Abstract
Shigellosis is the major cause of dysentery globally. It is mainly attributed to two Shigella species, Shigella sonnei and Shigella flexneri, which leads to approximately 165 million infections and 1.1 million deaths each year. Rapid increase and widening of spectrum in antibiotics resistance make Shigella hard to be adequately controlled through existing prevention and treatment measures. It has also been observed that enhanced virulence and advent of antibiotic resistance (AR) could arise almost simultaneously. However, genetic linkages between the two factors are missing or largely ignored, which hinders experimental verification of the relationship. In this study, we sequenced 15 clinically isolated S. flexneri strains. Genome assembly, annotation and comparison were performed through routine pipelines. Differential resistant profiles of all 15 S. flexneri strains to nine antibiotics were experimentally verified. Virulence factors (VFs) belonging to 4 categories and 31 functional groups from the Virulence Factor Database (VFDB) were used to screen all Shigella translated CDSs. Distribution patterns of virulence factors were analysed by correlating with the profiles of bacterial antibiotics resistance. In addition, multi-resistant S. flexneri strains were compared with antibiotic-sensitive strains by focusing on the abundance or scarcity of specific groups of VFs. By doing these, a clear view of the relationships between virulence factors and antibiotics resistance in Shigella could be achieved, which not only provides a set of genetic evidence to support the interactions between VFs and AR but could also be used as a guidance for further verification of the relationships through manipulating specific groups of virulence factors.
Keywords: Shigella, virulence factor, comparative genomics, antibiotics resistance, HMMER, Prokka
1. Introduction
Shigellosis is an acute gastroenteritis infection that leads to approximately 165 million infections and 1.1 million deaths per annum [1]. Most of the deaths are related to children in the age group of less than 5 years old [2]. Shigellosis is caused by facultatively anaerobic, non-motile Gram-negative, rod-shaped bacteria belonging to the genus Shigella. Shigella infection involves invasion and replication within the colonic epithelium, resulting in severe inflammation and epithelial destruction [3]. There are four groups of Shigella in the genus, which includes Shigella dysenteriae, S. boydii, S. flexneri and S. sonnei. Each group is further classified into serotypes and sub-serotypes based on their lipopolysaccharide O-antigen repeats [3]. Epidemiological studies showed that S. dysenteriae was a dominant cause of large epidemics in the past and is now rarely found while S. boydii is infrequently isolated [4]. On the other hand, S. flexneri and S. sonnei are the major causes for Shigellosis. However, the two Shigella strains show distinct geographic distribution patterns, which relies on the socioeconomic conditions of the area. Specifically, S. sonnei is more tightly linked to countries with higher human development index such as Europe and North America while S. flexneri dominates in the low-income regions such as Africa and some of the Asian countries [3]. With the improvement of socioeconomic conditions, transition of dominant strains from S. flexneri to S. sonnei was also observed [5].
S. flexneri and S. sonnei were earlier susceptible to a spectrum of antibiotics. New drug resistant phenotype normally develops within a decade of their release [3]. However, due to antibiotics abuse, drug or multi-drug resistant (MDR) strains emerge more frequently than ever [6]. In addition, international travellers and unprotected sex between men increase the dissemination of Shigella across countries and lead to potential increase of antibiotic resistance [3]. Historically, Shigella was treated with antibiotic drugs such as sulphonamides, tetracycline, and chloramphenicol successively [7]. Antibiotics such as ampicillin, co-trimoxazole, nalidixic acid, and fluoroquinolones were then introduced for combating the bug due to resistance to former drugs [7]. When fluoroquinolones resistant Shigella strains emerged, stronger drugs like ceftriaxone, pivmecillinam, and azithromycin were used for treating the infection [7]. Thus, MDR Shigella strains present a heavy burden and emerging threats to the society. Common strategies of bacterial antibiotic resistance include but not limited to reduced drug penetration, antibiotics efflux, target modification by mutation and antibiotics hydrolysis [7]. Currently, many shigellosis outbreaks are linked to resistant Shigella strains [7]. Rapid increase and widening of spectrum in antibiotics resistance makes Shigella hard to be adequately controlled by existing prevention and treatment measures [8]. Thus, endeavours have been tried to accelerate the development of Shigella vaccines. Vaccine antigens, Shigella subunit vaccines, live oral Shigella vaccines, and also killed whole-cell oral Shigella vaccines are currently under development [3].
Shigella spp. were evolved from non-pathogenic E. coli ancestors by acquisition of chromosomal pathogenicity islands and a large virulence plasmid while genes related with anti-virulence such as cadA and ompT, bacterial mobility like flagella and fimbriae, and catabolism were lost [9]. Pathogenicity of Shigella virulence mainly involves Type III secretion system (T3SS), adherence, invasion, intracellular mobility and spread, immune system manipulation and evasion, and toxin, etc [10]. Accepted paradigm indicated that increased antibiotic resistance is associated with fitness costs, resulting in reductions in in vivo virulence [11]. However, experimental validation of this accepted paradigm is modest. Recent studies suggested that there may be a complex interplay between bacterial virulence and resistance [12]. It has been observed that enhanced virulence and advent of antibiotic resistance often arise almost simultaneously [12]. In addition, a global sensory-transduction system BfmRS in Acinetobacter baumannii controls both enhanced virulence and resistance [13]. Accordingly, loss of aminoglycoside resistance regulator, AmgRS, was found to enhance aminoglycoside action against bacteria while reducing bacterial virulence [14]. Moreover, an experimental study found that increase in antibiotic resistance might be exacerbated by fitness advantages that enhance virulence in drug-resistant microbes, which was consistently verified in three pathogenic bacteria Pseudomonas aeruginosa, Acinetobacter baumannii and Vibrio cholerae [15]. Although recent progresses suggested that resistance and virulence might be coupled, genetic linkages between the two factors are still insufficient and largely ignored, which hinders further experimental verification of the relationship.
