Abstract
Myelin is synthesized about the time of birth. The Src-family tyrosine kinase Fyn is involved in the initial events of myelination. Fyn is present in myelin-forming cells and is activated through stimulation of cell surface receptors such as large myelin-associated glycoprotein (L-MAG). Here we show that Fyn stimulates transcription of the myelin basic protein (MBP) gene for myelination. MBP is a major component of the myelin membrane. In 4-week-old Fyn-deficient mice, MBP is significantly reduced, and electron microscopic analysis showed that myelination is delayed, compared with wild-type mice. The Fyn-deficient mice had thinner, more irregular myelin than the wild-type. We found that Fyn stimulates the promoter activity of the MBP gene by approximately sevenfold. The region responsible for the transactivation by Fyn is located between nucleotides −675 and −647 with respect to the transcription start site. Proteins binding to this region were found by gel shift study, and the binding activity correlates with Fyn activity during myelination. These results suggest that transactivation of the MBP gene by Fyn is important for myelination.
Keywords: myelination, Fyn tyrosine kinase, myelin basic protein, transactivation, developmental regulation, knock-out mouse
Myelin is the lipoprotein multimembrane that functions as an insulator preventing the flow of ion currents across the axonal membrane and facilitating the conduction of electrical nerve impulses. During brain development, myelin formation is one of the major events in maturation. It is synthesized by oligodendrocytes in the CNS soon after birth in mammals. For myelination, oligodendrocytes extend processes to recognize neuronal axons by cell surface receptors. The receptors generate intracellular signals in the oligodendrocytes, which enables them to wrap around the axon to form myelin. Thus, determining the signaling pathway in the oligodendrocytes is crucial to understanding the mechanism of myelination. We have shown that initial events of myelination involve Fyn protein tyrosine kinase (PTK) signaling (Umemori et al., 1994). Fyn, a member of the Src-family nonreceptor PTKs, is activated during the initial stages of myelination through stimulation of the large myelin-associated glycoprotein (L-MAG), an adhesion molecule that has been implicated in myelinogenesis.
Myelin is composed of a limited number of myelin proteins: proteolipid protein (PLP), myelin basic protein (MBP), 2′, 3′-cyclic nucleotide 3′-phosphohydrolase (CNPase), MAG, and several enzymes. Among them, MBP constitutes 30% of all myelin protein in the CNS. MBP, a functionally important structural protein of myelin, is localized at opposing cytoplasmic faces of the myelin lamellae, which form the major dense line in electron micrographs (Lemke, 1988). Five different forms of MBPs with molecular masses of 14–21.5 kDa are produced by alternative use of seven exons in mouse brain (for review, see Mikoshiba et al., 1991). In the CNS, MBP gene expression is differentially regulated during myelinogenesis (Okano et al., 1987). Cell- and stage-specific regulation of MBP gene expression in brain is regulated at the transcriptional level (Kamholz et al., 1988; Shiota et al., 1991). The 5′-flanking region of the MBP gene contains several regulatory elements that differentially contribute to the cell type-specific transcription (Miura et al., 1989; Tamura et al., 1989; Devine-Beach et al., 1990;Asipu and Blair, 1994). In addition, several MBP promoter-binding proteins have been identified (Inoue et al., 1990; Haque et al., 1994;Haas et al., 1995). However, biological signals from cell surface receptors that activate MBP gene transcription during myelination remain to be elucidated.
Here we show that signals through Fyn PTK stimulate transcription of the MBP gene. Fyn serves as a signaling molecule from cell surface receptors, such as MAG, to the nucleus to transactivate the MBP gene in the initial stages of myelination. The present study with Fyn-deficient mice shows that this signaling is important for myelin formation.
MATERIALS AND METHODS
Lysate preparation. Brains and spinal cords were removed from wild-type and Fyn-deficient mice and lysed in Tris-NP40-EDTA (TNE) buffer (Umemori et al., 1994), and equal amounts of lysates were subjected to immunoblotting.
