Abstract
Background/Aims
The epidemiology and pathogenesis of eosinophilic esophagitis (EoE) remain unclear in Asian countries. We investigated clinicopathological characteristics and diagnostic trends of EoE, and evaluated 3 tissue biomarkers for correlation with disease activity and treatment response in Korean patients with EoE.
Methods
We retrospectively reviewed 25 271 esophageal biopsies performed during upper endoscopies between 2006 and 2017. We diagnosed EoE based on ≥ 15 eosinophils/high-power field (HPF) and, symptoms of esophageal dysfunction. We performed immunohistochemical analysis for tryptase, eosinophilic derived neurotoxin (EDN), and eotaxin-3.
Results
We diagnosed EoE in 72 patients (53 men and 19 women; mean age, 46.2 years) with presenting symptoms of, dysphagia (15.3%), epigastric pain (31.9%), and heartburn (30.6%). The diagnostic rate of EoE considerably increased between 2006 and 2017, from 0.29 diagnoses to 7.99 diagnoses per 1000 esophageal biopsies (P < 0.001). The mean peak eosinophil count (PEC) was 56.0 (± 77.8)/HPF. Whereas the EDN (rho = 0.667, P < 0.001) and eotaxin-3 levels (rho = 0.465, P < 0.001) correlated with PEC, tryptase and PEC were weakly correlated (rho = 0.291, P = 0.013). EDN (rho = 0.279, P = 0.017), and tryptase (rho = 0.279, P = 0.033) correlated with the inflammatory score of Eosinophilic Esophagitis Endoscopic Reference Score. Immunohistochemical analysis and changes in tryptase, EDN, and eotaxin-3 levels were associated with histologic and endoscopic improvements.
Conclusions
EoE incidence considerably increased during the 12-year period, regardless of endoscopic esophageal biopsy rate. Tryptase, EDN, and eotaxin-3 levels in esophageal biopsy specimens could be promising biomarkers for disease activity, symptom, and endoscopic response in Korea.
Keywords: Endoscopy, Eosinophilic esophagitis, Esophagus
Introduction
Eosinophilic esophagitis (EoE) is now one of the most commonly diagnosed diseases during the assessment of feeding problems in children, and in the assessment of dysphagia and food impaction in adults in Western countries.1 One study in Korea showed an increasing trend in a limited number of patients during a short period of observation.2 However, the epidemiology and pathogenesis of EoE are not yet clearly defined among Asian populations, particularly in Korea.3
The current criteria for diagnosis of EoE require the presence of at least 15 eosinophils per high-power field (HPF) in esophageal biopsy, in the correct clinical setting and without comparing causes of esophageal eosinophilia.4–6 However, the eosinophil count alone is not specific and therefore insufficient as a sole biomarker.7 Moreover, EoE is a patchy disease with variations in both numbers of eosinophils and in histologic findings. Among biopsy specimens taken from the same site and even within a given biopsy specimen, the number of eosinophils can vary widely.8 The complex pathogenesis of EoE provides numerous biomarker candidates. Immunohistochemical (IHC) analysis is a viable option for predicting the disease activity and treatment response of EoE, and it has shown promising results in a few Western studies.9,10 In a previous study, a combination of 3 IHC stains (eotxain-3, major basic protein, and tryptase), distinguished EoE from control with 100% sensitivity and specificity.7 However, its utility in Korean patients with EoE has neither been tested nor validated. A set of predictors that are more specific to the disease activity and treatment response of EoE in the Korean population would be useful.
The objective of this study is to investigate the clinicopathologic characteristics and diagnostic trends of EoE in Korean patients. In addition, 3 tissue biomarkers (tryptase, eosinophil-derived neurotoxin [EDN], and eotaxin-3) were evaluated for their correlations with disease activity and treatment response.
Materials and Methods
Study Subjects and Clinical Data
We conducted a retrospective analysis of data obtained from a single center in Korea from 2006 to 2017. Adult patients (age > 18 years) who had undergone an endoscopic esophageal biopsy were enrolled in this study. The details of portions of our study design were as reported in a previous study.2 The clinical data were extracted: demographics, symptoms, endoscopy indications, endoscopic and histologic findings, and prescribed treatment. This study was approved by the Institutional Review Board of Asan Medical Center, University of Ulsan college of Medicine (IRB No. 2017-1232).
