Abstract
Eukaryotic transcription factors (TFs) from the same structural family tend to bind similar DNA sequences, despite the ability of these TFs to execute distinct functions in vivo. The cell partly resolves this specificity paradox through combinatorial strategies and the use of low-affinity binding sites, which are better able to distinguish between similar TFs. However, because these sites have low affinity, it is challenging to understand how TFs recognize them in vivo. Here, we summarize recent findings and technological advancements that allow for the quantification and mechanistic interpretation of TF recognition across a wide range of affinities. We propose a model that integrates insights from the fields of genetics and cell biology to provide further conceptual understanding of TF binding specificity. We argue that in eukaryotes, target specificity is driven by an inhomogeneous 3D nuclear distribution of TFs and by variation in DNA binding affinity such that locally elevated TF concentration allows low-affinity binding sites to be functional.
Keywords: transcription regulation, low-affinity binding sites, suboptimal binding sites, 3D genome architecture, transcriptional hubs, phase separation, local transcription factor concentration
INTRODUCTION
The study of gene regulation by transcription factors (TFs) dates back more than half a century, to the pioneering work by Jacob & Monod (1961), the isolation of Escherichia coli and phage repressors (Gilbert & Muller-Hill 1966, Ptashne 1967a), and the subsequent realization that these proteins bind DNA at specific sites (Gilbert & Maxam 1973, Ptashne 1967b). This last discovery raised a new fundamental question: How do these repressors recognize and bind to a specific stretch of DNA from among the millions of possible sites in the E. coli genome? Early X-ray crystallography structures for the Lac and λ repressors (Anderson et al. 1981, Ohlendorf et al. 1982, Steitz et al. 1982) suggested that TFs primarily recognize DNA sequences by forming hydrogen bonds with base pairs in the DNA major groove. Initially, it seemed that simple rules might predict which DNA sequences are bound by any TF (Pabo & Sauer 1984). However, with more structures solved, it became apparent that TFs use a variety of structural mechanisms to recognize DNA (Garvie & Wolberger 2001, Luscombe et al. 2001) and that a simple DNA recognition code might not exist (Pabo & Sauer 1992, Slattery et al. 2014).
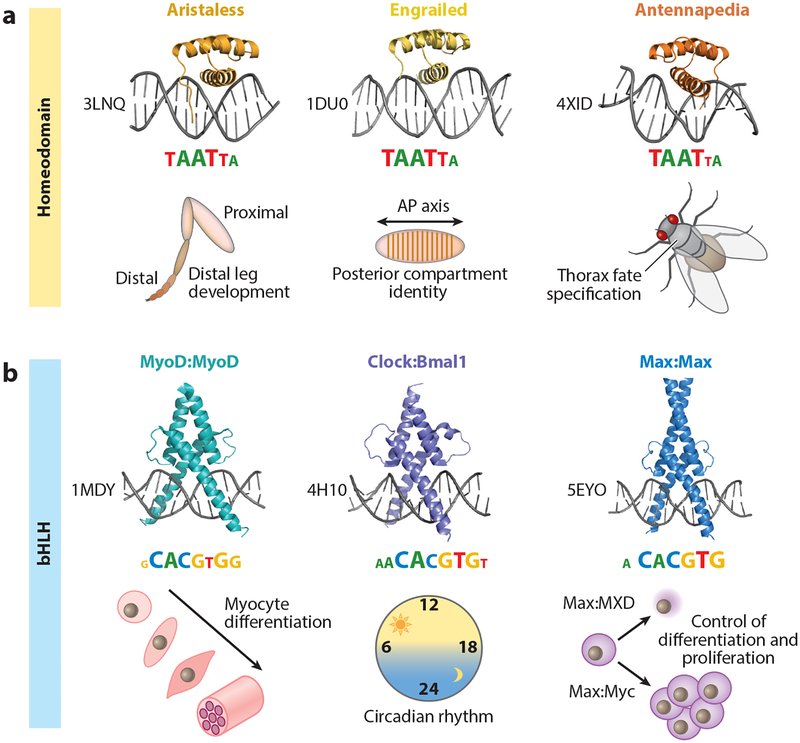
TFs can be grouped into distinct structural families on the basis of their DNA binding domains (DBDs). In eukaryotes, the largest of these are the zinc finger (ZF), homeodomain (HD), basic leucine zipper (bZIP), and basic helix-loop-helix (bHLH) families (Lambert et al. 2018). With the exception of ZF proteins, TF paralogs from the same DBD family tend to recognize similar DNA sequences in vitro (Jolma et al. 2013, Weirauch et al. 2014) (Figure 1a). Early genetic experiments that swapped closely related homeobox DBDs resulted in severe homeotic transformations (reviewed in Mann et al. 2009, Merabet & Mann 2016), indicating that TFs with very similar DBDs nevertheless execute distinct functions in vivo. Moreover, many TFs have crucial functions as master regulators of cell fate (Vierbuchen & Wernig 2012) and perform their functions reliably, despite highly similar sequence recognition. For instance, family members of the bHLH class of TFs control essential processes as different as myocyte differentiation [bHLH MyoD (Tapscott et al. 1988)], regulation of the circadian clock [bHLH Clock and BMAL1 (Dierickx et al. 2018)], and the decision to proliferate or differentiate [bHLH Max (Carroll et al. 2018)], despite recognizing very similar binding sites (Figure 1b).
Figure 1.
Shared binding mechanisms and sequence recognition among transcription factor (TF) paralogs with distinct in vivo functions. (a) Crystal structures, binding motifs, and broad in vivo functions of three examples from the Drosophila melanogaster homeodomain TF family: All three TFs—Aristaless (PDB ID: 3LNQ), Engrailed (PDB ID: 1DU0), and Antennapedia (PDB ID: 4XID)—share a conserved structural fold and recognize similar binding motifs, represented as information content logos (motif logos derived from Noyes et al. 2008). Despite the similarities in DNA recognition and sequence readout, Aristaless, Engrailed, and Antennapedia have distinct in vivo functions: distal leg development (Campbell & Tomlinson 1998), posterior compartment identity along the anterior-posterior (AP) axis (Nusslein-Volhard & Wieschaus 1980), and thoracic fate specification (Struhl 1982), respectively. (b) Crystal structures, binding motifs, and broad in vivo functions of three homo- and heterodimeric examples from the Mus musculus and Homo sapiens basic helix-loop-helix (bHLH) TF family: MyoD:MyoD (PDB ID: 1MDY), Clock:Bmal1 (PDB ID: 4H10), and Max:Max (PDB ID: 5EYO). Each of these TF dimers recognizes a highly similar, reverse-complement-symmetric motif [consensus = CACGTG; motifs taken from Jaspar (Khan et al. 2018)], yet has a distinct in vivo function: MyoD is a master regulator of myocyte fate specification (Tapscott et al. 1988); Clock:Bmal1 are central players in establishing a functional circadian clock (Dierickx et al. 2018); and Max is involved in either cell proliferation or differentiation, depending on its binding partners (e.g., Myc or TFs from the MXD class of bHLH TFs) (Carroll et al. 2018).
To solve this specificity paradox, it is necessary to develop a thorough understanding of the DNA binding mechanisms that TFs employ. In contrast to initial expectations, the majority (approximately two-thirds) of contacts between TFs and DNA are van der Waals (VdW) interactions as opposed to direct hydrogen bonding to bases; the latter occurs as frequently as water-mediated bonds (i.e., in approximately one-sixth of contacts) (Luscombe et al. 2001). This finding implies that sequence specificity emerges not only from specific hydrogen bonding patterns but also from DNA shape readout, which—through VdW, water-mediated, and DNA backbone bonds—contributes significantly to the total free energy of binding (Rohs et al. 2009, 2010).
Multiple studies have demonstrated the importance of DNA shape readout in conferring both binding affinity and DNA binding specificity (reviewed by Rohs et al. 2010, Slattery et al. 2014). For instance, analysis of the crystal structure and binding preferences of the Drosophila TALE (three-amino-acid loop extension) TF Extradenticle (Exd) in complex with the Hox HD factor Sex combs reduced (Scr) revealed that positively charged amino acids within the N-terminal arm of the Hox HD insert into the DNA minor groove, sensing its electrostatic potential (Abe et al. 2015, Joshi et al. 2007). Intrinsic geometric and physical properties of DNA, such as helical twist and electrostatic potential, can be readily predicted from the DNA sequence (Li et al. 2017, Yang et al. 2017) and used to analyze DNA recognition mechanisms (Rube et al. 2018, Zhou et al. 2015). Deviations from classical Watson-Crick base pairing (Nikolova et al. 2011) can also be critical for TF recognition, as exemplified by the presence of Hoogsteen base pairs in the structures of Matα (Aishima et al. 2002) and the p53 tetramer bound to DNA (Kitayner et al. 2010). These examples underscore the idea that, despite being constrained by particular DBD structures, TFs exploit a wide range of mechanisms to bind to a particular DNA sequence.
