Abstract
Background
It is important to explore the interaction between antibacterial nanoparticles and microbes for understanding bactericidal activity and developing novel applications. It is possible that the nanoparticulate size can govern the antibacterial potency.
Purpose
The purpose of this study was to evaluate the antimicrobial and antibiofilm properties of cetylpyridinium chloride (CPC)–decorated nanoemulsions against methicillin-resistant Staphylococcus aureus (MRSA).
Methods
The droplet size could be adjusted by varying the percentage of squalene, the main ingredient of the oily core.
Results
We fabricated cationic nanoemulsions of three different sizes, 55, 165, and 245 nm. The nanoemulsions showed greater storage stability than the self-assembled CPC micelles. The tested nanoemulsions exhibited more antimicrobial activity against Gram-positive bacteria than Gram-negative bacteria and fungi. The killing of MRSA was mainly induced by direct cell-membrane damage. This rupture led to the leakage of cytoplasmic DNA and proteins. The nanoemulsions might also degrade the DNA helix and disturb protein synthesis. The proteomic analysis indicated the significant downregulation of DNA-directed RNA polymerase (RNAP) subunits β and β’. The antibacterial effect of nanoemulsions increased with decreasing droplet size in the biofilm MRSA but not planktonic MRSA. The small-sized nanoemulsions had potent antibiofilm activity that showed a colony-forming unit (CFU) reduction of 10-fold compared with the control. The loss of total DNA concentration also negatively correlated with the nanoemulsion size.
Conclusion
The present report established a foundation for the development of squalene@CPC nanosystems against drug-resistant S. aureus.
Keywords: cetylpyridinium chloride, nanoemulsion, MRSA, biofilm, size effect
Introduction
Pathogenic infection is the second-highest cause of death worldwide, with 16 million victims each year.1 The growing resistance to antibiotics exacerbates the health problem of microbial infection. The emergence of methicillin-resistant Staphylococcus aureus (MRSA) is a huge threat to health with everything from superficial skin wounds to toxic shock syndrome.2 The development of novel approaches to solve this problem is urgent. Nanotechnology demonstrates a promising strategy for combating the antibacterial resistance. The large surface area to mass ratio, high reactivity, and surface functionalization of the nanostructures have led to the unique properties for efficient eradication of pathogens.3 Nanoemulsions are isotropic and thermodynamically stable nanosystems consisting of oil, water, and emulsifiers. They provide significant potential as functional additives in cosmetics, foods, and pharmaceutical products. Antimicrobial nanoemulsions show innate antibacterial activity against a broad species of pathogens.4,5 The surfactants in nanoemulsions are responsible for the strong bacteria-killing ability. These features make antimicrobial nanoemulsions feasible for application in wound treatment and surface decontamination.
The quaternary ammonium surfactants are often incorporated into nanoemulsions to form cationic nanosystems for antibacterial use. The cationic and amphiphilic characteristics of quaternary ammonium compounds can destabilize the bacterial membrane to exhibit the antimicrobial activity. Among these, cetylpyridinium chloride (CPC) as a cationic surfactant is approved by the USFDA as GRAS for extensive application in the food industry and hospitals as a disinfectant.6 It is found in many commercial products, including mouth rinses, wound dressings, nasal sprays, and cosmetics.7,8 CPC can form micelles in water via self-assembly in the amount of a higher than critical micellar concentration (CMC). However, the micelles usually reveal an unstable condition, resulting in the difficulty of quality control and application. Nanoemulsions can be ideal candidates as the vehicles of CPC. The size and composition of the nanoparticles can greatly influence the biological effects. The impact of the particulate size on the antimicrobial activity of metallic nanoparticles such as Ag, Pd, and ZnO has been widely studied.9–11 Nevertheless, no literature has been found pertaining to investigating the size effect of lipid nanosystems on the antibacterial effect. We intended to explore how the droplet size of cationic nanoemulsions affects the bacterial killing. Squalene was employed as the oil phase to produce CPC-coated nanoemulsions in the presence of blended emulsifiers (squalene@CPC nanoemulsions).
We fabricated three sizes of nanoemulsions: 55, 165, and 245 nm. MRSA was chosen as the main model microorganism in this study due to its prevalence and virulence in clinical infection. MRSA can exist in both planktonic and biofilm phenotypes, with the latter being predominant in the medical environment.12 A biofilm-forming capacity contributes to the unfavorable eradication of MRSA infection. Both phenotypes were used in this work to examine the anti-MRSA effect of the nanoemulsions. The proteomics of MRSA were established to understand the possible antibacterial mechanisms of squalene@CPC nanoemulsions. The size change of nanoemulsions under an accelerated stability test was evaluated to compare with self-assembled CPC micelles. It is possible to adjust the formulation and size of nanoemulsions in order to achieve optimized nanosystems based on the antibacterial profiles in the present study.
Materials And Methods
Preparation Of squalene@CPC Nanoemulsions
The oil and aqueous phases of nanoemulsions were fabricated separately. The oil phase consisted of soy phospatidylcholine (SPC, 150 mg) and squalene. The squalene amount was 200, 500, and 700 mg for preparation of cationic nanoemulsions of small (CN-S), medium (CN-M), and large (CN-L) size, respectively. The aqueous phase consisted of CPC (80 mg), Poloxamer 188 (150 mg), and water for all formulations. Both phases were heated in a water bath for 20 min. The aqueous phase was added into the oil phase in the presence of high-shear homogenization at 12,000 rpm for 20 min, followed by the agitation using a probe-type sonicator (VCX600, Sonics and Materials) for 20 min at 35 W. The total volume of the final product was 10 mL.
