Abstract
Background:
Zika virus (ZIKV) spread to Puerto Rico likely originated from southeast Brazil approximately 8.5 months earlier than blood donation screening for ZIKV was initiated, but the time of ZIKV introduction in the blood donor population remains unknown.
Methods:
To better understand when arboviral infections first appeared in the blood donor pool in Puerto Rico, we retrospectively screened for ZIKV-RNA (as well as chikungunya [CHIKV] and dengue [DENV] viral RNA) a repository of 1,186 linked blood donor and recipient samples collected from February 2015 – May 2016 as an end-point efficacy measure following the introduction of platelet pathogen reduction (PR). Phylogenetic analysis identified relatedness of donor strain to other circulating strains, and molecular clock analysis identified the estimated time of introduction.
Results:
An asymptomatic donor collected in December 2015 was ZIKV-RNA confirmed-positive, 4 months prior to investigational nucleic acid testing (NAT) implementation in April 2016; coincident and related to the first reported autochthonous cases. No CHIKV-RNA or DENV-RNA reactives were identified in donors or recipients, and no adverse events reported from PR use in recipients. Phylogenetic analysis confirmed the molecular relatedness of the donor ZIKV strain to the Puerto Rico lineage likely introduced approximately 4.5 months earlier.
Conclusion:
This study identified an asymptomatic ZIKV infection in a blood donor occurring prior to those previously recognized by blood donation screening. NAT and PR continue to be used as acceptable strategies to prevent transfusion-transmitted arboviral infections worldwide; however, repeated arboviral outbreaks warrant consideration of PR as a more proactive approach.
Keywords: Zika virus (ZIKV), Puerto Rico, viral outbreak, phylogenetic analysis, MSSPE, spiked primer enrichment, next-generation sequencing, metagenomic sequencing, viral epidemiology
INTRODUCTION
Zika virus (ZIKV) transmission in the Americas originated in Brazil in May 2015; however, phylogenetic, molecular clock analyses and computational modeling indicate that the virus was circulating at least one year prior7–9. The same studies inferred that ZIKV lineages disseminated from Brazil to Central America, Mexico, the Caribbean and the rest of South America7–9. ZIKV introduction to Puerto Rico likely originated from southeast Brazil approximately 4.5 months earlier than the first confirmed autochthonous case was published on December 31, 2015 and approximately 8.5 months earlier than ZIKV cases were identified by blood donation screening7–10. The index patient reportedly developed ZIKV-compatible clinical symptoms on November 23, 201510. ZIKV nucleic acid testing using an investigational blood donation screening assay, the cobas Zika test [Roche Molecular Systems, Inc. (RMS), Pleasanton, CA], was initiated in Puerto Rico on April 3, 2016, less than four months following the first autochthonous ZIKV case, resulting in the rapid identification of ZIKV-infected but asymptomatic blood donors11.
At the suspected time of ZIKV introduction, outbreaks of chikungunya (CHIKV) and dengue (DENV) viruses were dissipating in Puerto Rico. Due to the absence of licensed blood screening tests and the risk of CHIKV and DENV transfusion transmission, the American Red Cross (ARC) partnered with Cerus Corporation to distribute pathogen-reduced platelet concentrates starting February 2015 as part of Cerus’ investigational, prospective, open-label, “Treatment Use Study of Patient Safety Following Transfusion of INTERCEPT Blood-Platelet Components” clinical trial (TRUE). Given the temporal coincidence of the TRUE study and ZIKV introduction in Puerto Rico, we performed retrospective ZIKV RNA screening of repository samples collected between February 2015 and May 2016, following their screening for CHIKV and DENV RNA12, to better understand the timeline of ZIKV introduction to the blood donor population.
MATERIAL AND METHODS
TRUE clinical study:
The TRUE clinical study protocol was reviewed and approved by the ARC institutional review board and cleared by the US Food and Drug Administration (Investigational Device Exemption Study Number: BB IDE 6200). The study was registered on clinicaltrials.gov with the identification code . The goals of the study were to provide early access to pathogen-reduced apheresis platelets and evaluate their safety, including the prevention of transfusion-transmitted infections, particularly from arboviral agents that repeatedly occur in large epidemics in Puerto Rico and the Caribbean (e.g., CHIKV and DENV).