The notion that any one TF can bind different DNA sequences also dates to early work in prokaryotes. For instance, sequence degeneracies were identified within the λ operator of the E. coli promoter −10 element (Maniatis et al. 1975), and the binding affinity of the λ phage Cro and λ repressors was found to directly depend on the operator sequence (Hochschild et al. 1986). These observations were initially summarized using consensus sequences, which specify degenerate positions with IUPAC ambiguity symbols, and were later summarized using more refined position weight matrices (PWMs) (Stormo et al. 1982), which tabulate nucleotide base frequencies for each position within the TF-DNA interface. A theoretical framework in which competition between mutation and selection defines the information content of the PWM relative to a random genomic background (Berg & von Hippel 1987) in turn led to a widely used visualization known as DNA sequence logos (Schneider & Stephens 1990), in which letter height corresponds to the information gain measured in bits (Figure 1).
Today, TF binding preferences are best determined by performing high-throughput in vitro binding assays (see sidebar titled (Experimental Methods for Detecting Low-Affinity Binding Sites). The two most popular technologies are (a) methods such as protein binding microarrays (PBMs) (Badis et al. 2009, Berger et al. 2008, Bulyk 2007, Mukherjee et al. 2004, Weirauch et al. 2014) and cognate site identifier (Rodriguez-Martinez et al. 2017, Warren et al. 2006), in which binding is quantified for tens of thousands of immobilized DNA probes in parallel, and (b) methods such as high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) (Jolma et al. 2010, Zhao et al. 2009), SELEX-seq (Slattery et al. 2011), Spec-seq (Stormo et al. 2015), SMiLE-seq (Isakova et al. 2017), and DAP-seq (Bartlett et al. 2017), in which high-complexity libraries consisting of either random DNA or genomic fragments are subjected to one or more rounds of selection for TF binding followed by deep sequencing. PWMs derived from these data have been collected in databases such as CIS-BP (Weirauch et al. 2014), JASPAR (Khan et al. 2018), HOCOMOCO (Kulakovskiy et al. 2018), and UniPROBE (Hume et al. 2015).
EXPERIMENTAL METHODS FOR DETECTING LOW-AFFINITY BINDING SITES.
In recent years, high-throughput in vitro binding assays have been modified to improve the detection of low-affinity binding sites, providing an alternative strategy for the laborious trial-and-error process of identifying them in vivo; the latter typically requires enhancer bashing and a series of binding assays or mutational analyses to narrow down a small enhancer fragment (Crocker et al. 2015, Zandvakili et al. 2018). For instance, capturing specificity contributions from regions flanking the TF-DNA interface as observed in crystal structures (the so-called core binding site) can help resolve subtle but important binding preference differences in TF paralogs and TF complexes (Gordan et al. 2013, Shen et al. 2018) but requires targeted DNA probe designs. Assays such as BET-seq (Le et al. 2018), HiP-FA (Jung et al. 2018), MITOMI (Maerkl & Quake 2007), and Spec-seq (Stormo et al. 2015) yield quantitative affinity measurements that also cover the low-affinity range but require prior knowledge about a TF binding site to design the sequence pool. Complementary deep-sequencing-based assays such as HT-SELEX (Jolma et al. 2010), SELEX-seq (Slattery et al. 2011), and SMiLE-seq (Isakova et al. 2017) can measure a relative DNA binding affinity (the binding affinity of a transcription factor for any particular DNA sequence relative to the optimal sequence for the same transcription factor) and show new promise, given the recent emergence of highly accurate modeling approaches (for further details, see sidebar titled Sequence-Based Models of Low-Affinity Binding).
Direct comparison between in vitro DNA binding specificities and genome-wide in vivo TF binding profiles, as assayed using high-throughput methods such as ChIP-seq (Barski et al. 2007, Johnson et al. 2007, Mikkelsen et al. 2007) and related methods (Cheetham et al. 2018, He et al. 2015, Rhee & Pugh 2011, Skene & Henikoff 2017, Southall et al. 2013, van Steensel et al. 2001, Wang et al. 2007), has revealed that a majority of in vivo binding events are not accompanied by an obvious match to the corresponding PWM (Wang et al. 2012, Yang et al. 2006). This observation underscores the TF specificity paradox and suggests that current PWM models are missing a crucial aspect of TF–DNA sequence recognition. Binding sites alternatively referred to as low affinity (Crocker et al. 2015, 2016), suboptimal (Farley et al. 2015), or submaximal (Bhimsaria et al. 2018), which are poorly predicted by most PWM models, have been suggested as an important potential solution to this problem.
Here, we review evidence for the role played by low-affinity binding sites in eukaryotic gene regulation. We discuss how these sites are recognized by TFs and how recently developed computational tools can be used to distinguish them from nonspecific binding sites (see sidebar titled Sequence-Based Models of Low-Affinity Binding). We argue that low-affinity sites can help solve the problem of TF specificity but that this solution requires TF concentrations to be locally boosted to a sufficiently high level for low-affinity sites to be effectively bound. We suggest that eukaryotic nuclear architecture and transcriptional hubs may have coevolved with functional low-affinity binding sites to solve the eukaryotic TF specificity paradox.
SEQUENCE-BASED MODELS OF LOW-AFFINITY BINDING.
Despite the availability of high-quality data, the computational identification of relevant low-affinity sites from in vitro binding data has proven challenging. For example, researchers were unable to identify biologically functional low-affinity binding sites (Crocker et al. 2015) from oligomer enrichment tables derived from SELEX-seq data (Slattery et al. 2011). To be successful, computational approaches require an optimal statistical representation of the data generation process and a flexible representation of the features across the full TF-DNA binding interface that determine binding free energy.
A systematic comparison of methods for analyzing PBM data (Weirauch et al. 2013) revealed that algorithms that fit biophysically motivated models that use DNA sequence features as predictors (Riley et al. 2015, Zhao & Stormo 2011) achieve superior quantification of binding affinities. The resulting models can be represented as position-specific affinity matrices (Bussemaker et al. 2007) and visualized as energy logos (Foat et al. 2006), in which letter heights correspond directly to binding energy differences in bases at each position (Figure 4b, below).
Although technically more challenging due to the discrete nature of probe counts, analogous direct fitting of biophysically motivated feature-based models of protein-DNA interaction to SELEX data recently became possible and will help overcome the limitations of methods based on the oligomer enrichment tables initially used to analyze SELEX data. These feature-based algorithms include (a) an application of convolutional neural networks that treats binding site prediction as a classification problem in terms of bound and unbound probes (Alipanahi et al. 2015) and (b) two distinct maximum-likelihood algorithms, BEESEM (Ruan et al. 2017) and NRLB (Rastogi et al. 2018).
Of these, NRLB is currently the only algorithm capable of learning feature-based models of arbitrary length that accurately capture the entire range of binding affinity. When tested on Drosophila enhancers, NRLB not only successfully identified functional low-affinity Hox binding sites experimentally demonstrated to be >100-fold weaker than the best site in the fly genome, but also accurately predicted the quantitative loss of enhancer activity in vivo as individual sites were sequentially mutated (Rastogi et al. 2018).
If these algorithms can be further improved and leveraged to create a comprehensive resource of accurate sequence-to-affinity models for all TFs, this approach has the potential to transform the way in which enhancers are found and analyzed.
