Rapid cell divisions that occur during somatic embryogenesis in soybean are accompanied by hyperactive DNA methylation pathways to ensure the maintenance of genome integrity over the short term.
Abstract
Somatic embryogenesis is an important tissue culture technique that sometimes leads to phenotypic variation via genetic and/or epigenetic changes. To understand the genomic and epigenomic impacts of somatic embryogenesis, we characterized soybean (Glycine max) epigenomes sampled from embryos at 10 different stages ranging from 6 weeks to 13 years of continuous culture. We identified genome-wide increases in DNA methylation from cultured samples, especially at CHH sites. The hypermethylation almost exclusively occurred in regions previously possessing non-CG methylation and was accompanied by increases in the expression of genes encoding the RNA-directed DNA methylation (RdDM) machinery. The epigenomic changes were similar between somatic and zygotic embryogenesis. Following the initial global wave of hypermethylation, rare decay events of maintenance methylation were observed, and the extent of the decay increased with time in culture. These losses in DNA methylation were accompanied by downregulation of genes encoding the RdDM machinery and transcriptome reprogramming reminiscent of transcriptomes during late-stage seed development. These results reveal a process for reinforcing already silenced regions to maintain genome integrity during somatic embryogenesis over the short term, which eventually decays at certain loci over longer time scales.
INTRODUCTION
Genetic transformation and rapid clonal propagation of plant species are typically performed via tissue culture using various combinations of plant hormones. In general, the recovery of whole plants from undifferentiated cells is divided into organogenesis and somatic embryogenesis (Thorpe, 1990; Bhojwani and Dantu, 2013). Both methods induce plant regeneration using different combinations of hormones. The former induces the formation of mature organs such as shoots or roots from callus. By contrast, the latter induces embryo development via the use of an auxin. Although tissue culture has contributed to the development of modern agriculture, somaclonal variation, usually from induced mutations, is also introduced into the cultured plants. While some of these mutations have been useful, the accumulation of mutations can result in abnormal phenotypes and the loss of regenerative capability (Ahloowalia, 1986; Skirvin et al., 1994).
Tissue culture-induced somaclonal variation also results from epigenomic variants via altered DNA methylation levels, causing the introduction of novel epialleles in a variety of plant species (Stroud et al., 2013a; Vining et al., 2013; Stelpflug et al., 2014; Ong-Abdullah et al., 2015; Han et al., 2018). DNA methylation is one of the most extensively studied chromatin modifications and has been found in all plants studied to date (Niederhuth et al., 2016; Takuno et al., 2016; Bewick et al., 2017). This modification is associated with the silencing of transposable elements (TEs) and certain genes (Du et al., 2015).
In plants, DNA methylation is separated into three sequence contexts: CG, CHG, and CHH (H = A, T, and C), which are maintained by different enzymatic pathways (Law and Jacobsen, 2010; Stroud et al., 2013b; Matzke and Mosher, 2014; Du et al., 2015). In Arabidopsis (Arabidopsis thaliana), CG, CHG, and CHH are maintained by METHYLTRANSFERASE1 (MET1), CHROMOMETHYLASE3 (CMT3), and CMT2, respectively (Finnegan et al., 1996; Ronemus et al., 1996; Lindroth et al., 2001; Mathieu et al., 2007; Lister et al., 2008; Zemach et al., 2013; Stroud et al., 2014). CMT2 and CMT3 target regions by recognizing methylation of Lys-9 on histone H3 (Stroud et al., 2014; Du et al., 2015). The CMT2 pathway mediates CHH methylation across the entire length of long TEs in pericentromeric regions and has a sequence preference for the CAA/CTA contexts in Arabidopsis (Zemach et al., 2013; Stroud et al., 2014; Gouil and Baulcombe, 2016). Additionally, the RNA-directed DNA methylation (RdDM) pathway methylates DNA in all three sequence contexts, and its activity is often associated with repressed TEs, repeats, and certain genes (Law and Jacobsen, 2010). The canonical RdDM pathway starts with transcripts generated by the plant-specific RNA polymerase Pol IV (He et al., 2009). These transcripts are converted into double-stranded RNA by RNA-DEPENDENT RNA POLYMERASE2 (RDR2; Law et al., 2011; Haag et al., 2012) and cleaved into 24-nucleotide short interfering RNAs (24-nucleotide small interfering RNAs [siRNAs]) by DICER-LIKE3 (DCL3; Xie et al., 2004). Finally, these 24-nucleotide siRNAs are loaded into ARGONAUTE4 (AGO4; Zilberman et al., 2003) to target transcripts produced by another plant-specific RNA polymerase, Pol V (Huang et al., 2009; Lahmy et al., 2009). This serves to recruit the de novo DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) to methylate targeted regions in any cytosine context (Cao and Jacobsen, 2002; Cao et al., 2003). Additionally, there is a noncanonical RdDM pathway involved in the establishment of non-CG methylation that is mediated by RDR6 and DCL4 and guided by 21- or 22-nucleotide siRNAs (Cuerda-Gil and Slotkin, 2016). Lastly, DNA methylation specifically at CG sites also occurs in the gene bodies of a subset of transcribed genes (Tran et al., 2005; Zhang et al., 2006; Zilberman et al., 2007). This type of DNA methylation in flowering plants is referred to as gene body methylation (gbM) and is independently maintained by MET1 and CMT3 (Zhang et al., 2006; Bewick et al., 2016).
The topic of phenotypic defects resulting from changes in DNA methylation via tissue culture has gained agronomic attention. For instance, the Africa oil palm tree (Elaeis guineensis) is an important oil-bearing crop that can be clonally reproduced via somatic embryos. Occasionally, some of these palms have abnormal fruit shapes, resulting in low oil production or even oil-free fruits. These phenotypic defects result from hypomethylation of a retrotransposon located in the intronic region of a developmentally important transcription factor, DEFICIENS1 (EgDEF1, the oil palm ortholog of Antirrhinum majus DEFICIENS and Arabidopsis APETALA3). The loss of methylation results in the production of a novel transcript that leads to undesirable fruit shapes (Ong-Abdullah et al., 2015).
In addition to oil palm trees, changes in methylation associated with tissue culture have also been reported in other species, including maize (Zea mays) and rice (Oryza sativa) produced via somatic embryogenesis and poplar (Populus trichocarpa) produced via organogenesis. Although these studies did not find widespread DNA methylation changes in callus and regenerated plants, a combination of gains and losses in localized regions throughout the genome was observed. These methylation changes are inherited, as similar changes were observed in cultured tissues and regenerated plants (Stroud et al., 2013a; Vining et al., 2013; Stelpflug et al., 2014; Han et al., 2018). By contrast, little is known about epigenomic changes in eudicots during somatic embryogenesis throughout the tissue culture process. Moreover, the epigenome is crucial for the regulation of genome integrity. However, how the epigenome is maintained in embryonic tissue experiencing continuous rounds of cell division has not been fully explored. Given that sexual reproduction provides a channel to reinforce the integrity of the epigenome in plants (Teixeira et al., 2009; Walker et al., 2018), it is unclear whether continuous tissue culture over long time scales will affect the epigenome without the reinforcement that is experienced during sexual reproduction.
Exogenous auxin application will stimulate an embryogenic state in some plant tissues (Merkle et al., 1995; Nic-Can and Loyola-Vargas, 2016). The development of somatic embryos recapitulates the development of zygotic embryos and, for eudicots, proceeds through the globular, heart, torpedo, and cotyledonary stages. However, continued auxin exposure will hinder the development of normal somatic embryos. If the level of exogenous auxin is high enough, somatic embryo development is arrested at the globular stage. Instead of proceeding to the heart stage, new somatic embryos bud off the older one. This repetitive or cyclic embryogenesis will continue for years as long as the exogenous auxin level is high enough. Once the exogenous auxin is removed, the somatic embryos will resume normal histodifferentiation, reach physiological maturity, and germinate into plants.
In the case of soybean (Glycine max), plants regenerated after approximately 9 months of cell culture tend to be male sterile (Trick et al., 1997). Plants obtained from older cell lines have phenotypic abnormalities. All cell cultures eventually reach a point where somatic embryos from them are no longer able to germinate and convert into plants. In addition, at any age, cell lines are able to differentiate into amorphous structures incapable of forming more somatic embryos.
