoethanol (BME), heated to 70 °C for 10 min, and subjected to SDS–polyacrylamide gel electrophoresis. Proteins transferred to nitrocellulose membrane were then probed for anti-INO80 and anti-TRIM3.
TUBE affinity purification
TUBE affinity purification was performed as previously described (21) with minor modifications. Briefly, 200 μl of nickel-charged nitrilotriacetic acid (Ni-NTA) resin was added to 400 μl of equilibrium buffer (20 mM Na3PO4, 30 mM NaCl, and 10 mM imidazole). Anti–K48-TUBEs (15 μl; LifeSensors) and equilibrium buffer (385 μl) were added and rotated end-over-end for 4 hours at 4 °C. Anti–K48-TUBE slurry (10 μl) was removed, washed with extraction buffer, and centrifuged three times. Whole-cell NAc protein (100 μg) was added to anti–K48-TUBE slurry. Purification of bound proteins was performed using extraction buffer [2% NP-40 in 25 mM Hepes and 150 mM NaCl, (pH 7.2), 10 mM N-ethylmaleimide, 20 μM MG-132 (Tocris), and X protease inhibitor (Roche)]. Purified proteins were washed three times in extraction buffer, once in a high-salt buffer (25 mM Hepes and 500 mM NaCl), and once in a buffer without salt (25 mM Hepes). Proteins were then eluted in 30 μl of buffer containing 5 mM Hepes and analyzed using immunoblotting.
RNA extraction, isolation, and qPCR
NAc tissue punches were collected 24 hours after a cue-induced seeking test on AD30. Total RNA was extracted using TRIzol reagent (Ambion) and isolated using the E.Z.N.A. MicroElute Total RNA Kit (Omega Bio-Tek). RNA concentration was measured on a NanoDrop One spectrophotometer (Thermo Scientific) and then reverse-transcribed using SuperScript III First-Strand Synthesis System (Invitrogen). mRNA expression was measured using qPCR with IQ SYBR Green SuperMix (Bio-Rad Laboratories), and quantification of mRNA was conducted using an iQ5 system (Bio-Rad Laboratories). Reactions were run in triplicate and threshold cycles (Ct) were analyzed by a relative quantification method (ΔΔCt) using Gapdh as a housekeeping gene, as previously described. See table S1 for mRNA primer sequences.
Chromatin immunoprecipitation
Frozen rat NAc tissue punches were pooled (three samples per, and seven rats per sample), cross-linked, and quenched. Samples were washed before being subjected to lysis and sonication, as described (33, 54) with minor modification. After IP, samples were washed once with a low-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA (pH 8), 150 mM NaCl, and 20 mM tris-HCl (pH 8)], once with a high-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA (pH 8), 500 mM NaCl, and 20 mM tris-HCl (pH 8)], and finally once with a LiCl buffer [150 mM LiCl, 1% NP-40, 1% NaDOC, 1 mM EDTA (pH 8), and 10 mM tris-HCl (pH 8)] before tris-EDTA/NaCl washes and elution. Samples were then incubated with an INO80-specific antibody (7.5 μg per sample; Bethyl Laboratories) bound to M-280 Dynabeads on a rotator at 4 °C overnight. The next day, immunoprecipitates were washed, eluted, and reverse cross-linked. After RNA and protein digestions, DNA fragments were purified using a Qiagen PCR purification kit.
Library preparation
Following DNA purifications, ChIP-seq libraries were prepared according to Illumina protocols and sequenced with an Illumina HiSeq2500 Sequencing High Output mode v4.
ChIP-seq peak calling, differential analysis, and statistical analysis
Raw sequencing reads were aligned to the rat genome (rn6) using default settings of HISAT v0.1.7. Quality control was assessed using MultiQC/FastQC, and only uniquely mapped reads were kept. Alignments were filtered using SAMtools v0.1.19 to remove duplicates. Peak calling was performed using MACSv2.1.1 with default settings and filtered for P < 0.001; the window size was set as 300 base pairs. DNA inputs were used in all peak calling experiments to assess enrichment. Differential analysis of INO80 was performed using diffReps with a window size of 1 kb. A default P value cutoff of 0.001 was used with no fold change cutoffs. Peaks and differential sites were further annotated to nearby genes or intergenic regions using the region analysis tool from the diffReps package. To identify bona fide cocaine-mediated INO80 target genes for downstream validations, we used a highly conservative approach in which we overlapped those protein coding genes identified by MACS2 using replicates with MACS2 peak genes identified within each replicate to select those that displayed peaks in at least two-thirds of the samples per group (i.e., to eliminate sample biases). We then overlapped those genes with protein coding genes identified by diffReps to further examine target genes that displayed significant differential enrichment for INO80 comparing cocaine versus saline groups. While many additional target genes of INO80 are likely excluded using these strict criteria, our approach identified numerous high-confidence cocaine-regulated INO80 targets (112 genes), some of which we have further validated in our viral manipulation studies.