THE UNIQUE CHALLENGES OF EUKARYOTIC GENE REGULATION
Most early examples of TFs and their DNA binding sites came from prokaryotes such as E. coli and its phage. Consequently, theories regarding gene regulation were developed in the context of these organisms, which differ significantly from multicellular eukaryotes. Most prokaryotic TFs consist of only a DBD and an effector domain that mediates direct interactions with RNA polymerase (Perez-Rueda et al. 2018). The TFs of prokaryotes with larger genome sizes—and thus more genes to regulate—have additional DBD architectures, resulting in novel TF families and DNA recognition mechanisms (Minezaki et al. 2005). Consequently, a typical prokaryotic DBD binds with sufficient specificity to almost uniquely specify its target sites in the genome (Wunderlich & Mirny 2009). Use of the same strategy in eukaryotes would require DBDs with much larger binding sites and greater specificity to deal with much larger genomes. Surprisingly, however, the opposite phenomenon is observed: On average, eukaryotic DNA binding sites tend to be small, degenerate, and insufficiently specific to uniquely define a small number of genomic locations (Wunderlich & Mirny 2009).
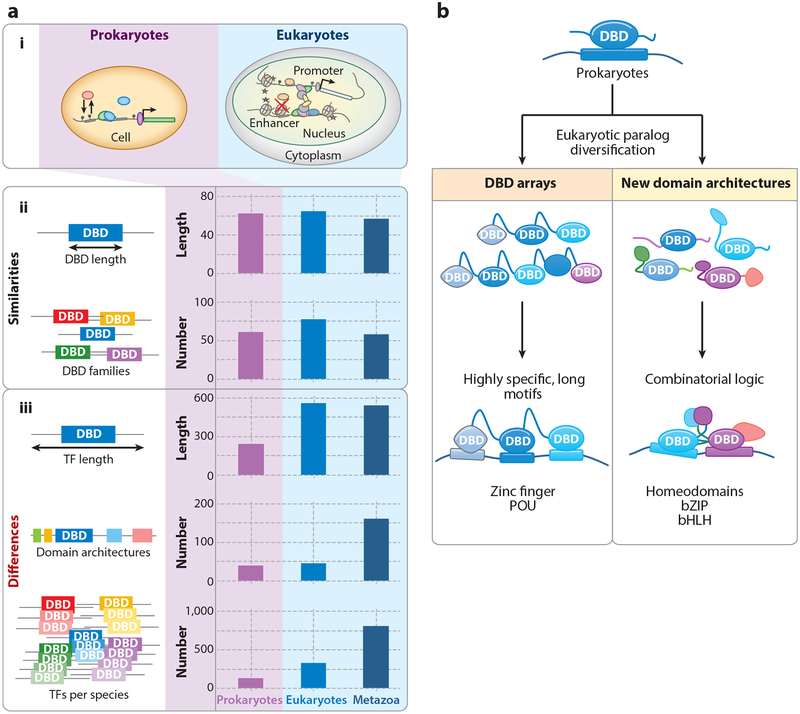
To begin to reconcile this discrepancy, it is informative to ask whether there are differences in TF composition in eukaryotes and prokaryotes that might play a role in diversifying DNA recognition beyond single, short motifs. Indeed, a systematic comparison of TFs across the kingdoms of life (Charoensawan et al. 2010) noted that the number of non-DBDs, the total number of unique domain architectures, the average number of DBDs per TF, and the average length of TFs increased in eukaryotes and, to an even greater extent, in metazoans (Figure 2a). In contrast, however, both the average length of DBDs and the number of DBD families remained largely constant (roughly 60 amino acids per DBD and approximately 60–70 DBD families per organism) (Figure 2a). These observations suggest that structural constraints may place an upper bound on family diversity and DBD size and, consequently, on the achievable specificity of TF binding. However, instead of increasing DBD diversity, eukaryotes dramatically increased the number of paralogs within individual TF families (Figure 2a). Most notably, the human TF repertoire is dominated by two major families: the C2H2 ZF proteins, with >700 members, and HD proteins, with roughly 200 members (Lambert et al. 2018). The existence of so many structurally related members of the same DBD family makes it even more challenging to understand how individual specificities and TF functions are achieved.
Figure 2.
Prokaryotic versus eukaryotic transcription factors (TFs): design principles and binding mechanisms (Charoensawan et al. 2010).(a) Comparison between prokaryotic and eukaryotic TF design principles. (i) Simplified cellular structure of prokaryotic and eukaryotic cells. (ii) Similarities in TFs. The average amino acid (aa) length of the DNA binding domain (DBD) (~60 aa) and the total number of distinct structural families per species are conserved across animal kingdoms. (iii) Differences in TFs. Compared to prokaryotes, eukaryotic and metazoan TFs are on average longer (in terms of total aa length) and have a higher number of unique domain architectures (i.e., combinations of DBDs and non-DBDs). In addition, eukaryotes have a much larger number of TF paralogs per structural family of DBDs. (b) Mechanisms of binding site diversification in eukaryotes beyond adding new DBDs. Prokaryotic binding sites are tailored to family-specific DBDs, with high information content and long binding sites. Following paralog expansion, eukaryotes diversified their binding site preferences by adding (left) tandem arrays of DBDs to specify longer or more complex sequence footprints and (right) new interaction domains that allowed for combinatorial binding logic. Other abbreviations: bHLH, basic helix-loop-helix; bZIP, basic leucine zipper; POU, domain shared by Pit-1, Oct-1, and Unc-86.
EUKARYOTES EXPLOIT COMPLEX DOMAIN STRUCTURES TO ENHANCE TRANSCRIPTION FACTOR DNA BINDING SPECIFICITY
Eukaryotes use several distinct combinatorial strategies to increase genomic target specificity. One strategy equips individual TFs with more than one DBD of the same family (Figure 2b) and, less frequently, with DBDs from different families (Lambert et al. 2018). In principle, this can lead to longer protein-DNA footprints with higher binding specificity, approaching that of prokaryotes. For example, human C2H2 ZF TFs can contain as many as 40 ZF domains, each recognizing 3 to 4 bp (Fedotova et al. 2017, Lambert et al. 2018). However, most ZF TFs have fewer fingers or use only subsets of fingers to recognize particular binding sites (Barazandeh et al. 2018, Fedotova et al. 2017, Najafabadi et al. 2015). Nevertheless, some ZF protein binding sites can be as long as 60 bp; an example is CTCF, which helps orchestrate chromosome architecture (Nakahashi et al. 2013). In a variant of this strategy, the human tumor suppressor p53 combines a DBD with a tetramerization domain (Wang et al. 1994, Weinberg et al. 2004), creating a four-subunit complex that recognizes an unusually long (>20-bp) binding site (Funk et al. 1992, Rastogi et al. 2018). It is noteworthy that both ZF TFs and p53 tetramers often exert highly specialized functions in the cell that are not restricted to specific cell types, thus allowing for a prokaryotic-type approach to recognize genomic target sites.
An alternative strategy combines DBDs with domains that mediate homo- or heterotypic interactions with other proteins (Figure 2b). This type of interaction is seen in some of the heavily expanded TF families in the human genome. For instance, bZIP proteins form a large variety of homo- and heterodimers (Miller 2009, Rodriguez-Martinez et al. 2017, Vinson et al. 2002). Cooperative TF complexes can also be formed by exploiting novel architectures beyond the DBD, such as the use of short linear motifs to mediate protein-protein interactions (Davey et al. 2012, Jolma et al. 2015, Merabet & Mann 2016, Slattery et al. 2011). Although new sequence specificities are created and footprints are extended by combining two TFs in this manner, this mechanism is typically not sufficient to bind a unique set of sites in vivo.
A few known examples of complexes consist of three or more cooperatively interacting TFs. The most prominent example is the IFN-β enhanceosome, in which eight factors are assembled on a DNA sequence with very stringent constraints on binding site position and orientation (Thanos & Maniatis 1995); given the paucity of protein-protein contacts seen in IFN-β crystal structures (Panne 2008, Panne et al. 2007), the assembly of the enhanceosome appears to be driven by a highly optimized DNA sequence. However, large-scale assemblies with strict constraints on binding site architecture seem to be the exception rather than the norm: While there are many examples of cooperatively bound TF pairs within enhancers (Jolma et al. 2015, Merabet & Mann 2016), most eukaryotic enhancers tolerate a much looser binding site arrangement (Arnosti & Kulkarni 2005, Panne 2008, Spitz & Furlong 2012).
