Key Points
Substantive familial risks are associated with each hematological malignancy, younger diagnosis age, and multiple affected relatives.
The familial relative risks provide evidence for shared etiology between the specific hematological malignancies.
Abstract
Estimating familial cancer risks is clinically important in being able to discriminate between individuals in the population at differing risk for malignancy. To gain insight into the familial risk for the different hematological malignancies and their possible inter-relationship, we analyzed data on more than 16 million individuals from the Swedish Family-Cancer Database. After identifying 153 115 patients diagnosed with a primary hematological malignancy, we quantified familial relative risks (FRRs) by calculating standardized incident ratios (SIRs) in 391 131 of their first-degree relatives. The majority of hematological malignancies showed increased FRRs for the same tumor type, with the highest FRRs being observed for mixed cellularity Hodgkin lymphoma (SIR, 16.7), lymphoplasmacytic lymphoma (SIR, 15.8), and mantle cell lymphoma (SIR, 13.3). There was evidence for pleiotropic relationships; notably, chronic lymphocytic leukemia was associated with an elevated familial risk for other B-cell tumors and myeloproliferative neoplasms. Collectively, these data provide evidence for shared etiological factors for many hematological malignancies and provide information for identifying individuals at increased risk, as well as informing future gene discovery initiatives.
Visual Abstract
Introduction
Each of the hematological malignancies is characterized by a distinctive clinical phenotype reflective of differences in its progenitor cell of origin and underling biology. The lymphomas, B- and T-cell leukemias, and myeloma are of lymphoid origin, arising at differing stages of maturation.1 Acute and chronic myeloid leukemia (CML), myelodysplastic syndrome (MDS), and the myeloproliferative diseases are all derived from a myeloid progenitor.2 Although many hematological malignancies are individually rare, collectively they contribute significantly to the overall cancer burden in the population.3
Aside from exposure to DNA-damaging agents and the association between Epstein-Barr virus (EBV), HIV, human T-cell lymphotropic virus (HTLV), and Helicobacter pylori with specific lymphoma subtypes, the etiological basis of most hematological malignancies is poorly understood.4-14
Epidemiological observational studies and reports of families segregating hematological malignancies over the years have supported the role of inherited factors in disease etiology.15-18 Direct evidence for predisposition to hematological tumors is provided by the increased risk associated with a number of rare inherited syndromes19 (eg, Fanconi anemia,20 Diamond-Blackfan anemia,21 and dyskeratosis congenita22), as well as rare germline mutations in a number of genes causing Mendelian susceptibility (ANKRD26,23 CEBPA,24 DDX41,25 ELANE,26 ETV6,27 GATA2,28 HAX1,29 RUNX1,30 SAMD9,31 SAMD9L,32 SRP72,33 and LSD134). Recently, genome-wide association studies have provided evidence for a heritable basis to sporadic forms of acute lymphoblastic leukemia (ALL),35 Hodgkin lymphoma (HL),36,37 diffuse large B-cell lymphoma (DLBCL),38 primary central nervous system lymphoma,39 follicular lymphoma (FL),40 marginal zone lymphoma,41 lymphoplasmacytic lymphoma/Waldenström macroglobulinemia (LPL/WM),42 chronic lymphocytic leukemia (CLL),43 multiple myeloma (MM),44 and the myeloproliferative neoplasms (MPNs).45
The increasing importance of recognizing inherited predisposition to hematological malignancy is underscored by the 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia, which has made recognition of a familial disease a component of diagnosing leukemia.2 Aside from precise estimates of familial risks for the hematological malignancies being clinically important in being able to discriminate between individuals at differing risk,46,47 such information is relevant to understanding the nature of genetic susceptibility (ie, contextualizing the effect of known syndromic forms of predisposition).19,48,49
Previous studies of familial risk have reported high disease-specific familial risks for ALL,17 HL,16 DLBCL,50 FL,50 MCL,15 LPL/WM,51 CLL,52 MM,53 polycythemia vera (PV),54 and essential thrombocythemia (ET).54 Furthermore, an increased risk for lymphoproliferative disorders has been reported in the relatives of patients with LPL/WM,51 DLBCL,15 FL,15 small lymphocytic lymphoma,15 CLL,52 and MM.55 However, these studies have limited power to quantify subtype-specific familial risks and to dissect familial risk based on age, sex, and familial relationship. In addition, these studies have not explored familial associations across the complete spectrum of hematological malignancies.
By analyzing the Swedish Family-Cancer Database, we have recently enumerated the familial risks for myeloid malignancies.18 To provide a more comprehensive analysis of the spectrum of all hematological malignancies, we have now extended our analysis of more than 16 million individuals to estimate risk for all lymphoid malignancies and define their inter-relationship. Our analysis provides evidence for shared familial risks consistent with a common etiological basis for a number of the hematological malignancies both within and extending across cell lineage of origin.
Materials and methods
Swedish Family-Cancer Database
We used the Swedish Family-Cancer Database to estimate familial relative risks (FRRs) of the major hematological malignancies. The Swedish Family-Cancer Database was created by linking information from the Multi-Generation Register, national censuses, the Swedish Cancer Registry, and death notifications.56 The Swedish Cancer Registry, established in 1958, is based on the compulsory reporting of all cancer diagnoses, providing near-complete coverage of all cancer registrations in Sweden.57 The 2015 update of these data includes a total population of more than 16.1 million individuals. The Swedish Family-Cancer Database is composed of all families from the Multigeneration Register since its inception.58 Since the Swedish Cancer Registry started in 1958, some individuals in the first generation will inevitably have been diagnosed with cancer before the registration was initiated. The possible influence of this left truncation on FRR estimation has, however, been analyzed and found not to cause bias.59
We considered all incident cases of hematological malignancies diagnosed between 1958 and 2015. Specifically, we analyzed all incident cases of all hematological malignancies, all myeloid malignancies, the MPNs, PV, ET, myelofibrosis, MPN not otherwise specified (MPN-NOS), CML, MDS, acute myeloid leukemia (AML), ALL, HL, nodular sclerosis HL, mixed cellularity HL, non-HL (NHL), DLBCL, FL, LPL/WM, mantle cell lymphoma (MCL), marginal zone lymphoma, Burkitt lymphoma, small lymphocytic lymphoma, hairy cell leukemia, CLL, MM, mature T-cell lymphoma, anaplastic T-cell lymphomas, and cutaneous T-cell lymphoma. The Swedish Cancer Register has implemented the International Classification of Disease (ICD)-7 since 1958, ICD-9 since 1987, ICD-O/2 and Systematized Nomenclature of Medicine (SNOMED) histopathological codes since 1993, and ICD-O/3 since 2005. We used a combination of ICD-7, ICD-O/2, and SNOMED codes to establish the diagnosis for most of the hematological malignancies. As MDS and subtypes of NHL and HL require ICD-O/2 and SNOMED codes, our analysis for these diseases was confined to data collected from 1993. Because SNOMED codes are based on the Kiel and Rappaport classifications, it was not possible to define all NHL subtypes on the basis of World Health Organization classification. The World Health Organization does, however, provide synonymous definitions across classifications, and these translations were used where possible. Individuals diagnosed with MDS before a diagnosis of AML only contributed the MDS analysis to mitigate against any possible confounding from treatment.