Size And Surface Charge Of Nanoemulsions
The mean diameter and zeta potential of the nanoemulsion droplets were recorded by a laser-scattering technique (Zetasizer ZS90, Malvern). The recordation was done on the nanodispersion concentration after a 100-fold dilution with water. The storage stability was carried out by size measurement under the condition at 50°C and 75% relative humidity. The duration for the accelerated stability test was 14 d.
Molecular Environment
The molecular environment of the nanoformulations was assessed by fluorescence spectrophotometry based on the solvatochromism of Nile red.13 The CPC formulations were loaded with Nile red (1 ppm). The emission spectra of dye-incorporated systems were scanned from 550 to 700 nm. The excitation wavelength was 546 nm.
Bacterial Strains And Culture Condition
MRSA (ATCC 33591), S. aureus (ATCC 6538), Escherichia coli (ATCC 8739), and Candida albicans (ATCC 10231) were purchased from American Type Culture Collection. The drug-resistant clinical isolates of MRSA (KM-1) and vancomycin-intermediate S. aureus (VISA, KV-2) were gifts provided by Dr. Po-Liang Lu (Kaohsiung Medical University Hospital). The strains were grown in tryptic soy broth (TSB) medium at 37°C and 150 rpm.
Minimum Inhibitory Concentration (MIC) And Minimum Bactericidal Concentration (MBC)
A broth twofold serial dilution method was utilized to measure MIC.14,15 Overnight culture of bacteria was diluted in TSB to achieve OD600 of 0.01 (about 2x106 CFU/mL). The bacterial population was exposed to several dilutions of CPC ranging from 0.03 to 62.5 μg/mL with TSB and incubated at 37°C for 20 h. The ELISA reader at 595 nm was used to detect the absorbance. The reading value of <0.09 was the level of MIC. For the MBC assay, the bacterial suspension was diluted in PBS and plated on TSB plates. The TSB plates were incubated at 37°C for 20 h. Subsequently the CFU was counted. MBC was defined as the lowest concentration that killed ≥99.9% of the bacteria.
Agar Diffusion Assay
This study was performed according to the previous investigation.16 This assay was performed by inoculating the bacteria (OD600=0.7) in the 0.75% TSB agar. The bacteria-agar mixture (5 mL) was poured into the dish for 15 min; then CPC (8 mg/mL) in micelles or nanoemulsions with a volume of 10 μL were pipetted onto the agar. The diameter of the inhibition zone was estimated after a 16-h incubation.
MRSA Morphology Visualized By Scanning Electron Microscopy (SEM)
MRSA at OD600=0.1 was treated with CPC (50 μg/mL) in micelles or nanoemulsions at 37°C for 24 h. The microbes were fixed with 3% glutaldehyde and 2% paraformaldehyde in cacodylate buffer several times. After dehydration in an ascending series of ethanol, the samples were coated with gold and observed under Hitachi SU8220 SEM.
Live/Dead MRSA Visualized By Fluorescence Microscopy
The viability and death of MRSA after treatment of CPC (10 and 100 μg/mL) for 4 h were observed using a Live/Dead BacLight® kit (Molecular Probes). The pellet of bacteria was obtained by centrifugation at 12,000 rpm before CPC treatment. The treated bacteria were stained with the kit for 15 min. The samples were monitored by Leica DMi8 fluorescence microscopy. The detailed procedure of this experiment is described by Manteca et al.17
MRSA Biofilm Determination
The biofilm was grown in a Cellview® dish by incubating MRSA (OD600=0.1) in TSB containing 1% glucose at 37°C for 24 h. The biofilm was treated by CPC (50 μg/mL) for 24 h. The culture medium was transferred to the microtubes. The biofilm was rinsed twice with PBS to remove the loosely adherent planktonic MRSA and suspended in PBS (200 μL). The recovered bacteria outside the biofilm and the bacteria inside the biofilm were serially diluted and plated in an agar plate for 24 h to count CFU. The biofilm was also stained by SYTO9 for 15 min. The biofilm was then gently rinsed with PBS. The three-dimensional structure and thickness of the biofilm were detected by Leica TSC SP2 confocal microscopy as described previously.18
Genomic DNA Analysis Of MRSA
MRSA at OD600=3 was treated by CPC (100 μg/mL) for 3 h. The genomic DNA was extracted using a Presto® Mini Bacteria kit based on the manufacturer’s instructions. The aliquot of purified genomic DNA (100 ng) was analyzed by electrophoresis on an 0.8% agarose gel.
Total Protein Analysis Of MRSA
MRSA was grown in TSB to OD600=6 and then treated with CPC (100 μg/mL) at 37°C for 3 h. The centrifuged pellet was resuspended with water. Following sonication for 20 min, the bacteria were centrifuged at 4°C and 10,000 rpm for 15 min. The total protein was quantified by a Bio-Rad protein assay kit in ELISA (595 nm).