Complex formation by pairs of TFs can reveal latent specificities (Slattery et al. 2011), whereby the binding preferences of the complex cannot simply be explained as a combination of the individual TF specificities (Ansari & Peterson-Kaufman 2011, Jolma et al. 2015, Mann & Chan 1996, Slattery et al. 2011). For instance, the Drosophila melanogaster Hox protein Scr is sensitive to variations in the DNA minor groove width only when bound in complex with its HD cofactor Exd (Abe et al. 2015, Zhou et al. 2015); Exd positions the N-terminal arm of Scr near the minor groove through a direct protein-protein interaction, which explains why this sensitivity is not observed when Scr binds as a monomer (Joshi et al. 2007). While latent specificity extends the classical concept of cooperative binding (Oehler et al. 1990) and thus contributes to the diversification of TF specificities, this gain in specificity is not sufficient to define a small subset of specific binding sites in eukaryotic genomes.
Recently, a direct link was made between DNA shape and TF affinity: By solving the X-ray crystal structures of the same Exd-Hox heterodimer bound to four different DNA binding sites of different affinity, a relationship between affinity and intrinsic DNA shape was revealed (Zeiske et al. 2018). All four ternary complexes had very similar protein and DNA structures, regardless of binding site affinity. However, although the predicted structures of unbound high-affinity binding sites were similar to the bound DNA shape, the shapes of lower-affinity sites were different, making binding energetically less favorable (Zeiske et al. 2018). Thus, in addition to intrinsic DNA shape playing a role in paralog binding specificity, differences in intrinsic DNA shape can also impact TF affinity.
Epigenetic DNA modifications also have the potential to modulate TF binding affinity and specificity. The most extensively studied case is the effect of CpG cytosine methylation on TF binding. Several studies have shown that methylation can influence TF binding both in vivo (Domcke et al. 2015) and in vitro (Kribelbauer et al. 2017, Mann et al. 2013, Yin et al. 2017, Zhu et al. 2016, Zuo et al. 2017). Depending on the TF, CpG methylation can both increase and decrease DNA binding affinity. Moreover, CpG methylation can alter DNA binding affinity in a paralog-specific manner, creating new low-affinity binding sites that are preferentially bound by specific members of the same TF family (Kribelbauer et al. 2017, Yin et al. 2017).
SPECIFICITY-AFFINITY TRADE-OFF: LOW-AFFINITY BINDING FACILITATES TRANSCRIPTION FACTOR PARALOG SPECIFICITY
The strategies outlined above help us understand how eukaryotes have been able to exploit cooperative binding to increase specificity from the perspective of an individual TF complex that binds across the genome. However, such strategies do not resolve the specificity problem from the point of view of an individual binding site that must recruit a particular TF. This paradox is especially striking for the eukaryote-specific family of HD TFs (Burglin & Affolter 2016, Charoensawan et al. 2010): Two large-scale studies comparing HD binding specificities found that most HD TFs bind relatively short (6–8-bp) sequences with a high degree of overlap between paralogs (Berger et al. 2008, Noyes et al. 2008). Given this degeneracy, the optimal site for a given HD will typically also be a high-affinity binding site for many other family members (Crocker et al. 2015). In addition, unless the available set of paralogs is restricted by cell type–specific TF expression, high-affinity binding sites appear to be useful only when neither paralog specificity nor tissue specificity is required (Mann et al. 2009). Therefore, the question is, how can a binding site be created that has high enough affinity for its target TF but also favors that TF over its close paralogs?
Recent work has demonstrated the importance of low-affinity sites in paralog-specific gene regulation. Two studies showed that replacing low-affinity sites with high-affinity ones resulted in ectopic gene activation (Crocker et al. 2015, Farley et al. 2015), suggesting that paralogous TFs in other tissues ignore the endogenous low-affinity site, yet occupy the artificial high-affinity one. In other work, the modification of Paired HD binding sites upstream of Drosophila Rhodopsin gene promoters resulted in altered spatial expression patterns, implying that subtle differences in affinity can cause a switch in sequence selectivity from one HD TF to another (Rister et al. 2015). More recently, low-affinity sites were shown to be important for distinguishing between the binding of and regulation by two different K50 HD proteins in Drosophila: Bicoid and Orthodenticle (Datta et al. 2018).
Low-affinity binding sites were also required for the correct interpretation of Hedgehog (Hh) signaling: Multiple low-affinity Cubitus interruptus (Ci) sites in Hh-regulated enhancers were required for activation in cells receiving low Hh signaling, whereas enhancers with artificially generated, high-affinity sites caused Ci to behave as a repressor (Ramos & Barolo 2013). Other studies have demonstrated the ability of low-affinity sites to distinguish between repression (by Senseless) and activation (by Pax2) in the Drosophila peripheral nervous system (Zandvakili et al. 2018) and to properly time the expression of important developmental genes (Gaudet & Mango 2002).
Another notable example is a Drosophila mesodermal enhancer controlled by the Ets domain paralogs Yan and Pnt. High-affinity Yan sites are required for repression, while low-affinity Pnt sites are required for activation (Boisclair Lachance et al. 2018). Because of Yan’s relative preference for the high-affinity sites, low levels of this TF are sufficient to prevent activation by Pnt, again suggesting that differences in binding site affinity of different TF paralogs can be exploited to fine-tune enhancer activities.
Taken together, these findings indicate that functional low-affinity binding sites (a) are common in eukaryotes, (b) contribute to paralog specificity, and (c) are capable of fine-tuning gene expression patterns both spatially and temporally. Low-affinity binding sites also have the advantage of requiring less stringent sequence conservation, as it is easier to recreate multiple low-affinity sites than to maintain a single high-affinity site. Consistent with this notion, low-affinity sites in functionally conserved enhancers can exhibit rapid turnover in closely related species (Crocker et al. 2015, 2016). More generally, weak cooperative interactions have been proposed to contribute to the large increase in the number of processes that eukaryotic cells need to execute (Gao et al. 2018). However, given that genomes contain many more low-affinity sites than high-affinity ones, it is still difficult to fathom how TFs can bind to their desired targets in a manner that is not only paralog specific but also locus specific.
THE CRUCIAL ROLE OF LOCAL TRANSCRIPTION FACTOR CONCENTRATION
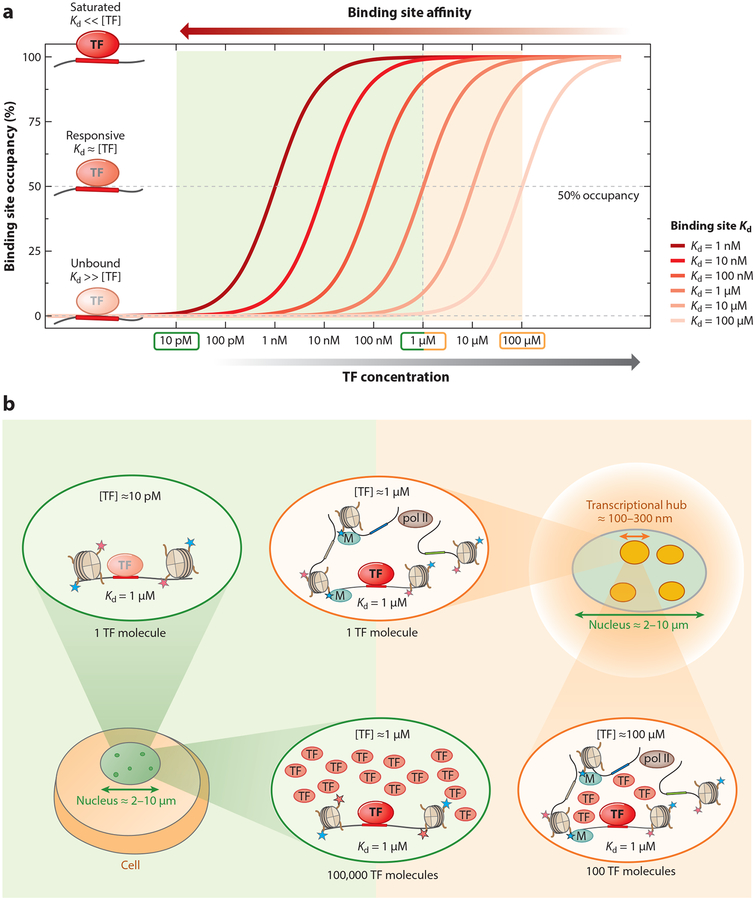
To understand how eukaryotic cells achieve significant binding at low-affinity binding sites, it is helpful to consider two parameters that are equally important in determining the average TF occupancy at a particular DNA binding site (Figure 3a). The first of these, [TF]free, is the local concentration of unbound, or free, DNA binding domains; the second, Kd, is the dissociation constant, which quantifies the strength of the protein-DNA interaction and is inversely proportional to affinity. The relationship between [TF]free and [TF]total is highly context dependent and difficult to predict. However, as long as thermodynamic equilibrium conditions hold on the relevant spatial and temporal scales, average TF occupancy depends only on the ratio of [TF]free to Kd (i.e., is proportional to both [TF]free and the affinity of the binding site). When [TF]free equals Kd, the bound state and the unbound state are equally probable, resulting in an average occupancy of 50%. At low TF concentrations, the occupancy is proportional to [TF]free; at high concentrations, occupancy saturates at the maximum value of 100%. To maintain significant occupancy at binding sites of lower affinity (higher Kd), the TF concentration needs to increase.