The study was undertaken with approval from the ethics committee at Lund University, Sweden, and was conducted in accordance with the tenets of the Declaration of Helsinki.
Statistical analysis
We define the FRR as the ratio of the number of observed cases of a given hematological malignancy in first-degree relatives (FDRs) of patients to the expected number of cases in FDRs of patients. As such, standardized incidence ratios (SIRs) as a measure of FRR were used to compare the risk for hematological malignancies in FDRs of patients (ie, parent, sibling, or child) with that of the general population.15,16,18,60,61 All FDRs were observed from the date of birth, immigration, or the start of cancer-specific registrations in the database: 1 January 1958, for MPNs, PV, ET, myelofibrosis, CML, AML, ALL, NHL, CLL, and MM; or 1 January 1993, for MPN-NOS, MDS, nodular sclerosis HL, mixed cellularity HL, DLBCL, FL, LPL/WM, MCL, marginal zone lymphoma, small lymphocytic lymphoma, Burkitt lymphoma, mature T-cell lymphoma, anaplastic T-cell lymphomas, and cutaneous T-cell lymphoma. Follow-up ended at diagnosis of cancer, date of death, emigration, or the end date of the registry (31 December 2015). The SIRs (indirect standardization) were calculated as the ratio of the observed number of affected individuals in FDRs (O) to the expected number of affected in FDRs (E):
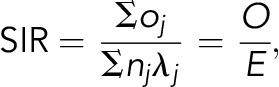 |
where λj is the strata-specific (cancer, age, sex, calendar year) incidence rate in the background population, nj is the number of strata-specific person-years in the FDRs, and oj is the number of observed cases in the FDRs. To provide an unbiased estimate of FRR, any individual who might potentially be both a case and an FDR was only counted once in the numerator.
A Poisson distribution was used to calculate the 95% confidence intervals (CIs). Tests for trend in SIRs were performed by evaluating the likelihood function in collapsed person-time additive Poisson regression models with and without the inclusion of the variable. In the stratified analyses where an FDR appears in both comparison groups as a result of being related to 2 or more incident cases, the individual is counted in each stratum, except in the age analysis, where an FDR of a younger incident case is given precedence. The age stratification was based on the first quartile of the age at diagnosis distribution of all incident cases for each hematological malignancy. Given the in utero origin of a subset of ALL,62 cases with a high probability of being monozygotic twins were excluded from the analysis.
The lifetime cumulative risk was calculated on the basis of the average life expectancy in Sweden in 2015 (82 years) and the following calculation63: lifelong cumulative rate = sum of all age-specific incident rates (0-82 years); lifelong cumulative risk = 1–e−Σλi. Exact values for person-years from individual data were used to calculate cumulative incidence.
To estimate the likely contribution of the bone marrow failure and leukemia susceptibility syndromes for which population-based effect sizes exist (ie, Fanconi anemia, Diamond-Blackfan anemia, Schwachman-Diamond syndrome, dyskeratosis congenita, Li Fraumeni, and ataxia telangiectasia) to the FRRs, we made use of published data on their population prevalence and the documented risk for respective hematological malignancies associated with each.64-69 Using this information and assuming the effect of all mutations being equal, the contribution of each mutated gene to the familial risk for each hematological malignancy was calculated from  , where λ0 is the familial risk to first-degree relatives of patients and λk is the familial relative risk associated with the gene mutations k, calculated as
, where λ0 is the familial risk to first-degree relatives of patients and λk is the familial relative risk associated with the gene mutations k, calculated as 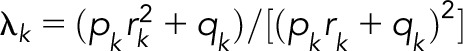 , where pk is the risk allele frequency k, qk = 1−pk, and rk is the published relative risk.70
, where pk is the risk allele frequency k, qk = 1−pk, and rk is the published relative risk.70
We estimated the power to demonstrate different relative risks as per Newman,71 based on the population incidence rate and number of person-years, stipulating a P value of .05 (2-sided).
Statistical analyses were performed using Stata version 14 (STATA, College State, TX) and R 3.3.3 software. A P value ≤ .05 (2-sided) was considered statistically significant.
Results
Overall estimates
Of the 16.1 million individuals registered in the Swedish Family-Cancer Database, 153 115 individuals were diagnosed with a major classifiable primary hematological malignancy between 1958 and 2015. Supplemental Table 1, available on the Blood Web site, shows the characteristics of cases and the FDRs. Familial cases represented 4.1% of all hematological malignancy diagnoses, which is higher than cancers of the nervous system (1.8%), kidney (2.8%), and pancreas (3.0%) ,but lower than those of the breast (8.5%), colorectum (10.1%), and prostate (15.3%).72
Familial aggregation of hematological malignancies
Almost all of the hematological malignancies showed statistically significant increased familial risks for the same tumor type (Figure 1; supplemental Table 2). We have reported a detailed analysis of the FRRs of the myeloid malignancies in this population.18 Briefly, we found 1.5-, 6.8-, 6.9-, and 7.7-fold increases in FRRs for AML, ET, MDS, and PV. For the B-cell tumors, a range of FRRs was observed, with 2-fold increases in FRRs for DLBCL, FL, and MM and 5.6-, 8.3-, 9.8-, 13.3-, 15.8-, and 16.7-fold increases in FRRs for CLL, hairy cell leukemia, nodular sclerosis HL, MCL, LPL/WM, and mixed cellularity HL. We were not able to provide evidence to support familial clustering of CML, myelofibrosis, and the T-cell neoplasms.