Proteomic Profiles Of MRSA
The treatment protocol of CPC was the same as the total protein analysis. The SDS-PAGE was carried out with a 5% stacking gel and a 10% separating gel followed by Coomassie blue staining. The bands were withdrawn and digested with trypsin overnight. The digested proteins were acidified with 0.5% trichloroacetic acid. The MALDI-TOF/TOF was detected by Bruker Ultraflex spectrometer. The detailed procedure was shown in the previous work.19 The fold change of the protein expression after CPC treatment was estimated by band quantification based on Prodigy Samespots analysis software. The protein masses were assigned and used for a database search with MASCOT search engine (www.matrixscience.com).
Statistical Analysis
The data presented as mean and standard deviation (S.D.). The difference in the data of the different experimental groups was analyzed using the Kruskal-Wallis test. The post hoc test for checking individual differences was Dunn’s test. The significance was demonstrated as * for p<0.05, ** for p<0.01, and *** for p<0.001 in the figures.
Results
Characterization Of Micelles And Nanoemulsions
The three nanoemulsions prepared by homogenization and sonication were depicted based on droplet size and zeta potential. CPC alone could self-assemble into micelles without the fabrication process. Table 1 compares the size, polydispersity index (PDI), and surface charge between micelles and squalene@CPC nanoemulsions. Self-assembly of CPC formed 1435 nm aggregates. The directed assembly of nanoemulsions showed much smaller droplets as compared to micelles. The droplet size increased from 55 nm (CN-S) to 165 (CN-M) and 245 nm (CN-L) with the increase of squalene from 2% to 7%. The micelles revealed a broad size distribution (PDI=1). The PDI of CN-S, CN-M, and CN-L was 0.40, 0.19, and 0.26, respectively. The surface of all formulations had cationic charges with zeta potential ranging from 9.3 to 51.1 mV, mainly due to the cationic feature of CPC. The positive charge of nanoemulsions was greater than that of micelles. CN-L showed a zeta potential slightly higher than that of the nanoemulsions with smaller droplets.
Table 1.
The Characterization Of Micelles And Cationic Nanoemulsions By Droplet Size, Polydispersity Index (PDI), And Zeta Potential
| Formulation | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| Micelles | 1434.67±102.47 | 1.0±0 | 9.27±2.23 |
| CN-S | 54.54±1.93 | 0.40±0.12 | 46.53±1.86 |
| CN-M | 164.73±1.44 | 0.19±0.05 | 46.37±2.35 |
| CN-L | 244.70±2.36 | 0.26±0.01 | 51.10±0.56 |
Note: Each value represents the mean±SD (n=3).
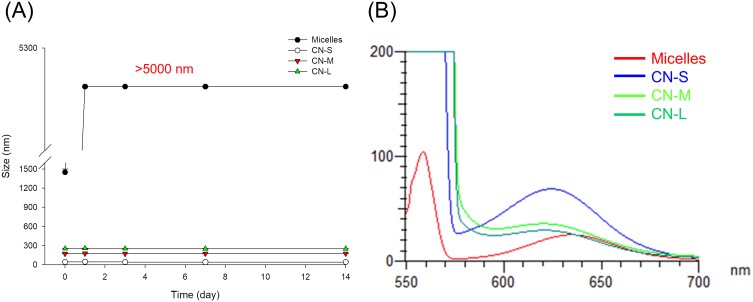
Figure 1A demonstrates the change in size as a function of storage time at 50°C. The micellar size increased very quickly during the first day of storage. The average diameter of micelles at Day-1 was >5,000 nm, which was the maximum detection limit of Zetasizer. All nanoemulsions retained their droplet size after 14 d. Figure 1B shows the emission spectrum of Nile red in micelles and nanoemulsions. Nile red fluorescence is quenched in the molecular environment with high hydrophilicity. The micelles exhibited the weakest fluorescence, implying the least lipophilicity. The smaller-sized nanoemulsions revealed a higher lipophilicity compared to the larger ones.
Figure 1.
The physicochemical characteristics of micelles and squalene@CPC nanoemulsions: (A) the size change of micelles and nanoemulsions at 50 °C and 75% relative humidity for 14 days; and (B) fluorescence emission of Nile red incorporation in micelles and nanoemulsions for determining molecular environment polarity. The stability data are presented as the mean of four experiments±S.D.
Antibacterial Effect Of Nanoemulsions Against Planktonic MRSA
To evaluate whether the response to micelles and nanoemulsions varies among different species of pathogens, we determined MIC and MBC against Gram-positive bacteria (MRSA, S. aureus, KM-1, and KV-2), Gram-negative bacteria (E. coli), and fungi (C. albicans). Table 2 presents the MIC of the CPC formulations. MIC is defined as the lowest CPC concentration that contributes to complete inhibition of visible growth. The results demonstrated that all formulations studied showed inhibitory activity for all pathogens investigated. Although a significant difference had been achieved in the droplet size by varying squalene content, the inhibitory effect was approximate. No inhibition was detected with the treatment of bulk materials (squalene, poloxamer 188, and SPC). This suggested that the inhibitory activity was mainly due to CPC in the formulations. The organisms of Gram-positive bacteria were found to be more susceptible to CPC than Gram-negative bacteria and fungi. The MIC assay indicated a superior inhibition toward Gram-positive bacteria compared to Gram-negative bacteria by about 8-fold. The CPC inhibition against MRSA was weaker than that against drug-sensitive S. aureus. The susceptibility of clinically isolated MRSA and VISA to CPC was analyzed. VISA is a strain highly prevalent in the epidemic MRSA species. CPC could significantly suppress the growth of both clinical isolates. MBC expressed the minimum bactericidal amount to kill >99.9% bacteria. As shown in Table 2, MBC values are slightly higher or comparable to the degrees of MIC. The same as MIC, Gram-positive bacteria were more sensitive to CPC, followed by E. coli and C. albicans. CPC at 1.95 μg/mL completely killed MRSA.