In this study, we collected 15 S. flexneri strains from 7 municipal Centers for Disease Control and Prevention (CDC) in Jiangsu province. Profiles of resistance to 9 antibiotics, that is, Amoxicillin/Clavulanic acid (AMC), Ceftiophene (CFT), Cefotaxime (CTX), Gentamicin (GEN), Nalidixic acid (NAL), Norfloxacin (NOR), Tetracycline (TBT), and compound Sulfamethoxazole (SMZ), were experimentally verified. All strains were sequenced via next-generation high-throughput sequencing platform, which were then analysed for core-/pan-genomes and phylogenomic relationships. Distribution patterns of virulence factors under 4 categories belonging to 31 functional groups were studied. Differential distribution patterns of virulence factors were observed in sequenced strains by comparing antibiotic sensitive and resistant strains. In addition, abundance of specific groups of virulence factors and extent of resistance were also correlated, which may provide genetic support for the positive relationship between virulence and resistance.
2. Methods and materials
2.1. Bacterial isolates, growth conditions and DNA extraction
Shigella is currently categorized as class B infectious disease in China. Pathogenic bacteria detected in local hospitals should be reported to the provincial CDC by municipal CDC. Through collaboration with provincial CDC, 15 Shigella flexneri strains from different patients with either diarrhea or dysentery in different hospitals of 7 municipal cities were isolated by using routine biochemical techniques. Resistance profile to 9 antibiotics (AMC, CFT, CTX, GEN, NAL, NOR, TBT and SMZ) as previously described for each strain was provided by CDC based on their routine screening procedures. Isolates of Shigella flexneri were plated on trypticase soya agar (TSA). Picked-up single colony was then inoculated in 5ml trypticase soya broth (TSB) and incubated overnight at 37 °C with shaking rate of 200 rpm. DNA isolation was performed using Easy-DNA™ Kit for genomic DNA isolation (Invitrogen Life Technologies, Carlsbad, CA, USA)
2.2. Genome sequencing, assembly, and annotation
Genomes of the 15 Shigella flexneri isolates were sequenced at Beijing Genome Institute (BGI) in Shenzhen, China by using next-generation sequencing platforms. Genomic DNA (1.5 µg) was fragmented in a microTube using M220 Focused Ultrasonicator (Covaris Inc., Woburn, MA, USA), which were then validated by average molecule length using the Agilent 2100 bioanalyzer instrument (Agilent DNA 1000 Reagents) and quantified by real-time quantitative PCR (qPCR). The qualified libraries were amplified within the flow cell on the cBot instrument for cluster generation (Hiseq 4000 PE Cluster Kit Illumina). The clustered flow cell was loaded onto the Hiseq 4000 Sequencer for paired-end sequencing (Hiseq 4000 SBS Kit, Illumina) with recommended read lengths 100bp or 150bp. Obtained sequences were assessed via FastQC, assembled via SPAdes [16], recorded and reordered via MAUVE [17] based on reference genome S. flexneri 2a str. 301 by following Edwards and Holt's beginner's guide to comparative bacterial genome analysis using next-generation sequence data (Version 2) [18]. For the annotation process, assembled DNA sequences of the new draft genomes from the 15 isolates were run through an automatic annotation pipeline via Prokka (rapid prokaryotic genome annotation), followed by manual curation in some cases [19]. Ten files were generated in the specified output directory, such as FASTA file of translated coding genes (protein), FASTA file of all genomic features (nucleotide), and Genbank file containing sequences and annotations, etc.
2.3. Orthologous gene prediction and genome sequence comparison
Core-/pan-genome analysis was performed by using standalone software Roary [20]. Core genes (99% ≤ strains ≤ 100%), soft core genes (95% ≤ strains < 99%), shell genes (15% ≤ strains < 95%) and cloud genes (0% ≤ strains < 15%) were calculated. Core and unique genes in the genomes were illustrated in Venn diagram. S. flexneri genomes were visualized in circular form genome by comparing to the reference genome S. flexneri 2a str. 301 via standalone software BRIG [21]. Bacterial analysis pipeline from Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/cge/index.php) was used to compare the genomes. Only pre-assembled contig files were submitted to the online server. Theoretical distributions of antibiotic resistance genes and virulence genes were identified, together with plasmid sequences. Multilocus sequence type (MLST) was also performed based on seven housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA.
2.4. Phylogenomic analysis and tree visualization
A Newick tree for 15 Shigella flexneri strains, together with another 12 Shigella sonnei strains (unpublished data), was generated based on 3052 core genes in each genome by the phylogenomic analysis with default 1000-time bootstrapping tests via FastTree incorporated in software Roary [22]. The tree was then visualized through online webserver interactive Tree of Life (iTOL) [23]. Genome size, number of MDR, and antibiotic resistance profiles for each strain were then added to the tree by using multi-bar and binary templates in iTOL server.
2.5. Identification and comparison of putative virulence factors in the Shigella genomes
31 groups of bacterial virulence factors that belong to four categories were downloaded from the Virulence Factor Database (VFDB) [24]. These virulence factors were then used to screen Shigella translated CDSs via phmmer command (E-value < 0.00001) in HMMER package [25]. For each group of virulence factors, multiple homologous sequences were found in corresponding proteomes. These homologous sequences were then processed to get rid of redundant sequences. MDR (resistance to more than 1 antibiotics) and sensitive S. flexneri strains (resistance to 0 antibiotics) were compared in terms of the abundance of specific groups of virulence factors via in-house Python scripts.