Immunoblotting. Equal amounts of lysates (20 μg protein) were separated on 15% SDS-PAGE and transferred to polyvinylidene difluoride membranes. After blocking, the filters were probed with anti-MBP antibody (Nichirei, Tokyo, Japan) followed by treatment with125I-labeled protein A (ICN, Costa Mesa, CA). The amount of MBP was measured using a Fujix Bio Image Analyzer, BAS 2000.
Plasmid construction. The deletions of the MBP promoter used in Figure 3 were described previously (Miura et al., 1989). These deletions were subcloned into pUC00 chloramphenicol acetyltransferase (CAT) (Uchiumi et al., 1992). Deletions used in Figure 4 were constructed by site-directed mutagenesis (Kunkel, 1985). ABamHI site was introduced at the position of bp −692, −675, −656, or −647 of pBG1b, and the more distal region was removed by digestion with BamHI. The deletion in Figure 5was constructed by PCR. The PCR product with primers 5′-GACCAAAGCTTATTCCTCACC-3′ and 5′-GGCACTGCAGAATCC-CTCTCC-3′ was subcloned into the HindIII-PstI site of pBG1b. Sense and antisense oligonucleotides for the Fyn response sequence withHindIII sites at both ends were synthesized, annealed, and cloned into TK-CAT (see Fig. 6) (Kadowaki et al., 1995). All constructs were confirmed by sequencing. The construction of the expression plasmid for the constitutively active form of Fyn has been described previously (Takeuchi et al., 1993).
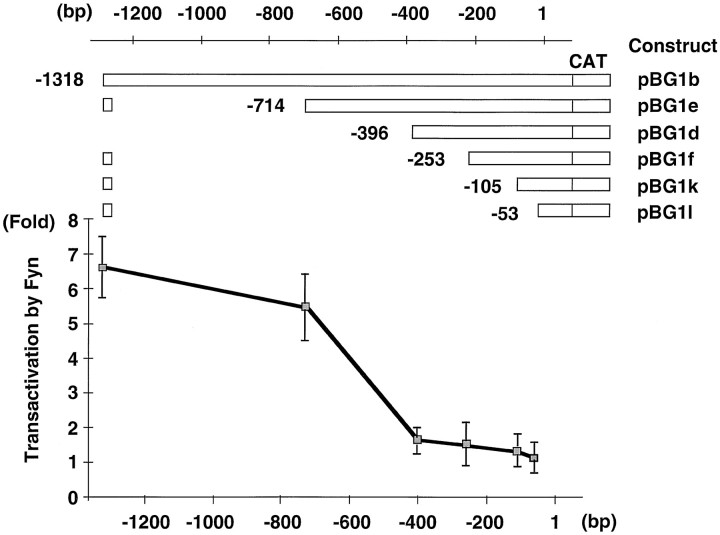
Fig. 3.
Activation of the MBP gene transcription by Fyn. Deletion constructs of the MBP promoter–CAT and the transactivation by Fyn (fold activation) are shown. The indicated base pair is from the transcription start site. CV1 cells were transfected with each MBP promoter–CAT plasmid and pME18S vector or Fyn expression plasmid. Enzyme activities in the presence of Fyn were normalized against the activities with pME18S vector. The results are expressed as means ± SD of five separate experiments.
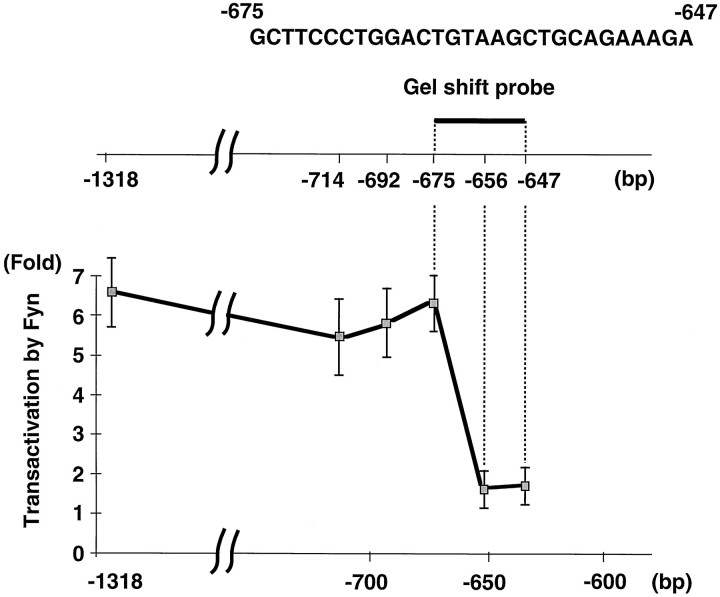
Fig. 4.