Sample collection:
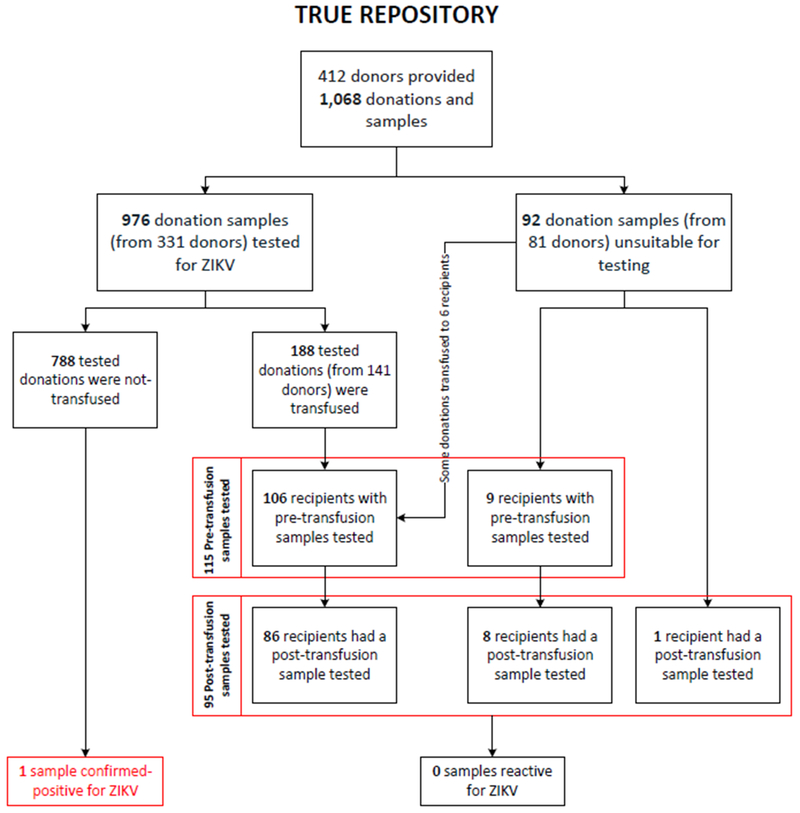
Serum and/or plasma samples from consenting donors were obtained prior to INTERCEPT treatment to create a repository as part of the TRUE study. Recipients who consented to receive INTERCEPT-treated platelets had pre- and post-transfusion samples collected and stored at the ARC Scientific Support Office (SSO). Post-transfusion samples were collected as early as 0 days after transfusion, range 0-16 days, average 4.9 days, median 4 days. Donor-recipient linkage information was retained. Repository samples from 412 apheresis platelet donors and 116 recipients consisted of 1,278 whole blood samples collected by venipuncture in paired EDTA, clot and plasma preparation tubes (Fig. 1). Sample tubes were kept at 4 ⁰C until spun; sample spinning occurred <72 h post collection; the plasma and serum were separated from the cell pellet prior to storing frozen at −18 ⁰C. Samples were subjected to two freeze/thaw cycles.
Fig. 1. Schematic of donor and recipient samples included in the TRUE repository.
Donors (412) enrolled in the TRUE clinical trial provided 1,068 plateletpheresis donations and matching donor samples (collected in paired EDTA, clot and plasma preparation tubes). Of these, 976 donor samples from 331 donors were suitable for retrospective ZIKV RNA screening. Only 188 donations from 141 donors whose donation samples were suitable for ZIKV RNA testing were transfused to 106 recipients who provided a suitable pre-transfusion sample; of these, 86 also provided a post-transfusion sample.
Donation samples (92) from 81 donors were unsuitable for ZIKV RNA testing including those transfused to nine recipients with pre-transfusion testing of whom eight also had their post-transfusion samples tested, and to 1 recipient whose pre-donation sample was unsuitable for testing but who provided a post-transfusion sample. Recipients (94) with pre-transfusion samples tested for ZIKV RNA also provided post-transfusion samples. No pre- or post-transfusion sample tested reactive for ZIKV (or CHIKV or DENV) RNA.
A total of 788 donations outdated and were discarded. The single ZIKV confirmed-positive sample came from a platelet unit that was not transfused.