Histologic Analysis
For each case, hematoxylin and eosin stained slides were reviewed for eosinophil counts. Eosinophils were counted in all available fields for each biopsy specimen. The total number of eosinophils in all fields was counted and the peak eosinophil count (PEC) per HPF was reported (HPF = 0.237 mm2).11
Treatment Outcomes
The treatment outcomes included symptom response (dichotomous patient-reported subjective improvement [yes/no]), endoscopic improvement according to the Eosinophilic Esophagitis Endoscopic Reference Score (EREFS),12,13 and post-treatment PEC. Total EREFS was composed of the sum of the maximal overall score for each individual sign. Inflammatory EREFS was derived by adding the 3 inflammatory signs (edema, exudate, and furrows), but excluding rings and stricture. All endoscopic findings concerning the study patients were reviewed by 3 experienced endoscopists (H.Y.J., K.W.J., and G.H.K.). Then the endoscopic findings were re-scored using the EREFS. Histologic response was defined as the presence of < 15 eosinophils/HPF on follow-up biopsy.14 Endoscopic response was defined as any improvement in EREFS or EREFS < 2.13
The treatment options were a topical steroid (fluticasone from a multidose inhaler 250–500 μg twice daily), proton pump inhibitors (standard dose once daily), or diet elimination which was consulted to an allergic physician for a period of 8 weeks.
Immunohistochemical Staining and Interpretation
Paraffin-embedded tissue specimens were stained with an anti-EDN (RNASE2; 1:50 dilution, rabbit polyclonal, PA5-60882; Thermo Fisher Scientific, Waltham, MA, USA), anti-mast cell tryptase (1:100 dilution, clone AA1, GTX22378; GeneTex, Irvine, CA, USA), and anti-eotaxin-3 (1:200 dilution, goat, #500-P156G; PeproTech, Rocky Hill, NJ, USA) antibodies, by using an automatic IHC staining device (Benchmark XT; Ventana Medical Systems, Tucson, AZ, USA). Briefly, 4 μm-thick whole tissue sections were transferred onto poly-L-lysine-coated adhesive slides and dried at 74°C for 30 minutes, followed by standard heat epitope retrieval for 1 hour in EDTA (pH 8.0) in the autostainer. After incubation with primary antibodies, the sections were incubated with the OptiView Universal DAB detection kit (Ventana Medical systems, Tucson, AZ, USA) and counterstained with Harris hematoxylin. Positive control slides were incubated with the primary antibody to confirm that the antibody was working as expected. Negative controls were stained with an antibody diluent and the secondary antibody to assess background staining. For these positive and negative controls, tissues expected to stain for the targeted antigen was used. Negative control tissue not expected to stain for the targeted antigen was used for stain optimization and control. Some pilot cases of EoE and normal esophagus were used as positive and negative controls. Cytoplasmic staining was considered positive. In case of EDN and tryptase, the maximum number of positively stained cells in the squamous epithelial layer was counted per HPF. Otherwise, eotaxin-3 immunohistochemistry results were evaluated semiquantitatively for both the intensity of staining and the percentage of stained cells, calculated as the H score. We divided the intensity level into 3 levels, namely negative, weak, and strong, and assigned scores of 0, 1, and 2 to these levels, respectively. H score was obtained through calculating the sum of individual multiplications of the staining intensity and the percentage of positive cells in each intensity level on a scale from 0 to 200 (staining intensity × percentage of positive cells). Either cytoplasmic or nuclear staining of squamous epithelial cells was considered positive.
Statistical Methods
The χ2 test or Fisher’s exact test was used to compare categorical variables, whereas continuous variables were assessed using the Mann-Whitney U test. Continuous variables were presented as mean ± standard deviation or numbers (%), and categorical variables were presented as numbers with percentages. A P-values of < 0.05 considered as statistical significance. Cochran-Armitage trend test was used for trend analysis according to the period.
All statistical tests were performed using SPSS version 21.0 (IBM Corp, Armonk, NY, USA).
Results
Clinical and Endoscopic Characteristics
A total of 25 271 endoscopic esophageal biopsies had been performed at Asan Medical Center in the study period. Among the biopsy reports, 602 reports included the term “eosinophil.” Finally, 72 patients were included our study (Supplementary Fig. 1). The baseline characteristics of the patients included in the study are summarized in Table 1. The study population consisted of 53 male and 19 female (mean age, 46.2 ± 14.4 years).
Table 1.