Figure 1.
The inter-relationship between familial relative risks for different hematological malignancies. The color corresponds to the magnitude of the familial risk as indicated. White indicates a familial risk that crosses unity (nonsignificant). HL comprises nodular sclerosis HL and mixed cellularity HL. NHL comprises DLBCL, FL, LPL, MCL, small lymphocytic lymphoma, hairy cell leukemia, and the T-cell leukemias/lymphomas. Myeloid malignancies comprise PV, ET, myelofibrosis, MPN-NOS, MDS, CML, and AML. Myeloproliferative neoplasms comprise PV, ET, myelofibrosis, MPN-NOS. Hematological malignancies with no significant familial associations are not shown.
We next examined FRRs of the lymphoid diseases by age at diagnosis, sex, and type of familial relationship. Familial risks were significantly higher for relatives of cases diagnosed young for all HL (5.76 vs 3.36) and CLL (6.99 vs 4.83; Table 1). The FRRs were significantly higher in siblings when compared with that of parent-offspring relationships for NHL (1.97 vs 1.69), HL (7.45 vs 3.09), and CLL (7.80 vs 5.36). In contrast, parent-offspring RRs were significantly higher for LPL/WM (21.88 vs 5.56; supplemental Table 3). We found no evidence for differences in FRR by sex (supplemental Tables 4 and 5). FRRs were significantly higher for relatives with 2 or more affected FDRs when compared with relatives with 1 affected FDR for all hematological malignancies (2.08 vs 1.31) and CLL (27.13 vs 5.36).
Table 1.
Familial relative risks for lymphoid malignancies and all hematological malignancy, stratified by the age at diagnosis
| Age groups, y* | N | SIR (95% CI) | P | |
|---|---|---|---|---|
| All hematological malignancies | ≤56 | 2070 | 1.38 (1.32-1.44) | .05 |
| >56 | 4150 | 1.31 (1.27-1.35) | ||
| ALL | ≤5 | 6 | 2.51 (0.60-5.66) | .69 |
| >5 | 8 | 0.36 (0.01-2.03) | ||
| HL | ≤28 | 45 | 5.76 (4.20-7.70) | 7.3 × 10−3 |
| >28 | 61 | 3.36 (2.57-4.32) | ||
| Nodular sclerosis HL | ≤22 | 6 | 14.29 (5.24-31.09) | .29 |
| >22 | 10 | 8.20 (3.93-15.07) | ||
| NHL | ≤57 | 392 | 1.76 (1.59-1.95) | .89 |
| >57 | 792 | 1.75 (1.63-1.87) | ||
| DLBCL | ≤59 | 18 | 2.65 (1.57-4.18) | .44 |
| >59 | 30 | 2.10 (1.42-3.00) | ||
| FL | ≤56 | 6 | 2.02 (0.74-4.40) | .78 |
| >56 | 12 | 1.76 (0.91-3.07) | ||
| LPL | ≤65 | 8 | 19.05 (8.22-37.53) | .53 |
| >65 | 8 | 13.79 (5.95-27.18) | ||
| MCL | ≤63 | 3 | 13.64 (2.81-39.85) | .93 |
| >63 | 5 | 13.16 (4.27-30.71) | ||
| CLL | ≤63 | 148 | 6.99 (5.91-8.21) | 1.1 × 10−3 |
| >63 | 169 | 4.83 (4.13-5.62) | ||
| MM | ≤63 | 92 | 2.24 (1.81-2.75) | .52 |
| >63 | 136 | 1.96 (1.72-2.43) | ||
| Myeloid malignancies | ≤56 | 167 | 2.19 (1.87-2.55) | .13 |
| >56 | 312 | 1.89 (1.69-2.11) | ||
| MPN | ≤59 | 80 | 6.46 (5.12-8.04) | 4.0 × 10−3 |
| >59 | 101 | 4.15 (3.38-5.04) | ||
| PV | ≤59 | 26 | 10.90 (7.12-15.97) | .03 |
| >59 | 27 | 5.96 (3.93-8.67) | ||
| ET | ≤56 | 10 | 9.76 (4.68-17.95) | .17 |
| >56 | 12 | 5.37 (2.77-9.38) | ||
| CML | ≤47 | 2 | 1.33 (0.16-4.80) | .39 |
| >47 | 2 | 0.55 (0.07-1.99) | ||
| MDS | ≤68 | 13 | 11.95 (6.36-20.43) | 8.8 × 10−3 |
| >68 | 5 | 3.27 (1.06-7.63) | ||
| AML | ≤52 | 13 | 1.48 (0.79-2.53) | .90 |
| >52 | 33 | 1.55 (1.07-2.18) |
Age group is defined by the lower quartile of the distribution of the diagnosis. P represents a trend test. The age stratification was based on the first quartile of the age at diagnosis distribution of all incident cases for each hematological malignancy. Where a first-degree relative appears in both age groups as a result of being related to 2 or more incident cases, the first-degree relative is counted in the younger age group. Data on the clonal myeloid neoplasms have previously been reported.18
N, number of cases.
As shown for the myeloid malignancies,18 familial risks were significantly higher for relatives of cases diagnosed young for all MPNs (6.46 vs 4.15), PV (10.91 vs 5.96), and MDS (11.95 vs 3.27; Table 1). Sibling relative risks were higher than parent-child relative risks for AML (3.08 vs 1.09), and all 53 familial cases of PV were of a parent-child relationship. Familial relative risks were significantly higher for relatives with 2 or more affected FDRs for all myeloid malignancies (4.55 vs 1.96) and all MPNs (17.82, 4.83).