ZIKV plasma sample screening:
Testing for ZIKV RNA was done at the SSO using the licensed cobas Zika nucleic acid test (cobas Zika), with a 95% limit of detection (LOD) of 8.1 copies per milliliter (c/mL) (95% CI: 6.1-13.6 c/mL) on the cobas 8800 System (RMS)13. Each sample was tested one-time individually. One of 1,278 samples tested initial ZIKV-reactive on the cobas 8800 System. This sample was retested once on the cobas 8800 System using the reactive sample tube. A paired serum sample was sent to the Wadsworth Center (New York State Department of Health) for ZIKV-IgM MAC-ELISA testing. Additionally, the amplification-detection (AD) plate for the initial cobas Zika reactive sample was sent to RMS for evaluation of amplicons by heminested (hn) PCR. The plasma sample was also tested using the Procleix Zika Virus Assay on the Procleix Panther System (Grifols Diagnostic Solutions Inc., San Diego, CA) with a 95% LOD of 3.9 c/mL (95% CI 3.2 – 4.8 c/mL)14. The donation was considered confirmed positive based on repeat cobas Zika reactivity, hnPCR positivity and Procleix reactivity.
Exploratory minipool testing:
Aliquots from the ZIKV confirmed-positive sample were further tested in duplicate in exploratory 6- and 16-sample minipools created by combining the positive sample aliquot with 5 or 15 equal volumes of negative samples, respectively, from routine NAT-nonreactive donation samples.
Estimation of Zika viral loads by standard curve analysis:
ZIKV RNA loads in ZIKV confirmed-positive samples were obtained by testing end-point dilutions of serum samples on the Procleix Zika Virus Assay on the Procleix Panther System as previously described15.
Phylogenetic analysis:
Using the metagenomic sequencing with spiked primer enrichment (MSSPE) method with short 13-nt ZIKV primers during the reverse transcription step as described previously9, 14 million shotgun metagenomic reads were generated, of which 601 corresponded to ZIKV. The consensus donor ZIKV genome along with 90 ZIKV genomes with a broad geographic representation of the 2014-2017 lineages from the Americas and the French Polynesian lineages from the 2013-2014 outbreak was aligned using the multiple alignment fast Fourier transform (MAFFT) algorithm, and phylogenetic trees were constructed using Phylogenies-Maximum Likelihood (PHYML) software16.
Molecular clock analysis:
Molecular clock analysis with inclusion of 12 fully sequenced ZIKV genomes from the National Center for Biotechnology Information (NCBI) GenBank database as of October 2018, including all Puerto Rico ZIKV genomes in GenBank, three representative Brazilian ZIKV strains from NCBI GenBank including a near neighbor to the Puerto Rico clade (KY785456 from Rio de Janeiro), and two French Polynesian strains as an outgroup was performed using Bayesian Evolutionary Analysis Sampling Trees (BEAST) software v1.8.117. Posterior probability distributions of the node corresponding to the lowest common ancestor of the Puerto Rico ZIKV strains were calculated and plotted using Tracer 1.7.118. The whole-genome donor ZIKV sequence has been deposited in NCBI GenBank (accession number 17421).
RESULTS
Of the 1,278 donor and recipient repository plasma samples, 1,186 were suitable for ZIKV screening on the cobas 8800 System. Of these, 976 samples were provided by 331 plateletpheresis donors at the time of each donation (donor samples), 115 samples were collected from 115 transfusion recipients prior to being transfused with INTERCEPT-treated platelets (pre-transfusion recipient samples), and 95 samples were collected from 95 recipients following transfusion (post-transfusion recipient samples). Among recipients, 94 provided a pre- and a post-transfusion sample, 21 provided a pre-transfusion sample and 1 had a post-transfusion sample that were suitable for ZIKV testing (Fig. 1). All samples tested for ZIKV RNA were non-reactive for CHIKV and DENV RNA12.