Baseline Characteristics and Endoscopic Findings of the Study Patients
| Variable | Value (N = 72) |
|---|---|
| Age (yr) | 46.2 ± 14.4 |
| Sex (male) | 53 (73.6) |
| Symptoms | |
| Dysphagia/food impaction | 11 (15.3) |
| Epigastric pain | 23 (31.9) |
| Heartburn | 22 (30.6) |
| Dyspepsia | 16 (22.2) |
| Any allergic disease | |
| Asthma | 9 (12.5) |
| Allergic rhinitis | 12 (16.7) |
| Food allergy | 3 (4.2) |
| Atopic dermatitis | 1 (1.4) |
| None | 47 (65.3) |
| Endoscopic findings based on EREFS | |
| Fixed rings | |
| Grade 0: none | 46 (63.9) |
| Grade 1: mild | 19 (26.4) |
| Grade 2: moderate | 7 (9.7) |
| Grade 3: severe | 0 (0.0) |
| Exudates | |
| Grade 0: none | 27 (37.5) |
| Grade 1: mild | 40 (55.6) |
| Grade 2: severe | 5 (6.9) |
| Furrows | |
| Grade 0: absent | 22 (30.6) |
| Grade 1: present | 50 (69.4) |
| Edema | |
| Grade 0: absent | 32 (44.4) |
| Grade 1: present | 40 (55.6) |
| Stricture | |
| Grade 0: absent | 69 (95.8) |
| Grade 1: present | 3 (4.2) |
| Dilation | 1 (1.48) |
| Tested for eosinophilia | 60 (83.3) |
| Eosinophilia (> 500 eosinophils/μL) | 9 (12.5) |
EREFS, eosinophilic esophagitis endoscopic reference score.
Data are presented as mean ± SD or n (%).
The clinical symptoms of the patients were dysphagia/food impaction (11 cases [15.3%]), epigastric pain (23 cases [31.9%]), heartburn (22 cases [30.6%]), and dyspepsia (16 cases [22.2%]). Twenty-five patients (34.7%) had a personal history of allergy.
The typical endoscopic appearance of EoE was observed in 68 patients (91.8%), including furrows in 50 (69.4%), rings in 26 (36.1%), white exudates in 45 (62.5%), and stricture in 3 (4.2%). The endoscopic features were classified using the EREFS system (Table 1).12
Among the 72 cases included in the study, 56 were treated with proton pump inhibitors and 4 were treated with steroids. Five patients had a spontaneous remission without any medication. Among the 27 patients who did not respond to proton pump inhibitors, 17 were treated with corticosteroids, which led to symptomatic relief or improvement of endoscopic findings. Despite treatment, 2 of 17 patients who underwent upper endoscopy still showed typical endoscopic findings and did not have symptomatic improvement (Supplementary Fig. 2). Moreover 1 patient underwent balloon dilation for stricture.
Diagnostic Trend
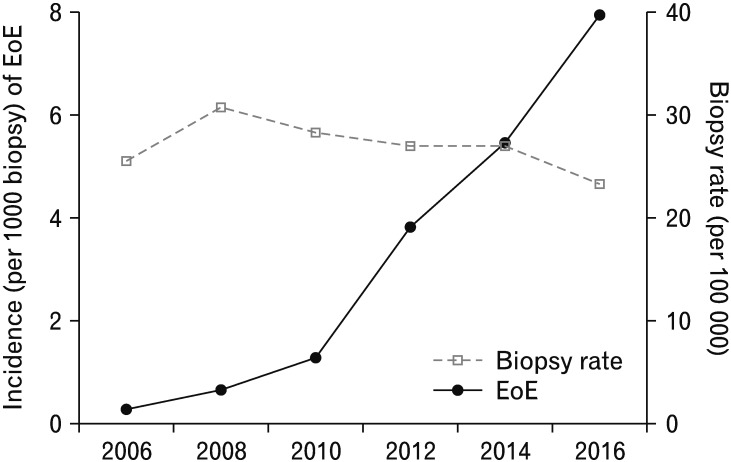
A comparison of the rate of esophageal biopsy performed during the study period with the rate of newly diagnosed EoE cases is shown in Figure 1. The number of esophageal biopsies appeared to have increased only slightly over the course of the study period. However, the number of patients with EoE seemed to have considerably increased over the 12-year period (P < 0.001).
Figure 1.