Cumulative risk
Supplemental Table 6 details the cumulative risk for each of the hematological malignancies. The lifetime cumulative risks for all primary hematological malignancies, NHL, and CLL in the Swedish population were 3.4%, 1.3%, and 0.4%, respectively (Table 2; supplemental Table 6). As the FRRs estimates for the hematological malignancies are relatively high compared with the FRRs of other cancers,73 the lifetime cumulative risk in FDRs of patients increased to 4.3%, 2.2%, and 2.3% for all primary hematological malignancies, NHL, and CLL, respectively (Figure 2). For individuals with 2 or more affected FDRs, markedly elevated cumulative risk estimates for all primary hematological malignancies (7.4%) and CLL (8.6%) were observed (Table 2).
Table 2.
Familial relative risks for all hematological malignancy, NHL, CLL, myeloid malignancy, and myeloproliferative neoplasm, stratified by number of affected first-degree relatives
| Population CR (%) | Number of FDRs | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 affected FDR | ≥2 affected FDR | P value | ||||||
| N | SIR (95% CI) | CR (%) | N | SIR (95% CI) | CR (%) | |||
| All hematological malignancies | 3.4 | 5941 | 1.31 (1.28-1.34) | 4.2 | 279 | 2.08 (1.84-2.33) | 7.4 | 2.1 × 10−12 |
| NHL | 1.3 | 1165 | 1.75 (1.65-1.85) | 2.1 | 19 | 2.06 (1.24-3.22) | 2.4 | .50 |
| CLL | 0.4 | 297 | 5.36 (4.77-6.00) | 2.1 | 20 | 27.13 (16.97-42.90) | 8.6 | 1.34 × 10−8 |
| Myeloid malignancies | 0.7 | 470 | 1.96 (1.79-2.15) | 1.4 | 9 | 4.55 (2.08-8.64) | 4.4 | .02 |
| MPN | 0.3 | 176 | 4.83 (4.14-5.60) | 1.2 | 5 | 17.82 (5.79-24.89) | 3.2 | .01 |
P represents a trend test. Data on the clonal myeloid neoplasms have previously been reported.18
CR, cumulative risk.
Figure 2.
Lifetime cumulative risk for all hematological malignancies, NHL, and CLL. (A) All hematological malignancies, (B) NHL, and (C) CLL.
Contribution of cancer syndromes to familial risk
A number of rare bone marrow failure and leukemia susceptibility syndromes caused by inheritance of germline mutations influence the risk for specific hematological malignancies.19 For example, ataxia telangiectasia is associated with an increased risk for ALL and AML.69 Others that are rarer include Fanconi anemia,64 Diamond-Blackfan syndrome,65 and dyskeratosis congenita.66 Thus far, no high-impact mutations have consistently been shown to influence the risk for the mature B-cell neoplasms in adults. The rare bone marrow failure and leukemia susceptibility syndromes contribute 17%, 7%, and 4% of the population-based familial risk estimates for MDS, ALL, and AML, respectively (supplemental Figure 1).
Shared familial risk across hematological malignancies
To explore the possibility of familial aggregation between different tumor types, we examined FRRs between all hematological malignancies. Although the strongest FRRs tended to be disease-specific, we also identified distinct patterns of familial risk (Figure 1; supplemental Table 2). These were predominantly cell-lineage specific; for example, CLL shared familial risks with nearly all the B-cell tumors. However, familial risks were also noted across cell lineage, with HL, DLBCL, MCL, CLL, and MM sharing risks with a number of myeloid malignancies. In contrast, no familial aggregation was observed for CML, Burkitt lymphoma, anaplastic T-cell lymphoma, and mature T-cell lymphoma (Figure 1; supplemental Table 2).
Discussion
This study provides further support for the significant familial aggregation of the major hematological malignancies. Because of the large sample size and long follow-up time, we have been able not only to demonstrate significantly elevated relative risks in FDRs of cases for the same tumor type but also to detect associations between the different hematological tumor types. Although we were able to capture the major histological subtypes of the hematological malignancy as defined by ICD-7 and ICD-O/2 codes, the study of FRRs using more recent developments to the classification system may further refine FRR estimates.1,2
Our analysis benefits greatly from being based on the Swedish Family-Cancer Database. This population-based family cancer registry possesses near-complete case registration for almost all hematological malignancies, thereby allowing familial relative risks to be derived while avoiding biases introduced by case-control study designs.74,75 Estimates of underreporting of cancer diagnoses are thought to be stable over periods and have been estimated as 4% in 1978, 3.7% in 1998, and 4% in 2014.74,76,77 Furthermore, our analysis is enhanced by the large number of cases when compared with earlier studies, which have previously exploited this resource, particularly in respect to the hematological diseases that have been classified from 1993.52,54,78 Although the strengths of our study are the large sample size and unbiased assessment of cancer status in relatives, we did not have information concerning other possible risk factors for malignancies that may be correlated within families.
The familial risk estimates provided in this study are broadly comparable to those previously published from the Swedish population.15-17,51-55 One notable difference is the discrepancy in familial risk for DLBCL.50 This difference may be a result of previous analyses based on a case-control design with a consequential inflation of risk estimates, definition of affected relatives, and greater case numbers with longer follow-up in this study.
Although elevated familial risks cluster with diseases from the same hematopoietic cell lineage, a number of associations extend across hematopoietic cell lineages. These findings suggest that etiological factors for hematological malignancies are shared not only between diseases arising from cells at different stages of differentiation of the same hematopoietic lineage but also between diseases originating from different hematopoietic lineages. It is likely that there is heterogeneity in the mechanisms by which these factors exert their effects on different phenotypes, as evidenced by the observation that the majority of FRRs are highest for concordant disease, and that some of the risks differed depending on the relationship of the FDR.