Quantitative ChIP
Chromatin flow through from the INO80 ChIP-seq experiment described above was incubated with an Egr1-specific antibody (7.5 μg per sample; Cell Signaling Technology, 4153) bound to M-280 Dynabeads on a rotator at 4 °C overnight. The next day, immunoprecipitates were washed, eluted, and reverse cross-linked. After RNA and protein digestions, DNA fragments were purified using a Qiagen PCR purification kit. Following DNA purification, levels of Egr1 binding at an Egr1 binding site of Trim3 promoter were quantified through qPCR (iQ5 system, Bio-Rad Laboratories) using custom-made primers for amplifying promoter regions (table S2). Input and immunoprecipitated DNA amplifications were run in duplicates, and fold changes were determined as cocaine relative to saline controls.
Statistical analysis
Sample sizes were similar to those reported in previous publications (20, 33). All statistical analyses were performed using Prism (GraphPad Software). Self-administration training was analyzed using two-way repeated-measures within-subject analyses of variance (ANOVAs). Immunoblotting, co-IP, TUBE affinity purification, and qPCR studies were analyzed using Student’s t tests. Cue-induced seeking behavior, locomotor activity, and ChIP studies were analyzed using one-way ANOVA, followed by planned comparisons. Tests were two-tailed except where directional hypotheses were deduced. The criterion for statistical significance was set at P < 0.05. All data are presented as means ± SEM.
Supplementary Material
Acknowledgments
We thank R. E. Van Kesteren (VU Amsterdam) for gifting the pcDNA-myc-TRIM3-ΔRBCC plasmid and the NIDA Drug Supply Program for providing the cocaine used in these studies. Funding: This work was supported by NIDA (R01DA037257, S1-R01DA037257, and R21DA044486 to D.M.D.; DP1DA042078 and R21DA044767 to I.M.; and F31DA045428 to A.E.L.), NIMH (R01MH116900 to I.M.), NIGMS (R25GM09545902 to University at Buffalo), and NINDS (F99NS108543 to J.A.M.). P.H.G. was supported by FAPESP 2017/19284-0. Author contributions: C.T.W., S.M., A.F.S., I.M., and D.M.D. designed experiments. C.T.W., S.M., J.A.M., A.F.S., P.H.G., Z.-J.W., and A.M.G. performed behavioral experiments. C.T.W., S.M., J.A.M., A.F.S., A.E.L., and P.H.G. conducted biochemical experiments. R.L.N. generated and provided custom viral constructs. C.T.W. and S.M. performed statistical analyses. A.R., L.S., and I.M. performed ChIP-seq analyses. C.T.W. wrote the manuscript with review from D.M.D., I.M., S.M., J.A.M., A.F.S., and P.H.G. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The accession number for all sequencing data described in this paper is GEO: GSE119627.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaay0351/DC1
Fig. S1. Incubated cocaine craving during prolonged abstinence following extended-access cocaine self-administration.
Fig. S2. Experimental timeline and tissue collection.
Fig. S3. Custom HSV vectors made to manipulate INO80.
Fig. S4. Cue-induced food seeking and locomotor activity following food self-administration.
Fig. S5. Custom HSV vectors made to manipulate TRIM3.
Fig. S6. Expression of TRIM3 and synaptic substrates during abstinence following extended-access self-administration.
Fig. S7. Similar cocaine-seeking behavior in rats that received intra-NAc injections of HSV-TRIM3 or HSV-NLS-TRIM3.
Fig. S8. Schematic of INO80 signaling in the NAc.
Table S1. Primer list for qPCR.
Table S2. Primer list for ChIP.
Data S1. INO80 ChIP-seq data tables.