Comparison of esophageal biopsy rate and number of diagnosed eosinophilic esophagitis (EoE) cases. P for trend < 0.001.
Histologic and Immunohistochemical Staining Evaluations
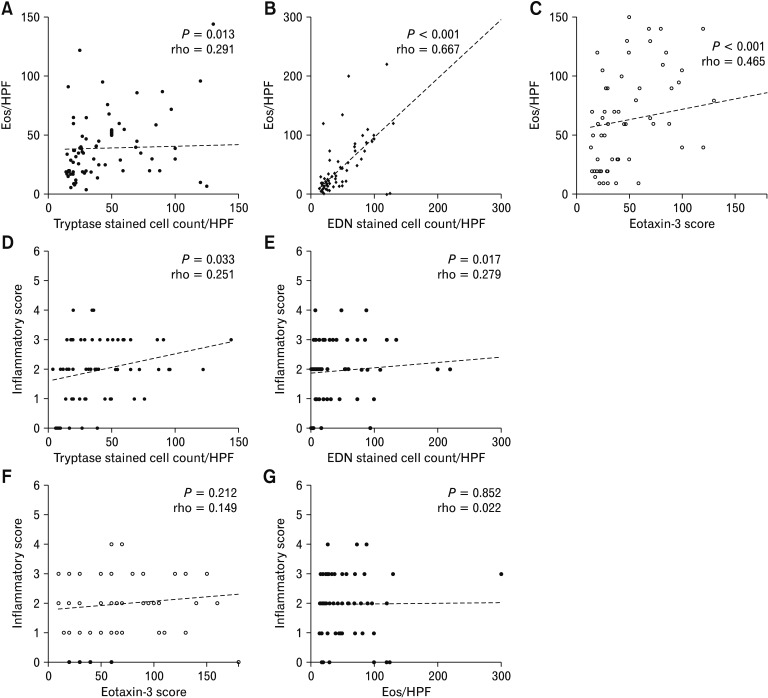
Histopathologic examination revealed eosinophilic infiltration of the esophageal epithelium (≥ 15 eosinophils/HPF) in all patients. The mean PEC was 56.0 ± 77.8. The mean counts of tryptase- and EDN-stained cells/HPF were 39.4 ± 27.8 and 53.1 ± 85.3, respectively. The mean score of eotaxin-3 stained cells/HPF was 39.4 ± 27.8 (Table 2). A significant correlation was found between the PEC and the levels of EDN (rho = 0.667, P < 0.001) and eotaxin-3 (rho = 0.465, P < 0.001; Fig. 2B and 2C). However, the tryptase levels showed a weak correlation with PEC (rho = 0.291, P = 0.013; Fig. 2A). EDN (rho = 0.251, P = 0.033), and tryptase (rho = 0.279, P = 0.017) levels correlated with the inflammatory score of EREFS in the patients with EoE (Fig. 2D and 2E). However, the eotaxin-3 levels did not correlate with the inflammatory score of EREFS (rho = 0.149, P = 0.212; Fig. 2F). Moreover, the total EREFS did not correlate with any biomarker (EDN: rho = 0.227, P = 0.055; tryptase: rho = 0.194, P = 0.155; and eotaxin-3: rho = 0.152, P = 0.201). In addition, there was no correlation between EREFS and PEC (rho = 0.022, P = 0.852; Fig. 2G).
Table 2.