The association between familial risk and age of onset for many of the hematological malignancies provides support for the role of genetic predisposition influencing disease risk.49 This was especially marked for PV, MDS, HL, and CLL, which displayed high FRRs for relatives of cases and for those related to patients diagnosed young. The genetic architecture of susceptibility to hematological malignancy is not fully defined. Rare high-impact mutations in a number of genes have been identified, conferring elevated risks for myeloid and/or lymphoproliferative malignancies.19-21,23-25,28,31-33,79-88 Our analysis indicates that the known genetic predisposition syndromes for which population-based effect estimates exist are unlikely to account for the observed FRRs. Although the existence of additional major genes, as well as more recently described mutations, may account for part of the observed FRR, it is likely that some of the FRR is enshrined in common genetic variation.49 Indeed, although few published studies have investigated the role of such variation in MDS and AML, common genetic variants have already been shown to contribute significantly to the FRR of the other hematological malignancies.36,43,44,89,90 Albeit preliminary, evidence for a shared susceptibility between different hematological malignancies consistent with our observations has been proposed from recent analyses of GWAS data.91,92 A component of the FRR may also be explained by shared environmental risk factors, although the magnitude of their role in explaining familial aggregation has been debated.93-96 To date, few robustly identified environmental risk factors for hematological malignancy have been described, with examples including infection with EBV, HIV, HTLV, and Helicobacter pylori, as wel as benzene, cytotoxic therapy, and ionizing radiation.9-12 Of note, there are some, albeit nonconclusive, data, to suggest ionizing radiation may increase the risk for lymphoid as well as myeloid malignancies.11,97-99
Although the FRRs associated with some of these hematological malignancies are high compared with those associated with the solid cancers,73 the absolute risk is small (Table 2). For example, given that the population lifetime risk for CLL is only 0.4%, an FRR of 6.99 translates to a risk of 2.2% for a first-degree relative of a CLL case. Such familial risks may therefore be of limited clinical significance, with a few notable caveats. First, a diagnosis of CLL represents a proportion of the observable CLL-related phenotype. Monoclonal B-lymphocytosis, the precursor of CLL, is present in 3% to 4% of adults older than 40 years and 14% to 18% of FDRs of CLL.100 An increase in prevalence of such precursor diseases in relatives is therefore likely to be observed for myeloma and the myeloid malignancies with respect to the precursor lesion monoclonal gammopathy of unknown significance and clonal hematopoiesis, respectively.101-107 Second, FRRs reflect the influence of all forms of family history on risk. Inevitably, this may include a subset with clear Mendelian inheritance. This is reflected in the greater lifetime risk associated with having multiple affected relatives of more than 4% and 8% for the myeloid malignancies and CLL, respectively, and is consistent with the existence of genes with stronger effects such as those predisposing to AML, MDS, and CLL.19,79 The significant familial aggregation shown here therefore justifies the continued application of gene-mapping approaches in high-risk families and suggests that within families, this may lead to identification of additional high-penetrance mutations.
There are a number of limitations to our analysis. First, our findings may only apply to Western countries and may not therefore be applicable to economically developing countries that have different tumor incidence rates.108-111 Second, although we had good power to demonstrate modest FRRs (ie, greater than 80% power for an FRR of 1.5) associated with the major classes of hematological malignancies (ALL, CLL, AML, CML), for the malignancies only cataloged post-1993, we have had limited power (eg, only 50% power for an FRS of 2.5 in the case of LPL/WM; supplemental Figure 2). Finally, although we have sought to reduce the effect of misclassification of cancer outcomes by applying ICD-O/2 and SNOMED codes and the use of synonymous classification terms across time, there remains a potential for misclassification to affect estimates.
Accepting these caveats, the results of our analyses have important implications. First, these results can inform initiatives to identify and characterize etiological risk factors for hematological malignancy. Such efforts offer the prospect of providing insight into disease biology.49,112-116 Furthermore, given the presence of disease nonconcordant FRR, it may be appropriate, with careful study design, to expand such initiatives to include multiple hematological phenotypes. Second, given that even moderate familial associations indicate large differences in risk between individuals in the population,47,117 such risks can improve the management of patients with hematological malignancy and their relatives through counseling and genetic testing.19,118
In summary, we have performed a comprehensive analysis of familial risks of the major hematological malignancies in the Swedish population. These results have the potential to assist in the management of patients with hematological malignancy and their relatives and to inform future studies investigating the etiological basis of these hematological malignancies.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This project was supported by grants from the German Cancer Aid, the Swedish Research Council (2014-2517, 2014-10134 and 2016-01176), ALF funding from Region Skåne. A.S. is the recipient of a guest scientist Fellowship from DKFZ. The work of R.S.H. is supported by funding from Bloodwise.
Footnotes
These data are not publicly available because of restrictions (information that could compromise participant privacy).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.S., R.S.H., and K.H. designed the study; K.S., J.S., and K.H. provided the data; A.S., S.C., and H.T. performed data extraction and statistical analysis; A.S., R.S.H., and K.H. drafted the manuscript; and all authors contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Amit Sud, Division of Genetics and Epidemiology, The Institute of Cancer Research, 15 Cotswold Rd, London SM2 5NG, United Kingdom; e-mail: amit.sud@icr.ac.uk.