REFERENCES AND NOTES
- 1.Parvaz M. A., Moeller S. J., Goldstein R. Z., Incubation of cue-induced craving in adults addicted to cocaine measured by electroencephalography. JAMA Psychiat. 73, 1127–1134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu L., Grimm J. W., Dempsey J., Shaham Y., Cocaine seeking over extended withdrawal periods in rats: Different time courses of responding induced by cocaine cues versus cocaine priming over the first 6 months. Psychopharmacology 176, 101–108 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Sesack S. R., Grace A. A., Cortico-basal ganglia reward network: Microcircuitry. Neuropsychopharmacology 35, 27–47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf M. E., Synaptic mechanisms underlying persistent cocaine craving. Nat. Rev. Neurosci. 17, 351–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman W. M., Patel K. M., Brucklacher R. M., Lull M. E., Erwin M., Morgan D., Roberts D. C. S., Vrana K. E., Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology 33, 1807–1817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freeman W. M., Lull M. E., Patel K. M., Brucklacher R. M., Morgan D., Roberts D. C. S., Vrana K. E., Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 11, 29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massart R., Barnea R., Dikshtein Y., Suderman M., Meir O., Hallett M., Kennedy P., Nestler E. J., Szyf M., Yadid G., Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J. Neurosci. 35, 8042–8058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X., Mizuguchi G., Hamiche A., Wu C., A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L., Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., Hopfner K.-P., Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udugama M., Sabri A., Bartholomew B., The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 31, 662–673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayala R., Willhoft O., Aramayo R. J., Wilkinson M., McCormack E. A., Ocloo L., Wigley D. B., Zhang X., Structure and regulation of the human INO80-nucleosome complex. Nature 556, 391–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCann T. S., Tansey W. P., Functions of the proteasome on chromatin. Biomolecules 4, 1026–1044 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bach S. V., Hegde A. N., The proteasome and epigenetics: Zooming in on histone modifications. Biomol. Concepts 7, 215–227 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Jarome T. J., Devulapalli R. K., The ubiquitin-proteasome system and memory: Moving beyond protein degradation. Neuroscientist 24, 639–651 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Hegde A. N., Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 138, 98–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neklesa T. K., Winkler J. D., Crews C. M., Targeted protein degradation by PROTACs. Pharmacol. Ther. 174, 138–144 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Ren Z.-Y., Liu M.-M., Xue Y.-X., Ding Z.-B., Xue L.-F., Zhai S.-D., Lu L., A critical role for protein degradation in the nucleus accumbens core in cocaine reward memory. Neuropsychopharmacology 38, 778–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner C. T., Milovanovic M., Christian D. T., Loweth J. A., Wolf M. E., Response of the ubiquitin proteasome system to memory retrieval after extended-access cocaine or saline self-administration. Neuropsychopharmacology 40, 3006–3014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner C. T., Viswanathan R., Martin J. A., Gobira P. H., Mitra S., Thomas S. A., Wang Z.-J., Liu J.-F., Stewart A. F., Neve R. L., Li J.-X., Gancarz A. M., Dietz D. M., E3 ubiquitin-protein ligase Smurf1 in the nucleus accumbens mediates cocaine seeking. Biol. Psychiatry 84, 881–892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber J., Végh M. J., Dawitz J., Kroon T., Loos M., Labonté D., Li K. W., van Nierop P., van Diepen M. T., De Zeeuw C. I., Kneussel M., Meredith R. M., Smit A. B., van Kesteren R. E., Ubiquitin ligase TRIM3 controls hippocampal plasticity and learning by regulating synaptic γ-actin levels. J. Cell Biol. 211, 569–586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf M. E., The Bermuda Triangle of cocaine-induced neuroadaptations. Trends Neurosci. 