In this study, we performed a comprehensive genome-wide investigation of the epigenome during somatic embryogenesis in soybean. Samples were collected and profiled from nine different developmental stages from various time points throughout the process of tissue culture. The results revealed a genome-wide increase in DNA methylation from cultured samples, which was most apparent for CHH methylation. Interestingly, regions already silenced by DNA methylation were preferred targets of the observed hypermethylation, which was coupled with the upregulation of genes encoding the RdDM machinery. These results reveal that there is a built-in methylome reinforcement program present during somatic embryogenesis. Additionally, the molecular changes observed were similar to previously documented epigenomic changes that occur during zygotic embryogenesis (Bouyer et al., 2017; Kawakatsu et al., 2017; Lin et al., 2017; Narsai et al., 2017). Relative to the reference methylome, the most significant amount of local-specific DNA methylation loss occurred in samples that experienced long-term tissue culture, revealing that epigenomic stability can decay over time, which is perhaps connected to the absence of sexual reproduction. The instability is coupled with the upregulation of genes that function in the late stage of seed development, indicative of the occurrence of a developmental transition, potentially explaining the inability of these somatic embryos to germinate into plants.
RESULTS
Genome-Wide Hypermethylation Occurs during Somatic Embryogenesis
To investigate the soybean epigenome during somatic embryogenesis, we collected replicated samples (see Methods) from 10 different developmental stages (Figure 1A; Supplemental Tables 1 to 3). Immature cotyledons were collected after 24 h on hormone-free medium (24 h-2,4-D) or on medium containing 40 mg/L (equivalent to 182 µM/L) 2,4-D (24 h+2,4-D). A 24-h exposure to exogenous auxins is enough to induce the formation of somatic embryos, so 24 h-2,4-D was used as the reference epigenome to assess changes during somatic embryogenesis and its induction. The next sampling was at 6 weeks, when repetitive embryogenesis is well established and able to continue in the presence of reduced exogenous auxin levels. Sampling 3 (6 week globular) and 4 (6 week differentiated) represented embryogenic and differentiated tissues at this stage, respectively. Sampling 5 (4 d subculture) was performed 4 weeks and 4 d later. Four weeks is the frequency of transfer onto fresh medium, making this the first cycle of repetitive globular-stage embryos. Four days after transfer is the time when cells are most mitotically active and, hence, most transformable (Hazel et al., 1998). Sampling 6 (6 month globular) took place when cultures were 6 months old, which is when somaclonal variation becomes more frequent. Sampling 7 (12 month globular) and 8 (12 month differentiated) represented embryogenic and differentiated tissue after 1 year in culture. Sampling 9 (8 year globular) represented an old cell line that was still embryogenic after 8 years of culture, although the embryos would no longer convert into plants. The last sampling (13 year globular) was propagated from the 8 year globular line, adding another 5 years of continuous embryogenic tissue culture. Therefore, the globular-stage embryo samples cultured for 8 and 13 years provided an opportunity to explore the maintenance of methylome integrity under continuous rounds of cell division in the absence of epigenome reinforcement programs that occur during sexual reproduction. Samples derived from the globular stage or the differentiated stage represent the maintenance or loss of capacity to form more somatic embryos, respectively (Parrott et al., 1988). It should be noted that a recent study observed that tissue used as explant can have an effect on the epigenomic profiles of plants propagated from somatic embryos (Wibowo et al., 2018); however, all samples from our study were regenerated from the same explant (immature cotyledons) using the same tissue culture method.
Figure 1.
Overview of Genome-Wide DNA Methylation Changes during Somatic Embryogenesis.
(A) Diagram of collected soybean samples and their developmental stages. Samples are denoted by different numbers, followed by a detailed description. + and – indicate treatment with synthetic auxin (2,4-D). 24 h-2,4-D was used as the reference epigenome to assess changes during somatic embryogenesis. Samples from 4 weeks plus 4 d were used, as this is a common method of transfer to induce mitotically active and transformable cells. Somaclonal variation starts to appear after 6 months of tissue culture. The auxin concentration is labeled under each sample. Samples from the globular stages and the differentiated stages result from germination of somatic embryos and indicate the maintenance or loss of the capacity to form more somatic embryos, respectively. 24 h-2,4-D has three independent biological replicates for MethylC-seq and RNA-seq. 24 h+2,4-D, 6 wk globular, 6 wk diff, 4-d subculture, and 6 mo globular have three independent biological replicates for MethylC-seq, RNA-seq, and small RNA-seq. The 13-year globular only has MethylC-seq data.
(B) to (D) Bar graphs of genome-wide weighted methylation levels in the CG (B), CHG (C), and CHH (D) contexts. Error bars indicate sd. Asterisks denote P value cutoffs 0.01 (**) and 0.05 (*).
(E) to (G) Heatmaps of weighted CG (E), CHG (F), and CHH (G) methylation levels across chromosome I (500-kb windows). Averaged methylation levels are displayed, if biological replicates exist in that stage. Gene and TE densities are plotted using 500-kb windows.
To investigate the impact of somatic embryogenesis on DNA methylation states, we generated replicated single-base resolution DNA methylomes for each sample using whole-genome bisulfite sequencing (Supplemental Table 1). First, genome-wide methylation levels were computed for each sequenced sample. The sample (24 h-2,4-D) that did not undergo auxin induction was used as the baseline to assess changes to other stages, which revealed methylation levels of 45.0, 25.8, and 1.8% for CG, CHG, and CHH, respectively. In all other samples, increases in DNA methylation were observed in all three contexts, with the greatest change occurring in CHH methylation (Figures 1B to 1D; Supplemental Figures 1A to 1C). For example, in cultured embryos, CG, CHG, and CHH methylation levels increased to maxima of 53.2, 36.3, and 10.5%, respectively (Figures 1B to 1D). The CG and CHG contexts are methylated and maintained in a symmetrical manner with high fidelity, potentially explaining the relative stability of CG and CHG methylated sites compared with CHH methylated sites. Taken together, these results indicate that DNA methylation increases during somatic embryo development. The increases in CHH methylation could be related to an active transcriptional silencing program to reinforce DNA methylation genome-wide during this process.
Exogenous auxin is necessary for the successful induction of somatic embryos (Jenik and Barton, 2005). To explore the role of auxin in the observed hypermethylation, immature cotyledons were treated with auxin (24 h+2,4-D) for 24 h. A subtle increase in DNA methylation was observed upon auxin induction, with mCHH increasing to 2.8% (Figures 1B to 1D). We also observed a decrease in CHH methylation in the 4-d subculture samples. This could be due to the increased cell division rate that occurs after 4 d of being on fresh medium at that stage (Figure 1D).
CHH Hypermethylation Occurs Genome-Wide at Previously Silenced Regions
The soybean genome contains domains of heterochromatin in pericentromeric regions of the chromosomes (Schmutz et al., 2010). These regions have a higher density of TEs and non-CG methylation compared with euchromatin (Schmitz et al., 2013). Therefore, to determine the genome-wide impact of hypermethylation observed in the tissue-cultured samples, we calculated DNA methylation levels at a chromosome-wide scale (Figures 1E to 1G; Supplemental Figure 2). The results revealed that the genome-wide CHH hypermethylation occurred throughout the entire genome, with the greatest increases in TE-enriched regions accompanied by moderate increases in gene-enriched regions (Figure 1G). Collectively, these results indicate that increases in CHH methylation during somatic embryogenesis occur across the entire genome and correlate with the abundance of TE and non-CG methylation, suggesting that hypermethylation might occur in regions previously targeted by DNA methylation pathways.
To create an unbiased assessment of the changes to DNA methylation during induction and globular-stage embryogenesis, we first pooled all methylome samples together to conduct an undirected identification of differentially methylated regions (DMRs) across all stages in the CG, CHG, and CHH contexts (Figures 2A to 2C). In total, 36,169 CG DMRs, 41,416 CHG DMRs, and 189,339 CHH DMRs were identified throughout the entire genome. Further analysis revealed that 69.6% of CG DMRs, 68.1% of CHG DMRs, and 74.4% of CHH DMRs are located in intergenic regions (Figure 2D). Among the CG, CHG, and CHH DMRs occurring in intergenic regions, 84.2, 86.7, and 97.3% of them overlapped with TEs, respectively. In addition, 8.7% of CG DMRs, 7.9% of CHG DMRs, and 9.7% of CHH DMRs are located in promoter regions (Figure 2D). Among the CG, CHG, and CHH DMRs occurring in promoter regions, 76.7, 86.1, and 93.5% of them overlapped with TEs, respectively. In at least one cultured embryo stage relative to the initial stage (24 h-2,4-D), 419 (1.2%) CG DMRs gained at least 50% methylation (all gains and losses were measured in absolute values; see Methods), 4992 (12.1%) CHG DMRs gained at least 30% methylation, and 173,555 (91.7%) CHH DMRs gained at least 10% methylation. In samples undergoing tissue culture for more than 24 h, anywhere from 38.1 to 240.0 megabases (Mb) (3.9–24.5%) of sequences showed evidence of hypermethylated CHH DMRs (Figure 2E).