Figure 3.
Transcription factor (TF) occupancy is a function of both binding site affinity and TF concentration. (a) TF binding to a specific site depends on both the free TF concentration ([TF]free) and the Kd (the inverse of affinity) of the binding site. TF occupancy as a function of free TF concentration is shown for six different binding site affinities (with Kd from 1 nM to 100 μM). Depending on the concentration and Kd, TFs are either unbound ([TF]free ≪ Kd) in a responsive regime, where occupancy varies with concentration ([TF]free ~ Kd), or saturated ([TF]free ≫ Kd). (b) The influence of nuclear compartment size on TF concentration and on the ability of sites of different affinity to be bound by the TF. Three different concentration regimes (low, intermediate, and high) are highlighted. On the low-concentration end, having only a single TF molecule inside a nucleus with a diameter of 2–10 μm (green) yields [TF]total ~ 10 pM, which represents an upper bound on [TF]free. At such a low concentration, a weak binding site (Kd ~ 1 μM) would essentially be unbound; to achieve significant occupancy, 100,000 TF molecules would be required, resulting in a nuclear concentration of ~1 μM, similar to the Kd. Eukaryotic nuclear architecture brings groups of regulatory elements into proximity in 3D, forming transcriptional hubs of ~100 nm in diameter (orange). Restricting a single TF molecule to a subnuclear compartment of this size also corresponds to a TF concentration of 1 μM. Recruiting a larger number of free TF molecules to the hub would allow even ultralow-affinity sites with Kds of up to 100 μM to be occupied, while saturation would no longer be limited to just the near-optimal sites (Kd ~ 10 nM).
It is informative to consider how many molecules of a particular TF would be required to reach ~50% occupancy for a given Kd range (Figure 3b). Having one TF molecule in a typical eukaryotic nucleus with a diameter of ~6 μm corresponds to a total nuclear TF concentration of ~10 pM, which is an upper bound on [TF]free. This value is far lower than the typical Kd values (~10 nM) for optimal TF binding sites, which would therefore rarely be bound under these conditions. Having 100,000 TF molecules in the nucleus would correspond to a concentration of ~1 μM, at which even low-affinity sites with a Kd of ~1 μM (i.e., 100-fold weaker than a typical high-affinity site) are significantly bound. However, if the distribution of TF molecules within the nucleus is uniform, all binding sites will experience the same [TF]free, implying that binding at all high-affinity sites will be saturated and that therefore these sites will no longer be responsive to variation in TF concentration. That eukaryotes would choose to have their TFs constitutively occupy a large number of sites in the genome seems implausible, and indeed TF concentration levels may be tuned to the Kd values of their top target sites (Brewster et al. 2014, Gerland et al. 2002). However, functionally relevant low-affinity binding sites will not be sufficiently occupied to impact gene expression at TF concentrations at which saturated binding is avoided for the strongest binding sites in the genome.
PHASE SEPARATION, TRANSCRIPTIONAL HUBS, AND SUBNUCLEAR COMPARTMENTS
In any given cell type, only a portion of the genome is accessible for TF binding (Guertin & Lis 2013), as shown in both DNase-seq (Hesselberth et al. 2009) and ATAC-seq (Buenrostro et al. 2013) datasets. While nonuniform chromatin accessibility may appear to address the binding site selection problem, there are still far too many binding sites within accessible regions for most TFs. Moreover, accessible regions in chromatin are themselves a consequence of prior TF binding and activity, which further raises the question of how these regions are selected to begin with.
A more complete answer to the target specificity conundrum may lie in the highly compartmentalized structure of eukaryotic nuclei (Dixon et al. 2012, Furlong & Levine 2018, Nora et al. 2012). During the past decade, chromosome conformation capture (3C) and derived high-throughput assays (Dekker et al. 2002, Lieberman-Aiden et al. 2009) as well as DNA imaging studies (Boettiger et al. 2016, Chen et al. 2014, Chen et al. 2015, Fraser et al. 2015, Giorgetti & Heard 2016) have revolutionized our understanding of 3D genome architecture. Imaging studies with tagged TFs have shown that they are nonrandomly distributed within both fixed and unfixed nuclei and form clusters with high protein and polymerase content (Cisse et al. 2013, Liu et al. 2014, Mir et al. 2018, Tsai et al. 2017; see Furlong & Levine 2018 and Liu & Tjian 2018 for recent reviews).
Importantly, these studies suggest the existence of spatial clusters of regulatory elements that are potentially coregulated by the same set of TFs. An intriguing advantage of such subnuclear compartments, or transcriptional hubs, is that TF concentration is likely to differ greatly from the nuclear average. For example, if one restricts a single TF molecule to a compartment with a diameter 10% that of the nucleus, its concentration increases 1,000-fold. A single TF molecule in a typical nuclear hub with a diameter of ~100 nm (Furlong & Levine 2018) corresponds to a local [TF]free of ~1 μM, equivalent to having 100,000 TF molecules distributed uniformly throughout the nucleus (Figure 3b).
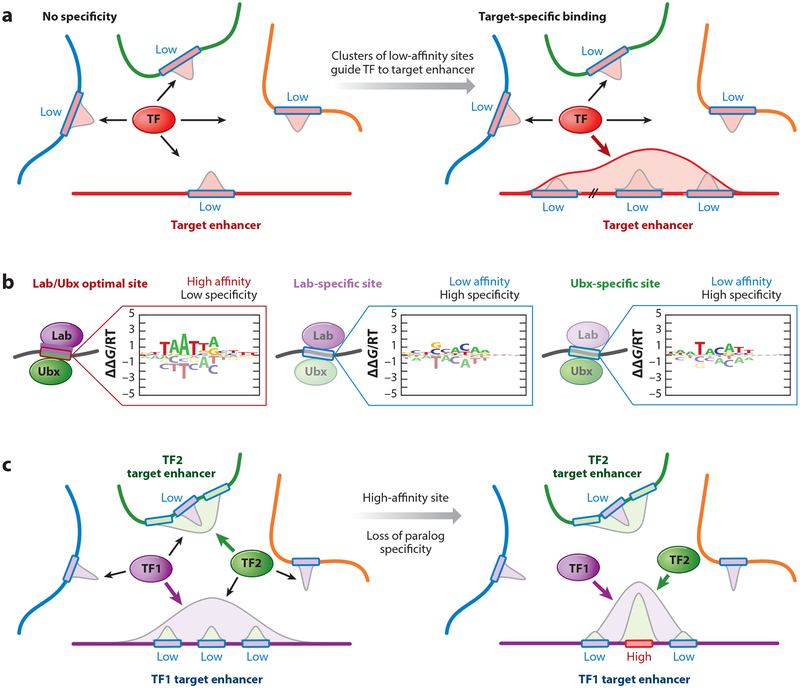
The 3D compartmentalization of the nucleus thus provides a powerful mechanism for increasing local [TF]free for specific regulatory elements inside a hub. It also immediately provides a rationale for the pervasiveness of functional low-affinity binding sites: Any binding site with a Kd below 1 μM would be fully saturated inside a hub, a quality not desirable for tunable gene expression. In contrast, the Kds of low-affinity targets are likely within a tunable range, even at the high TF concentrations occurring within hubs. Control of specific enhancers inside a hub (Figure 4) can be further refined using latent specificity (Slattery et al. 2011), TF cooperativity (Jolma et al. 2015), binding site syntax (i.e., the relative order, orientation, and spacing of binding sites) (Farley et al. 2016), and clustering of low-affinity sites (also referred to as homotypic clusters of TF binding sites) (Ezer et al. 2014, Gotea et al. 2010) (Figure 4a). Moreover, the high TF concentration within transcriptional hubs allows low-affinity sites to be bound, which in turn provides the flexibility for TF paralogs to functionally diverge (Figure 4b,c).