33, 391–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Cai Y., Jin J., Florens L., Swanson S. K., Washburn M. P., Conaway J. W., Conaway R. C., Subunit organization of the human INO80 chromatin remodeling complex: An evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11283–11289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve R. L., Neve K. A., Nestler E. J., Carlezon W. A. J., Use of herpes virus amplicon vectors to study brain disorders. Biotechniques 39, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Hung A. Y., Sung C. C., Brito I. L., Sheng M., Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLOS ONE 5, e9842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M. S., Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valjent E., Aubier B., Corbillé A.-G., Brami-Cherrier K., Caboche J., Topilko P., Girault J.-A., Hervé D., Plasticity-associated gene Krox24/Zif268 is required for long-lasting behavioral effects of cocaine. J. Neurosci. 26, 4956–4960 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad K. L., Tseng K. Y., Uejima J. L., Reimers J. M., Heng L.-J., Shaham Y., Marinelli M., Wolf M. E., Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature 454, 118–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purgianto A., Scheyer A. F., Loweth J. A., Ford K. A., Tseng K. Y., Wolf M. E., Different adaptations in AMPA receptor transmission in the nucleus accumbens after short vs long access cocaine self-administration regimens. Neuropsychopharmacology 38, 1789–1797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker D. M., Nestler E. J., Neuroepigenetics and addiction. Handb. Clin. Neurol. 148, 747–765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar A., Choi K.-H., Renthal W., Tsankova N. M., Theobald D. E. H., Truong H.-T., Russo S. J., LaPlant Q., Sasaki T. S., Whistler K. N., Neve R. L., Self D. W., Nestler E. J., Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Sun H., Martin J. A., Werner C. T., Wang Z.-J., Damez-Werno D. M., Scobie K. N., Shao N.-Y., Dias C., Rabkin J., Koo J. W., Gancarz A. M., Mouzon E. A., Neve R. L., Shen L., Dietz D. M., Nestler E. J., BAZ1B in nucleus accumbens regulates reward-related behaviors in response to distinct emotional stimuli. J. Neurosci. 36, 3954–3961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z.-J., Martin J. A., Mueller L. E., Caccamise A., Werner C. T., Neve R. L., Gancarz A. M., Li J.-X., Dietz D. M., BRG1 in the nucleus accumbens regulates cocaine-seeking behavior. Biol. Psychiatry 80, 652–660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun H., Damez-Werno D. M., Scobie K. N., Shao N.-Y., Dias C., Rabkin J., Wright K. N., Mouzon E., Kabbaj M., Neve R., Turecki G., Shen L., Nestler E. J., Regulation of BAZ1A and nucleosome positioning in the nucleus accumbens in response to cocaine. Neuroscience 353, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopfner K.-P., Gerhold C.-B., Lakomek K., Wollmann P., Swi2/Snf2 remodelers: Hybrid views on hybrid molecular machines. Curr. Opin. Struct. Biol. 22, 225–233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cannella N., Oliveira A. M. M., Hemstedt T., Lissek T., Buechler E., Bading H., Spanagel R., Dnmt3a2 in the nucleus accumbens shell is required for reinstatement of cocaine seeking. J. Neurosci. 38, 7516–7528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belyantseva I. A., Perrin B. J., Sonnemann K. J., Zhu M., Stepanyan R., McGee J., Frolenkov G. I., Walsh E. J., Friderici K. H., Friedman T. B., Ervasti J. M., γ-Actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. U.S.A. 106, 9703–9708 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson E. M., Self D. W., It's only a matter of time: Longevity of cocaine-induced changes in dendritic spine density in the nucleus accumbens. Curr. Opin. Behav. Sci. 13, 117–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammer R. P. Jr., Cooke E. S., Sensitization of neuronal response to cocaine during withdrawal following chronic treatment. Neuroreport 7, 2041–2045 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Li X., DeJoseph M. R., Urban J. H., Bahi A., Dreyer J.-L., Meredith G. E., Ford K. A., Ferrario C. R., Loweth J. A., Wolf M. E., Different roles of BDNF in nucleus accumbens core versus shell during the incubation of cue-induced cocaine craving and its long-term maintenance. J. Neurosci. 