Figure 2.
DMRs during Somatic Embryogenesis.
(A) to (C) Heatmaps of CG methylation levels of CG DMRs (A), CHG methylation levels of CHG DMRs (B), and CHH methylation levels of CHH DMRs (C) across different developmental stages. The total number of DMRs is labeled above each heatmap.
(D) Distribution of identified DMRs in genomic features. Promoter regions and downstream regions were defined as 1 kb upstream of the transcription start site and 1 kb downstream of the transcription termination site, respectively. The distribution of identified DMRs in each genomic feature was compared with genomic distribution using a χ2 test. *, P < 0.05. UTR, untranslated region.
(E) Summation of all DMRs genome-wide across individual samples. DMRs were defined relative to 24 h-2,4-D. Absolute methylation differences of +/−50% for CG, +/−30% for CHG, and +/−10% for CHH were defined as hypermethylation/hypomethylation, respectively.
Further analysis revealed that 99.2% of CHH DMRs also possessed at least 5% CHG methylation in the 24 h-2,4-D sample. Only 122 loci lacked any preexisting methylation, which could represent spontaneous epimutations (Becker et al., 2011; Schmitz et al., 2011). By contrast, 24,645 (68.1%) of the CG DMRs, 27,664 (66.8%) of the CHG DMRs, and 1187 (0.6%) of the CHH DMRs gained at least 50% mCG, 30% mCHG, and 10% mCHH, respectively. The CHH methylation levels are very low in the initial stage, which could explain the low proportion of hypomethylated CHH DMRs. Additionally, in samples undergoing tissue culture for more than 24 h, the loss of CG and CHG methylation in cultured embryos accumulated over time (Supplemental Figure 3), ranging from 104.4 kb to 13.8 Mb (up to 1.4%) and 450.4 kb to 27.1 Mb (up to 2.8%) of the total sequences that had reduced DNA methylation, respectively (Figure 2E). These results indicate that a substantial amount of DNA methylation is reduced over time, depending on the sample profiled. Therefore, over the course of somatic embryogenesis, there is an initial, marked increase in predominantly CHH methylation, followed by a reduction in DNA methylation in all contexts. Taken together, these results confirm the notion that the most extensive changes in DNA methylation occurred in the CHH context and almost exclusively in regions that were already methylated in non-CG contexts.
In most flowering plant genomes, the majority of genes are unmethylated (UM), followed by two small groups that possess gbM and TE-like methylation (TEM), respectively (Tran et al., 2005; Kawakatsu et al., 2016a; Niederhuth et al., 2016; Takuno et al., 2016; Bewick et al., 2017). The observed hypermethylation appeared limited to regions previously possessing non-CG methylation as opposed to UM genes or genes possessing gbM (Figure 3A). Therefore, to quantitatively assess if hypermethylation is restricted to specific genomic features, we grouped genes into one of three categories (UM = 26,685 genes, gbM = 6834 genes, and TEM = 3563 genes; Supplemental Data Set 1). Increases to CHH methylation primarily occurred on TEs/TEM genes and not on gbM/UM genes (Figures 3B to 3E). Quantifying changes to DNA methylation as a result of tissue culture revealed that 87.0% of TEs, 0.1% of UM genes, 0.1% of gbM genes, and 55.0% of TEM genes gained at least 10% CHH methylation in at least one cultured embryo stage. The proportions of gain of methylation in TEs and TEM genes are significantly higher than the numbers in UM and gbM genes (Supplemental Table 4). Collectively, these results indicate that the increase in DNA methylation is associated with preexisting non-CG methylation and occurs on both TEs and TEM genes.
Figure 3.
CHH Hypermethylation Targets Previously Methylated Regions.
(A) Genome browser view of the DNA methylation profile of a representative region of the soybean genome (purple vertical lines = CG methylation, blue vertical lines = CHG methylation, and gold vertical lines = CHH methylation). The horizontal lines represent different categories of genes.
(B) to (E) Heatmaps of methylation levels across TE (B), UM (C), gbM (D), and TEM (E) genes and 1-kb flanking regions in different contexts. Averaged methylation levels are displayed, if biological replicates exist in that stage. TSS, transcription start site; TTS, transcription termination site.
To investigate if the observed CHH hypermethylation leads to changes to transcription in TEs and TEM genes, we generated replicated RNA sequencing (RNA-seq) data for samples isolated from different stages of tissue culture. Of the 642,428 TEs and 1601 TEM genes gaining over 10% mCHH in 6 mo globular embryos, only 109 of the TEs and 11 of the TEM genes were differentially expressed in 6 mo globular embryos relative to the initial stage (Supplemental Table 5). Elevated CHH methylation in the promoter regions and an earlier stage (6 wk globular embryos) also did not result in a wide range of transcriptional changes (Supplemental Table 5). These results suggest that the hypermethylation did not lead to a widespread change in TE or TEM gene expression. Collectively, the low correlation between hypermethylation and the expression of TEs and TEM genes indicates that the observed hypermethylation is primarily used to reinforce regions that are already methylated. This conclusion is further supported by the lack of hypermethylation associated with gbM loci, where DNA methylation is restricted to CG sites due to a lack of RdDM activity.
RdDM Drives CHH Hypermethylation during Somatic Embryogenesis
In plants, CHH methylation is catalyzed by CMT2 and/or DRM2 (Cao et al., 2000; Zemach et al., 2013; Matzke and Mosher, 2014; Stroud et al., 2014). CHH hypermethylation in Arabidopsis has been identified in root columella cells, during seed development, and in the vegetative nucleus of pollen (Calarco et al., 2012; Kawakatsu et al., 2016b, 2017; Lin et al., 2017). In some cases, hypermethylation was identified as a combinatorial effect of the canonical RdDM pathway and the CMT2 pathway (Kawakatsu et al., 2016b, 2017; Lin et al., 2017).
To investigate the potential causes for the observed changes in CHH methylation during embryo induction and globular-stage somatic embryogenesis, we identified enzymes involved in the RdDM pathway and determined the expression levels of their underlying genes using RNA-seq data (Supplemental Table 2; Supplemental Data Set 2). Genes for numerous canonical RdDM pathway components as well as other enzymes required for the maintenance of DNA methylation were immediately downregulated at the stage when immature cotyledons were treated with auxin (Figure 4A; Supplemental Table 6). The expression of the canonical RdDM pathway components subsequently increased in cultured somatic embryos. Notably, genes encoding two noncanonical RdDM pathway enzymes, RDR6 and DCL4, were highly expressed and did not experience reduced expression under initial auxin treatment (Figure 4A). Taken together, these findings suggest that enzymes from both the canonical and noncanonical RdDM pathways are involved in the observed increase in DNA methylation. The only exception was the 24 h+2,4-D material, which showed downregulation of many genes encoding canonical RdDM pathway enzymes yet still exhibited minor increases in DNA methylation, suggesting that other factors might lead to methylation at this stage (Figures 1B to 1D).
Figure 4.
High Activity of the RdDM Pathway.
(A) Expression profile of genes involved in the regulation of DNA methylation throughout somatic embryogenesis using fragments per kilobase per million (FPKM) for quantification. The expression levels were normalized by the values in 24 h-2,4-D.
(B) Bar plots of the size distribution of 21-, 22-, and 24-nt small RNAs. Error bars indicate sd. Size distribution of small RNAs was compared with 24 h-2,4-D using χ2 test. *, P < −105.
(C) and (D) Metaplots of CHH methylation level (C) and normalized 24-nt read density (D) across four different TE classes and 1-kb flanking regions. TE classes are as follows: LINEs (long interspersed nuclear elements), LTRs (long terminal repeats), TIRs (terminal inverted repeats), and helitrons. Averaged methylation levels are displayed, if biological replicates exist for that stage.
(E) Distribution of methylated sites in each subcontext across different developmental stages. Raw numbers were normalized by the values in 24 h-2,4-D. Error bars indicate sd.