REFERENCES
- 1.Swerdlow SH, Campo E, Pileri SA, et al. . The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, et al. . The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 3.Sant M, Allemani C, Tereanu C, et al. ; HAEMACARE Working Group . Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116(19):3724-3734. [DOI] [PubMed] [Google Scholar]
- 4.Hjalgrim H, Askling J, Rostgaard K, et al. . Characteristics of Hodgkin’s lymphoma after infectious mononucleosis. N Engl J Med. 2003;349(14):1324-1332. [DOI] [PubMed] [Google Scholar]
- 5.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1(7335):702-703. [DOI] [PubMed] [Google Scholar]
- 6.Wyatt JI, Rathbone BJ. Immune Response of the Gastric Mucosa to Campylobacter pylori. Scand J Gastroenterol Suppl. 1988;142:44-49. [PubMed] [Google Scholar]
- 7.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77(12):7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziegler JL, Beckstead JA, Volberding PA, et al. . Non-Hodgkin’s lymphoma in 90 homosexual men. Relation to generalized lymphadenopathy and the acquired immunodeficiency syndrome. N Engl J Med. 1984;311(9):565-570. [DOI] [PubMed] [Google Scholar]
- 9.Grogg KL, Miller RF, Dogan A. HIV infection and lymphoma. J Clin Pathol. 2007;60(12):1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allan JM, Travis LB. Mechanisms of therapy-related carcinogenesis. Nat Rev Cancer. 2005;5(12):943-955. [DOI] [PubMed] [Google Scholar]
- 11.Hsu W-L, Preston DL, Soda M, et al. . The incidence of leukemia, lymphoma and multiple myeloma among atomic bomb survivors: 1950-2001. Radiat Res. 2013;179(3):361-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fircanis S, Merriam P, Khan N, Castillo JJ. The relation between cigarette smoking and risk of acute myeloid leukemia: an updated meta-analysis of epidemiological studies. Am J Hematol. 2014;89(8):E125-E132. [DOI] [PubMed] [Google Scholar]
- 13.Cocco P, Vermeulen R, Flore V, et al. . Occupational exposure to trichloroethylene and risk of non-Hodgkin lymphoma and its major subtypes: a pooled InterLymph analysis. Occup Environ Med. 2013;70(11):795-802. [DOI] [PubMed] [Google Scholar]
- 14.’t Mannetje A, De Roos AJ, Boffetta P, et al. Occupation and risk of non-Hodgkin lymphoma and its subtypes: a pooled analysis from the InterLymph Consortium. Environ Health Perspect 2016;124(4):396-405. [DOI] [PMC free article] [PubMed]
- 15.Fallah M, Kharazmi E, Pukkala E, et al. . Familial risk of non-Hodgkin lymphoma by sex, relationship, age at diagnosis and histology: a joint study from five Nordic countries. Leukemia. 2016;30(2):373-378. [DOI] [PubMed] [Google Scholar]
- 16.Kharazmi E, Fallah M, Pukkala E, et al. . Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: a joint study from five Nordic countries. Blood. 2015;126(17):1990-1995. [DOI] [PubMed] [Google Scholar]
- 17.Kharazmi E, da Silva Filho MI, Pukkala E, Sundquist K, Thomsen H, Hemminki K. Familial risks for childhood acute lymphocytic leukaemia in Sweden and Finland: far exceeding the effects of known germline variants. Br J Haematol. 2012;159(5):585-588. [DOI] [PubMed] [Google Scholar]
- 18.Sud A, Chattopadhyay S, Thomsen H, et al. . Familial risks of acute myeloid leukemia, myelodysplastic syndromes, and myeloproliferative neoplasms. Blood. 2018;132(9):973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godley LA, Shimamura A. Genetic predisposition to hematologic malignancies: management and surveillance. Blood. 2017;130(4):424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337-349. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto KM, Narla A. Perspective on Diamond-Blackfan anemia: lessons from a rare congenital bone marrow failure syndrome. Leukemia. 2018;32(2):249-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dokal I. Dyskeratosis congenita. Hematology Am Soc Hematol Educ Program. 2011;2011:480-486. [DOI] [PubMed] [Google Scholar]
- 23.Noris P, Perrotta S, Seri M, et al. . Mutations in ANKRD26 are responsible for a frequent form of inherited thrombocytopenia: analysis of 78 patients from 21 families. Blood. 2011;117(24):6673-6680. [DOI] [PubMed] [Google Scholar]
- 24.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351(23):2403-2407. [DOI] [PubMed] [Google Scholar]
- 25.Polprasert C, Schulze I, Sekeres MA, et al. . Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23(4):433-436. [DOI] [PubMed] [Google Scholar]
- 27.Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol. 2017;106(2):189-195. [DOI] [PubMed] [Google Scholar]
- 28.Hahn CN, Chong C-E, Carmichael CL, et al. . Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein C, Grudzien M, Appaswamy G, et al. . HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease). Nat Genet. 2007;39(1):86-92. [DOI] [PubMed] [Google Scholar]
- 30.Antony-Debré I, Manchev VT, Balayn N, et al. . Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood. 2015;125(6):930-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narumi S, Amano N, Ishii T, et al. . SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792-797. [DOI] [PubMed] [Google Scholar]
- 32.Tesi B, Davidsson J, Voss M, et al. . Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirwan M, Walne AJ, Plagnol V, et al. . Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am J Hum Genet. 2012;90(5):888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X, Calvo-Vidal MN, Chen S, et al. . Germline lysine-specific demethylase 1 (LSD1/KDM1A) mutations confer susceptibility to multiple myeloma. Cancer Res. 2018;78(10):2747-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayakrishnan J, Kumar R, Henrion MYR, et al. . A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31(3):573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sud A, Thomsen H, Law PJ, et al. ; PRACTICAL consortium . Genome-wide association study of classical Hodgkin lymphoma identifies key regulators of disease susceptibility [published correction appears in Nat Commun. 2019;10(1):157]. Nat Commun. 2017;8(1):1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sud A, Thomsen H, Orlando G, et al. ; PRACTICAL Consortium . Genome-wide association study implicates immune dysfunction in the development of Hodgkin lymphoma. Blood. 2018;132(19):2040-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerhan JR, Berndt SI, Vijai J, et al. . Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014;46(11):1233-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labreche K, Daniau M, Sud A, et al. ; LOC Network . A genome-wide association study identifies susceptibility loci for primary central nervous system lymphoma at 6p25.3 and 3p22.1: a LOC network study. Neuro-oncol. 2019;noz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skibola CF, Berndt SI, Vijai J, et al. . Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet. 2014;95(4):462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijai J, Wang Z, Berndt SI, et al. . A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6(1):5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMaster ML, Berndt SI, Zhang J, et al. . Two high-risk susceptibility loci at 6p25.3 and 14q32.13 for Waldenström macroglobulinemia. Nat Commun. 