Biomarkers in Atopic and Non-atopic Patients With Eosinophilic Esophagitis at Baseline
| Variable | Total (n = 72) | Atopic (n = 25) | Non-atopic (n = 47) | P-valuea |
|---|---|---|---|---|
| Age (yr) | 46.2 ± 14.4 | 41.8 ± 10.0 | 48.4 ± 15.8 | 0.106 |
| Sex (male) | 53 (73.6) | 19 (76.0) | 34 (72.3) | 0.575 |
| Peak eosinophil countb | 56.2 ± 77.8 | 78.6 ± 124.1 | 44.3 ± 29.5 | 0.523 |
| EDN stained cell countb | 53.1 ± 85.3 | 74.6 ± 132.0 | 41.6 ± 41.9 | 0.986 |
| Tryptase stained cell countb | 39.4 ± 27.8 | 39.6 ± 30.5 | 39.4 ± 26.7 | 0.937 |
| Eotaxin-3 stained score | 64.6 ± 43.1 | 70 ± 48.4 | 61.7 ± 40.3 | 0.491 |
| EREFS | 1.9 ± 1.2 | 2.1 ± 1.2 | 1.9 ± 1.2 | 0.441 |
| Endoscopic findings | ||||
| Normal | 8 (11.1) | 2 (8.3) | 7 (14.6) | 0.317 |
| Rings | 26 (36.1) | 10 (41.7) | 16 (33.3) | 0.637 |
| Exudates | 45 (62.5) | 16 (66.7) | 29 (60.4) | 0.712 |
| Furrows | 47 (65.3) | 15 (62.5) | 32 (66.7) | 0.795 |
| Edema | 8 (11.1) | 2 (8.3) | 6 (12.5) | 0.710 |
| Stricture | 2 (2.7) | 1 (4.2) | 1 (2.7) | > 0.999 |
| Symptomatic response | 55/59 (93.2) | 20/23 (86.9) | 35/36 (97.2) | |
| Endoscopic response | 26/31 (66.7) | 9/11 (81.8) | 17/20 (85.0) | |
Obtained using the chi-square test and analyzed using the Fisher exact test for categorical variables and, whereas were assessed using the Mann-Whitney U test for continuous variables.
Per high-power field.
EDN, eosinophil-derived neurotoxin; EREFS, eosinophilic esophagitis endoscopic reference score.
Data are presented as mean ± SD or n (%).
Figure 2.
Scatter plot comparing (A) tryptase levels and peak eosinophil count (PEC), (B) eosinophil-derived neurotoxin (EDN) levels and PEC, (C) Eotaxin-3 levels and PEC, (D) tryptase levels and inflammatory scores of eosinophilic esophagitis endoscopic reference score (EREFS), (E) EDN levels and inflammatory scores of EREFS, (F) eotaxin-3 levels and inflammatory scores of EREFS, and (G) PEC and inflammatory scores of EREFS. Eos, eosinophil; HPF, high-power field. P-values were analyzed by Spearman rank-order correlation.
We compared the tissue biomarker levels between atopic (n = 24) and non-atopic (n = 48) patients; however, no significant differences were observed for any biomarkers (Table 2).
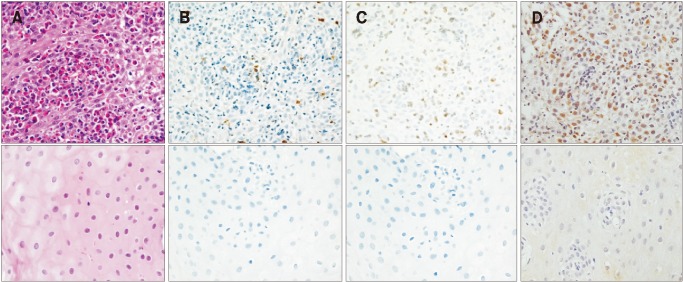
Paired esophageal tissue samples before and after treatment of 18 patients with EoE were available and stained for tryptase and EDN (Fig. 3). There were 12 (61.0%) histologic responders with < 15 eosinophils/HPF, and 6 (39.0%) nonresponders. The histologic responders did not differ from the non-responders in age, sex, EREFS, and baseline eosinophil count, with the exception of the post-treatment eosinophil count (2.3 ± 4.6 vs 35.4 ± 12.3; P < 0.001; Table 3).
Figure 3.
Histologic findings before and after treatment in a patient with eosinophilic esophagitis (EoE). (A) Hematoxylin and eosin staining shows an elevated eosinophil counts (×400) and normalization after therapy (×400). (B) The tryptase staining shows elevated number of stained cells and normalization after therapy (×400). (C) The eosinophil-derived neurotoxin staining shows an elevated number of stained cells (×400) and normalization after therapy (×400). (D) Eotaxin-3 staining shows elevated number of stained cells (×400) and normalization after therapy (×400).
Table 3.