Table 2.
The Minimum Inhibitory Concentration (MIC, µg/mL) And Minimum Bactericidal Concentration (MBC, µg/mL) Of Cetylpyridinium Chloride (CPC) In Micelles And Cationic Nanoemulsions Against Different Strains Of Bacteria
| Strain | Micelles | CN-S | CN-M | CN-L | |
|---|---|---|---|---|---|
| MIC | MRSA | 0.98 | 0.98 | 0.98 | 0.98 |
| S. aureus | 0.49~0.98 | 0.49 | 0.49 | 0.49 | |
| KM-1 | 0.49~0.98 | 0.49 | 0.49 | 0.49 | |
| KV-2 | 0.49~0.98 | 0.49 | 0.49 | 0.49 | |
| E. coli | 3.91 | 3.91~7.81 | 3.91~7.81 | 7.81 | |
| C. albicans | 1.95~3.91 | 3.91 | 1.95~3.91 | 1.95~3.91 | |
| MBC | MRSA | 1.95 | 1.95 | 1.95 | 0.98~1.95 |
| S. aureus | 0.49~0.98 | 0.98~1.95 | 0.98 | 0.49~0.98 | |
| KM-1 | 0.98 | 0.98 | 0.98 | 0.98 | |
| KV-2 | 0.98~1.95 | 1.95 | 0.98~1.95 | 0.98 | |
| E. coli | 3.91 | 7.81 | 7.81 | 7.81~15.63 | |
| C. albicans | 3.91~7.81 | 7.81~15.63 | 7.81~15.63 | 7.81~15.63 |
Note: KM-1 and KV-2 are the clinical isolates of MRSA and VISA, respectively. Each value represents the mean±S.D. (n=3).
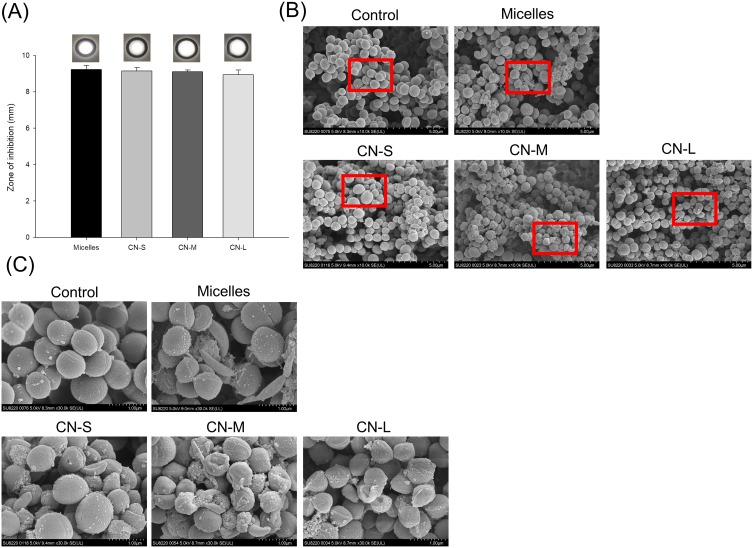
The MRSA inhibition by CPC formulations was recognized by the agar diffusion study as shown in Figure 2A. CPC displayed a clear inhibition zone on the agar media against planktonic MRSA. We observed no difference in the effect of any of the droplet sizes on MRSA growth. Application of CPC provided the inhibition zone of about 9 mm for all formulations tested. SEM allowed us to directly visualize MRSA integrity and morphology after CPC treatment. As shown in Figure 2B, the untreated MRSA reveals a smooth and bright surface without debris. The CPC treatment disrupted the intact morphology, leading to the shrinkage of some of the MRSA membrane. The enlarged image showed surface blebbing and roughening after CPC treatment (Figure 2C). The formation of vacuoles and debris was detected. The bacterial membrane damage resulted in the cytoplasmic material leakage. The different droplet sizes did not significantly affect the level of interference.
Figure 2.
Determination of the antibacterial activity of micelles and squalene@CPC nanoemulsions against planktonic MRSA: (A) zone of inhibition measured from agar diffusion assay; (B) morphological changes of MRSA viewed under SEM; and (C) the enlarged images of B. The agar diffusion data are presented as the mean of three experiments±S.D.
The live/dead staining was conducted to further assess the planktonic MRSA killing by micelles and nanoemulsions. Live MRSA with intact membrane would be stained green by SYTO9-DNA interaction. PI is a molecule that diffuses to the bacteria with compromised membrane. The PI uptake is an implication of cellular injury and death. Figure 3A and B represent the live/dead images of MRSA at a CPC dose of 10 and 100 μg/mL, respectively. The PI signal was increased by nanoemulsion treatment as compared to the untreated control. SYTO9 covered the same area with PI in the nanoemulsion-treated groups. This was because SYTO9 stained all cells while PI stained only dead cells. PI would cover the entire visual field of SYTO9. An accumulation of red signal was observed in the nanoemulsion-treated MRSA. The accumulation was more obvious following the increase of droplet size. This could be due to the massive loss of cell integrity causing DNA release from the bacteria. The red signal aggregation was not visualized in MRSA treated with micelles. The SYTO9 staining was also absent in the group of micelles. This could be because micelles prompted complete MRSA damage, leaving the entire DNA disruption. The detailed explanation is explored in the Discussion section.