2.6. Correlational analysis between virulence and antibiotics resistance
The total number of non-redundant putative virulence factors in each group for each proteome was calculated. Distinct distribution patterns of virulence factors were observed. Correlation between virulence and resistance were further studied by principal component analysis (PCA), which clustered sensitive and resistant strains separately based on the differences of virulence factors.
3. Results
3.1. Collection of clinically isolated Shigella strains
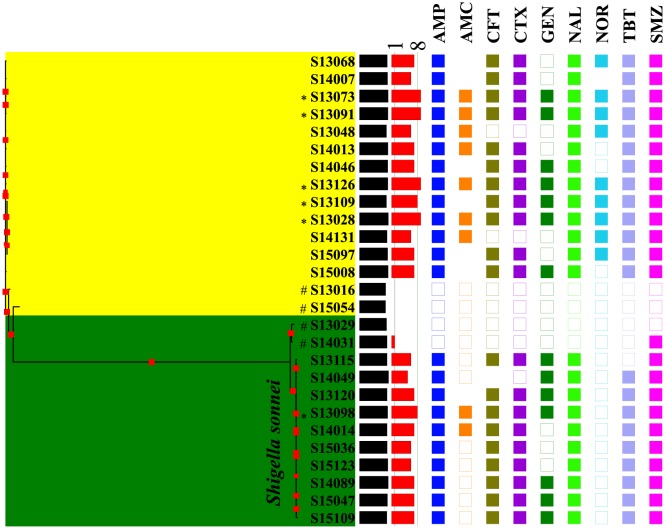
15 Shigella flexneri strains with different antibiotic resistance profiles were isolated from 7 cities in Jiangsu province of China. Another 12 strains belonging to S. sonnei were also collected and used here only as a comparison for phylogenomic study. Four serotypes of S. flexneri were experimentally verified, which includes F1a (4 strains), F1b (1 strain), F2a (8 strains), and F2b (2 strains). Sequence type ST245 was identified in these Shigella isolates according to MLST based on seven housekeeping genes. As for antibiotic resistant profiles of the 27 strains, MDR strains ranges from 5 to 9 drug resistance while no or single resistance strains were considered as sensitive. A phylogenomic tree was constructed via core genomes of studies strains, which was incorporated with genome sizes and resistance profiles (Figure 1). Distinct features were observed between the two bacterial strains. A clear genome reduction was identified in sensitive strains when compared with MDR strains (P-value < 0.05). In addition, all S. sonnei strains are sensitive to Norfloxacin (NOR) and more labile toward Amoxicillin/Clavulanic acid (AMC) when compared with S. flexneri.
Figure 1. Phylogenomic tree generated via core genes of 27 Shigella strains, which divided S. sonnei and S. flexneri into two branches. Genome size (black bar) and degree of antibiotic resistance (red bar) were incorporated, accordingly. Bootstrapping values (1000 times) were visualized through red square symbols of varying size on the branches. The presence (filled squares) and absence (empty squares) of nine antibiotics that were tested in this study, which included Amoxicillin/Clavulanic acid (AMC), Ceftiophene (CFT), Cefotaxime (CTX), Gentamicin (GEN), Nalidixic acid (NAL), Norfloxacin (NOR), Tetracycline (TBT), and compound Sulfamethoxazole (SMZ), were presented. No square means intermittent level of resistance. AMC and NOR shows the most apparent resistance difference between S. flexneri and S. sonnei. # Most sensitive strains with MDR value of 0 and 1 (vertical line 1). *Most resistant strains with MDR value of 8 and 9 (vertical line 8).
3.2. Genome assembly, annotation, and comparison
General features of the 15 S. flexneri genomes are presented in Table 1, which were obtained by integrating genome assembly and annotation results. Genome size ranges from 4.21 Mbps to 4.63 Mbps. The number of predicted protein-encoding open reading frames (ORFs) in the 15 isolates varied from 4160 (S15054) to 4608 (S13028). The total GC content ranges from 50.38% to 50.78% and is relatively consistent among isolates. All strains have a single tmRNA coding gene. The number of ribosome RNA (rRNA) and transfer RNA (tRNA) coding genes among strains varies slightly with no significant difference.
Table 1. Comparison of 15 S. flexneri strains based on key genome assembly and annotation parameters.
| ID | Serotype | BPs# | N50 | Contigs | CG%# | CDS# | tmRNA# | tRNA# | rRNA# |
| S13016 | F2a | 4253276 | 24106 | 379 | 50.75 | 4173 | 1 | 80 | 6 |
| S13028 | F2b | 4633307 | 28586 | 418 | 50.38 | 4608 | 1 | 79 | 5 |
| S13048 | F1a | 4354182 | 30242 | 320 | 50.7 | 4305 | 1 | 81 | 6 |
| S13068 | F2b | 4480462 | 30192 | 356 | 50.5 | 4434 | 1 | 81 | 6 |
| S13073 | F1a | 4608113 | 29656 | 382 | 50.5 | 4577 | 1 | 79 | 5 |
| S13091 | F2a | 4586630 | 28583 | 403 | 50.47 | 4538 | 1 | 81 | 5 |
| S13109 | F2a | 4387425 | 22591 | 472 | 50.66 | 4309 | 1 | 78 | 6 |
| S13126 | F2a | 4616160 | 19223 | 600 | 50.52 | 4532 | 1 | 81 | 4 |
| S14007 | F2a | 4542085 | 29286 | 391 | 50.45 | 4487 | 1 | 77 | 5 |
| S14013 | F2a | 4601323 | 29308 | 398 | 50.47 | 4554 | 1 | 80 | 5 |
| S14046 | F1a | 4557758 | 29656 | 394 | 50.46 | 4520 | 1 | 81 | 5 |
| S14131 | F2a | 4475708 | 29656 | 360 | 50.42 | 4417 | 1 | 79 | 6 |
| S15008 | F1a | 4608066 | 30073 | 381 | 50.48 | 4581 | 1 | 80 | 5 |
| S15054 | F1b | 4212908 | 31762 | 292 | 50.78 | 4160 | 1 | 79 | 6 |
| S15097 | F2a | 4542196 | 30475 | 371 | 50.4 | 4507 | 1 | 79 | 5 |
#BPs: Base pairs; CDS: coding sequences; GC%: Percentage of GC pairs. tmRNA: transfer-messenger RNA gene. tRNA: transfer RNA gene. rRNA: ribosomal RNA gene.