Histologic Response After Treatment in 18 Patients With Eosinophilic Esophagitis
| Variable | Responders (n = 12) | Non-responders (n = 6) | P-valuea |
|---|---|---|---|
| Age (yr) | 46.3 ± 13.3 | 46.1 ± 11.9 | 0.972 |
| Sex (male) | 0 (0.0) | 4 (66.7) | 0.137 |
| Peak eosinophil countb | 87.9 ± 172.6 | 42.7 ± 40.6 | > 0.999 |
| EDN stained cell countb | 83.6 ± 172.7 | 45.6 ± 56.5 | 0.084 |
| Tryptase stained cell countb | 48.0 ± 31.3 | 54.4 ± 45.4 | 0.784 |
| Eotaxin-3 stained score | 53.3 ± 35.8 | 78.3 ± 43.5 | 0.131 |
| Post eosinophil countb | 2.3 ± 4.6 | 35.4 ± 12.3 | < 0.001 |
| Post EDN stained cell countb | 6.7 ± 15.2 | 45.6 ± 56.5 | 0.047 |
| Post tryptase stained cell countb | 21.5 ± 27.1 | 34.3 ± 31.5 | 0.219 |
| Post Eotaxin-3 stained score | 43.8 ± 26.4 | 69.2 ± 62.6 | 0.538 |
| EREFS | 1.2 ± 1.3 | 1.4 ± 1.6 | 0.441 |
| Symptomatic response | 12 (100) | 5 (83.3) | 0.137 |
| Endoscopic response | 9 (75) | 5 (83.3) | > 0.999 |
Obtained using the chi-square test and analyzed using the Fisher exact test for categorical variables and, whereas were assessed using the Mann-Whitney U test for continuous variables.
Per high-power field.
EDN, eosinophil-derived neurotoxin; EREFS, eosinophilic esophagitis endoscopic reference score.
Data are presented as mean ± SD or n (%).
In the IHC staining analysis, the mean changes in tryptase, EDN, and eotaxin-3 levels were 34.5, 76.8, and 9.6, respectively, in histologic responders. In the case of histologic non-responders, the mean changes in tryptase, EDN, and eotaxin-3 levels were 2.8, 23.5, and 9.2, respectively (Supplementary Fig. 3); however, the difference did not reach statistical significance. On the basis of the EREF score, the mean changes in the tryptase, EDN, and eotaxin-3 levels were 25.7, 71.1, and 15.0 in endoscopic responders. In the case of the endoscopic non-responders, the mean changes in tryptase, EDN, and eotaxin-3 levels were 17.7, 16.7, and −10.0, respectively (Supplementary Fig. 4).
Discussion
This study evaluated the clinicopathological characteristics and diagnostic trends of EoE in Korea, and assessed 3 potential tissue biomarkers for diagnosing the disease and monitoring the treatment efficacy in patients. EoE is a chronic inflammatory disease, with a rapidly increasing incidence reported in Western countries.15–19 However, this has not been fully investigated in Asian countries. We found that, the incidence of EoE considerably increased during the 12-year period, even after accounting for the endoscopic esophageal biopsy rate. The EDN, eotaxin-3, and tryptase levels correlated with PEC. Moreover, the EDN and tryptase levels correlated with the inflammatory score of EREFS, and post treatment changes in tryptase, EDN, and eotaxin-3 levels were associated with histologic and endoscopic improvements.
The diagnosis of EoE is based on the correct presentation of clinical, endoscopic, and histologic findings.4–6,20 A PEC, of ≥ 15 intraepithelial eosinophils in at least one HPF, in an esophageal biopsy is the gold standard parameter for the histologic aspect of the diagnosis. Reduced PEC constitutes an endpoint in clinical trials of therapies for EoE and is a common goal in clinical management. However, re-review of slides with PECs of 1-14/HPF yields counts at or above the diagnostic threshold value in > 20% of such biopsies, which is a limitation of using PEC as the sole pathologic diagnostic parameter.21 No guidelines have set firm response thresholds for any outcome measures.
IHC analysis is an attractive option because it is commercially available, relatively inexpensive, and reproducible between laboratories. While it has been investigated in EoE in Western countries,22,23 its usefulness has not been validated in Korea. The tissue biomarkers evaluated in this study (EDN, eotaxin-3, and tryptase) were selected on the basis of their role in the pathogenesis of EoE. Eosinophil granule proteins such as EDN are believed to play a functional role in the gastrointestinal tract by increasing permeability, but elevated levels result in chronic inflammation.24 Furthermore, plasma EDN level has been proposed as a non-invasive biomarker for diagnosing or monitoring EoE.25 Our findings agree with those of a previous study,10 as we found a significant correlation between EDN and eosinophil counts. Improvements in endoscopic appearance and PEC are associated with changes in EDN levels. Eotaxins are chemoattractants that recruit eosinophils.26 Tissue cytokine levels and eotaxin-3 expression levels are elevated in EoE.27,28 In a previous study, decreased levels of eotaxin-3 at baseline predicted which patients would not respond to therapy.29
Mast cells have been implicated in the pathogenesis of EoE in several studies, and they have potential as a specific tissue marker of EoE.20,30 Mast cell-associated genes are upregulated in EoE,31 and mast cells produce interleukin-13 and transforming growth factor-β, which have been implicated in eosinophil migration and fibrosis, respectively; however, the potential role of mast cells in diagnosing EoE is unknown, particularly in an Korean cohort. In this study, by comparing tryptase-positive mast cells between before and after treatment, we found that tryptase levels correlated with endoscopic and symptomatic responses, but not with tissue eosinophil counts. This result suggests that analysis of mast cells could add diagnostic information not gained from simple eosinophil counts.