Figure 3.
The live/dead imaging by treatment of micelles and squalene@CPC nanoemulsions against planktonic MRSA: (A) treatment with CPC at 10 μg/mL; and (B) treatment with CPC at 100 μg/mL.
Antibacterial Effect Of Nanoemulsions Against Biofilm MRSA
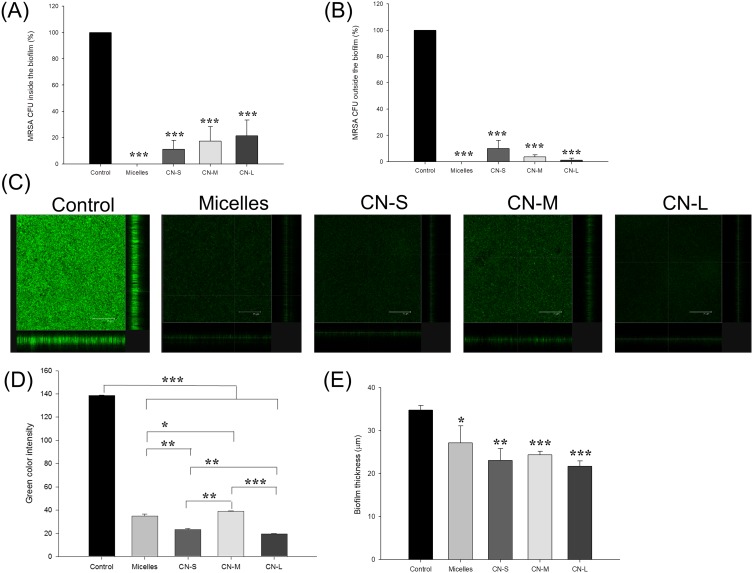
Figure 4 presents the antibiofilm impact of micelles and nanoemulsions against one-day-old MRSA biofilm cells. Figure 4A shows the corresponding percentage of MRSA inhibition inside biofilm measured as CFU. Similar to the strong killing on planktonic cells, CPC could eliminate MRSA in biofilm. The biofilm formation was inhibited up to 89%, 82%, and 78% after application of CN-S, CN-M, and CN-L, respectively. This suggested that the size reduction led to an improvement in the nanoemulsion penetration into the biofilm matrix. The antimicrobial activity towards planktonic MRSA outside the biofilm was also examined as depicted in Figure 4B. A complete planktonic cell killing was observed by micelle treatment. Contrary to the result of MRSA inside the biofilm, treatment by nanoemulsions with smaller droplets showed less CFU reduction compared to the larger ones. CN-M and CN-L consistently produced a lower CFU percentage against MRSA outside the biofilm than that against bacteria inside the biofilm. The antibiofilm effect was seen by SYTO9 staining as illustrated in Figure 4C. The viable MRSA in the intact biofilm (control) displayed a dense and thick architecture. CPC was vital to diminishing the viability and thickness of biofilm MRSA. The SYTO9 intensity and biomass thickness was calculated as shown in Figure 4D and E, respectively. There was a 3.5-fold decrease of green fluorescence by micelle treatment. CN-S and CN-L further reduced the green signal as compared to micelles. The mid-sized nanoemulsions are less toxic to biofilm MRSA based on green intensity. The treatment of CPC reduced the average thickness of the biofilm. The biofilm treated with micelles, CN-S, CN-M, and CN-L was 27.2, 23.1, 24.4, and 21.7 μm, respectively. The trend of thickness inhibition was similar to that of SYTO9 intensity.
Figure 4.
Determination of the antibacterial activity of micelles and squalene@CPC nanoemulsions against biofilm MRSA: (A) MRSA CFU inside the biofilm; (B) MRSA CFU outside the biofilm; (C) the three-dimensional images of biofilm visualized by confocal microscopy; (D) the corresponding biofilm green intensity; and (E) the corresponding biofilm thickness. All data are presented as the mean of three experiments±S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
The Genomic And Proteomic Profiles Of MRSA Treated By Nanoemulsions
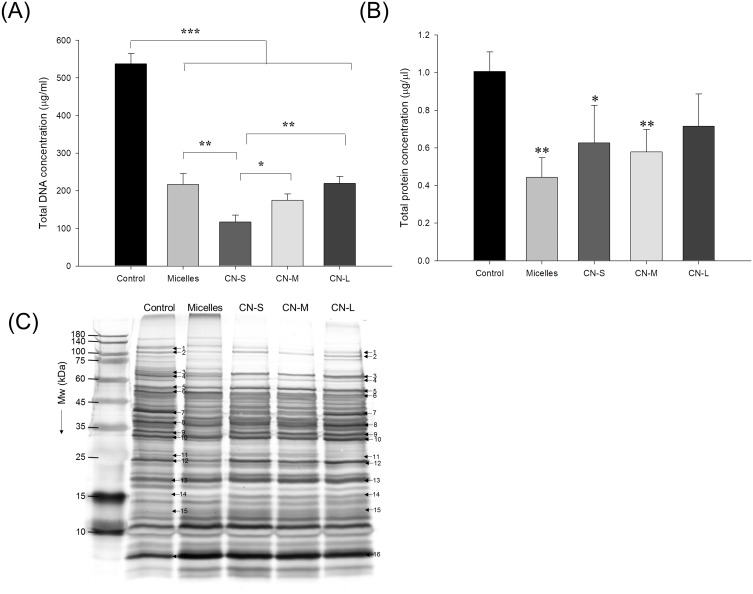
The genomic DNA detection was conducted to evaluate the anti-MRSA activity of CPC as shown in Figure 5A. The micelles exerted a significant 2.5-fold reduction of DNA concentration compared to the untreated control. The DNA elimination by nanoemulsions increased with lowering the droplet size. The application of CN-S caused a 5-fold decrease in the DNA concentration compared to the control. Figure 5B expresses that micelles and nanoemulsions decrease the cellular protein content. This reduction could be due to the bacterial membrane injury and/or the inhibition of protein synthesis after DNA denaturalization. The protein reduction by the nanoemulsions was not size-dependent. The changes in the MRSA proteins by CPC were further examined by SDS-PAGE. The control MRSA exhibited apparently clear and intense protein bands as shown in Figure 5C. The protein bands of >75 kDa had faded after treatment with CPC formulations. No significant change of protein bands of <75 kDa was observed before and after CPC treatment. The qualitative and quantitative analysis of the proteins is conducted by mass as summarized in Table 3. The protein change of >±2-fold can be regarded as significant. DNA-directed RNA polymerase (RNAP) subunits β and β’ (band No. 1 and 2) were significantly downregulated by CPC. The other proteins (band No. 3~16) generally showed negligible change after CPC application.