2018;9(1):4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law PJ, Berndt SI, Speedy HE, et al. . Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;8(1):14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Went M, Sud A, Försti A, et al. ; PRACTICAL consortium . Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma [published correction appears in Nat Commun. 2019;10(1):213]. Nat Commun. 2018;9(1):3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapper W, Jones AV, Kralovics R, et al. . Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat Commun. 2015;6(1):6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemminki K, Eng C. Clinical genetic counselling for familial cancers requires reliable data on familial cancer risks and general action plans. J Med Genet. 2004;41(11):801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valberg M, Stensrud MJ, Aalen OO. The surprising implications of familial association in disease risk. BMC Public Health. 2018;18(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohlmann W, Schiffman JD. Discussing and managing hematologic germ line variants. Hematology Am Soc Hematol Educ Program. 2016;2016:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017;17(11):692-704. [DOI] [PubMed] [Google Scholar]
- 50.Goldin LR, Björkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009;146(1):91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kristinsson SY, Björkholm M, Goldin LR, McMaster ML, Turesson I, Landgren O. Risk of lymphoproliferative disorders among first-degree relatives of lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia patients: a population-based study in Sweden. Blood. 2008;112(8):3052-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104(6):1850-1854. [DOI] [PubMed] [Google Scholar]
- 53.Landgren O, Linet MS, McMaster ML, Gridley G, Hemminki K, Goldin LR. Familial characteristics of autoimmune and hematologic disorders in 8,406 multiple myeloma patients: a population-based case-control study. Int J Cancer. 2006;118(12):3095-3098. [DOI] [PubMed] [Google Scholar]
- 54.Landgren O, Goldin LR, Kristinsson SY, Helgadottir EA, Samuelsson J, Björkholm M. Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24,577 first-degree relatives of 11,039 patients with myeloproliferative neoplasms in Sweden. Blood. 2008;112(6):2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank C, Fallah M, Chen T, et al. . Search for familial clustering of multiple myeloma with any cancer. Leukemia. 2016;30(3):627-632. [DOI] [PubMed] [Google Scholar]
- 56.Hemminki K, Li X, Plna K, Granström C, Vaittinen P. The nation-wide Swedish family-cancer database--updated structure and familial rates. Acta Oncol. 2001;40(6):772-777. [DOI] [PubMed] [Google Scholar]
- 57.Ji J, Sundquist K, Sundquist J, Hemminki K. Comparability of cancer identification among Death Registry, Cancer Registry and Hospital Discharge Registry. Int J Cancer. 2012;131(9):2085-2093. [DOI] [PubMed] [Google Scholar]
- 58.Hemminki K, Ji J, Brandt A, Mousavi SM, Sundquist J. The Swedish Family-Cancer Database 2009: prospects for histology-specific and immigrant studies. Int J Cancer. 2010;126(10):2259-2267. [DOI] [PubMed] [Google Scholar]
- 59.Leu M, Reilly M, Czene K. Evaluation of bias in familial risk estimates: a study of common cancers using Swedish population-based registers. J Natl Cancer Inst. 2008;100(18):1318-1325. [DOI] [PubMed] [Google Scholar]
- 60.Andrieu N, Launoy G, Guillois R, Ory-Paoletti C, Gignoux M. Estimation of the familial relative risk of cancer by site from a French population based family study on colorectal cancer (CCREF study). Gut. 2004;53(9):1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czene K, Adami H-O, Chang ET. Sex- and kindred-specific familial risk of non-Hodgkin’s lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2496-2499. [DOI] [PubMed] [Google Scholar]
- 62.Ford AM, Ridge SA, Cabrera ME, et al. . In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363(6427):358-360. [DOI] [PubMed] [Google Scholar]
- 63.Statistics Sweden Life expectancy 1751–2017. https://www.scb.se/en/finding-statistics/statistics-by-subject-area/population/population-composition/population-statistics/pong/tables-and-graphs/yearly-statistics--the-whole-country/life-expectancy. Accessed 14 November 2018. [Google Scholar]
- 64.Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27(3-4):214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mai PL, Best AF, Peters JA, et al. . Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122(23):3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suarez F, Mahlaoui N, Canioni D, et al. . Incidence, presentation, and prognosis of malignancies in ataxia-telangiectasia: a report from the French national registry of primary immune deficiencies. J Clin Oncol. 2015;33(2):202-208. [DOI] [PubMed] [Google Scholar]
- 70.Houlston RS, Ford D. Genetics of coeliac disease. QJM. 1996;89(10):737-743. [DOI] [PubMed] [Google Scholar]
- 71.Newman SC. Sample Size and Power. Biostatistical Methods in Epidemiology. New York: John Wiley & Sons, Inc.; 2002:281-294. [Google Scholar]
- 72.Hemminki K, Li X, Czene K. Familial risk of cancer: data for clinical counseling and cancer genetics. Int J Cancer. 2004;108(1):109-114. [DOI] [PubMed] [Google Scholar]
- 73.Frank C, Fallah M, Sundquist J, Hemminki A, Hemminki K. Population landscape of familial cancer. Sci Rep. 2015;5(1):12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barlow L, Westergren K, Holmberg L, Talbäck M. The completeness of the Swedish Cancer Register: a sample survey for year 1998. Acta Oncol. 2009;48(1):27-33. [DOI] [PubMed] [Google Scholar]
- 75.Chang ET, Smedby KE, Hjalgrim H, Glimelius B, Adami H-O. Reliability of self-reported family history of cancer in a large case-control study of lymphoma. J Natl Cancer Inst. 2006;98(1):61-68. [DOI] [PubMed] [Google Scholar]
- 76.Mattsson B, Wallgren A. Completeness of the Swedish Cancer Register. Non-notified cancer cases recorded on death certificates in 1978. Acta Radiol Oncol. 1984;23(5):305-313. [DOI] [PubMed] [Google Scholar]
- 77.Pukkala E, Engholm G, Højsgaard Schmidt LK, et al. . Nordic Cancer Registries - an overview of their procedures and data comparability. Acta Oncol. 2018;57(4):440-455. [DOI] [PubMed] [Google Scholar]
- 78.Goldin LR, Kristinsson SY, Liang XS, Derolf ÅR, Landgren O, Björkholm M. Familial aggregation of acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol. 2012;30(2):179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Speedy HE, Kinnersley B, Chubb D, et al. . Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood. 2016;128(19):2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dokal I, Vulliamy T, Mason P, Bessler M. Clinical utility gene card for: dyskeratosis congenita - update 2015. Eur J Hum Genet. 2015;23(4):558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boocock GRB, Morrison JA, Popovic M, et al. . Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97-101. [DOI] [PubMed] [Google Scholar]