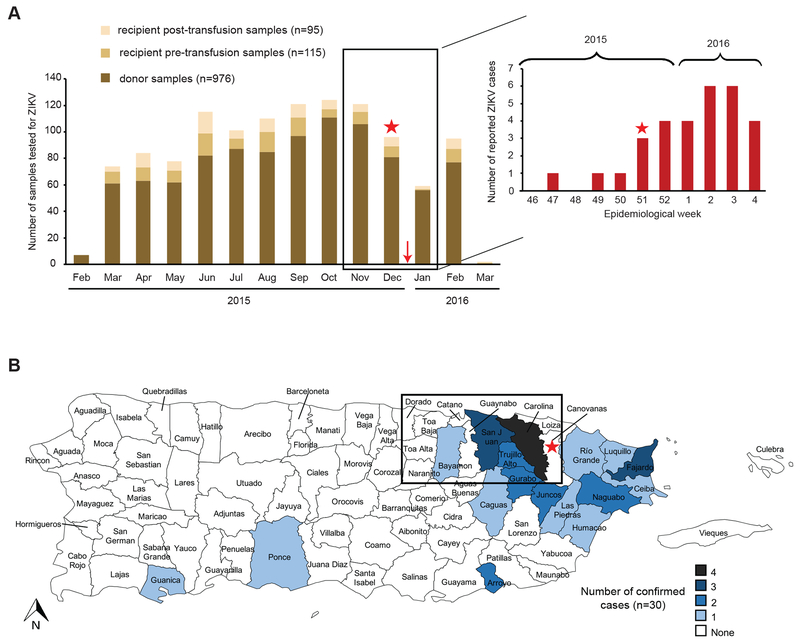
Of 976 donor samples tested, one (0.1%) was ZIKV-reactive and ZIKV confirmed positive by repeat cobas Zika (cycle threshold 36) and hn-PCR testing. The donation was collected on December 21, 2015, near the time of onset of the ZIKV outbreak on the island, and prior to the first publication of autochthonous cases (Fig. 2A, box). ZIKV RNA reactivity was also demonstrated by means of an alternate assay, Procleix Zika Virus Assay. The Procleix reactive result showed a signal-to-cutoff ratio of 32.30 against a cutoff for reactivity of 1.00. The donation was ZIKV IgM non-reactive, therefore classified as a presumptive window-period donation. ZIKV RNA was detected in 100% of reactions tested in duplicate on the Procleix Panther System when a donation sample was mixed with ZIKV-negative samples mimicking a 6- or 16-sample pool, relevant given the transition of US blood donor ZIKV screening to mini-pool testing and the potential infectivity of window-period donations15. By standard curve analysis, ZIKV RNA loads in the donor sample at the time of donation were estimated at 2,000 c/mL, consistent with those previously reported for window-period donations15.
Fig. 2. Geographic and temporal localization of the ZIKV confirmed-positive donor. (A) Donor and recipient blood donations tested for ZIKV from February 2015 to March 2016 (left bar graph) and reported ZIKV cases in Puerto Rico during the same time period (right bar graph).
Recipients that consented to receive INTERCEPT-treated platelets had pre- and/or post-transfusion serum and plasma samples collected, frozen and retained for research relating to blood-borne pathogens. Surplus plasma and serum samples from consenting platelet donors, prior to INTERCEPT treatment, were also stored frozen and retained for future research following routine blood donation screening. Donor and recipient frozen repository samples were stored by the ARC with linkage of recipient to donor samples available pre- and/or post-transfusion of INTERCEPT-treated platelets to investigate possible infectious disease transfusion transmissions. The number of donor and pre- and post-transfusion recipient samples tested are shown grouped by collection month. Each sample was tested one-time individually. The red stars denote the time period during which the sample from the ZIKV confirmed-positive donor was collected. The red arrow refers to the date of the first reported autochthonous ZIKV case in Puerto Rico, December 31, 2015. (B) Municipalities of residence corresponding to 30 laboratory-confirmed cases of ZIKV disease from November 23, 2015 to February 11, 2016. The municipality of the ZIKV confirmed-positive donor identified in this study is denoted on the map with a red star. The box highlights the Metro/San Juan region; the area of Puerto Rico most severely impacted by the outbreak within the timeframe of this donation, with four cases identified in Carolina, three cases in San Juan, two cases each in Trujillo alto and Gurabo, and one case each in Bayamón and Caguas10.
All 1,068 apheresis platelet donations collected in the study were subject to INTERCEPT treatment as part of the TRUE clinical trial prior to transfusion, including those of the reactive donor; donor repository samples were collected prior to INTERCEPT treatment. Of the 976 donations screened for ZIKV, a total of 188 INTERCEPT-treated plateletpheresis donations from 141 donors were transfused to 106 of 115 recipients; the remaining 9 recipients received INTERCEPT-treated plateletpheresis donation(s) from donors whose samples were not screened due to sample suitability issues (Fig. 1). None of the 115 pre- or 95 post-transfusion samples tested reactive for ZIKV, including 9 post-transfusion samples from recipients of INTERCEPT-treated plateletpheresis transfusions from donors with unknown ZIKV infection status due to their donation samples being unsuitable for testing (Fig. 1).