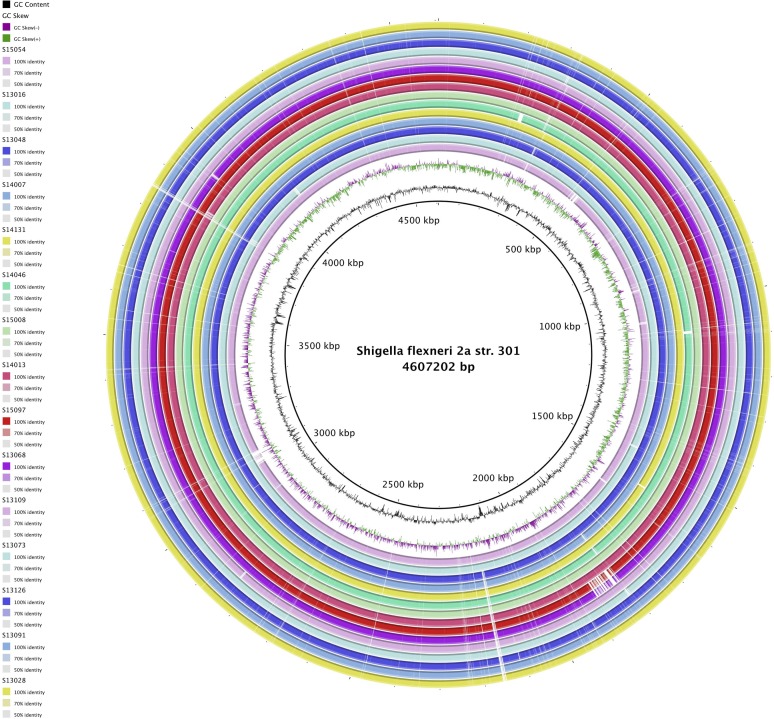
By using progressive Mauve from the Mauve software, we compared the ordered genome assembly of S. flexneri with S. flexneri 2a str. 301 as a reference genome. It seems that the chromosomal alignments of these strains are approximately identical. Additional genomic features of the 15 S. flexneri strains against reference genome S. flexneri 2a str. 301, such as sequence similarity and distribution of GC content were also analyzed and presented in Figure 2, which indicated that S. flexneri genomes are comparatively well reserved among strains.
Figure 2. Genome comparison of 15 isolated S. flexneri strains against reference genome S. flexneri 2a str. 301 generated by BRIG 0.95. The inner cycle (black) represents the complete genome of the reference strain and the shade of each colors denote the similarities between each strain with reference strain. GC content and GC skew (+/-) were illustrated in-between.
3.3. Core- and pan-genome of 15 Shigella flexneri isolates
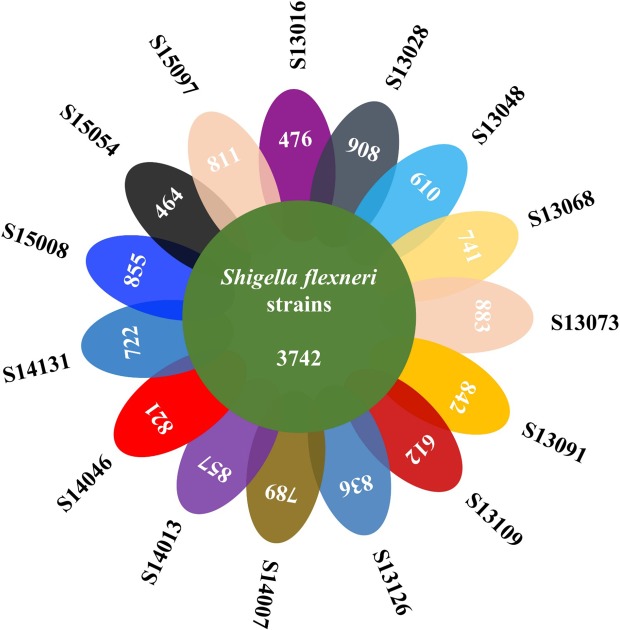
Core and pan-genome analyses for 15 S. flexneri isolates were determined by Roary through comparison of the translated CDS set, followed by clustering of orthologous proteins and the representatives of each orthologous cluster and strain-specific CDS in the total pan-genome. The total pan-genome for the 15 compared S. flexneri strains encompasses 5626 CDS. Of these, 3742 (66.51% of total CDS) are core conserved genes across all 15 Shigella genomes. A total of 1884 protein CDS (33.49% of the pan-genome total) constitute the accessory fraction, which are unique to each genome. The lowest numbers of specific genes were encoded by S. flexneri strains S13016 and S15054, with 476 and 464, respectively. The highest numbers of specific genes belong to S. flexneri strains S13028 and S13073, with 908 and 883, respectively (Figure 3). Interestingly, the former two strains are completely sensitive to the nine antibiotics while the latter two strains are resistant to all the tested antibiotics. This is consistent with theoretical prediction of antibiotic resistance genes (ARGs) via Bacterial Analysis Pipeline from Center for Genomic Epidemiology, in which S13016 has no resistance genes and S15054 was found to harbour a single resistance gene sul2 only. In contrast, other MDR strains have abundant ARGs. In addition, MDR S. flexneri strains are commonly equipped with virulence factors such as capU, gad, ipaD, lpfA, pic, sepA, sigA, and virF. In contrast, sensitive strains only have partial set of these genes, that is, gad, lpfA, pic, and sigA. Theoretical analysis found that S13016 has no known plasmid while S15054 harbors plasmid replicons of Col(MG828) and ColRNAI with no known typing. On the other hand, MDR strains have replicon typing IncN, IncI, and IncF except for S13048. For details, please refer to Table S1.