The canonical and noncanonical RdDM pathways in plants are mediated by 24-nucleotide and 21- or 22-nucleotide siRNA, respectively (Law and Jacobsen, 2010; Cuerda-Gil and Slotkin, 2016). To determine if the canonical RdDM pathway is the primary mechanism coordinating CHH hypermethylation during somatic embryogenesis, we generated small RNA-seq data with independent biological replicates (Supplemental Table 3) and profiled the distribution of aligned sequences (Figure 4B). The distribution of small RNA fragments was similar between the 24 h-2,4-D and 24 h+2,4-D samples, with abundant 21nucleotide and 24-nucleotide RNAs, which are mostly microRNAs and siRNAs, respectively. After the initial stages, the 24-nucleotide siRNAs were the dominant size class, representing over 70.7% of all small RNAs (Figure 4B). These results provide one line of evidence that the RdDM pathway is active at its targeted regions.
The CMT2 pathway mediates CHH methylation throughout long TEs, which are often located in pericentromeric regions, whereas the RdDM pathway is responsible for directing CHH methylation at TE edges (Zemach et al., 2013; Stroud et al., 2014). Therefore, to determine the relative contribution of the CMT2 pathway to the observed CHH hypermethylation, we assessed DNA methylation levels of four major soybean TE classes: long interspersed nuclear elements, long terminal repeats, terminal inverted repeats, and helitrons (Figure 4C). The increases in CHH methylation occurred evenly at the edges and bodies of all four families, along with an increase in abundance of 24-nucleotide small RNAs throughout these same regions (Figure 4D). Similar trends were observed in TEs longer than 3 kb (Supplemental Figure 4), suggesting that the processivity of the RdDM pathway is functioning to methylate these regions instead of CMT2. In support of this result, no increase in the steady state levels of CMT2 mRNA was observed throughout the soybean somatic embryogenesis stages that were evaluated (Figure 4A).
Previous studies have revealed trinucleotide preferences of CMT2 in different species (Gouil and Baulcombe, 2016). To further understand the role of CMT2 during somatic embryogenesis, we split the methylated cytosines into 16 trinucleotide combinations (CNN, N = A, T, C, and G; Figure 4E). The number of methylated sites was calculated from each sample and normalized by values in the 24 h-2,4-D sample. The changes at CG and CHG sites were minor (Figure 4E). By contrast, all nine CHH combinations were highly hypermethylated, without a specific site preference. These results indicate that the RdDM pathway is likely acting as the main driver of genome-wide CHH hypermethylation and that CMT2 is providing a supporting role. Alternatively, CMT2 specificity could be masked by the higher activity of the RdDM machinery, or perhaps CMT2 in soybean does not have specific site preferences like CMT2 in Arabidopsis (Gouil and Baulcombe, 2016).
DNA Methylation Is Lost at Certain Loci during Tissue Culture
DMR analysis revealed rare losses of DNA methylation after 12 months of continued tissue culture, with even greater losses detected in the 8 year globular-stage sample (Figures 2A to 2C). Therefore, to explore the potential cause of the observed hypomethylation in CG and CHG DMRs, we examined the underlying contexts to estimate the involvement of different methylation pathways. Accordingly, 12,744 (35.2%) CG DMRs and 16,899 (40.8%) CHG DMRs lost over 50% CG methylation and 30% CHG methylation in the 8 yr globular-stage sample, respectively (Supplemental Figures 5A and 5B). Moreover, 91.34% of hypomethylated CG DMRs contained at least 5% CHG methylation in the 24 h-2,4-D sample and 99.97% of hypomethylated CHG DMRs contained at least 5% CG methylation in the 24 h-2,4-D sample. These results suggest that the vast majority of hypomethylated regions are targeted by non-CG methylation pathways.
The loss of a hyperactive RdDM pathway in the 12 month differentiated and 8 year globular stages could explain the reduction observed at CHH sites (Figure 1D). However, the absence of highly active RdDM machinery should not lead to any losses of CG and CHG methylation, as these methylation types are likely maintained independently by MET1 and CMT3, once methylated. Furthermore, the reduction of methylation at CG and CHG in DMRs was almost indistinguishable between 12 month globular and 12 month differentiated embryos (Supplemental Figures 5A and 5B), but the latter stage had much lower transcript abundance for the RdDM machinery (Figure 4A). These results suggest that the decay of methylation at some regions is unlikely due directly to the loss of RdDM activity and might instead be due to the stochastic loss of maintenance of DNA methylation.
In plants, CG and CHG are symmetrically maintained by MET1 and CMT3, with high fidelity levels on both strands of the DNA (Niederhuth et al., 2016). Therefore, to assess the functionality of soybean MET1 and CMT3 activities throughout the tissue culture stage examined, we measured the methylation states of symmetrically methylated CG and CHG pairs. The results demonstrated that both methylated CG and CHG contexts were faithfully maintained in a symmetrical manner (Supplemental Figures 6 and 7), indicating that these enzymes were still functional in all of the samples tested.
To further test for a decay in the maintenance of methylation at a subset of loci in the genome, we evaluated gbM loci, as they represent non-RdDM-targeted loci (Stroud et al., 2013b; Bewick et al., 2016). DNA methylation states were stable in most gbM genes during continuous tissue culture, and a maximum of 66 gbM genes gained more than 20% CG methylation during somatic embryogenesis. However, many more gbM loci experienced losses of DNA methylation over longer time scales (Figure 5A). In total, 280 (4.1%), 285 (4.2%), 543 (7.9%), and 838 (12.3%) gbM genes lost more than 20% CG methylation in 12 mo globular embryos, 12 month diff embryos, 8 year globular embryos, and 13 year globular embryos, respectively (Figure 5B). By contrast, a maximum of 70 gbM genes lost similar amounts of CG methylation during other stages. These results further support the notion that long-term somatic embryogenesis leads to the loss of maintenance methylation at a subset of loci.
Figure 5.
Losses of Maintenance of DNA Methylation Occur in gbM Loci.
(A) Representative gbM locus of the soybean genome that lost maintenance methylation (purple vertical lines = CG methylation, blue vertical lines = CHG methylation, and gold vertical lines = CHH methylation). The black and red horizontal lines represent a hypermethylated region and a gbM locus, respectively.
(B) Heatmap of CG methylation levels of gbM genes throughout somatic embryogenesis. The number of gbM genes is labeled on the left side.
Given the loss of maintenance of DNA methylation at some regions, we hypothesized that TE reactivation and transposition might occur. Using the RNA-seq data, we examined the transcriptional changes of TEs that lost maintenance methylation in the 8 yr globular-stage embryo sample. Of the 50,339 TEs that lost over 50% CG methylation, only 284 (0.6%) were differentially expressed in 8 year globular embryos relative to the initial stage. To assess if any of the loss in methylation leads to novel TE insertions, we used epiTEome (Daron and Slotkin, 2017) and identified a subtle increase in TE insertion events over time (Supplemental Figure 8). However, different integration events were found at all stages compared with the 24 h-2,4-D stage, suggesting that the TEs present in this genotype do not accumulate during tissue culture. In summary, the loss of maintenance methylation at rare loci occurs over time and with cell divisions. Further studies are needed to determine if some of these rare loci are associated with TE reactivation and transposition.
Reduced DNA Methylation Leads to the Reactivation of Silenced Genes
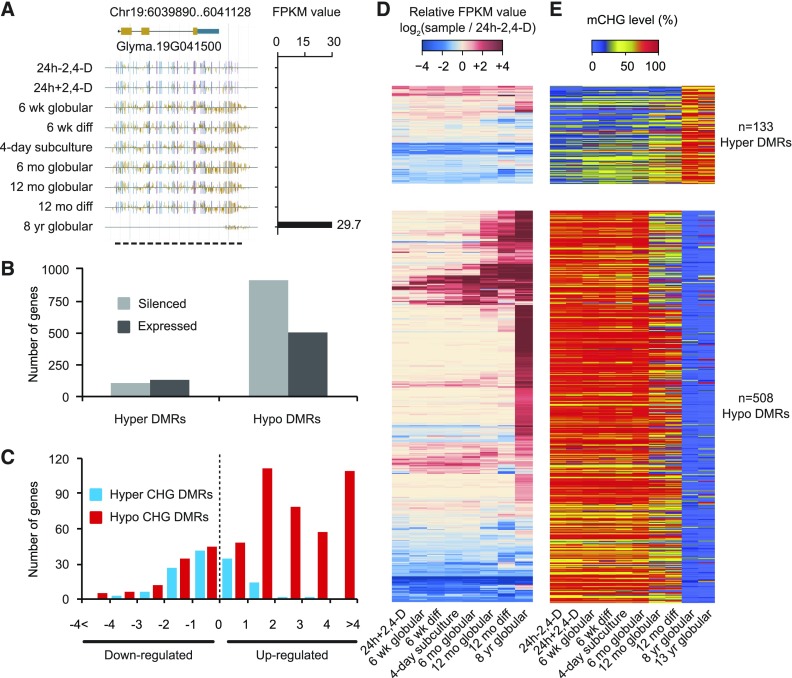
Our high-resolution investigation uncovered a subset of genes that lose DNA methylation and become transcriptionally active in the 8 year globular embryo sample (Figure 6A). Therefore, to quantify the changes in gene expression associated with the loss of DNA methylation, two categories of genes were defined based on the type of CHG DMRs they overlapped with in the 8 year globular sample. In total, we identified 241 genes that overlapped with CHG DMRs (gaining at least 10% CHG methylation), 133 of which were expressed in at least one sample. By contrast, 1418 genes overlapped with CHG DMRs (losing at least 10% CHG methylation) and 508 were expressed in at least one sample (Figure 6B). Using the RNA-seq data, we found slight bias in the expression profiles of genes that overlapped with hypermethylated CHG DMRs (t test, P = 0.013). However, 356 (70.1%) genes overlapping hypomethylated CHG DMRs were upregulated (t test, P = 6.6E-52; Figure 6C).