Identification of the sequence required for Fyn-dependent transactivation of the MBP promoter. The deletion constructs of the MBP promoter contain positions indicated on thetop line. Enzyme activities in the presence of Fyn were normalized against the activities with pME18S vector (fold activation). The results are expressed as means ± SD of three separate experiments. The sequence used for the experiments depicted in Figures6 and 7 is shown at the top.
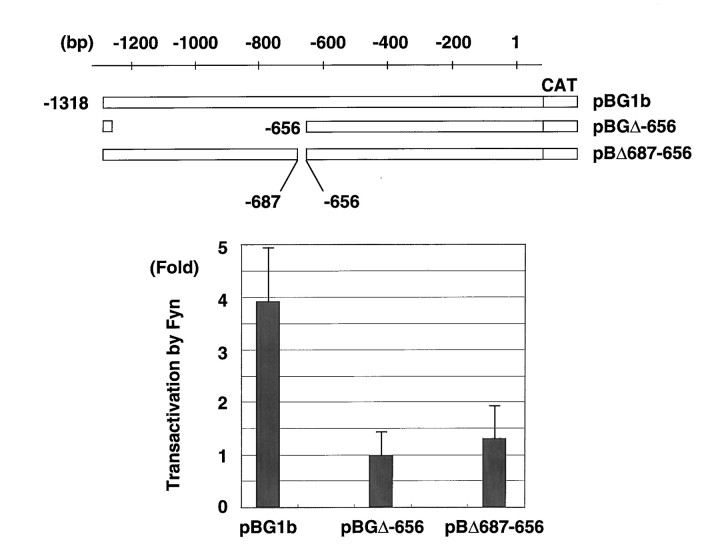
Fig. 5.
Deletion of the MBP promoter between −687 and –656 eliminates transactivation by Fyn. The deletion constructs of the MBP promoter contain positions indicated on the top line. Enzyme activities in the presence of Fyn were normalized against the activities with pME18S vector (fold activation). The results are expressed as means ± SD of three separate experiments.
Fig. 6.

The sequence between bp –675 and –647 of the MBP promoter is responsible for Fyn-dependent transactivation. The sequence between bp –675 and –647 of the MBP promoter was fused to the CAT reporter gene with TK promoter (A,TK-FRS-CAT) and transfected with pME18S vector (−) or Fyn expression plasmid (+) into CV1 cells. Results of the CAT assay are shown as fold activation by Fyn (B). Means ± SD.
Chloramphenicol acetyltransferase assay. CV1 cells were transfected with 5 μg of each MBP promoter–CAT plasmid and 5 μg of pME18S vector (Takebe et al., 1988) or Fyn expression plasmid by the calcium phosphate precipitation method. After 48 hr incubation, the cell extracts were prepared as described previously (Kadowaki et al., 1995), and protein concentrations were normalized. CAT assay was performed as described previously (Umesono et al., 1988). CAT activities were quantified by a Fujix Bio Image Analyzer, BAS 2000.
Gel shift assay. The gel shift assay was performed as described previously (Kadowaki et al., 1992). Briefly, 5 μg of nuclear extract from CV1 cells or brains in a binding buffer was incubated at 4°C for 20 min. Unlabeled competitors were added at this time. Then, 30 fmol of [32P]-labeled Fyn response sequence probe (1 × 105 cpm) was added, and the reaction mixture was incubated again for 30 min at room temperature. The bound complex was then resolved by 5% PAGE.
Electron microscopic analysis. Spinal cords were removed from 4-week-old Fyn-deficient and wild-type mice and processed for electron microscopy. Electron microscopic analysis was performed as described previously (Yoon et al., 1996).