Figure 3. Venn diagram of core- and pan-genome of 15 S. flexneri strains. 3742 core genes were shared by all strains while varied number of genes were present in each strain as unique genes. The two sensitive strains (S13106 and S15054) have the lowest number of unique genes (476 and 464) while the two most resistant strains (S13028 and S13073) have the highest number of unique genes (908 and 993).
3.4. Distribution of virulence factors among S. flexneri strains
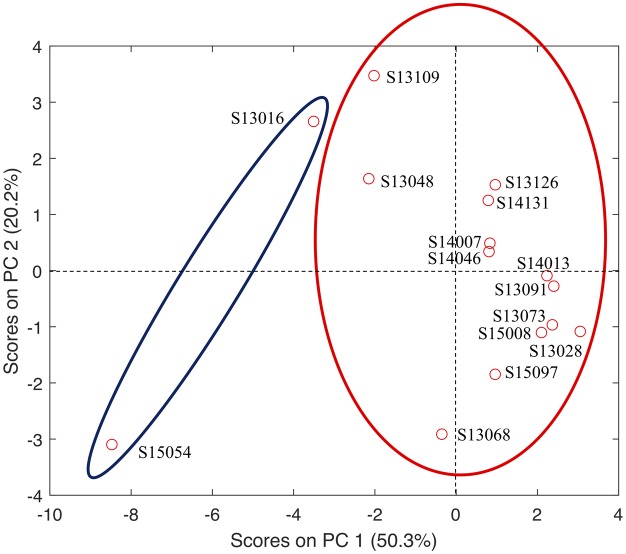
In silico identification of the putative virulence genes were performed on the translated CDSs of all isolated S. flexneri strains. All the putative virulence factors were classified into 4 major categories, that is, adhesion and invasion, secretion system and effectors, toxin, and iron acquisition, which were further divided into 31 functional groups. Distribution patterns of the virulence factors and their abundance in each strain were presented in Table 2. Nine groups of virulence factors are completely missing in all S. flexneri strains, which are β-PFTs (pore-forming toxin), superantigens and superantigen like protein, surface acting enzymes, glucosyltransferase, guanylate adenylate cyclase, deamidase, rRNA N-glycosidase, metalloprotease, intracellular PFTs. Five groups of virulence factors, that is, sortase assembled pili, fibrinogen-binding protein, collagen-binding protein, T7SS, and ADP Ribosyltransferase, are highly conserved and equally distributed in these strains. For the rest of the 17 functional groups, most of them were skewedly distributed in highly resistant strains (MDR = 9) when compared with sensitive strains, especially for Chaperone usher and T3SS. In order to better understand the relationship between resistance and virulence, principal component analysis was performed. Although many factors interfere, an apparent cluster could be observed for sensitive and resistant strains in terms of abundance of functional groups of virulence factors (Figure 4).
Table 2. Distribution patterns of 4 categories of virulence factors that belong to 31 groups among 15 Shigella flexneri strains in terms of antibiotic resistance. Sensitive strains have no resistance or only resist to one antibiotics. The four categories of VFs are Adhesion & Invasion, Secretion system & Effectors, Toxin, Iron acquisition. MDR strains have more than one resistance. PTS: Pore-forming toxins. It is noteworthy that the number of virulence factors represents the number of hits identified in bacterial proteomes via blastp.