Figure 6.
Reactivation of Genes Is Accompanied by Losses of Maintenance Methylation.
(A) DNA methylation and corresponding transcript abundance of a reactivated gene throughout somatic embryogenesis (purple vertical lines = CG methylation, blue vertical lines = CHG methylation, and gold vertical lines = CHH methylation).
(B) Bar plot of correlation between genes and hypermethylated/hypomethylated CHG DMRs. Fragments per kilobase per million (FPKM) = 1 was used as the cutoff to determine absolute expression.
(C) Fold change distribution of expressed genes associated with CHG DMRs. Fold change was defined relative to the values in 24 h-2,4-D.
(D) Heatmaps of transcription levels in expressed genes that overlapped with defined hypermethylated/hypomethylated CHG DMRs. The expression levels were normalized by the values in 24 h-2,4-D.
(E) Heatmaps of CHG methylation levels in corresponding overlapping CHG DMRs. The numbers of expressed genes are labeled on the right side.
We also tracked the changes in mRNA expression levels and the corresponding methylation levels for these genes at other stages (Figures 6D and 6E). The reactivation of these silenced genes is highly correlated to the loss of DNA methylation. Additionally, of the 508 hypomethylated DMRs found in the 8-year sample, 96.3% were present in the 13-year sample, suggesting that once the loss of DNA methylation occurs, it is rarely restored in embryogenic tissue culture. Gene Ontology enrichment analysis did not find any significant categories. These results suggest that long-term tissue culture leads to the loss of non-CG methylation in the body of a subset of silenced genes. The observed loss of DNA methylation might be associated with the reactivation of genes that were previously silenced. It is also possible that the expression differences may precede the reduction in DNA methylation (Secco et al., 2015), which will require additional studies to validate.
Continuous Tissue Culture Leads to a Developmental Transition
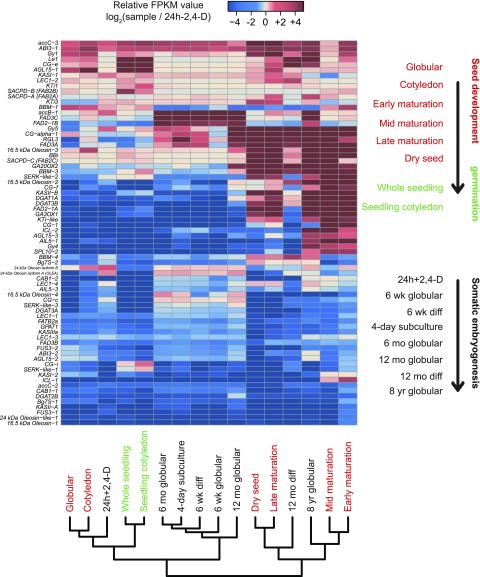
A recent study revealed 75 genes that are important for seed development in soybean (Lin et al., 2017). To determine if a developmental transition occurs in embryos that experience long-term tissue culture, we used the expression of these genes to conduct a comparative analysis across our samples. The analysis indicated that cultured globular-stage embryos (except 8 year globular) have an expression profile most similar to that of zygotic globular-stage embryos and tissues from seed germination (Figure 7). A few exceptions are genes for accB-1 (BIOTIN CARBOXYL CARRIER PROTEIN OF ACETYL-COA CARBOXYLASE), FAD3C (FATTY ACID DESATURASE3C), and FAD2-1B (FATTY ACID DESATURASE2-1B), which are all involved in oil biosynthesis (Lin et al., 2017). Interestingly, the expression profiles of 6 week globular-stage, 6 week differentiated, and 12 month globular-stage embryos were closer to those of early-stage zygotic embryos during seed development. By contrast, the 12 month diff and 8 year globular-stage embryos had strong similarities to zygotic embryos from late stages of seed development (Figure 7). Collectively, these results suggest that long-term tissue culture leads to a developmental transition in cell fate. Further studies are needed to understand whether the early expression of these genes is an active or passive process during somatic embryogenesis and to determine if this developmental transition is what prevents successful plant regeneration from these embryos.
Figure 7.
Expression Profile of Genes Known To Be Important for Seed Development and Germination.
The gene list was presented in a recent publication (Lin et al., 2017), and gene names are labeled on the left side. The expression levels were normalized by the values in 24 h-2,4-D using fragments per kilobase per million (FPKM). Hierarchical clustering relationships are illustrated at the bottom. The red and black colors denote samples obtained during seed development and somatic embryogenesis, respectively. The arrows indicate the orientation of development of these processes.
DISCUSSION
In this study, we discovered a genome-wide increase in DNA methylation during induction and globular-stage somatic embryogenesis in soybean (Figures 1B to 1D). Other genome-wide tissue culture studies have revealed a combination of gains and losses in localized regions, but the extent of changes was not as widespread as those observed in this more-detailed study (Stroud et al., 2013a; Vining et al., 2013; Stelpflug et al., 2014; Ong-Abdullah et al., 2015). Late-stage embryos are hypermethylated in Arabidopsis (Gehring et al., 2009; Hsieh et al., 2009). Therefore, one possible explanation for the observed hypermethylation during somatic embryogenesis is that the genome-wide increase in DNA methylation is an inherent property of embryogenesis. This idea is partly supported by recent studies that profiled epigenomic changes during zygotic embryogenesis in soybean and Arabidopsis (Bouyer et al., 2017; Kawakatsu et al., 2017; Lin et al., 2017; Narsai et al., 2017). These studies revealed striking similarities, including CHH hypermethylation primarily targeting non-CG methylation as well as upregulation of RdDM machinery genes during seed development. Perhaps this mechanism is conserved during embryogenesis to maintain the integrity of the genome given that a subset of these cells give rise to germ cells.
However, there are also differences between somatic embryos and zygotic embryos in developing seeds. Hypermethylation occurs in globular-stage embryos during somatic embryogenesis (Figures 1B to 1D), but it does not occur until the early embryo maturation stage of seed development (Kawakatsu et al., 2017; Lin et al., 2017). Auxin is essential for precise embryonic development. Endogenous auxin levels are low in globular-stage embryos and progressively increase during seed development (Jenik and Barton, 2005; Robert et al., 2015). By contrast, globular-stage embryos are cultured with a very high concentration of exogenous auxin (Figure 1A). Thus, auxin could be associated with the observed early wave of hypermethylation. This idea is partly supported by a previous study focusing on carrot (Daucus carota) somatic embryogenesis showing a positive correlation between the concentration of auxin and hypermethylation (Loschiavo et al., 1989). Moreover, the columella is a hypermethylated cell type in Arabidopsis that has enriched auxin signaling (Ottenschläger et al., 2003; Kawakatsu et al., 2016b). Additionally, in our study, we found that the immature cotyledon showed a slight increase in DNA methylation after 24 h of auxin treatment (Figures 1A to 1D). We speculate that auxin could be involved in the reinforcement of DNA methylation at previously non-CG methylated loci. However, how auxin participates in hypermethylation and leads to hyperactivity of the RdDM pathway requires further exploration.