This study has several potential limitations. First, as this study was conducted at a single center, the results may not be generalizable. However, as our data are focused on the Korean population, they are novel, and some findings related to the IHC are similar to those previously described in non-Asian populations. Second, as this was a retrospective study, not all outcomes are available in all patients. Third, esophageal biopsy specimens were not obtained from each upper and lower levels of the esophagus.
Despite these limitations, the study has multiple strengths. This is the first study to investigate tissue biomarkers of EoE in Korea. Moreover, we analyzed multiple treatment response definitions to demonstrate the meaning of tissue biomarkers. The findings of this study may provide useful information on the epidemiology and pathogenesis of EoE in Asian countries
Our findings suggest that the number of patients with EoE considerably increased over the 12-year study period, regardless of the endoscopic esophageal biopsy rate. The tryptase, EDN, and eotaxin-3 levels in esophageal biopsy specimens could be promising biomarkers of disease activity, symptom and endoscopic response in Korea, and EDN level is correlated most with PEC and treatment response. Large-scale, prospective studies are required for these biomarkers to be adopted in the diagnosis and treatment of EoE.
Supplementary Material
Footnotes
Supplementary Materials
Note: To access the supplementary table mentioned in this article, visit the online version of Journal of Neurogastroenterology and Motility at http://www.jnmjournal.org/, and at https://doi.org/10.5056/jnm19066.
Conflicts of interest: None.
Author contributions: Ga Hee Kim, Kee Wook Jung, and Hwoon-Yong Jung designed the study; Young Soo Park and Kee Don Choi analyzed the raw data and performed statistical analyses; Mi mi Kim, Hee Kyong Na, Do Hoon Kim, and Jeong Hoon Lee contributed to data collection; Ji Yong Ahn, Ho June Song, and Gin Hyug Lee interpreted the data; Evan S Dellon contributed to data interpretation and critical revision of the manuscript; and Ga Hee Kim and Young Soo Park drafted the manuscript and prepared for submission. All authors revised the manuscript critically for important intellectual content and approved the version to be published. Hwoon-Yong Jung is guarantor of the article.
Financial support: This work was partly supported by the Korean College of Helicobacter and Upper Gastrointestinal Research Foundation Grant.
References
- 1.Straumann A, Bussmann C, Zuber M, Vannini S, Simon HU, Schoepfer A. Eosinophilic esophagitis: analysis of food impaction and perforation in 251 adolescent and adult patients. Clin Gastroenterol Hepatol. 2008;6:598–600. doi: 10.1016/j.cgh.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Kim GH, Jung KW, Jung HY, et al. Diagnostic trends and clinical characteristics of eosinophilic esophagitis: a Korean, single-center database study. J Neurogastroenterol Motil. 2018;24:248–254. doi: 10.5056/jnm17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, Kim MJ, Kim JH, et al. Clinical analysis of primary eosinophilic esophagitis. J Neurogastroenterol Motil. 2013;19:204–209. doi: 10.5056/jnm.2013.19.2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 5.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG clinical guideline: evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE) Am J Gastroenterol. 2013;108:679–692. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 6.Dellon ES, Liacouras CA, Molina-Infante J, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology. 2018;155:1022–1033. e10. doi: 10.1053/j.gastro.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellon ES, Speck O, Woodward K, et al. Markers of eosinophilic inflammation for diagnosis of eosinophilic esophagitis and proton pump inhibitor-responsive esophageal eosinophilia: a prospective study. Clin Gastroenterol Hepatol. 2014;12:2015–2022. doi: 10.1016/j.cgh.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins MH. Histopathologic features of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. 2008;18:59–71. viii–ix. doi: 10.1016/j.giec.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Wolf WA, Cotton CC, Green DJ, et al. Evaluation of histologic cutpoints for treatment response in eosinophilic esophagitis. J Gastroenterol Hepatol Res. 2015;4:1780–1787. doi: 10.17554/j.issn.2224-3992.2015.04.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kephart GM, Alexander JA, Arora AS, et al. Marked deposition of eosinophil-derived neurotoxin in adult patients with eosinophilic esophagitis. Am J Gastroenterol. 2010;105:298–307. doi: 10.1038/ajg.2009.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meuten DJ, Moore FM, George JW. Mitotic count and the field of view area: time to standardize. Vet Pathol. 2016;53:7–9. doi: 10.1177/0300985815593349. [DOI] [PubMed] [Google Scholar]