| Shigella flexneri Strains | S13016 | S15054 | S13048 | S14007 | S14131 | S13068 | S14013 | S14046 | S15008 | S15097 | S13109 | S13028 | S13073 | S13091 | S13126 | |
| MDR | 0 | 0 | 6 | 6 | 6 | 7 | 7 | 7 | 7 | 7 | 8 | 9 | 9 | 9 | 9 | |
| Adhesion & Invasion | Chaperone usher | 129 | 131 | 135 | 137 | 135 | 134 | 136 | 137 | 138 | 137 | 135 | 138 | 138 | 137 | 135 |
| Extracellular nucleation precipitation | 14 | 13 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 13 | |
| Type 4 pili | 122 | 130 | 125 | 129 | 127 | 135 | 142 | 130 | 140 | 135 | 124 | 138 | 140 | 139 | 129 | |
| Flagella | 192 | 192 | 191 | 197 | 198 | 197 | 198 | 197 | 197 | 196 | 192 | 198 | 197 | 197 | 197 | |
| Autotransporter | 17 | 13 | 17 | 19 | 19 | 19 | 19 | 19 | 19 | 19 | 18 | 19 | 19 | 19 | 19 | |
| Fibronectin-binding protein | 51 | 45 | 51 | 53 | 53 | 49 | 53 | 53 | 53 | 50 | 53 | 53 | 53 | 53 | 53 | |
| Other adherence invasion related VFs | 62 | 59 | 60 | 61 | 61 | 57 | 62 | 61 | 61 | 58 | 63 | 61 | 61 | 61 | 60 | |
| Secretion system & Effectors | T2SS | 8 | 8 | 8 | 9 | 9 | 13 | 12 | 9 | 13 | 13 | 8 | 14 | 13 | 13 | 9 |
| T3SS | 159 | 157 | 161 | 214 | 211 | 213 | 214 | 217 | 218 | 210 | 168 | 218 | 217 | 215 | 212 | |
| T4SS | 32 | 33 | 31 | 33 | 32 | 40 | 41 | 35 | 42 | 41 | 32 | 53 | 41 | 41 | 34 | |
| T5SS | 18 | 14 | 18 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 19 | 20 | 20 | 20 | 20 | |
| T6SS | 152 | 152 | 153 | 154 | 154 | 152 | 154 | 153 | 154 | 154 | 152 | 154 | 154 | 154 | 155 | |
| Toxin | α-PFTs | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 1 |
| Dnase1 genotoxin | 29 | 27 | 29 | 28 | 29 | 28 | 29 | 28 | 28 | 28 | 29 | 29 | 28 | 29 | 29 | |
| MDR | 0 | 0 | 6 | 6 | 6 | 7 | 7 | 7 | 7 | 7 | 8 | 9 | 9 | 9 | 9 | |
| Iron acquisition | Siderophore mediated iron uptake | 272 | 259 | 275 | 277 | 277 | 273 | 279 | 277 | 277 | 274 | 275 | 278 | 279 | 279 | 280 |
| Heme-mediated iron uptake | 109 | 105 | 108 | 108 | 108 | 106 | 108 | 108 | 108 | 108 | 108 | 108 | 108 | 108 | 108 | |
| Transferrin and lactoferrin mediated iron uptake | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 |
#Nine groups of virulence factors are not present in all S. flexneri strains, which are β-PFTs, Superantigens and superantigen like protein, Surface acting enzymes, Glucosyltransferase, Guanylate adenylate cyclase, Deamidase, rRNA N-glycosidase, Metalloprotease, and Intracellular PFTs. *Five groups of virulence factors have equal number of virulence factors in all strains, which are Sortase assembled pili (n = 1), Fibrinogen-binding protein (n = 3), Collagen-binding protein (n = 8), T7SS (n = 1) and ADP Ribosyltransferase (n = 1).
Figure 4. Principal component analysis (PCA) of the relationship between antibiotic resistance and virulence factors in 15 sequenced Shigella flexneri strains. S13016 and S15054 are antibiotics-sensitive strains while the rest strains are multi-drug resistant. The two sensitive strains have comparatively less virulence factors in a majority of functional groups while resistant strains have comparatively higher number of certain groups of virulence factors. Thus, resistance and virulence could have a mutual improvement relationship, although exceptions do exist.
3.5. Genes exclusively associated with antibiotic resistant and sensitive strains
The gene presence and absence matrix for all S. flexneri strains were produced by Roary. A complete list of total genes for all strains were listed against 15 S. flexneri strains with 0 as absence and 1 as presence (Table S2). Function for translated protein is also annotated except for hypothetical proteins. By using filter function in Excel table, genes that are exclusively associated with resistant strains (12 genes) or sensitive strains (9 genes) were selected and presented in Table 3.
Table 3. Genes exclusively associated with antibiotic resistant and sensitive strains, respectively. Corresponding functions were obtained from UniProt database. Each gene was assigned into functional groups of virulence factors if there is any match.
| S. flexneri | Gene | Functions | VF Groups |
| Resistant strains | aidB_2 | Putative acyl-CoA dehydrogenase | DNase I genotoxin |
| bisC | Biotin sulfoxide reductase | - | |
| dhfrI | Trimethoprim resistance protein | - | |
| flgE | Flagellar hook protein | Flagella | |
| tnsA | Transposon Tn7 transposition protein | - | |
| tnsB | |||
| tnsC | |||
| tnsE | |||
| wecD_2 | dTDP-fucosamine acetyltransferase | - | |
| xerC_4 | Tyrosine recombinase | Chaperone usher pathway | |
| xerD_3 | |||
| ydiN_1 | Amino acid/amine transport protein | - | |
| Sensitive Strains | alkA | DNA-3-methyladenine glycosylase | - |
| bcsC_1 | Cellulose synthase subunit | - | |
| dgcE_1 | Putative diguanylate cyclase | Type IV pill | |
| dgcE_2 | |||
| dgcE_3 | |||
| dnaK_2 | Putative chaperone | Adherence and invasion | |
| dnaK_3 | |||
| pgrR_3 | HTH-type transcriptional regulator | Siderophore mediated iron uptake | |
| ycaM | Inner membrane transporter | - |
#Genes with unknown functions are not included.
4. Discussion
15 newly isolated and completely sequenced S. flexneri strains were thoroughly analysed in terms of distributions of virulence factors. Although classical thoughts support that virulence and resistance are negatively related, more evidence suggested that virulence and resistance could be enhanced simultaneously [11],[12],[15]. Initial phylogenomic analysis separated sensitive and resistant strains into different clusters (Figure 1), which reflected the intrinsic differences of evolutionary pathways between the two groups. Genome sizes of sensitive and resistant groups also show apparent difference, that is, smaller genomes (4167 CDSs on average) associated with sensitive strains and larger genome (4490 CDSs on average) linked to resistant strains. Similarly, another study focusing on sensitive and resistant E. coli isolates also found that more antibiotic sensitive Sudanese strain have smaller genome size while the genome of the resistant Chinese strain is larger [26]. Physically, it is rather difficult for bacteria to develop genetic systems with small genomes. In fact, it has been observed that multidrug resistance phenotype is a function of genome size based on comparative analysis of 22 bacterial species, which is also known as the ‘size matters’ hypothesis [27].