Figure 4.
Low-affinity binding sites and their importance in guiding transcription factors (TFs) to their cognate target sites. (a) The role of clusters of low-affinity sites in guiding TF binding within transcriptional hubs. Since local TF concentration inside a hub is >1 μM (see Figure 3b), the preferred Kd for responsive TF binding sites is also 1 μM or higher, which is low relative to optimal sites. Since many different DNA sequences can be used to realize low-affinity binding, each enhancer inside the hub has a high probability of harboring at least one low-affinity binding site, resulting in ambiguity. (Left) An extreme case in which each of the four enhancers has one low-affinity binding site within the appropriate Kd regime. Averaged over time or a population of cells, the TF will occupy each enhancer equally, resulting in a uniform distribution with no specificity for the target enhancer. (Right) Clusters of low-affinity binding sites confer target enhancer specificity by increasing its share in TF binding. On average, the TF will now predominantly occupy its target enhancer, which has the largest cumulative binding affinity. (b,c) The importance of low-affinity sites in paralog-specific binding. (b) Sequence specificities of the closely related Drosophila melanogaster TF paralogs Labial (Lab) and Ultrabithorax (Ubx), represented as energy logos (Foat et al. 2006). The optimal, high-affinity site for Lab is strongly bound by both Lab and Ubx (left logo) due to their shared DNA recognition strategy. The sequences that maximize specificity for Lab and Ubx, respectively (middle and right logos), are low affinity (~10,000-fold weaker than the optimal site) and, unlike high-affinity sites, have equal contributions across nucleotide positions within the binding interface. (c) Replacing a low-affinity, yet highly specific, binding site within the target enhancer for one paralog (TF1, purple) with a high-affinity site will cause loss of paralog-specific binding. TF2 (green) will no longer predominantly occupy its target enhancer; instead, both TF1 and TF2 will be directed to the same enhancer.
While nuclear hub assembly provides a plausible explanation for how low-affinity binding sites can effectively be occupied in cells, it raises the question of how these microenvironments form in the first place. Low-complexity protein domains, also termed intrinsically disordered regions (IDRs), are present in many nuclear proteins (Shin & Brangwynne 2017, Zhu & Brangwynne 2015) and TFs (Chong et al. 2018; see Alberti et al. 2019 for a recent review). Recent studies suggest that IDRs can promote the creation of distinct microenvironments via a process referred to as phase separation (Banani et al. 2016, Erdel & Rippe 2018, Hyman et al. 2014, Wheeler & Hyman 2018). Phase separation is thought to underlie the formation of heterochromatin via the biophysical properties of HP1 (Strom et al. 2017) and the 3D segregation of superenhancers into hubs, driven by specific combinations of TFs and mediator proteins (Hnisz et al. 2017, Sabari et al. 2018). Another notable example is the regulation of olfactory receptor (OR) gene expression, in which phase separation has been proposed to play a role in mediating long-range interchromosomal interactions that silence all but one of >1,000 OR genes in olfactory sensory neurons (Monahan et al. 2019). Finally, activation domains of several TFs phase-separate together with transcriptional coactivator proteins, and the formation of these condensates is involved in gene activation (Boija et al. 2018).
Despite these examples, how phase-separated TF hubs are formed is currently unknown. Their formation may emerge from a combination of multiple weak protein-protein and protein-DNA interactions (Figure 5). Regardless of the mechanism, we suggest that the ability of phase-separated hubs to form in higher eukaryotes enabled closely related TFs to regulate distinct target genes by taking advantage of paralog-specific low-affinity binding sites (Figure 5).
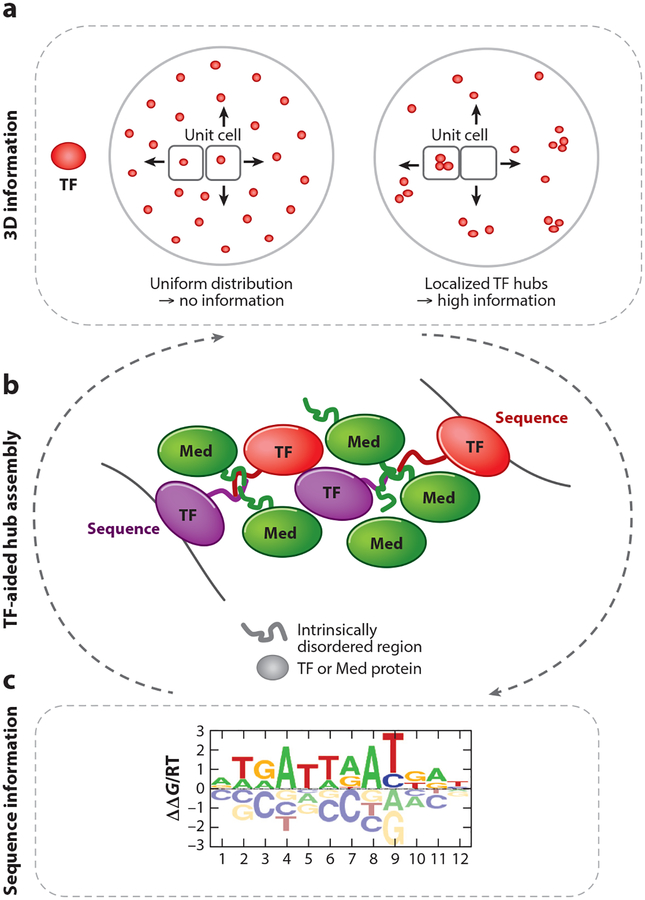
Figure 5.
Weak protein-protein interactions as a means to concentrate transcription factors (TFs) and assemble nuclear hubs. (a) The inhomogeneous 3D nuclear distribution of TFs provides a regulatory layer that complements the information encoded in the DNA sequence, by limiting TFs to distinct nuclear compartments and boosting their local concentration. (b) The assembly of transient, phase-separated protein condensates is mediated by weak protein-protein interactions between intrinsically disordered regions (IDRs) of both TFs and mediator (Med) proteins. (c) These microenvironments allow TFs to bind to paralog-specific low-affinity binding sites. A combination of weak protein-protein and protein-DNA interactions likely drive hub formation. Whether TF-DNA binding precedes phase separation, or vice versa, and what role chromatin plays remain to be determined.
CONCLUDING REMARKS
We discuss above how the occupancy by a particular TF along the genome may be shaped as much by the nonuniform 3D distribution of TF molecules within the nucleus as by the 1D landscape of DNA binding affinity (Figure 5). A key remaining challenge is to find ways to characterize and represent transcriptional hubs and the resulting profile of local TF concentration along the genome to enable quantitative predictions of TF occupancy. A convergence between genetics and cell biology, two fields that are historically distinct, will likely be required to achieve this goal.
SUMMARY POINTS.
There is a trade-off between a DNA sequence (a) having a high binding affinity for a particular TF and (b) being able to preferentially recruit that TF in favor of closely related, paralogous TFs from the same structural family.
The emergence of large TF families in metazoans through duplication and divergence has made it necessary for the cell to find ways to make use of low-affinity binding sites.
To what degree a particular binding site in the genome is occupied by a TF molecule depends on the ratio of [TF] to Kd, i.e., between the (effective, local, free) TF concentration [TF] and the dissociation constant Kd that quantifies the protein-TF interaction.
When a TF is restricted to a subnuclear volume with a diameter 10% that of the entire nucleus, its concentration goes up 1,000-fold, and thus the formation of phase-separated subnuclear compartments, or hubs, is a powerful strategy for making use of low-affinity sites.
Recent advances in high-throughput genomics and statistical learning are starting to make it feasible to build highly accurate models of protein-DNA interaction that can reliably detect low-affinity binding sites in genomic DNA.
To predict which promoters and enhancers are regulated by a particular TF, it is not sufficient to know its full genomic affinity profile; information about the TF’s 3D distribution within the cell nucleus is also important.
FUTURE ISSUES.
New methods are required for estimating local TF concentration, either by direct experimental observation or by computational inference.
Better strategies are needed to predict which TFs from among a set of close paralogs will preferentially bind to a given DNA sequence.
Methods are required to quantify the nonuniform distribution of TF hubs within eukaryotic nuclei.
We need a better understanding of how hubs emerge from a combination of weak protein-protein and protein-DNA interactions.