RESULTS
Electron microscopic analysis of myelin in Fyn-deficient mice
Because Fyn is activated through L-MAG in the initial stages of myelination (Umemori et al., 1994), we searched for target genes downstream of the Fyn-mediated signaling pathway. Because the amount of myelin in 4-week-old Fyn-deficient mice (Yagi et al., 1993) is ∼50–60% of that in wild-type mice (Umemori et al., 1994), we first examined the structure of myelin in 4-week-old Fyn-deficient and wild-type mice by electron microscopy. As shown in Figure1, myelin in Fyn-deficient mice was thinner than that in wild-type mice. Quantitative analysis showed that myelin thickness in Fyn-deficient mice was ∼70% of that in wild-type mice. In addition, myelin in Fyn-deficient mice was irregular in form relative to that in wild-type mice. These observations suggest that myelin in 4-week-old Fyn-deficient mice is immature. When we examined myelin in 12-week old mice, no obvious difference was detected between Fyn-deficient and wild-type mice (data not shown). Therefore, we conclude that myelination is delayed in Fyn-deficient mice.
Fig. 1.
Electron microscopic analysis of myelin in 4-week-old Fyn-deficient and wild-type mice. Typical electron micrographs of spinal cord sections of 4-week-old Fyn-deficient (Fyn−/−) and wild-type mice are shown. Scale bars, 1 μm. 5000× magnification.
Amount of MBP is reduced in Fyn-deficient mice
On the basis of the result from electron microscopic analysis, we assumed that the expression of structural proteins of myelin might be a target of Fyn signaling. In wild-type mice, MBP expression is increased during postnatal days 4–12, which is just after Fyn activation (Umemori et al., 1994). To examine whether MBP expression is a downstream event of Fyn signaling, we measured the amount of MBP in 4-week-old mice. Equal amounts of brain or spinal cord lysates obtained from wild-type and Fyn-deficient mice were subjected to immunoblotting with anti-MBP antibody. As shown in Figure2, the amount of MBP was significantly decreased in Fyn-deficient mice compared with that in wild-type mice (56.7% of wild-type in Fyn-deficient brain and 31.9% in spinal cord).
Fig. 2.
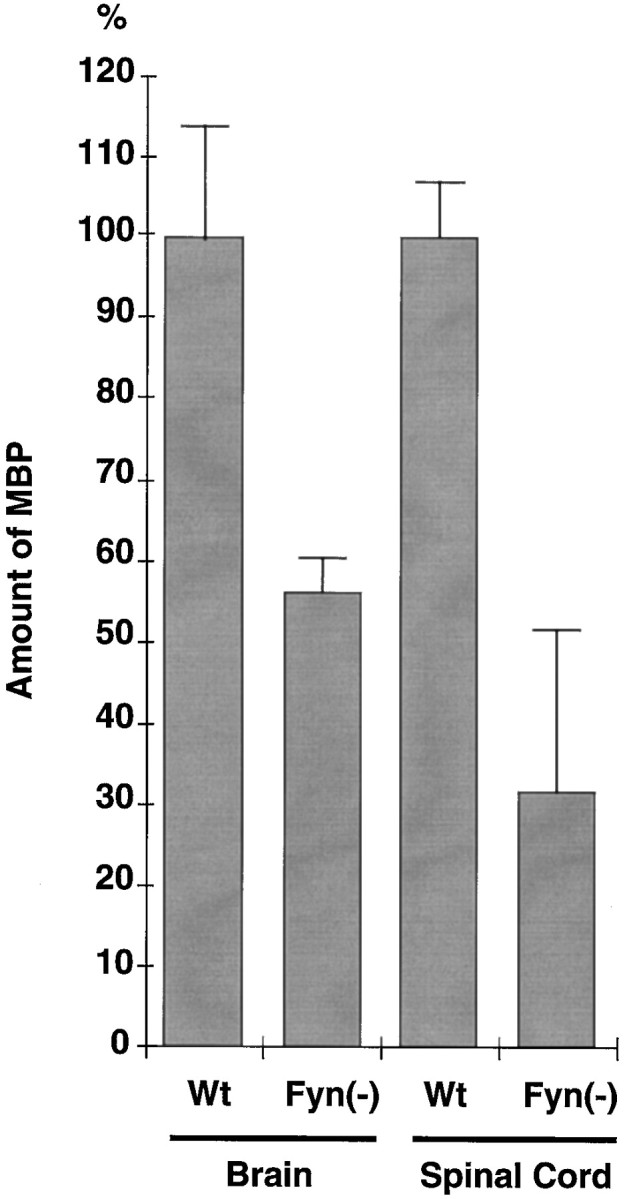
Amount of MBP in wild-type and Fyn-deficient mice. Equal amounts of brain or spinal cord lysates obtained from 4-week-old wild-type and Fyn-deficient mice were subjected to immunoblotting with anti-MBP. The amounts of MBP were determined by densitometric scanning of the corresponding bands. Results are presented as densitometric units normalized to the value for the wild type. Error bars represent SEM (n = 6).