Core-/pan-genome analysis identified that S. flexneri strains have different number of unique genes except for the 3742 shared core genes, which reflects the heterogeneity within the same strains. In addition, the two sensitive strains, S13106 and S15054, have the lowest number of unique genes (476 and 464) while bacteria with the highest number of unique genes (908 and 993) are two most resistant strains, S13028 and S13073 (Figure 3). Specific genes in these strains could reflect bacterial characteristics and dynamics, leading to a better understanding of epidemiological features of S. flexneri [28]. Insights into these genes are out of the scope of this study and will be explored for future studies.
As for the distribution patterns of functional groups of virulence factors in S. flexneri strains, specific patterns were observed, which may provide evidence to support the positive correlation between increased virulence and enhanced antibiotic resistance. It was clear and consistent that nine groups of virulence factors are not present, and another five groups of virulence factors are equally distributed in all S. flexneri strains. Among the 17 virulence factors that are differentially distributed in the studied strains, chaperon usher, type 4 pili, flagella, T3SS, T6SS, siderophore mediated iron uptake, and heme-mediated iron uptake are abundantly present in the genomes of all strains. As for these seven groups of virulence factors, T3SS shows most distinct differences between sensitive strains (158 VFs) and resistant strains (207 VFs) on average. As previously reported, T3SS is a group of specialized protein export systems utilized by bacteria to effectively exploit eukaryotic hosts and contributes to bacterial adherence, invasion, and manipulation of the host's intracellular trafficking and immune systems [10]. Thus, in the multi-resistant Shigella strains, high number of virulence factors in T3SS groups could be very likely to occur at the same time. In fact, virulence mechanisms functioning to overcome host defence systems and antibiotic resistance are necessary for bacteria to survive antimicrobial treatments. Their collaborative work facilitates the MDR S. flexneri strains to adapt to and survive in competitive and demanding environments [29]. In addition, principal component analysis incorporating antibiotic resistance profile and 17 functional groups of virulence factors also showed that sensitive strain S. flexneri strain S15054 is isolated from other strains and most closely related to another sensitive strain S13016. All other resistant strains were all closely clustered together. Thus, from statistical point of view, it was also shown that virulence and antibiotic resistance are closely and positively correlated [12]. However, it should be notified that the spread of the two low resistant isolates in Figure 4 is a bit large. More S. flexneri strains should be included in future to further validate the claim.
Unique genes associated with sensitive and resistant strains were also identified based on the gene presence and absence table generated by core-/pan-genome analysis. It was found that all multi-drug resistant S. flexneri strains uniquely harbors four Tn7 transposon genes (tnsA, tnsB, tnsC, tnsE) and a trimethoprim resistance protein, which reflects that these strains are probably and comparatively more plastic and versatile at genome level and are more capable of acquiring resistance [30]. On the other hand, sensitive strains uniquely have type IV pill related genes (dgcE_1, dgcE_2, dgcE_3), genes involved in adherence and invasion (dnaK_2, dnaK_3), and the HTH-type transcriptional regulator gene pgrR_3 that is responsible for iron uptake. All these features emphasize that sensitive strains are more likely have specialized tools to exploit cells for reproduction. Studies confirmed that bacterial strains acquiring antibiotic resistance have a lower growth rate and are less transmissible than their susceptible counterparts [31].
Except for the interplay between virulence and resistance, several studies also proposed that antibiotic resistance is linked with bacterial intracellular and environmental persistence. In specificity, antibiotic-resistant strains such as Escherichia coli have been reported to survive longer in macrophages [32]. Further study confirmed that resistance to antibiotics and to immune system are interconnected [33]. Moreover, Vogwill et al. showed that survival of antibiotic and environmental stressors is positively correlated while specific mechanisms are unrelated in Pseudomonas strains [34]. Thus, survival and resistance could have potential interactions in bacteria. However, it was also reported that antibiotic-resistant fecal enterococci did not survive longer than antibiotic sensitive strains [35]. The possession of the antibiotic resistance plasmids in E. coli did not promote bacterial survival under starvation conditions, neither [36]. Considering the controversial conclusions, it would also be interesting to investigate this relationship via theoretical analysis, which could provide more insights into this issue and support for experimental studies in future.
5. Conclusions
15 newly sequenced S. flexneri genomes isolated from clinical samples were assembled, annotated and compared by following several standardized genome analysis pipelines [16],[19],[20]. We then identified strain-specific differences in the gain and loss of putative virulence factors in this preliminary study. In addition, abundance of certain functional groups of virulence factors is positively correlated with the extent of antibiotic resistance based on the comparison of the highly resistant and susceptible strains. Several groups of virulence factors were highlighted due to their tight relationships with strong resistant phenotypes, such as chaperone usher and T3SS, etc. Although virulence and resistance develop on different timescales and share no much common mechanisms, they may share some common characteristics [29]. Thus, antibiotic resistance and virulence are likely to have synergistic effects toward efficiently exploiting host cells in order to reproduce and transmit extensively. However, association between virulence and resistance is an increasing problem and the answer to this question is becoming more beneficial for pathogenic bacteria [29]. This study provides a starting point to address the question of how virulence and antibiotic resistance may interplay in Shigella flexneri by looking into the subtle classification of virulence factors into 31 functional groups. Although the result would be much more convincing if we can incorporate other Shigella flexneri genomes from the public database (1121 sub-strains in PATRIC database version 3.5.39) into the study, antibiotic resistance and susceptibility phenotype data for these strains are largely missing, which greatly hinders the understanding of the interactions between the two factors. Thus, in further studies, more antibiotic resistance phenotypes should be deposited into database, together with virulence phenotypes and genomic data. In addition, fitness costs should also be incorporated to tackle the intriguing relationship among virulence, stress resistance, and antibiotic resistance from the bioinformatics point of view.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81871734, 81471994), Jiangsu Provincial Natural Science Foundation (BK20151154, BK20180997), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), Advanced Health Talent of Six-one Project of Jiangsu Province (LGY2016042), and Jiangsu Provincial Commission of Health and Family Planning Research Project (H201631), Startup Foundation for Excellent Researchers at Xuzhou Medical University (No. D2016007), The Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 17KJB360014, No. 16KJB180028), and Innovative and Entrepreneurial Talent Scheme of Jiangsu Province (2017). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Footnotes
Author contribution: BG, LW, and ZZ proposed the core ideas of the study and designed the experiment. HQ, YL, and YC did all the experiments for bacterial sequencing. BG, LW, ZZ, and PM performed all the data analysis. All authors contribute to the writing of the manuscript.