Figure 5.
The genomic and proteomic profiles of MRSA treated by micelles and squalene@CPC nanoemulsions: (A) analysis of the quality of MRSA genomic DNA by agarose gel electrophoresis; (B) total protein concentration in MRSA; and (C) the protein change of MRSA analyzed by SDS-PAGE and MALDI-TOF/TOF mass. All data are presented as the mean of three experiments±S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
Table 3.
Differentially Expressed Proteins In MRSA Following The Treatments Of Micelles And Cationic Nanoemulsions
| Band No. | Protein | Accession No. | Mw (Da) | Matched Peptides | Coverage % (SCORE) |
Ratios To MRSAa,b | Biological Function | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Micelles | CN-S | CN-M | CM-L | |||||||
| 1 | DNA-directed RNA polymerase subunit β | P60279 | 133,409 | 19 | 16% (100) | −2.33 | −3.23 | −8.34 | −3.45 | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. |
| 2 | DNA-directed RNA polymerase subunit β’ | P60286 | 134,327 | 24 | 24% (115) | −2.78 | −0.71 | −3.57 | −0.64 | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. |
| 3 | Transketolase | Q6GH64 | 72,235 | 15 | 28% (136) | 0.64 | 2.07 | 1.16 | 2.44 | Catalyzes the transfer of a two-carbon ketol group from a ketose donor to an aldose acceptor, via a covalent intermediate with the cofactor thiamine pyrophosphate. |
| 4 | Phosphoenolpyruvate carboxykinase (ATP) | Q8NVZ8 | 59,599 | 26 | 56% (168) | −0.59 | −0.21 | −0.96 | −0.54 | Involved in the gluconeogenesis. Catalyzes the conversion of oxaloacetate (OAA) to phosphoenolpyruvate (PEP) through direct phosphoryl transfer between the nucleoside triphosphate and OAA. |
| 5 | Catalase | Q8NWV5 | 51,221 | 14 | 47% (141) | −0.28 | 0.95 | 0.97 | 2.20 | Decomposes hydrogen peroxide into water and oxygen; serves to protect cells from the toxic effects of hydrogen peroxide. |
| 6 | Enolase | P64079 | 46,036 | 14 | 47% (90) | 0.91 | −1.59 | 1.82 | 1.18 | Catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. It is essential for the degradation of carbohydrates via glycolysis. |
| 7 | Pyruvate dehydrogenase E1 component subunit alpha | P60090 | 36,459 | 7 | 34% (84) | −3.03 | −2.50 | −3.57 | −1.94 | The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2. |
| 8 | L-lactate dehydrogenase | P65257 | 34,650 | 15 | 58% (96) | −2.33 | −1.87 | −2.22 | −2.21 | Catalyzes the conversion of lactate to pyruvate. Appears to be the primary factor that allows S.aureus growth during nitrosative stress in both aerobically and anaerobically cultured cells. |
| 9 | Cysteine synthase | P63872 | 33,013 | 9 | 45% (84) | −2.86 | −0.27 | −0.31 | −1.85 | O-acetyl-L-serine + hydrogen sulfide = L-cysteine + acetate. |
| 10 | Pyridoxal 5ʹ-phosphate synthase subunit PdxS | P60799 | 32,114 | 8 | 42% (95) | −0.07 | 1.13 | 0.99 | 2.34 | Catalyzes the formation of pyridoxal 5ʹ-phosphate from ribose 5-phosphate (RBP), glyceraldehyde 3-phosphate (G3P) and ammonia. |
| 11 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | P65709 | 26,697 | 24 | 86% (204) | 1.08 | 1.19 | −0.12 | −0.21 | Catalyzes the interconversion of 2-phosphoglycerate and 3-phosphoglycerate. |
| 12 | DUF1642 domain-containing protein | PF07852.6 | 22,691 | 9 | 40% (82) | −1.79 | 1.22 | −0.95 | 1.25 | Protein of unknown function (DUF1642) |
| 13 | Superoxide dismutase [Mn/Fe] 2 | P66832 | 23,098 | 10 | 72% (92) | 1.06 | 1.09 | 1.99 | 1.82 | Destroys superoxide anion radicals which are normally produced within the cells and which are toxic to biological systems. |
| 14 | Ferritin, partial | Q7A0H5 | 16,627 | 7 | 52% (112) | 1.04 | 1.78 | 1.55 | 2.17 | Iron-storage protein. |
| 15 | DNA protection during starvation protein 2 | Q8RPQ2 | 16,749 | 9 | 76% (88) | 1.23 | 1.16 | 0.93 | 1.58 | Protects DNA from oxidative damage by sequestering intracellular Fe2+ ion and storing it in the form of Fe3+ oxyhydroxide mineral. |
| 16 | DNA-binding protein HU | Q7A0U9 | 9,606 | 7 | 72% (96) | 1.14 | 1.25 | 1.28 | 1.61 | Histone-like DNA-binding protein which is capable of wrapping DNA to stabilize it, and thus to prevent its denaturation under extreme environmental conditions. |
Notes: aRatios to MRSA indicated the fold changes in protein volume among micelles, CN-S, CN-M, and CN-L versus MRSA samples, respectively. The ratios > 1.0 mean the proteins whose expression levels were increased upon treatments of compounds, while ratios < −1.0 indicate the proteins were downregulated under the exposure to compounds. bAnalyzing the gel images using GeneTools software.