It remains unclear what leads to the losses of DNA methylation observed in cultured cells after many years of consecutive tissue culture. It is possible that the loss of DNA methylation simply results from a mutation in a gene important for the maintenance of DNA methylation. However, if this were the case, we would expect a genome-wide loss in these samples rather than the locus-specific loss that accumulates gradually over time. Notably, we found an even more severe reduction of DNA methylation in 13 year globular-stage embryos relative to the 8 year sample (Figures 2A and 2B). This was apparent at gbM loci, where over 12% of them lost gbM (Figure 5B). Additionally, 96.3% of the hypomethylated DMRs overlapping genes identified in the 8-year sample were found in the 13-year sample. Lastly, the decreases in DNA methylation also occurred in two independent 1-year-old samples. Collectively, we conclude that the process of tissue culture results in rare failures to maintain DNA methylation and that these demethylated regions accumulate over time.
One possible explanation is that the maintenance of DNA methylation pathways cannot keep up with the continuous and rapid rate of cell division in the absence of the epigenome reinforcement program provided by sexual reproduction (Teixeira et al., 2009; Walker et al., 2018). Over time, this lack of reinforcement would lead to the occasional failure to maintain methylation during DNA replication. However, this does not fully explain the depletion of non-CG methylation observed at certain loci. Alternatively, data from studies of seed development have revealed an active non-CG demethylation process during late-stage embryogenesis (Kawakatsu et al., 2017). Interestingly, our results indicate that the reduction in DNA methylation is coupled with a developmental transition. Therefore, active demethylation pathways may also play an important role in somatic embryos, resulting in the active removal of maintenance DNA methylation over long time scales. Further studies are needed to explore the mechanisms for epigenome maintenance in cultured cells.
METHODS
Plant Preparation and Collection
Soybean (Glycine max cv ‘Jack’) plants were grown under greenhouse conditions, which consisted of 10 to 14 h of daylight, depending on time of year, and supplemented with 12 h under 400-W high-pressure sodium lighting. The temperature in the greenhouse ranged from 20 to 27°C. Plants were placed in C2100 pots (Nursery Supplies) with a 42% sand and 58% Fafard 3B soil mix (Conrad Fafard), watered daily, and fertilized weekly with 0.453 kg of Peters Professional All Purpose Plant Food per 380 liters of water. Pods containing immature seeds 3 to 5 mm long were harvested. At this stage, the zygotic embryo is still growing via cell division rather than cell enlargement. The pods were surface-sterilized, and the seeds were removed. The embryonic axis was cut off from each seed, and the two cotyledons were extracted from the seed coat and placed on medium adaxial side up. The medium consisted of Murashige and Skoog basal salts, B5 vitamins (100 mg/mL myo-inositol, 1 mg/mL nicotinic acid [free acid], 1 mg/mL pyridoxine HCl, and 10 mg/mL thiamine hydrochloride), 3% (w/v) Suc, and 40 mg/L 2,4-D, adjusted to pH 5.8 and hardened with 0.2% (w/v) gellan gum. All embryogenic cell lines/cultures were maintained at 24°C at 5 to 10 µmol m−2 s−1 photosynthetically active radiation provided by cool-white fluorescent tubes and a 16-h-light/1-h-dark photoperiod (Trick et al., 1997). Figure 1 contains a diagram of each sampling tissue and stage. The number of replicates per sample/experiment is listed in Supplemental Tables 1 to 3. Briefly, 24 h-2,4-D had three independent biological replicates (experiments) for MethylC-seq and RNA-seq. The 24 h+2,4-D, 6 wk globular, 6 wk diff, 4-d subculture, and 6 mo globular samples had three independent biological replicates for MethylC-seq, RNA-seq, and small RNA-seq. Only MethylC-seq data were obtained for the 13-year globular sample.
DNA, RNA, and Small RNA Isolation
Collected cotyledons/somatic embryos were flash-frozen and finely ground to a powder using a mortar and pestle. DNA extraction was performed on all samples using a DNeasy Plant Mini Kit (Qiagen), and the DNA was sheered to ∼200 bp by sonication. Total RNA was isolated using the PureLink Plant RNA Reagent (Thermo Fisher) following the manufacturer’s instructions. Total RNA quality and quantity were assessed before library construction. Small RNAs (20–30 nucleotide) were size selected on a 15% polyacrylamide/urea gel and used for small RNA library preparation as described (Mathioni et al., 2017). A 3-µg aliquot of total RNA was used for size selection. For RNA-seq library preparation, total RNA was treated with DNase I (New England Biolabs [NEB]) and cleaned using RNA Clean and Concentrator-5 (Zymo Research).
Library Preparation
MethylC-seq libraries were prepared as described (Urich et al., 2015). Briefly, genomic DNA was sonicated to 200 bp using a Covaris S-series focused ultrasonicator and end-repaired using an End-It DNA end-repair kit (Epicentre). The end-repaired DNA was subjected to A-tailing using Klenow 3′-5′ exo− (NEB) and ligated to methylated adapters using T4 DNA ligase (NEB). The ligated DNA was subsequently bisulfite converted using an EZ DNA Methylation-Gold kit as per the manufacturer’s instructions and amplified using KAPA HiFi Uracil + Readymix Polymerase. Non-stranded RNA-seq libraries were constructed using a TruSeq RNA Library Preparation Kit v2 (Illumina catalog number RS-122-2001) following the manufacturer’s instructions. Small RNA-seq libraries were constructed using a TruSeq Small RNA Library Preparation Kit (Illumina catalog number RS-200-0012) following the manufacturer’s instructions and as described (Mathioni et al., 2017). Briefly, 20- to 30-nt small RNAs were size selected using PAGE gels. Then, 3′ and 5′ adapters were ligated, followed by reverse transcription and PAGE purification. Finally, a PCR amplification step was performed, and the libraries were quantified, diluted, and submitted for sequencing.
High-Throughput Sequencing
Illumina sequencing was performed on an Illumina HiSeq 2500 instrument at the University of Delaware Sequencing and Genotyping Center in the Delaware Biotechnology Institute and an Illumina NextSeq 500 instrument at the University of Georgia Genomics Facility. For MethylC-seq and small RNA-seq data, raw reads were trimmed for adapters and preprocessed to remove low-quality reads using cutadapt 1.9.dev1 (Martin, 2011). For small RNA-seq, only 18- to 30-nucleotide-long reads were kept for subsequent analysis. For RNA-seq data, these processes were performed with Trimmomatic v0.32 (Bolger et al., 2014).
MethylC-Seq Data Processing
Qualified reads were aligned to the G. max Wm82.a2.v1 assembly as described (Schmitz et al., 2013). Chloroplast DNA (which is fully UM) was used as a control to calculate the sodium bisulfite reaction nonconversion rate of unmodified cytosines (Supplemental Table 1). A binomial test coupled with Benjamini-Hochberg correction was used to determine the methylation status of each cytosine. All gains and losses of DNA methylation were measured in absolute values, including defining DMRs and calculating hypermethylated/hypomethylated regions. CG/CHG DMRs containing at least 10 mappable (with at least three reads) CG sites and at least 10 mappable CHG sites were used to determine the number of loci losing methylation in 8 year globular-stage embryos and methylation levels in the 24 h-2,4-D stage. CHH DMRs containing at least 10 mappable CHG sites were used to determine if the observed hypermethylation occurs in previously methylated regions. Loci without preexisting methylation were defined as follows: mCG < 1%, mCHG < 1%, mCHH < 1%. The CG methylation level in the coding regions of a gbM gene lower than 1% was defined as “completely lost.” Only TEs with at least 5% CHG methylation were used to compute the percentage of TEs gaining over 10% CHH methylation.
RNA-Seq Data Processing
Qualified reads were aligned to the G. max Wm82.a2.v1 assembly using TopHat v2.0.13 (Kim et al., 2013; Supplemental Table 2). Gene expression values were computed using Cufflinks v2.2.1 (Trapnell et al., 2010). Fragments per kilobase per million (FPKM) = 1 was used as the cutoff to determine expression. Genes with at least 2 log2-fold expression change and significant statistical difference (false discovery rate cutoff = 0.05) were identified as differentially expressed genes. Gene Ontology enrichment analysis was performed in the SoyBase database (https://www.soybase.org/; Morales et al., 2013). To estimate the expression of TEs, only TEs that did not overlap with genes and that were not expressed in the 24 h-2,4-D stage were analyzed. TEs with at least 2 log2-fold expression change and significant statistical differences were identified as differentially expressed TEs. The raw soybean seed development transcriptomes were downloaded from the Gene Expression Omnibus database under accession number GSE29163 and processed using the same procedure.