The ZIKV confirmed-positive donor was a 50-year-old male who donated twice prior to the reactive sample, on March 9, 2015 and November 9, 2015; samples from both donations tested non-reactive by cobas Zika. No additional donations were made by this donor after December 2015 (Fig. 2A). The donor was a resident of Canovanas, Puerto Rico, a municipality with no confirmed cases of ZIKV infection at the time of donation10, but located in the Metro/San Juan Region (Fig. 2B, box), the area of Puerto Rico most severely impacted by the outbreak with 13 of the 30 (43%) ZIKV-confirmed population-based cases within the timeframe of this donation10. The proximity of this donor residence to Carolina, the municipality with the most cases on the island (n=4), supports the probable autochthonous origin of this case. Attempts to contact and interview the ZIKV confirmed-positive donor to investigate symptom development compatible with ZIKV infection following the December 21, 2015 donation, and to obtain additional samples to evaluate donor seroconversion were unsuccessful. Likewise, we could not determine whether ZIKV infection in this donor was travel related.
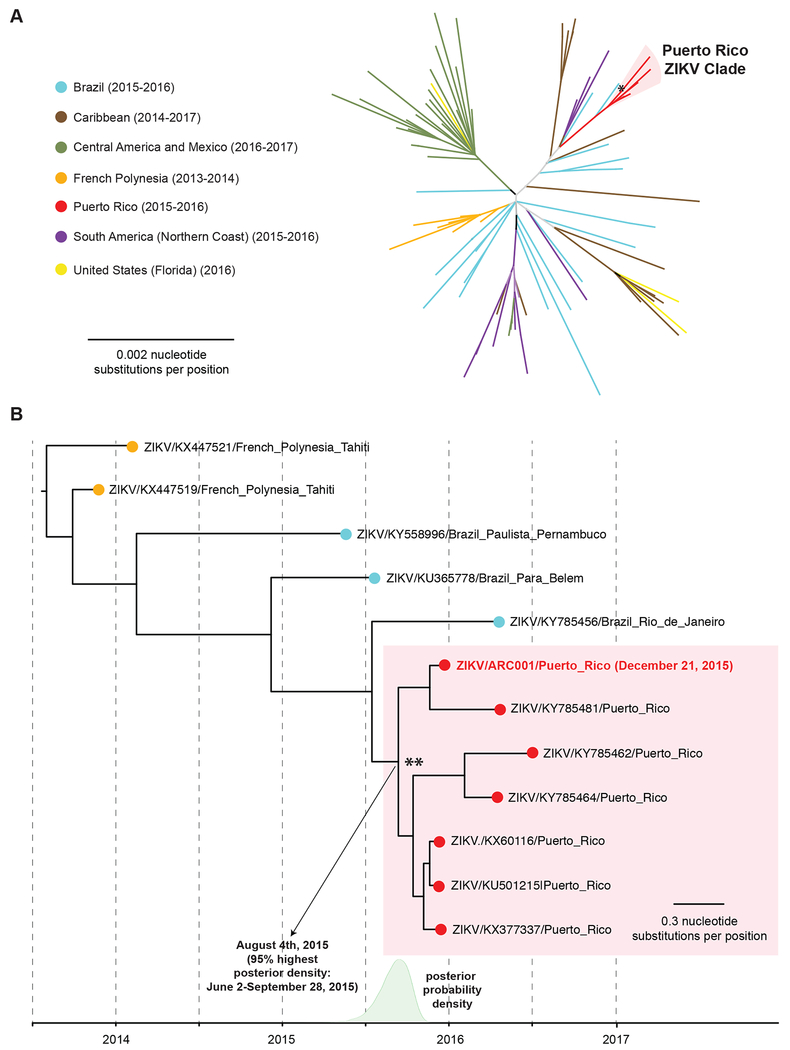
To further investigate the phylogenetic origin and the molecular relatedness of the donor ZIKV strain to the Puerto Rico lineage, we performed phylogenetic and molecular clock analyses of the ZIKV sequence recovered from the donor’s sample. The phylogenetic tree positioned the donor ZIKV strain in the Puerto Rico clade (Fig. 3A, asterisk). Molecular clock analysis revealed that the donor ZIKV strain belonged to a subclade shared with strain KY785481 (Fig. 3B). The donor was found to be an early branching offshoot of the most common ancestral node giving rise of the 10Puerto Rico clade (Fig. 3B, double asterisk), which was predicted to diverge on August 4, 2015, approximately 4.5 months prior to the time of donation (95% highest posterior density interval, June 2 – September 28, 2015).