Transcriptional activation of the MBP gene by Fyn
To determine the mechanism of the MBP expression through Fyn signaling, we examined the effect of Fyn on MBP gene transcription. We fused the 1318 bp 5′-flanking sequence of the MBP gene (Miura et al., 1989) to the CAT reporter gene. This construct was transfected into CV1 cells, and the CAT activity was measured in the presence or absence of a constitutively active form of Fyn (Takeuchi et al., 1993). As shown in Figure 3 (pBG1b), Fyn stimulated promoter activity of the MBP gene by approximately sevenfold.
Identification of Fyn response sequence in the MBP promoter
To identify the critical regions responsible for Fyn-mediated transactivation of the MBP gene, we used a series of deletions in the MBP promoter as shown in Figure 3 (Miura et al., 1989). The transactivation effect of Fyn on the MBP promoter disappeared when the promoter sequence was eliminated up to bp –396 (pBG1d). Therefore, the Fyn response region is located between bp –714 and –396 with respect to the transcription start site.
To further narrow the Fyn response sequence on the MBP promoter, we constructed another series of more minute deletions between bp –714 and –396. As shown in Figure 4, Fyn stimulated the deletion of up to bp –675. However, the promoter with deletion of up to bp –656 was not transactivated by Fyn. Thus, Fyn-induced activation of the MBP promoter requires the sequence between bp –675 and –656.
To prove that this is the only Fyn response sequence, we constructed a mutant promoter with deletion between bp –687 and –656 (Fig.5,pBΔ687–656). This deleted promoter was not transactivated by Fyn (Fig. 5). Therefore, the sequence between bp –687 and –656 is essential for transactivation of the MBP promoter by Fyn, suggesting that this region is the only sequence responsive to Fyn in the MBP promoter.
To show that the sequence above is sufficient for activation by Fyn, we fused the sequence between bp –675 and –647 with CAT reporter gene containing thymidine kinase promoter (Fig.6A); we then transfected the construct with Fyn expression plasmid into CV1 cells. The CAT assay revealed that the sequence between bp –675 and –647 was sufficient for transactivation by Fyn (approximately four- to fivefold) (Fig. 6B). Thus, we conclude that the sequence between bp –675 and –647 of the MBP promoter includes the Fyn response sequence.
Binding of nuclear proteins to the Fyn response sequence is developmentally regulated
We next examined whether nuclear proteins can interact with the sequence required for Fyn-dependent transactivation. As a gel shift probe, we prepared a synthetic oligonucleotide corresponding to the sequence between bp –675 and –647 (Fig. 4). Nuclear extracts were prepared from CV1 cells and subjected to the gel shift assay. As shown in Figure 7A, proteins interacting with the Fyn response sequence were found in the nuclear extracts prepared from CV1 cells. The competition assays using specific and nonspecific oligonucleotide competitors suggested that the complex formation was sequence specific.
Fig. 7.
Developmental regulation of protein binding to the Fyn response element. A, Nuclear extracts were prepared from CV1 cells and incubated with end-labeled Fyn response element (−) in the presence of unlabeled specific (S) or nonspecific (N) competitors. Thearrow indicates a band specific for the Fyn response element. B, Nuclear extracts were prepared from brains of 4-d-old (P4) and 30-d-old (P30) mice and incubated with Fyn response element. The arrowindicates a developmentally regulated band specific for the Fyn response element.