33, 1130–1142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B. T., Bowers M. S., Martin M., Hopf F. W., Guillory A. M., Carelli R. M., Chou J. K., Bonci A., Cocaine but not natural reward self-administration nor passive cocaine infusion produces persistent LTP in the VTA. Neuron 59, 288–297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai Y., Jin J., Yao T., Gottschalk A. J., Swanson S. K., Wu S., Shi Y., Washburn M. P., Florens L., Conaway R. C., Conaway J. W., YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Chang B., Chen Y., Zhao Y., Bruick R. K., JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wen C., Xu M., Mo C., Cheng Z., Guo Q., Zhu X., JMJD6 exerts function in neuropathic pain by regulating NF‑κB following peripheral nerve injury in rats. Int. J. Mol. Med. 42, 633–642 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Russo S. J., Wilkinson M. B., Mazei-Robison M. S., Dietz D. M., Maze I., Krishnan V., Renthal W., Graham A., Birnbaum S. G., Green T. A., Robison B., Lesselyong A., Perrotti L. I., Bolanos C. A., Kumar A., Clark M. S., Neumaier J. F., Neve R. L., Bhakar A. L., Barker P. A., Nestler E. J., Nuclear factor κB signaling regulates neuronal morphology and cocaine reward. J. Neurosci. 29, 3529–3537 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maze I., Covington H. E. III, Dietz D. M., LaPlant Q., Renthal W., Russo S. J., Mechanic M., Mouzon E., Neve R. L., Haggarty S. J., Ren Y., Sampath S. C., Hurd Y. L., Greengard P., Tarakhovsky A., Schaefer A., Nestler E. J., Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Awasthi S., Chakrapani B., Mahesh A., Chavali P. L., Chavali S., Dhayalan A., DDX39B promotes translation through regulation of pre-ribosomal RNA levels. RNA Biol. 15, 1157–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gancarz A. M., Wang Z.-J., Schroeder G. L., Damez-Werno D., Braunscheidel K. M., Mueller L. E., Humby M. S., Caccamise A., Martin J. A., Dietz K. C., Neve R. L., Dietz D. M., Activin receptor signaling regulates cocaine-primed behavioral and morphological plasticity. Nat. Neurosci. 18, 959–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gancarz A. M., Kausch M. A., Lloyd D. R., Richards J. B., Between-session progressive ratio performance in rats responding for cocaine and water reinforcers. Psychopharmacology 222, 215–223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gancarz-Kausch A. M., Adank D. N., Dietz D. M., Prolonged withdrawal following cocaine self-administration increases resistance to punishment in a cocaine binge. Sci. Rep. 4, 6876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunah A. W., Standaert D. G., Dopamine D1 receptor-dependent trafficking of striatal NMDA glutamate receptors to the postsynaptic membrane. J. Neurosci. 21, 5546–5558 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorn D. A., Jing L., Qiu Y., Gancarz-Kausch A. M., Galuska C. M., Dietz D. M., Zhang Y., Li J.-X., Effects of the trace amine-associated receptor 1 agonist RO5263397 on abuse-related effects of cocaine in rats. Neuropsychopharmacology 39, 2309–2316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loweth J. A., Scheyer A. F., Milovanovic M., LaCrosse A. L., Flores-Barrera E., Werner C. T., Li X., Ford K. A., Le T., Olive M. F., Szumlinski K. K., Tseng K. Y., Wolf M. E., Synaptic depression via mGluR1 positive allosteric modulation suppresses cue-induced cocaine craving. Nat. Neurosci. 17, 73–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maze I., Wenderski W., Noh K.-M., Bagot R. C., Tzavaras N., Purushothaman I., Elsässer S. J., Guo Y., Ionete C., Hurd Y. L., Tamminga C. A., Halene T., Farrelly L., Soshnev A. A., Wen D., Rafii S., Birtwistle M. R., Akbarian S., Buchholz B. A., Blitzer R. D., Nestler E. J., Yuan Z.-F., Garcia B. A., Shen L., Molina H., Allis C. D., Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaay0351/DC1
Fig. S1. Incubated cocaine craving during prolonged abstinence following extended-access cocaine self-administration.
Fig. S2. Experimental timeline and tissue collection.
Fig. S3. Custom HSV vectors made to manipulate INO80.
Fig. S4. Cue-induced food seeking and locomotor activity following food self-administration.
Fig. S5. Custom HSV vectors made to manipulate TRIM3.
Fig. S6. Expression of TRIM3 and synaptic substrates during abstinence following extended-access self-administration.
Fig. S7. Similar cocaine-seeking behavior in rats that received intra-NAc injections of HSV-TRIM3 or HSV-NLS-TRIM3.
Fig. S8. Schematic of INO80 signaling in the NAc.
Table S1. Primer list for qPCR.
Table S2. Primer list for ChIP.
Data S1. INO80 ChIP-seq data tables.