Small RNA-Seq Data Processing
Qualified reads were aligned to the G. max Wm82.a2.v1 assembly using bowtie1 v1.1.1 with the parameters “-n 0 -l 18 -M 1” (Langmead et al., 2009). Reads with multiple locations were randomly assigned to one location (Supplemental Table 3).
Gene Category Identification
First, genes with significant similarity to TEs were identified by BLAST search against RepeatMasker libraries version 4.0.5 (http://www.repeatmasker.org/) as described (El Baidouri et al., 2018). These genes were defined as TE-like genes and removed from subsequent analyses. Qualified genes were then classified into different categories as described (Niederhuth et al., 2016). Briefly, CG, CHG, and CHH methylation levels of the coding regions of all genes (primary transcripts) were calculated as the background methylation levels. A binomial test coupled with a Benjamini-Hochberg correction was used to determine the significance of the tested gene and an adjusted P of 0.05 was used as the cutoff. Only the 24 h-2,4-D methylome (first biological replicate) was used to perform these tests. The detailed criteria used to categorize annotated genes based on their methylation states were as follows: UM genes, (1) not significant mCG, mCHG, and mCHH levels, (2) at least 20 mapped CHH sites, (3) mCHG, mCHH, and mCHH levels less than 1%; GbM genes, (1) significant mCG level, (2) at least 20 mapped CG sites, (3) not significant mCHG and mCHH levels, (4) both mCHG and mCHH levels less than 1%; TEM genes, (1) significant mCHH level, (2) at least 20 mapped CHH sites, (3) not expressed in the 24 h-2,4-D stage.
TE Annotation and Identification of Novel TE Insertions
Soybean TE annotation was performed by TASR (El Baidouri et al., 2015) in addition to public TE annotation from SoyBase (http://soybase.org/soytedb/) as described (Kim et al., 2015). TE classification was performed using PASTEClassifier (https://urgi.versailles.inra.fr/Tools/PASTEClassifier; Hoede et al., 2014).
Identification of nonreference TE insertions was performed using epiTEome as described (Daron and Slotkin, 2017). Only TEs that were not present in the 24 h-2,4-D were outputted. Briefly, trimmed FASTQ reads were mapped to the G. max Wm82.a2.v1 assembly using Bismark (Krueger and Andrews, 2011) with the following parameters: –bowtie2–ambiguous–unmapped –R 10 -score_min L,0,-0.6 -N 1. New TE insertion sites were predicted using the nonmapped reads by epiTEome using the soybean TE annotation and reference genome.
To discard putative false positives and validate the TE insertions, we developed a postprocessing strategy downstream of epiTEome analysis. For each TE insertion predicted by epiTEome, both TE edges were concatenated with the genomic DNA flanking the insertion site, creating a pseudosequence of the TE insertion that is not present in the reference genome. Using Bismark, reads that failed to map to the reference genome (unmapped reads) were mapped to pseudosequences. The presence of a TE insertion was validated only if a minimum of five unmapped reads spanned through the TE insertion pseudosequence for at least one of the two TE edges. Only reads that mapped with a minimum of 25 bp on either side of the TE insertion breakpoint were considered.
Metaplot Analysis
For metaplot analyses, 20 50-bp bins were created for both the upstream and downstream regions of gene bodies/TEs. Gene bodies/TE regions were evenly divided into 20 bins. Weighted methylation levels were computed for each bin as described previously (Schultz et al., 2012).
DMR Analysis
Identification of DMRs was performed as described (Schultz et al., 2015). Only DMRs with at least 10 differential methylated sites and a 10% methylation level difference within each DMR were reported and used for subsequent analysis. Absolute methylation differences of +/−50% for CG, +/−30% for CHG, and +/−10% for CHH were defined as hypermethylation/hypomethylation.
Phylogenetic Analysis of 5mC-Related Genes in Soybean
A phylogenetic approach, guided by annotations in Arabidopsis (Arabidopsis thaliana), was used to identify 5mC-related proteins in soybean. Annotations of 5mC-related proteins in A. thaliana v10 were used as queries in BLASTp searches for homologous sequences against itself and protein annotations of G. max Wm82.a1.v1.1, Populus trichocarpa v3.0, Phaseolus vulgaris v1.0, and Vitis vinifera v12 (Camacho et al., 2009). Best-hit proteins (e-value < 1E-25 and bit score > 100) were extracted and aligned using PASTA (Mirarab et al., 2015). Following alignment, phylogenies were estimated using RAxML with 1000 bootstrap replicates and the PROTGAMMAGTR model of amino acid substitution (Stamatakis, 2014). The resulting phylogenies identified 12 orthogroups: (1) AGO1/2/3/4/5/6/7/8/9/10, (2) CMT1/2/3, (3) DCL1/2/3/4 and others, (4) DML1/2/3 and others, (5) DRM1/2/3, (6) IBM1 and others, (7) MET1/2 and others, (8) NRPA/B/C/D1, (9) NRPE1, (10) RDR1/2/6 and others, (11) SUVH1/2/3/4/5/6/7/8/9/10, and (12) VIM1/2/3/4 and others. Hence, each orthogroup contains one or more monophyletic clades of 5mC-related proteins based on annotations in Arabidopsis and also contains protein sequences from soybean and other species. Homologs of 5mC-related proteins in soybean, and the other species, are found within each clade. Thus, homologous proteins in soybean were reduced to a single representative clade using the top Arabidopsis BLASTp hit.
Accession Numbers
The data generated from this study has been uploaded to the Gene Expression Omnibus database and can be retrieved through accession number GSE94299.
Supplemental Data
Supplemental Figure 1. Bar graphs of genome-wide DNA methylation changes during somatic embryogenesis.
Supplemental Figure 2. Heatmaps of genome-wide DNA methylation changes during somatic embryogenesis.
Supplemental Figure 3. Pairwise comparison of DNA methylation differences.
Supplemental Figure 4. DNA methylation and small RNA profiles of four major TE classes longer than 3 kb.
Supplemental Figure 5. Overview of methylation profiles in identified CG and CHG DMRs.
Supplemental Figure 6. Methylation states of methylated CG pairs.
Supplemental Figure 7. Methylation states of methylated CHG pairs.
Supplemental Figure 8. Profile of novel TE insertions during somatic embryogenesis.
Supplemental Table 1. Methylome sequencing summary statistics.
Supplemental Table 2. RNA-seq summary statistics.
Supplemental Table 3. Small RNA-seq summary statistics.
Supplemental Table 4. Pairwise X2 test of different genomic features.
Supplemental Table 5. Differentially expressed TEM genes in 6 wk and 6 mo globular embryos.
Supplemental Table 6. Adjusted P values of genes compared with the reference stage.
Supplemental Data Set 1. Identified UM, gbM, and TEM genes in soybean.
Supplemental Data Set 2. Unaligned and aligned 5mC-related gene protein sequences in FASTA format, and corresponding phylogenetic trees in nexus format.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jered Wendte, Brigitte Hofmeister, and William Jordan for comments and discussions. This work was supported by the United Soybean Board (award 1420-532-5645 to B.C.M and W.A.P.) and by the National Science Foundation (grant IOS-1649424 to B.C.M.; MCB-1252370 to R.K.S. and J.D.; MCB-1339194, IOS-1546867, and IOS-1856627 to R.J.S and S.A.J.; and IOS-1844427 to R.J.S.).
AUTHOR CONTRIBUTIONS
W.A.P., B.C.M., and R.J.S. conceived the project and designed the experiments. S.J., D.T., and W.A.P. performed the experiments and collected tissues. S.M.M. and B.C.M. performed deep sequencing and prepared the small RNA and RNA-seq data. A.J.B. identified 5mC-related genes in soybean. K.D.K. and S.A.J. identified TE genes and generated the TE annotation. J.D. and R.K.S. performed the analysis to identify novel TE insertions. L.J. and R.J.S. analyzed the data. L.J. wrote the article. All authors discussed the results and commented on the article.