Fig. 3. Phylogeny and molecular clock analysis of the donor ZIKV strain. (A) Radial phylogeny of representative ZIKV genomes from the 2015-2016 epidemic.
The consensus donor ZIKV genome was assembled using Geneious software version 11.1.321 by mapping the reads to the Puerto Rico strain KY785481. The consensus donor ZIKV genome (red boldface) along with 90 ZIKV genomes with a broad geographic representation of the 2014-2017 lineages from the Americas and the French Polynesian lineages from the 2013-2014 Yap Island outbreak were aligned using the MAFFT algorithm22, and phylogenetic trees were constructed using PHYML software16. Branch lengths are drawn proportionally to the number of nucleotide substitutions per position. The asterisk denotes the donor ZIKV strain. (B) Molecular clock analysis of ZIKV genomes from the Puerto Rico clade. Molecular clock analysis with inclusion of all 7 fully sequenced ZIKV genomes from Puerto Rico, 3 Brazilian ZIKV strains including a near neighbor (KY785456 from Rio de Janeiro), and 2 French Polynesian strains as an outgroup was performed using BEAST software17. Branch lengths are drawn proportionally to the number of years. The more recent common ancestral node to the Puerto Rico clade is denoted with a double asterisk. The posterior probability density for that node is plotted on the timeline in green, and the mean (corresponding to the predicted divergence date of the ancestral node) and 95% highest posterior density interval are shown.
DISCUSSION
Retrospective screening of plasma samples collected at the time of ZIKV spread to Puerto Rico identified acute ZIKV infection in a plateletpheresis donor at the time autochthonous infections were recognized in the general population. Platelet donations collected at the ARC Puerto Rico during this period were subject to INTERCEPT pathogen reduction. No post-transfusion recipient sample tested ZIKV-reactive, including nine obtained from recipients of plateletpheresis transfusions from donors with unknown ZIKV status due to their donation samples being unsuitable for testing. Unfortunately, sample aliquots were not obtained from platelet collections following INTERCEPT treatment that would have allowed the clinical assessment of the effectiveness of ZIKV reduction; the implicated donation outdated and was discarded. Nonetheless, infectivity studies demonstrate that ZIKV is highly sensitive to INTERCEPT treatment19. No INTERCEPT-related adverse reactions, including any transfusion-transmitted infection occurred as part of this study20. All donor and recipient samples tested non-reactive for CHIKV and DENV RNA consistent with the end of these concurrent outbreaks during the time of sample collection12. The total number of recipients in the repository from whom pre-transfusion samples were tested was 115 (Fig. 1), although only 90 of the 115 recipients exclusively received PR-platelet transfusions with 256 INTERCEPT-treated platelets. Thus, 25 additional recipients were represented in the repository receiving both PR-treated and imported platelets from the continental US20; however, this discrepancy has no impact on our testing results since no recipient was found to have ZIKV (or CHIKV or DENV) RNA.
The temporal and spatial coincidence of the ZIKV confirmed-positive donation with the beginning of the ZIKV outbreak and its occurrence in the Puerto Rico region with higher numbers of ZIKV cases suggest that this is the first case of autochthonous ZIKV infection in a blood donor. This observation, albeit supported by phylogenetic and molecular clock analyses that position the donor ZIKV strain in a subclade that was predicted to diverge from the most recent ancestral node giving rise to the Puerto Rico clade nearly 5 months prior to the report of the first autochthonous case in the general population, is limited by the absence of donor follow-up and travel history. Nevertheless, this study identified an asymptomatic ZIKV infection in a blood donor occurring at least 4 months prior to the implementation of blood donor screening11.
Nucleic acid testing and pathogen reduction continue to be used as acceptable strategies to prevent transfusion-transmitted arboviral infections worldwide; however, repeated arboviral outbreaks warrant consideration of PR as a more proactive approach.
Acknowledgments
Funding:
This study was supported in part by grants from the National Institutes of Health (NIH) R01-HL105704 from the National Heart, Lung, and Blood Institute. R33-AI129455 (to CYC) from the National Institute of Allergy and Infectious Diseases, and Abbott Laboratories (CYC). Laboratory support for this study was provided to the American Red Cross by Roche Molecular Diagnostics. The TRUE study and sample repository used in this study were funded by Cerus Corporation.