ACKNOWLEDGMENTS
We thank H. Tomas Rube and other members of the Bussemaker and Mann labs for valuable discussions. This work was supported by an HHMI International Student Research Fellowship (to J.F.K.) and by NIH grants R35GM118336 to R.S.M. and R01HG003008 to H.J.B.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Glossary
- Transcription factor (TF)
a protein containing at least one DNA binding domain along with domains that mediate interactions with cofactors and transcriptional machinery
- Paralogs
related transcription factors in the same organism that belong to the same family; paralogs typically have similar DNA binding domains, yet different biological functions
- DNA binding specificity
a quantification of the relative binding affinity of a transcription factor (or transcription factor complex) across all possible DNA sequences
- Consensus sequence
a DNA sequence pattern specifying the frequently observed nucleotides at each position of a transcription factor binding site
- IUPAC
International Union of Pure and Applied Chemistry (https://iupac.org/)
- Position weight matrix (PWM)
a mathematical model that predicts binding affinity by quantifying the effect of base substitution at each position in the binding site
- Protein binding microarray (PBM)
a method to characterize the DNA binding specificity of a transcription factor by using <105 immobilized DNA probes
- Systematic evolution of ligands by exponential enrichment (SELEX)
a method for characterizing the DNA binding specificity of a transcription factor by using a random DNA library
- Low-affinity binding site
a DNA site bound up to 1,000-fold more weakly than the optimal DNA sequence, but still more strongly than the immediately surrounding sequence
- Transcriptional hub
a type of subnuclear compartment characterized by a high concentration of RNA polymerase and the site of active transcription
- Intrinsic DNA shape
the structure of a DNA binding site prior to binding of a transcription factor; this shape may be different from the DNA shape in the transcription factor–bound complex
- Transcription factor paralog specificity
the ability of a DNA ligand to recruit a particular paralog among multiple available transcription factors from the same structural famil
- Transcription factor locus specificity
the ability of a specific genomic site to preferentially bind a transcription factor as opposed to other potential sites in the genome
- Binding saturation
a situation that arises whenever the local free transcription factor concentration is much larger than the dissociation constant of the DNA binding site
- Subnuclear compartment
a three-dimensional membraneless volume within a nucleus with distinct molecular composition; this compartment is transiently created by a complex network of protein-protein and protein-DNA interactions
LITERATURE CITED
- Abe N, Dror I, Yang L, Slattery M, Zhou T, et al. 2015. Deconvolving the recognition of DNA shape from sequence. Cell 161:307–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. 2002. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 30:5244–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T. 2019. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176:419–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol 33:831–38 [DOI] [PubMed] [Google Scholar]
- Anderson WF, Ohlendorf DH, Takeda Y, Matthews BW. 1981. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290:754–58 [DOI] [PubMed] [Google Scholar]
- Ansari AZ, Peterson-Kaufman KJ. 2011. A partner evokes latent differences between Hox proteins. Cell 147:1220–21 [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell Biochem 94:890–98 [DOI] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324:1720–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, et al. 2016. Compositional control of phase-separated cellular bodies. Cell 166:651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazandeh M, Lambert SA, Albu M, Hughes TR. 2018. Comparison of ChIP-seq data and a reference motif set for human KRAB C2H2 zinc finger proteins. G3 8:219–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–37 [DOI] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, et al. 2017. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc 12:1659–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. 1987. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol 193:723–50 [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133:1266–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhimsaria D, Rodriguez-Martinez JA, Pan J, Roston D, Korkmaz EN, et al. 2018. Specificity landscapes unmask submaximal binding site preferences of transcription factors. PNAS 115:E10586–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, et al. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529:418–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175:1842–55.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisclair Lachance JF, Webber JL, Hong L, Dinner AR, Rebay I. 2018. Cooperative recruitment of Yan via a high-affinity ETS supersite organizes repression to confer specificity and robustness to cardiac cell fate specification. Genes Dev. 32:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster RC, Weinert FM, Garcia HG, Song D, Rydenfelt M, Phillips R. 2014. The transcription factor titration effect dictates level of gene expression. Cell 156:1312–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10:1213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk ML. 2007. Protein binding microarrays for the characterization of DNA-protein interactions. Adv. Biochem. Eng. Biotechnol 104:65–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR, Affolter M. 2016. Homeodomain proteins: an update. Chromosoma 125:497–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, Foat BC, Ward LD. 2007. Predictive modeling of genome-wide mRNA expression: from modules to molecules. Annu. Rev. Biophys. Biomol. Struct 36:329–47 [DOI] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. 1998. The roles of the homeobox genes aristaless and Distal-less in patterning the legs and wings of Drosophila. Development 125:4483–93 [DOI] [PubMed] [Google Scholar]
- Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. 2018. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front. Med 12:412–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoensawan V, Wilson D, Teichmann SA. 2010. Genomic repertoires of DNA-binding transcription factors across the tree of life. Nucleic Acids Res. 38:7364–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham SW, Gruhn WH, van den Ameele J, Krautz R, Southall TD, et al. 2018. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development 145:dev170209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156:1274–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. 2015. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, et al. 2018. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, et al. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341:664–67 [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160:191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Noon EP, Stern DL. 2016. The soft touch: low-affinity transcription factor binding sites in development and evolution. Curr. Top. Dev. Biol 117:455–69 [DOI] [PubMed] [Google Scholar]
- Datta RR, Ling J, Kurland J, Ren X, Xu Z, et al. 2018. A feed-forward relay integrates the regulatory activities of Bicoid and Orthodenticle via sequential binding to suboptimal sites. Genes Dev. 32:723–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, et al. 2012. Attributes of short linear motifs. Mol. Biosyst 8:268–81 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–11 [DOI] [PubMed] [Google Scholar]
- Dierickx P, Van Laake LW, Geijsen N. 2018. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 19:18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Ginno PA, Hartl D, Burger L, Schubeler D. 2015. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528:575–79 [DOI] [PubMed] [Google Scholar]
- Erdel F, Rippe K. 2018. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J 114:2262–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Zabet NR, Adryan B. 2014. Homotypic clusters of transcription factor binding sites: a model system for understanding the physical mechanics of gene expression. Comput. Struct. Biotechnol. J 10:63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. 2015. Suboptimization of developmental enhancers. Science 350:325–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. 2016. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. PNAS 113:6508–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotova AA, Bonchuk AN, Mogila VA, Georgiev PG. 2017. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Nat. 9:47–58 [PMC free article] [PubMed] [Google Scholar]
- Foat BC, Morozov AV, Bussemaker HJ. 2006. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics 22:e141–49 [DOI] [PubMed] [Google Scholar]
- Fraser J, Williamson I, Bickmore WA, Dostie J. 2015. An overview of genome organization and how we got there: from FISH to Hi-C. Microbiol. Mol. Biol. Rev 79:347–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol 12:2866–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, Levine M. 2018. Developmental enhancers and chromosome topology. Science 361:1341–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Shrinivas K, Lepeudry P, Suzuki HI, Sharp PA, Chakraborty AK. 2018. Evolution of weak cooperative interactions for biological specificity. PNAS 115:E11053–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C. 2001. Recognition of specific DNA sequences. Mol. Cell 8:937–46 [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295:821–25 [DOI] [PubMed] [Google Scholar]
- Gerland U, Moroz JD, Hwa T. 2002. Physical constraints and functional characteristics of transcription factor–DNA interaction. PNAS 99:12015–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Maxam A. 1973. The nucleotide sequence of the lac operator. PNAS 70:3581–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Muller-Hill B. 1966. Isolation of the lac repressor. PNAS 56:1891–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Heard E. 2016. Closing the loop: 3C versus DNA FISH. Genome Biol. 17:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Shen N, Dror I, Zhou T, Horton J, et al. 2013. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 3:1093–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. 2010. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 20:565–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. 2013. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr. Opin. Genet. Dev 23:116–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. 2015. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol 33:395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6:283–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A, Douhan J 3rd, Ptashne M. 1986. How lambda repressor and lambda Cro distinguish between OR1 and OR3. Cell 47:807–16 [DOI] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. 2015. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 43:D117–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30:39–58 [DOI] [PubMed] [Google Scholar]