- 12.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system. Gut. 2013;62:489–495. doi: 10.1136/gutjnl-2011-301817. [DOI] [PubMed] [Google Scholar]
- 13.Dellon ES, Cotton CC, Gebhart JH, et al. Accuracy of the eosinophilic esophagitis endoscopic reference score in diagnosis and determining response to treatment. Clin Gastroenterol Hepatol. 2016;14:31–39. doi: 10.1016/j.cgh.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed CC, Wolf WA, Cotton CC, et al. Optimal histologic cutpoints for treatment response in patients with eosinophilic esophagitis: analysis of data from a prospective cohort study. Clin Gastroenterol Hepatol. 2018;16:226–233. e2. doi: 10.1016/j.cgh.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sealock RJ, Rendon G, El-Serag HB. Systematic review: the epidemiology of eosinophilic oesophagitis in adults. Aliment Pharmacol Ther. 2010;32:712–719. doi: 10.1111/j.1365-2036.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 16.Dellon ES, Hirano I. Epidemiology and natural history of eosinophilic esophagitis. Gastroenterology. 2018;154:319–332. e3. doi: 10.1053/j.gastro.2017.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rhijn BD, Verheij J, Smout AJ, Bredenoord AJ. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013;25:47–52.e5. doi: 10.1111/nmo.12009. [DOI] [PubMed] [Google Scholar]
- 18.Molina-Infante J, Gonzalez-Cordero PL, Ferreira-Nossa HC, Mata-Romero P, Lucendo AJ, Arias A. Rising incidence and prevalence of adult eosinophilic esophagitis in midwestern Spain (2007–2016) United European Gastroenterol J. 2018;6:29–37. doi: 10.1177/2050640617705913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellon ES, Erichsen R, Baron JA, et al. The increasing incidence and prevalence of eosinophilic oesophagitis outpaces changes in endoscopic and biopsy practice: national population-based estimates from Denmark. Aliment Pharmacol Ther. 2015;41:662–670. doi: 10.1111/apt.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Stucke EM, Clarridge KE, Collins MH, Henderson CJ, Martin LJ, Rothenberg ME. Value of an additional review for eosinophil quantification in esophageal biopsies. J Pediatr Gastroenterol Nutr. 2015;61:65–68. doi: 10.1097/MPG.0000000000000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellon ES, Chen X, Miller CR, et al. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–271. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis. Am J Gastroenterol. 2012;107:1503–1511. doi: 10.1038/ajg.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abu-Ghazaleh RI, Fujisawa T, Mestecky J, Kyle RA, Gleich GJ. IgA-induced eosinophil degranulation. J Immunol. 1989;142:2393–2400. [PubMed] [Google Scholar]
- 25.Konikoff MR, Blanchard C, Kirby C, et al. Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4:1328–1336. doi: 10.1016/j.cgh.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Rankin SM, Conroy DM, Williams TJ. Eotaxin and eosinophil recruitment: implications for human disease. Mol Med Today. 2000;6:20–27. doi: 10.1016/S1357-4310(99)01635-4. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya B, Carlsten J, Sabo E, et al. Increased expression of eotax-in-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38:1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf WA, Cotton CC, Green DJ, et al. Predictors of response to steroid therapy for eosinophilic esophagitis and treatment of steroid-refractory patients. Clin Gastroenterol Hepatol. 2015;13:452–458. doi: 10.1016/j.cgh.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta SK, Fitzgerald JF, Kondratyuk T, HogenEsch H. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:22–26. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 31.Abonia JP, Blanchard C, Butz BB, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–149. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.