We then examined whether the complex formation is developmentally regulated. For this, we prepared nuclear extracts from the brains of 4- and 30-d-old mice and subjected them to the gel shift assay. As shown in Figure 7B, strong interaction of the Fyn response sequence with binding proteins was detected with the P4 extract, whereas much weaker interaction was observed with the P30 extract. Therefore, the complex formation is regulated developmentally, which correlates with Fyn kinase activity (Umemori et al., 1994). From these results, we conclude that the sequence between bp –675 and –647 of the MBP promoter includes the Fyn response element with which transcription factors stimulated by Fyn interact during myelination.
DISCUSSION
This study shows that Fyn stimulates MBP gene transcription for myelination. Despite recent progress in the identification and characterization of genes encoding myelin components, the molecular mechanisms controlling the process of myelination have remained elusive. Here we have demonstrated a signaling pathway from the cell surface to a gene expression that is important in the early phase of myelination. The putative signaling pathway is schematically illustrated in Figure 8. The signal from neuron–oligodendrocyte interaction activates Fyn PTK. Fyn phosphorylates its substrates and may stimulate transcription factors. Consequently, MBP gene transcription is stimulated, and the amount of MBP increases, which is necessary for myelin formation. The study with Fyn-deficient mice supports the importance of this signaling pathway for myelination.
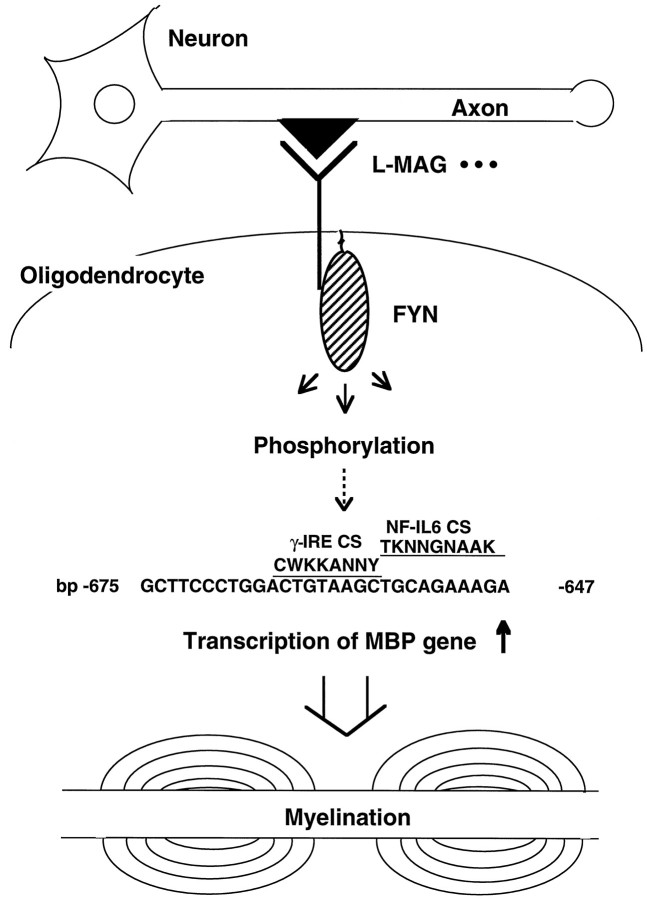
Fig. 8.
Schematic illustration for the role of the signaling through Fyn PTK in myelination. The signal from neuron–oligodendrocyte interaction (such as from L-MAG) activates Fyn PTK. Fyn phosphorylates its substrates and stimulates transcription factors. Thus, MBP gene transcription is stimulated and the amount of MBP increases, which is necessary for forming myelin. The Fyn response element and the positions of the interferon-γ response element core sequence (γ-IRE CS) and NF-IL6 core sequence (NF-IL6 CS) are indicated with their consensus sequences. Single letter code: W = Aor T, K = G orT, Y = C orT, N = A orG or C or T.