References
- Ahloowalia B.S. (1986). Limitations to the use of somaclonal variation in crop improvement In Somaclonal Variations and Crop Improvement, Semal J., ed (Dordrecht: Springer; ), pp. 14–27. [Google Scholar]
- Becker C., Hagmann J., Müller J., Koenig D., Stegle O., Borgwardt K., Weigel D. (2011). Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249. [DOI] [PubMed] [Google Scholar]
- Bewick A.J., et al. (2016). On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA 113: 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A.J., Niederhuth C.E., Ji L., Rohr N.A., Griffin P.T., Leebens-Mack J., Schmitz R.J. (2017). The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhojwani S.S., Dantu P.K. (2013). Plant Tissue Culture: An Introductory Text. (India: Springer; ). [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Kramdi A., Kassam M., Heese M., Schnittger A., Roudier F., Colot V. (2017). DNA methylation dynamics during early plant life. Genome Biol. 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J.P., Borges F., Donoghue M.T., Van Ex F., Jullien P.E., Lopes T., Gardner R., Berger F., Feijó J.A., Becker J.D., Martienssen R.A. (2012). Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cao X., Springer N.M., Muszynski M.G., Phillips R.L., Kaeppler S., Jacobsen S.E. (2000). Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. USA 97: 4979–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., Slotkin R.K. (2016). Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163. [DOI] [PubMed] [Google Scholar]
- Daron J., Slotkin R.K. (2017). EpiTEome: Simultaneous detection of transposable element insertion sites and their DNA methylation levels. Genome Biol. 18: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Johnson L.M., Jacobsen S.E., Patel D.J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M., Kim K.D., Abernathy B., Arikit S., Maumus F., Panaud O., Meyers B.C., Jackson S.A. (2015). A new approach for annotation of transposable elements using small RNA mapping. Nucleic Acids Res. 43: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M., Kim K.D., Abernathy B., Li Y.-H., Qiu L.-J., Jackson S.A. (2018). Genic c-methylation in soybean is associated with gene paralogs relocated to transposable element-rich pericentromeres. Mol. Plant 11: 485–495. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Peacock W.J., Dennis E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93: 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q., Baulcombe D.C. (2016). DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Ream T.S., Marasco M., Nicora C.D., Norbeck A.D., Pasa-Tolic L., Pikaard C.S. (2012). In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 48: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Crisp P.A., Stelpflug S., Kaeppler S.M., Li Q., Springer N.M. (2018). Heritable epigenomic changes to the maize methylome resulting from tissue culture. Genetics 209: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel C.B., Klein T.M., Anis M., Wilde H.D., Parrott W.A. (1998). Growth characteristics and transformability of soybean embryogenic cultures. Plant Cell Rep. 17: 765–772. [DOI] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.-S., Zhu J.-K. (2009). NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 23: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoede C., Arnoux S., Moisset M., Chaumier T., Inizan O., Jamilloux V., Quesneville H. (2014). PASTEC: An automatic transposable element classification tool. PLoS One 9: e91929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Jones A.M., Searle I., Patel K., Vogler H., Hubner N.C., Baulcombe D.C. (2009). An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 16: 91–93. [DOI] [PubMed] [Google Scholar]
- Jenik P.D., Barton M.K. (2005). Surge and destroy: The role of auxin in plant embryogenesis. Development 132: 3577–3585. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., et al. ; 1001 Genomes Consortium (2016a). Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Nery J.R., Castanon R., Ecker J.R. (2017). Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Stuart T., Valdes M., Breakfield N., Schmitz R.J., Nery J.R., Urich M.A., Han X., Lister R., Benfey P.N., Ecker J.R. (2016b). Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2: 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.D., El Baidouri M., Abernathy B., Iwata-Otsubo A., Chavarro C., Gonzales M., Libault M., Grimwood J., Jackson S.A. (2015). A Comparative Epigenomic Analysis of Polyploidy-Derived Genes in Soybean and Common Bean. Plant Physiol. 168 (4): 1433–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S., Pontier D., Cavel E., Vega D., El-Shami M., Kanno T., Lagrange T. (2009). PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc. Natl. Acad. Sci. USA 106: 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Vashisht A.A., Wohlschlegel J.A., Jacobsen S.E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Le B.H., Chen M., Henry K.F., Hur J., Hsieh T.-F., Chen P.-Y., Pelletier J.M., Pellegrini M., Fischer R.L., Harada J.J., Goldberg R.B. (2017). Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. USA 114: E9730–E9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschiavo F., Pitto L., Giuliano G., Torti G., Nuti-Ronchi V., Marazziti D., Vergara R., Orselli S., Terzi M. (1989). DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones and hypomethylating drugs. Theor. Appl. Genet. 77: 325–331. [DOI] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12.
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862. [DOI] [PubMed] [Google Scholar]
- Mathioni S.M., Kakrana A., Meyers B.C. (2017). Characterization of plant small RNAs by next generation sequencing. Curr. Protoc. Plant Biol. 2: 39–63. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Merkle S.A., Parrott W.A., Flinn B.S. (1995). Morphogenic aspects of somatic embryogenesis In In Vitro Embryogenesis in Plants, Thorpe T.A., ed (Dordrecht: Springer; ), pp. 155–203. [Google Scholar]
- Mirarab S., Nguyen N., Guo S., Wang L.S., Kim J., Warnow T. (2015). PASTA: Ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 22: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales A.M., O’Rourke J.A., Van De Mortel M., Scheider K.T., Bancroft T.J., Borém A., Nelson R.T., Nettleton D., Baum T.J., Shoemaker R.C. (2013). Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct. Plant Biol. 40: 1029–1047. [DOI] [PubMed] [Google Scholar]
- Narsai R., Gouil Q., Secco D., Srivastava A., Karpievitch Y.V., Liew L.C., Lister R., Lewsey M.G., Whelan J. (2017). Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nic-Can G.I., Loyola-Vargas V.M. (2016). The role of the auxins during somatic embryogenesis In Somatic Embryogenesis: Fundamental Aspects and Applications, Loyola-Vargas V., and Ochoa-Alejo N., eds (Cham: Springer; ), pp. 171–182. [Google Scholar]
- Niederhuth C.E., et al. (2016). Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M., et al. (2015). Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott W.A., Dryden G., Vogt S., Hildebrand D.F., Collins G.B., Williams E.G. (1988). Optimization of somatic embryogenesis and embryo germination in soybean. In Vitro Cell. Dev. Biol. 24: 817–820. [Google Scholar]
- Robert H.S., Grunewald W., Sauer M., Cannoot B., Soriano M., Swarup R., Weijers D., Bennett M., Boutilier K., Friml J. (2015). Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142: 702–711. [DOI] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J., Dellaporta S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657. [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., He Y., Valdés-López O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G., Ecker J.R. (2013). Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Schultz M.D., Lewsey M.G., O’Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. (2011). Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schultz M.D., et al. (2015). Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M.D., Schmitz R.J., Ecker J.R. (2012). ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28: 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., Schultz M.D., Chiarenza S., Nussaume L., Ecker J.R., Whelan J., Lister R. (2015). Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: 9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirvin R.M., McPheeters K.D., Norton M. (1994). Sources and frequency of somaclonal variation. HortScience 29: 1232–1237. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug S.C., Eichten S.R., Hermanson P.J., Springer N.M., Kaeppler S.M. (2014). Consistent and heritable alterations of DNA methylation are induced by tissue culture in maize. Genetics 198: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Ding B., Simon S.A., Feng S., Bellizzi M., Pellegrini M., Wang G.L., Meyers B.C., Jacobsen S.E. (2013a). Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013b). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S., Ran J.H., Gaut B.S. (2016). Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants 2: 15222. [DOI] [PubMed] [Google Scholar]
- Teixeira F.K., et al. (2009). A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604. [DOI] [PubMed] [Google Scholar]
- Thorpe T.A. (1990). The current status of plant tissue culture In Plant Tissue Culture: Applications and Limitations, Bhojwani S.S., ed (Amsterdam: Elsevier; ), pp. 1–33. [Google Scholar]
- Tran R.K., Henikoff J.G., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S. (2005). DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15: 154–159. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick H.N., Dinkins R.D., Santarem E.R., Di R., Samoylov V., Meurer C.A., Walker D.R., Parrott W.A., Finer J.J., Collins G.B. (1997). Recent advances in soybean transformation. Plant Tissue Cult. Biotechnol. 3: 9–26. [Google Scholar]
- Urich M.A., Nery J.R., Lister R., Schmitz R.J., Ecker J.R. (2015). MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protoc. 10: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining K., Pomraning K.R., Wilhelm L.J., Ma C., Pellegrini M., Di Y., Mockler T.C., Freitag M., Strauss S.H. (2013). Methylome reorganization during in vitro dedifferentiation and regeneration of Populus trichocarpa. BMC Plant Biol. 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J., Gao H., Zhang J., Aldridge B., Vickers M., Higgins J.D., Feng X. (2018). Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 50: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A., et al. (2018). Partial maintenance of organ-specific epigenetic marks during plant asexual reproduction leads to heritable phenotypic variation. Proc. Natl. Acad. Sci. USA 115: E9145–E9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W.-L., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69. [DOI] [PubMed] [Google Scholar]