Discussion
Most of the investigations involved in the antimicrobial nanoemulsions have used susceptible bacteria as the model pathogens.20 Further study of the capability of nanoemulsions to eradicate drug-resistant microorganisms is required. In the present study, we employed CPC to form cationic nanoemulsions, and we attempted to optimize the formulations for achieving anti-MRSA activity. Our ability to control the droplet size gave the benefit of showing how specific sizes influence bacteria killing. We demonstrate here that squalene@CPC nanosystems could significantly inhibit the growth of MRSA and VISA. Most of the evaluation platforms showed no significant effect of size on anti-MRSA activity. In the experiment of biofilm suppression, the smaller-sized nanostructures more facilely restrained MRSA viability than did the bigger ones. A similar trend (CN-S>CN-M>CN-L) was observed when detecting the total genomic DNA loss in MRSA. The nanoemulsions potentially represented the capacity to treat MRSA and those that developed an impermeable biofilm.
The surface charge of cationic nanomaterials depends upon the number of cationic surfactants on the particulate surface and the associated degree of the counterions. Poloxamer 188 and SPC as the emulsifiers in lipid nanoparticles offer negative charge to the surface.21 The high positive charge in squalene@CPC nanoemulsions reported here verified the CPC association with the droplet shell. Besides the aggregates of CMC to create micelles, some monomers existed in the aqueous dispersion. This is the reason for the low zeta potential obtained from the micelles. The size of the micelles was much larger than that of the nanoemulsions. The micelles with CPC alone might produce electrostatic repulsion, resulting in the loose structure with large size. The blend of CPC with anionic emulsifiers caused electrostatic attraction, leading to an enhancement of particulate cohesion and condensation. Previous study22 also suggests the formation of a close surface by the introduction of phospholipids into the cationic nanostructures. The nanoemulsion size was directly proportional to the squalene percentage. Compared to micelles, squalene@CPC nanoemulsions were more stable. Zeta potential serves as an important fact governing the stability as it provides electrostatic repulsion among the charged droplets.23 The nanoemulsions processed greater zeta potential, signifying a high-energy barrier to maintain the storage stability. The metastable solid form of CPC in the micellar dispersion suggested that the stability of micelles is easily disturbed by the environmental change.24
Our results suggested a stronger growth inhibition of CPC against Gram-positive than Gram-negative microbes. Gram-negative bacteria possess an outer membrane in the cell wall, forming an extra barrier against molecular penetration. On the other hand, there are plenty of pores in the cell wall of S. aureus comprising teichoic acid and peptidoglycan.25 The greater barrier function of Gram-negative bacteria than Gram-positive bacteria had led to the poor delivery of the nanoemulsions into Gram-negative microbes. Our findings highlighted the potential of nanoemulsions for treatment of S. aureus that are resistant to methicillin and vancomycin. VISA is S. aureus that can transform the cell wall and become less susceptible to vancomycin in the presence of some pressures.26 This is becoming a public health problem. The antibacterial mechanism of cationic surfactants is predominantly based on the membrane damage. Cationic surfactants facilely attach to the MRSA surface with negative charge via electrostatic affinity.27 In addition, the long alkyl chain of CPC can partition into the lipophilic region of the membrane. The increased total surface area of nanoemulsions improves the interaction with the site of action.28 The evidence of MRSA membrane impairment was supported by SEM and live/dead imaging. The distortion involved the pore formation and the subsequent loss of cytoplasmic materials. Besides the leakage of molecules and ions, it is possible that DNA, proteins, and lipids in MRSA would be denatured by nanoemulsion treatment.