Search for previously known consensus transcription factor response elements revealed that the Fyn response region contains the interferon-γ responsive element core sequence and the NF-IL6 core sequence (NF-IL6 CS) (Fig. 8). Interferon-γ signaling involves Janus kinase (JAK) family PTKs (Ihle et al., 1994). Therefore Fyn and JAK family PTKs may share downstream signals. Indeed, both PTK families are known to activate the signal transducers and activators of transcription family of transcription factors (Cao et al., 1996). NF-IL6 belongs to the CCAAT/enhancer binding protein (C/EBP) family of transcription factors, the activity of which is regulated by phosphorylation by several kinases including MAP kinase (Nakajima et al., 1993) and PKC (Mahoney et al., 1992). Fyn may directly phosphorylate C/EBP transcription factors or may regulate them through MAP kinase or PKC. In addition, C/EBP family transcription factor is known to interact with NFκB (Stein et al., 1993). We have shown previously that Fyn can activate NFκB-like proteins (Houhashi et al., 1995). Thus, C/EBP–NFκB interaction may be regulated by Fyn PTK. Myelination signaling through Fyn might stimulate such transcription factors.
Because the cell type-specific transcription of the MBP gene is regulated by a more proximal region of the MBP promoter than the Fyn response region (Tamura et al., 1989; Devine-Beach et al., 1990), signaling through Fyn would not regulate cell specificity but rather regulates stage-specific transcription as suggested in Figure7B. Fyn signaling might be required for initiating myelination. Identification of both cell- and stage-specific transcription signaling will further clarify the molecular mechanism of myelination and will be a clue to other biological events in development and differentiation.
Footnotes
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan. We thank S. Aizawa for providing Fyn-deficient mice, and M. Miura, K. Mikoshiba, and T. Tanaka for their encouragement.
Correspondence should be addressed to Dr. T. Yamamoto, Department of Oncology, Institute of Medical Science, The University of Tokyo, Minato-ku, Tokyo 108-8639, Japan.
Dr. Umemori’s present address: Department of Anatomy and Neurobiology, Washington University School of Medicine, St. Louis, MO 63110.
Dr. Kadowaki’s present address: Department of Internal Medicine (1), Daisan Hospital, The Jikei University School of Medicine, Izumihonchou Komae City, Tokyo 201, Japan.
REFERENCES
- 1.Asipu A, Blair GE. Regulation of myelin basic protein-encoding gene transcription in rat oligodendrocytes. Gene. 1994;150:227–234. doi: 10.1016/0378-1119(94)90431-6. [DOI] [PubMed] [Google Scholar]
- 2.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devine-Beach K, Lashgari MS, Khalili K. Myelin basic protein gene transcription. Identification of proximal and distal cis-acting regulatory elements. J Biol Chem. 1990;265:13830–13835. [PubMed] [Google Scholar]
- 4.Haas S, Thatikunta P, Steplewski A, Johnson EM, Khalili K, Amini S. A 39-kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J Cell Biol. 1995;130:1171–1179. doi: 10.1083/jcb.130.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haque NS, Buchberg AM, Khalili K. Isolation and characterization of MRF-1, a brain-derived DNA-binding protein with a capacity to regulate expression of myelin basic protein gene. J Biol Chem. 1994;269:31149–31156. [PubMed] [Google Scholar]
- 6.Houhashi N, Hayashi T, Fusaki N, Takeuchi M, Higurashi M, Okamoto T, Semba K, Yamamoto T. The protein tyrosine kinase Fyn activates transcription from the HIV promoter via activation of NFκB-like DNA-binding proteins. Int Immunol. 1995;7:1851–1859. doi: 10.1093/intimm/7.11.1851. [DOI] [PubMed] [Google Scholar]
- 7.Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Thierfelder WE, Kreider B, Silvennoinen O. Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem Sci. 1994;19:222–227. doi: 10.1016/0968-0004(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 8.Inoue T, Tamura T, Furuichi T, Mikoshiba K. Isolation of complementary DNAs encoding a cerebellum-enriched nuclear factor I family that activates transcription from the mouse myelin basic protein promoter. J Biol Chem. 1990;265:19065–19070. [PubMed] [Google Scholar]