Conflict of interest: The authors declare that they have no conflict of interest.
Ethical statement: All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation at Xuzhou Medical University and its 2nd Affiliated Hospital, China. This article does not contain any studies with animal subjects performed by the any of the authors.
References
- 1.Lima IFN, Havt A, Lima AAM. Update on molecular epidemiology of Shigella infection. Cur Opin Gastroenterol. 2015;31:30–37. doi: 10.1097/MOG.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 2.Jennison AV, Verma NK. Shigella flexneriinfection: pathogenesis and vaccine development. FEMS Microbiol Rev. 2004;28:43–58. doi: 10.1016/j.femsre.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Kotloff KL, Riddle MS, Platts-Mills JA, et al. Shigellosis. The Lancet. 2018;391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 4.Clements ACA, Thompson CN, Duy PT, et al. The rising dominance of Shigella sonnei: An intercontinental shift in the etiology of bacillary dysentery. PLOS Negl Trop Dis. 2015;9:e0003708. doi: 10.1371/journal.pntd.0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson J, Jenkins C, Clements A, et al. Shigella sonnei does not use amoebae as protective hosts. Appl Environ Microb. 2018;84:e02679–17. doi: 10.1128/AEM.02679-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 7.Puzari M, Sharma M, Chetia P. Emergence of antibiotic resistant Shigella species: A matter of concern. J Infect Public Health. 2018;11:451–454. doi: 10.1016/j.jiph.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya D, Sugunan AP, Bhattacharjee H, et al. Antimicrobial resistance in Shigella--rapid increase & widening of spectrum in Andaman Islands, India. Indian J Med Res. 2012;135:365–370. [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: Controlling host cell signaling, invasion, and death by Type III secretion. Clin Microbiol Rev. 2008;21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bliven KA, Lampel KA. Shigella: Virulence Factors and Pathogenicity. In: Gurtler JB, Doyle MP, Kornacki JL, editors. Foodborne Pathogens, 7Eds. Springer; 2017. pp. 169–208. [Google Scholar]
- 11.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder M, Brooks B, Brooks A. The complex relationship between virulence and antibiotic resistance. Genes. 2017;8:1–23. doi: 10.3390/genes8010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonomo R, Geisinger E, Mortman NJ, et al. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLOS Pathog. 2018;14:e1007030c. doi: 10.1371/journal.ppat.1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Hinz A, Bauerle E, et al. Targeting a bacterial stress response to enhance antibiotic action. Proc Nati Acad Sci. 2009;106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roux D, Danilchanka O, Guillard T, et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci Transl Med. 2015;7:297ra114–297ra114. doi: 10.1126/scitranslmed.aab1621. [DOI] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darling ACE. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards DJ, Holt KE. Beginner's guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp. 2013;3:1–9. doi: 10.1186/2042-5783-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [Google Scholar]
- 20.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alikhan N-F, Petty NK, Ben Zakour NL, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2004;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:1–8. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelgader SA, Shi D, Chen M, et al. Antibiotics resistance genes screening and comparative genomics analysis of commensal Escherichia coli isolated from poultry farms between China and Sudan. BioMed Res Int. 2018;2018:1–9. doi: 10.1155/2018/5327450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Projan SJ. (Genome) Size Matters. Antimicrob Agents Ch. 2007;51:1133–1134. doi: 10.1128/AAC.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks AR, Peters JE. Tn7 elements: Engendering diversity from chromosomes to episomes. Plasmid. 2009;61:1–14. doi: 10.1016/j.plasmid.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz zur Wiesch P, Engelstadter J, Bonhoeffer S. Compensation of fitness costs and reversibility of antibiotic resistance mutations. Antimicrob Agents Ch. 2010;54:2085–2095. doi: 10.1128/AAC.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miskinyte M, Gordo I. Increased survival of antibiotic-resistant Escherichia coli inside macrophages. Antimicrob Agents Ch. 2013;57:189–195. doi: 10.1128/AAC.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramiro RS, Costa H, Gordo I. Macrophage adaptation leads to parallel evolution of genetically diverse Escherichia coli small-colony variants with increased fitness in vivo and antibiotic collateral sensitivity. Evol Appl. 2016;9:994–1004. doi: 10.1111/eva.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabral D, Wurster J, Belenky P. Antibiotic persistence as a metabolic adaptation: stress, metabolism, the host, and new directions. Pharmaceuticals. 2018;11:1–19. doi: 10.3390/ph11010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettibone GW, Sullivan SA, Shiaris MP. Comparative survival of antibiotic-resistant and -sensitive fecal indicator bacteria in estuarine water. Appl Environ Microbiol. 1987;53:1241–1245. doi: 10.1128/aem.53.6.1241-1245.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flint KP. The long-term survival of Escherichia coliin river water. J Appl Bacteriol. 1987;63:261–270. doi: 10.1111/j.1365-2672.1987.tb04945.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.