- Isakova A, Groux R, Imbeault M, Rainer P, Alpern D, et al. 2017. SMiLE-seq identifies binding motifs of single and dimeric transcription factors. Nat. Methods 14:316–22 [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3:318–56 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–502 [DOI] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, et al. 2010. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20:861–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152:327–39 [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, et al. 2015. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384–38 [DOI] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, et al. 2007. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131:530–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Bandilla P, von Reutern M, Schnepf M, Rieder S, et al. 2018. True equilibrium measurement of transcription factor–DNA binding affinities using automated polarization microscopy. Nat. Commun 9:1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, et al. 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46:D260–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, et al. 2010. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol 17:423–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kribelbauer JF, Laptenko O, Chen S, Martini GD, Freed-Pastor WA, et al. 2017. Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 19:2383–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, et al. 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46:D252–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, et al. 2018. The human transcription factors. Cell 172:650–65 [DOI] [PubMed] [Google Scholar]
- Le DD, Shimko TC, Aditham AK, Keys AM, Longwell SA, et al. 2018. Comprehensive, high-resolution binding energy landscapes reveal context dependencies of transcription factor binding. PNAS 115:E3702–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sagendorf JM, Chiu TP, Pasi M, Perez A, Rohs R. 2017. Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 45:12877–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, et al. 2014. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3:e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tjian R. 2018. Visualizing transcription factor dynamics in living cells. J. Cell Biol 217:1181–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. 2001. Amino acid–base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29:2860–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR. 2007. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315:233–37 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Ptashne M, Backman K, Kield D, Flashman S, et al. 1975. Recognition sequences of repressor and polymerase in the operators of bacteriophage lambda. Cell 5:109–13 [DOI] [PubMed] [Google Scholar]
- Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, et al. 2013. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 23:988–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Chan SK. 1996. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 12:258–62 [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. 2009. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol 88:63–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Mann RS. 2016. To be specific or not: the critical relationship between Hox and TALE proteins. Trends Genet. 32:334–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M 2009. The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci 10:244–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Nishikawa K. 2005. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res. 12:269–80 [DOI] [PubMed] [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, et al. 2018. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7:e40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Horta A, Lomvardas S. 2019. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565:448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, et al. 2004. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet 36:1331–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Albu M, Hughes TR. 2015. Identification of C2H2-ZF binding preferences from ChIP-seq data using RCADE. Bioinformatics 31:2879–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi H, Kieffer Kwon KR, Resch W, Vian L, Dose M, et al. 2013. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3:1678–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova EN, Kim E, Wise AA, O’Brien PJ, Andricioaei I, Al-Hashimi HM. 2011. Transient Hoogsteen base pairs in canonical duplex DNA. Nature 470:498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801 [DOI] [PubMed] [Google Scholar]
- Oehler S, Eismann ER, Kramer H, Muller-Hill B. 1990. The three operators of the lac operon cooperate in repression. EMBO J 9:973–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf DH, Anderson WF, Fisher RG, Takeda Y, Matthews BW. 1982. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature 298:718–23 [DOI] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. 1984. Protein-DNA recognition. Annu. Rev. Biochem 53:293–321 [DOI] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem 61:1053–95 [DOI] [PubMed] [Google Scholar]
- Panne D 2008. The enhanceosome. Curr. Opin. Struct. Biol 18:236–42 [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. 2007. An atomic model of the interferon-beta enhanceosome. Cell 129:1111–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rueda E, Hernandez-Guerrero R, Martinez-Nunez MA, Armenta-Medina D, Sanchez I, Ibarra JA. 2018. Abundance, diversity and domain architecture variability in prokaryotic DNA-binding transcription factors. PLOS ONE 13:e0195332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M 1967a. Isolation of the lambda phage repressor. PNAS 57:306–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M 1967b. Specific binding of the lambda phage repressor to lambda DNA. Nature 214: 232–34 [DOI] [PubMed] [Google Scholar]
- Ramos AI, Barolo S. 2013. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R. Soc. B Biol. Sci 368:20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi C, Rube HT, Kribelbauer JF, Crocker J, Loker RE, et al. 2018. Accurate and sensitive quantification of protein-DNA binding affinity. PNAS 115:E3692–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147:1408–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley TR, Lazarovici A, Mann RS, Bussemaker HJ. 2015. Building accurate sequence-to-affinity models from high-throughput in vitro protein-DNA binding data using FeatureREDUCE. eLife 4:e06397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rister J, Razzaq A, Boodram P, Desai N, Tsanis C, et al. 2015. Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science 350:1258–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martinez JA, Reinke AW, Bhimsaria D, Keating AE, Ansari AZ. 2017. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. eLife 6:e19272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. 2010. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem 79:233–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. 2009. The role of DNA shape in protein-DNA recognition. Nature 461:1248–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan S, Swamidass SJ, Stormo GD. 2017. BEESEM: estimation of binding energy models using HT-SELEX data. Bioinformatics 33:2288–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube HT, Rastogi C, Kribelbauer JF, Bussemaker HJ. 2018. A unified approach for quantifying and interpreting DNA shape readout by transcription factors. Mol. Syst. Biol 14:e7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Zhao J, Schipper JL, Zhang Y, Bepler T, et al. 2018. Divergence in DNA specificity among paralogous transcription factors contributes to their differential in vivo binding. Cell Syst. 6:470–83.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6:e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, et al. 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147:1270–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordan R, Rohs R. 2014. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci 39:381–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, et al. 2013. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 26:101–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13:613–26 [DOI] [PubMed] [Google Scholar]
- Steitz TA, Ohlendorf DH, McKay DB, Anderson WF, Matthews BW. 1982. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. PNAS 79:3097–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold L, Ehrenfeucht A. 1982. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 10:2997–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Zuo Z, Chang YK. 2015. Spec-seq: determining protein-DNA-binding specificity by sequencing. Brief Funct. Genom 14:30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547:241–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G 1982. Genes controlling segmental specification in the Drosophila thorax. PNAS 79:7380–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242:405–11 [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091–100 [DOI] [PubMed] [Google Scholar]
- Tsai A, Muthusamy AK, Alves MR, Lavis LD, Singer RH, et al. 2017. Nuclear microenvironments modulate transcription from low-affinity enhancers. eLife 6:e28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet 27:304–8 [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. 2012. Molecular roadblocks for cellular reprogramming. Mol. Cell 47:827–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol 22:6321–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Johnston M, Mitra RD. 2007. Calling cards for DNA-binding proteins. Genome Res. 17:1202–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22:1798–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Reed M, Wang Y, Mayr G, Stenger JE, et al. 1994. p53 domains: structure, oligomerization, and transformation. Mol. Cell. Biol 14:5182–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, et al. 2006. Defining the sequence-recognition profile of DNA-binding molecules. PNAS 103:867–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Fersht AR. 2004. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol 341:1145–59 [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, et al. 2013. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol 31:126–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158:1431–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RJ, Hyman AA. 2018. Controlling compartmentalization by non-membrane-bound organelles. Philos. Trans. R. Soc. B Biol. Sci 373:20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA. 2009. Different gene regulation strategies revealed by analysis of binding motifs.Trends Genet. 25:434–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, et al. 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24:593–602 [DOI] [PubMed] [Google Scholar]
- Yang L, Orenstein Y, Jolma A, Yin Y, Taipale J, et al. 2017. Transcription factor family–specific DNA shape readout revealed by quantitative specificity models. Mol. Syst. Biol 13:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandvakili A, Campbell I, Gutzwiller LM, Weirauch MT, Gebelein B. 2018. Degenerate Pax2 and Senseless binding motifs improve detection of low-affinity sites required for enhancer specificity. PLOS Genet. 14:e1007289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiske T, Baburajendran N, Kaczynska A, Brasch J, Palmer AG 3rd, et al. 2018. Intrinsic DNA shape accounts for affinity differences between Hox-cofactor binding sites. Cell Rep. 24:2221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Granas D, Stormo GD. 2009. Inferring binding energies from selected binding sites. PLOS Comput. Biol 5:e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Stormo GD. 2011. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol 29:480–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Shen N, Yang L, Abe N, Horton J, et al. 2015. Quantitative modeling of transcription factor binding specificities using DNA shape. PNAS 112:4654–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. 2016. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet 17:551–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Brangwynne CP. 2015. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol 34:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Roy B, Chang YK, Granas D, Stormo GD. 2017. Measuring quantitative effects of methylation on transcription factor–DNA binding affinity. Sci. Adv 3:eaao1799. [DOI] [PMC free article] [PubMed] [Google Scholar]