- 9.Kadowaki Y, Toyoshima K, Yamamoto T. Ear3/COUP-TF binds most tightly to a response element with tandem repeat separated by one nucleotide. Biochem Biophys Res Commun. 1992;183:492–498. doi: 10.1016/0006-291x(92)90509-j. [DOI] [PubMed] [Google Scholar]
- 10.Kadowaki Y, Toyoshima K, Yamamoto T. Dual transcriptional control by Ear3/COUP: negative regulation through the DR1 direct repeat and positive regulation through a sequence downstream of the transcriptional start site of the mouse mammary tumor virus promoter. Proc Natl Acad Sci USA. 1995;92:4432–4436. doi: 10.1073/pnas.92.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamholz J, Toffenetti J, Lazzarini RA. Organization and expression of the human myelin basic protein gene. J Neurosci Res. 1988;21:62–70. doi: 10.1002/jnr.490210110. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemke G. Unwrapping the genes of myelin. Neuron. 1988;1:535–543. doi: 10.1016/0896-6273(88)90103-1. [DOI] [PubMed] [Google Scholar]
- 14.Mahoney CW, Shuman J, McKnight SL, Chen H-C, Huang K-P. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J Biol Chem. 1992;267:19396–19403. [PubMed] [Google Scholar]
- 15.Mikoshiba K, Okano H, Tamura T, Ikenaka K. Structure and function of myelin protein genes. Annu Rev Neurosci. 1991;14:201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- 16.Miura M, Tamura T, Aoyama A, Mikoshiba K. The promoter elements of the mouse myelin basic protein gene function efficiently in NG108–15 neuronal/glial cells. Gene. 1989;75:31–38. doi: 10.1016/0378-1119(89)90380-6. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano H, Miura M, Moriguchi A, Ikenaka K, Tsukada Y, Mikoshiba K. Inefficient transcription of the myelin basic protein gene possibly causes hypomyelination in myelin deficient mutant mice. J Neurochem. 1987;48:470–477. doi: 10.1111/j.1471-4159.1987.tb04116.x. [DOI] [PubMed] [Google Scholar]
- 19.Shiota C, Ikenaka K, Mikoshiba K. Developmental expression of myelin protein genes in dysmyelinating mutant mice: analysis by nuclear run-off transcription assay, in situ hybridization, and immunohistochemistry. J Neurochem. 1991;56:818–826. doi: 10.1111/j.1471-4159.1991.tb01997.x. [DOI] [PubMed] [Google Scholar]
- 20.Stein B, Cogswell PC, Baldwin AS., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi M, Kuramochi S, Fusaki N, Nada S, Kawamura-Tsuzuku J, Matsuda S, Semba K, Toyoshima K, Okada M, Yamamoto T. Functional and physical interaction of protein-tyrosine kinases Fyn and Csk in the T-cell signaling system. J Biol Chem. 1993;268:27413–27419. [PubMed] [Google Scholar]
- 23.Tamura T, Aoyama A, Inoue T, Miura M, Okano H, Mikoshiba K. Tissue-specific in vitro transcription from the mouse myelin basic protein promoter. Mol Cell Biol. 1989;9:3122–3126. doi: 10.1128/mcb.9.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchiumi F, Semba K, Yamanashi Y, Fujisawa J, Yoshida M, Inoue K, Toyoshima K, Yamamoto T. Characterization of the promoter region of the src family gene lyn and its transactivation by human T-cell leukemia virus type I-encoded p40tax. Mol Cell Biol. 1992;12:3784–3795. doi: 10.1128/mcb.12.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T. Initial events of myelination involve Fyn tyrosine kinase signalling. Nature. 1994;367:572–576. doi: 10.1038/367572a0. [DOI] [PubMed] [Google Scholar]
- 26.Umesono K, Giguere V, Glass SK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 27.Yagi T, Aizawa S, Tokunaga T, Shigetani Y, Takeda N, Ikawa Y. A role for Fyn tyrosine kinase in the suckling behaviour of neonatal mice. Nature. 1993;366:742–745. doi: 10.1038/366742a0. [DOI] [PubMed] [Google Scholar]
- 28.Yoon C-S, Hirosawa K, Suzuki E. Studies on the structure of ocellar photoreceptor cells of Drosophila melanogaster with special reference to subrhabdomeric cisternae. Cell Tissue Res. 1996;284:77–85. doi: 10.1007/s004410050568. [DOI] [PubMed] [Google Scholar]