Footnotes
Conflicts of Interest: Charles Chiu is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center and receives research support from Abbott Laboratories, Inc. Laurence Corash and Richard Benjamin are employees and stockholders in Cerus Corporation. The American Red Cross Scientific Affairs department receives laboratory testing support from Abbott Laboratories, Grifols Diagnostics, Roche Molecular Diagnostics, Terumo BCT and Cerus Corporation.
REFERENCES
- 1.Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev 2016;29: 487–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krauer F, Riesen M, Reveiz L, Oladapo OT, Martinez-Vega R, Porgo TV, Haefliger A, Broutet NJ, Low N, Group WHOZCW. Zika Virus Infection as a Cause of Congenital Brain Abnormalities and Guillain-Barre Syndrome: Systematic Review. PLoS Med 2017;14: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessler J, Ott CT, Carcelen AC, Konikoff JM, Williamson J, Bi Q, Kucirka LM, Cummings DA, Reich NG, Chaisson LH. Times to key events in Zika virus infection and implications for blood donation: a systematic review. Bull World Health Organ 2016;94: 841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnard M, Lastere S, Teissier A, Cao-Lormeau V, Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill 2014;19. [PubMed] [Google Scholar]
- 5.Motta IJ, Spencer BR, Cordeiro da Silva SG, Arruda MB, Dobbin JA, Gonzaga YB, Arcuri IP, Tavares RC, Atta EH, Fernandes RF, Costa DA, Ribeiro LJ, Limonte F, Higa LM, Voloch CM, Brindeiro RM, Tanuri A, Ferreira OC Jr., Evidence for transmission of Zika virus by platelet transfusion. N Engl J Med 2016;375: 1101–3. [DOI] [PubMed] [Google Scholar]
- 6.Kim CR, Counotte M, Bernstein K, Deal C, Mayaud P, Low N, Broutet N, Sexual Transmission of Zika virus Expert Meeting p. Investigating the sexual transmission of Zika virus. Lancet Glob Health 2018;6: e24–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faria NR, Quick J, Claro IM, Theze J, de Jesus JG, Giovanetti M, Kraemer MUG, Hill SC, Black A, da Costa AC, Franco LC, Silva SP, Wu CH, Raghwani J, Cauchemez S, du Plessis L, Verotti MP, de Oliveira WK, Carmo EH, Coelho GE, Santelli A, Vinhal LC, Henriques CM, Simpson JT, Loose M, Andersen KG, Grubaugh ND, Somasekar S, Chiu CY, Munoz-Medina JE, Gonzalez-Bonilla CR, Arias CF, Lewis-Ximenez LL, Baylis SA, Chieppe AO, Aguiar SF, Fernandes CA, Lemos PS, Nascimento BLS, Monteiro HAO, Siqueira IC, de Queiroz MG, de Souza TR, Bezerra JF, Lemos MR, Pereira GF, Loudal D, Moura LC, Dhalia R, Franca RF, Magalhaes T, Marques ET Jr., Jaenisch T, Wallau GL, de Lima MC, Nascimento V, de Cerqueira EM, de Lima MM, Mascarenhas DL, Neto JPM, Levin AS, Tozetto-Mendoza TR, Fonseca SN, Mendes-Correa MC, Milagres FP, Segurado A, Holmes EC, Rambaut A, Bedford T, Nunes MRT, Sabino EC, Alcantara LCJ, Loman NJ, Pybus OG . Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 2017;546: 406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metsky HC, Matranga CB, Wohl S, Schaffner SF, Freije CA, Winnicki SM, West K, Qu J, Baniecki ML, Gladden-Young A, Lin AE, Tomkins-Tinch CH, Ye SH, Park DJ, Luo CY, Barnes KG, Shah RR, Chak B, Barbosa-Lima G, Delatorre E, Vieira YR, Paul LM, Tan AL, Barcellona CM, Porcelli MC, Vasquez C, Cannons AC, Cone MR, Hogan KN, Kopp EW, Anzinger JJ, Garcia KF, Parham LA, Ramirez RMG, Montoya MCM, Rojas DP, Brown CM, Hennigan S, Sabina B, Scotland S, Gangavarapu K, Grubaugh ND, Oliveira G, Robles-Sikisaka R, Rambaut A, Gehrke L, Smole S, Halloran ME, Villar L, Mattar S, Lorenzana I, Cerbino-Neto J, Valim C, Degrave