- 82.Skokowa J, Dale DC, Touw IP, Zeidler C, Welte K. Severe congenital neutropenias. Nat Rev Dis Primers. 2017;3(1):17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song W-J, Sullivan MG, Legare RD, et al. . Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166-175. [DOI] [PubMed] [Google Scholar]
- 84.Zhang MY, Churpek JE, Keel SB, et al. . Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47(2):180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malkin D, Li FP, Strong LC, et al. . Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233-1238. [DOI] [PubMed] [Google Scholar]
- 86.Shah S, Schrader KA, Waanders E, et al. . A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45(10):1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de la Chapelle A, Träskelin AL, Juvonen E. Truncated erythropoietin receptor causes dominantly inherited benign human erythrocytosis. Proc Natl Acad Sci USA. 1993;90(10):4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Churchman ML, Qian M, Te Kronnie G, et al. . Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell. 2018;33(5):937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vijayakrishnan J, Studd J, Broderick P, et al. ; PRACTICAL Consortium . Genome-wide association study identifies susceptibility loci for B-cell childhood acute lymphoblastic leukemia [published correction appears in Nat Commun. 2019;10(1):419]. Nat Commun. 2018;9(1):1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Halvarsson B-M, Wihlborg A-K, Ali M, et al. . Direct evidence for a polygenic etiology in familial multiple myeloma. Blood Adv. 2017;1(10):619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Law PJ, Sud A, Mitchell JS, et al. . Genome-wide association analysis of chronic lymphocytic leukaemia, Hodgkin lymphoma and multiple myeloma identifies pleiotropic risk loci. Sci Rep. 2017;7(1):41071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Went M, Sud A, Speedy H, et al. . Genetic correlation between multiple myeloma and chronic lymphocytic leukaemia provides evidence for shared aetiology. Blood Cancer J. 2018;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khoury MJ, Beaty TH, Liang KY. Can familial aggregation of disease be explained by familial aggregation of environmental risk factors? Am J Epidemiol. 1988;127(3):674-683. [DOI] [PubMed] [Google Scholar]
- 94.Aalen OO. Modelling the influence of risk factors on familial aggregation of disease. Biometrics. 1991;47(3):933-945. [PubMed] [Google Scholar]
- 95.Guo SW. Familial aggregation of environmental risk factors and familial aggregation of disease. Am J Epidemiol. 2000;151(11):1121-1131. [DOI] [PubMed] [Google Scholar]
- 96.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10(7):733-741. [PubMed] [Google Scholar]
- 97.Little MP, Wakeford R, Borrego D, et al. . Leukaemia and myeloid malignancy among people exposed to low doses (<100 mSv) of ionising radiation during childhood: a pooled analysis of nine historical cohort studies. Lancet Haematol. 2018;5(8):e346-e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vrijheid M, Cardis E, Ashmore P, et al. ; 15-Country Study Group . Ionizing radiation and risk of chronic lymphocytic leukemia in the 15-country study of nuclear industry workers. Radiat Res. 2008;170(5):661-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leuraud K, Richardson DB, Cardis E, et al. . Ionising radiation and risk of death from leukaemia and lymphoma in radiation-monitored workers (INWORKS): an international cohort study. Lancet Haematol. 2015;2(7):e276-e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Tute R, Yuille M, Catovsky D, Houlston RS, Hillmen P, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL) in CLL families: substantial increase in relative risk for young adults. Leukemia. 2006;20(4):728-729. [DOI] [PubMed] [Google Scholar]
- 101.Hinds DA, Barnholt KE, Mesa RA, et al. . Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood. 2016;128(8):1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kralovics R, Stockton DW, Prchal JT. Clonal hematopoiesis in familial polycythemia vera suggests the involvement of multiple mutational events in the early pathogenesis of the disease. Blood. 2003;102(10):3793-3796. [DOI] [PubMed] [Google Scholar]
- 104.Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomsen H, Chattopadhyay S, Weinhold N, et al. . Genome-wide association study of monoclonal gammopathy of unknown significance (MGUS): comparison with multiple myeloma. Leukemia. 2019;33(7):1817-1821. [DOI] [PubMed] [Google Scholar]
- 106.Landgren O, Kristinsson SY, Goldin LR, et al. . Risk of plasma cell and lymphoproliferative disorders among 14621 first-degree relatives of 4458 patients with monoclonal gammopathy of undetermined significance in Sweden. Blood. 2009;114(4):791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vachon CM, Kyle RA, Therneau TM, et al. . Increased risk of monoclonal gammopathy in first-degree relatives of patients with multiple myeloma or monoclonal gammopathy of undetermined significance. Blood. 2009;114(4):785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Humphreys K, Grankvist A, Leu M, et al. . The genetic structure of the Swedish population. PLoS One. 2011;6(8):e22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ameur A, Dahlberg J, Olason P, et al. . SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet. 2017;25(11):1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hemminki K, Granström C, Chen B. The Swedish family-cancer database: update, application to colorectal cancer and clinical relevance. Hered Cancer Clin Pract. 2005;3(1):7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 112.Li N, Johnson DC, Weinhold N, et al. . Genetic predisposition to multiple myeloma at 5q15 Is mediated by an ELL2 enhancer polymorphism. Cell Reports. 2017;20(11):2556-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Studd JB, Vijayakrishnan J, Yang M, Migliorini G, Paulsson K, Houlston RS. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2 [published correction appears in Nat Commun. 2018;9:16204]. Nat Commun. 2017;8(1):14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kandaswamy R, Sava GP, Speedy HE, et al. . Genetic predisposition to chronic lymphocytic leukemia is mediated by a BMF super-enhancer polymorphism. Cell Reports. 2016;16(8):2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li N, Johnson DC, Weinhold N, et al. . Multiple myeloma risk variant at 7p15.3 creates an IRF4-binding site and interferes with CDCA7L expression. Nat Commun. 2016;7(1):13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Studd JB, Yang M, Li Z, et al. . Genetic predisposition to B-cell acute lymphoblastic leukemia at 14q11.2 is mediated by a CEBPE promoter polymorphism. Leukemia. 2019;33(1):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stensrud MJ, Valberg M. Inequality in genetic cancer risk suggests bad genes rather than bad luck. Nat Commun. 2017;8(1):1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Turnbull C, Sud A, Houlston RS. Cancer genetics, precision prevention and a call to action [published correction appears in Nat Genet. 2019;51(1):196]. Nat Genet. 2018;50(9):1212-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