W, Bozza PT, Gnirke A, Andersen KG, Isern S, Michael SF, Bozza FA, Souza TML, Bosch I, Yozwiak NL, MacInnis BL, Sabeti PC. Zika virus evolution and spread in the Americas. Nature 2017;546: 411–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theze J, Li T, du Plessis L, Bouquet J, Kraemer MUG, Somasekar S, Yu G, de Cesare M, Balmaseda A, Kuan G, Harris E, Wu CH, Ansari MA, Bowden R, Faria NR, Yagi S, Messenger S, Brooks T, Stone M, Bloch EM, Busch M, Munoz-Medina JE, Gonzalez-Bonilla CR, Wolinsky S, Lopez S, Arias CF, Bonsall D, Chiu CY, Pybus OG. Genomic Epidemiology Reconstructs the Introduction and Spread of Zika Virus in Central America and Mexico. Cell Host Microbe 2018;23: 855–64 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas DL, Sharp TM, Torres J, Armstrong PA, Munoz-Jordan J, Ryff KR, Martinez-Quinones A, Arias-Berrios J, Mayshack M, Garayalde GJ, Saavedra S, Luciano CA, Valencia-Prado M, Waterman S, Rivera-Garcia B. Local Transmission of Zika Virus--Puerto Rico, November 23, 2015-January 28, 2016. MMWR Morb Mortal Wkly Rep 2016;65: 154–8. [DOI] [PubMed] [Google Scholar]
- 11.Kuehnert MJ, Basavaraju SV, Moseley RR, Pate LL, Galel SA, Williamson PC, Busch MP, Alsina JO, Climent-Peris C, Marks PW, Epstein JS, Nakhasi HL, Hobson JP, Leiby DA, Akolkar PN, Petersen LR, Rivera-Garcia B. Screening of Blood Donations for Zika Virus Infection - Puerto Rico, April 3-June 11, 2016. MMWR Morb Mortal Wkly Rep 2016;65: 627–8. [DOI] [PubMed] [Google Scholar]
- 12.Stramer SL, Stanley J, Nguyen ML, Bertuzis R, Huynh N, Duncan JR, Albrecht P, Pate LL, Galel SA. Duplex nucleic acid test for the detection of chikungunya and dengue RNA viruses in blood donations. Transfusion 2019;0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.cobas® Zika. Nucleic acid test for use on the cobas® 6800/8800 Systems 08082871001–01EN: Roche Diagnostics GmbH, 2017. [Google Scholar]
- 14.Gao K, James A, Green T, Le T, Do D, Knight J, Peling T, Proctor M, Grimm K, Stramer S, Linnen J. Performance and evaluation of Procleix® Zika Virus Assay on the fully automated Procleix Panther System 27th Regional Congress of the ISBT. Copenhagen, Denmark, 2017. [Google Scholar]
- 15.Saa P, Proctor M, Foster G, Krysztof D, Winton C, Linnen JM, Gao K, Brodsky JP, Limberger RJ, Dodd RY, Stramer SL. Investigational Testing for Zika Virus among U.S. Blood Donors. N Engl J Med 2018;378: 1778–88. [DOI] [PubMed] [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59: 307–21. [DOI] [PubMed] [Google Scholar]
- 17.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007;7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 2018;67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santa Maria F, Laughhunn A, Lanteri MC, Aubry M, Musso D, Stassinopoulos A. Inactivation of Zika virus in platelet components using amotosalen and ultraviolet A illumination. Transfusion 2017;57: 2016–25. [DOI] [PubMed] [Google Scholar]
- 20.Rico S, Stramer SL, Benjamin RJ, Cartagena E, Arroyo A, Robles N, Carlo S, Vera L, Solivan-Ortiz P, Huang N, Koontz C, Berry T, Corash L. Treatment Use Study of Platelet Components Treated with Amotosalen and Ultraviolet a Light in Response to Emerging Arboviruses in Puerto Rico - True Study Final Results. Blood 2016;128: 3844. [Google Scholar]
- 21.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28: 1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Asimenos G, Toh H. Multiple Alignment of DNA Sequences with MAFFT. In: Posada D, ed. Bioinformatics for DNA Sequence Analysis: Humana Press, 2009:39–64. [DOI] [PubMed] [Google Scholar]