The aggregates of DNA were found after treatment of squalene@CPC nanoemulsions in the live/dead visualization, especially the bigger droplets. In addition to the DNA leakage from the damaged MRSA, cationic nanoemulsions acted as a crosslinker of DNA to develop the aggregates. CPC provided positive surface charge and long alkyl chains, which were capable of winding with negatively charged DNA. It has been demonstrated that cationic lipid nanoparticles can efficiently adsorb DNA in the biological medium.21 The higher positive charge of CN-L might increase this effect to form the larger aggregates. The limited zeta potential and loose structure of micelles contributed to the absence of DNA aggregates. The bactericidal behavior of nanoparticles is suggested with respect to particulate size, surface charge, and lipophilicity. As the size decreases, the surface of bacteria covered by the nanoparticles increases. The stronger nanoparticle-bacteria interaction is observed for the smaller size. This can be the pivotal factor in promoting antibacterial activity. This phenomenon can be found in the nanoparticles made of polymers and metals.29–31 This was not found in our case since no significant size effect was detected in the assays of MIC/MBC, agar diffusion, SEM, and live/dead imaging. This may suggest that the droplet diameter of cationic nanoemulsions was not the major cause of regulating the battle against planktonic MRSA.
Biofilm is a sessile structure with clusters of bacteria enclosed in extracellular polymeric substance (EPS). Biofilm formation is an essential factor contributing to the infection of S. aureus.32 Because of the rigid architecture, most of the antibiotics cannot freely diffuse into the biofilm. Biofilm also prefers gene transfer between bacteria, spreading the antibiotic resistance for virulence.33 Although CPC has been well documented to reveal antibacterial activity against planktonic culture, the investigation of biofilm is limited.34 Our results demonstrated an efficient prevention of biofilm development by squalene@CPC nanoemulsions. The nanoparticles are highly beneficial in disassembling biofilm due to the proficient penetration into the porous matrix and close contact with the biofilm surface offering a high concentration of antibacterial agents.35 The lipophilic nature of nanoemulsions can produce interaction with EPS, leading to the disruption and disengagement of the lipid layer.36 Biofilm is composed of polysaccharides, proteins, and DNA. The cationic CPC can be a chelator to interact with extracellular DNA. The CFU determination inside the biofilm showed that the matrix was not a major barrier for the entry of nanoemulsions, especially the smaller droplets. The ability of nanoparticles to penetrate into the biofilm can be affected by their size. The smaller droplets easily penetrated the biofilm mesh. The anti-MRSA effect was enhanced because of the larger contact surface of the smaller droplets. The cationic ammonium surfactants can make lipophilic interaction with S. aureus.37 The incrementation of lipophilicity may promote the nanoparticle diffusion in the biofilm. The lipophilicity of the smaller-sized nanoemulsions was greater than that of the larger-sized nanoemulsions based on the molecular environment assay. This allowed the higher biofilm deterioration of the smaller droplets.
Some of the bacteria can be dispersed from the biofilm to the perimeter in the final stage of EPS development.38 The tendency of bacterial killing outside the biofilm was quite different from that inside the biofilm. Since it was more difficult for the bigger droplets to diffuse into the EPS, these large-sized droplets remained outside the biofilm to eradicate the planktonic cells derived from the matrix. Different from the CFU determination in biofilm, the trend of SYTO9 intensity was CN-M>CN-S≥CN-L. CFU evaluation is simply an indicator of bacterial viability. The SYTO9 staining manifests both the cell viability and the ability to interact with DNA. The mid-sized nanoemulsions might exhibit a strong binding with extracellular DNA in biofilm. Further study is needed to show the evidence. Overall, the smaller-sized nanoemulsions can be an ideal antibiofilm agent to deliver into the matrix and exhibit a bactericidal effect. The micelles also showed a strong activity towards biofilm MRSA. There was almost no viable MRSA inside and outside the biofilm after micelle treatment. Both monomers and micelles coexisted in the aqueous CPC dispersion. The monomers of cationic surfactants quickly bind to the bacterial surface, disturbing the membrane structure.22,39 The tiny volume of monomers is also advantageous for diffusing into the biofilm mesh.
It was apparent that the DNA concentration reduction was magnified when the droplet size decreased. However, the size effect on the total protein concentration was not apparent. Both DNA and protein could be lost by the capability of CPC to damage the cellular membrane. According to SEM, some bacteria maintained surface integrity after CPC treatment. The mechanisms other than DNA and protein leakage should be discussed. The intercalation of antibacterial agents or nanoparticles into the DNA helix can block DNA replication to exert anti-MRSA activity.3,40 CPC is known to bind to and precipitate double-stranded DNA.41 This interaction results in the bacterial DNA degradation and denaturing. This effect may be more significant for the small-sized nanoemulsions. The DNA degradation can interfere with the DNA-protein binding in cytoplasm, inhibiting the enzymatic and metabolic activity. It is possible that CPC produced both direct and indirect membrane collapse. In order to understand the possible mechanism, proteomic profiles were established in CPC-treated MRSA. CPC significantly reduced the expression of RNAP subunits β and β’. Transcription is an important process for tight control of gene expression for S. aureus survival. All transcriptional activity is maintained by RNAP.42 Bacteria consist of four subunits of RNAP: α, β, β’, and ω.43 The downregulation of this pathway suggested that squalene@CPC nanoemulsions had a profound effect on the transcription of DNA to RNA. A similar example is rifamycin binding to the β subunit of RNAP as the antibacterial mechanism of action.44,45 Compared to RNAP, we did not find any significant concordance with the proteins involving oxidative stress, Fenton reaction, and the Krebs cycle. In-depth work on the anti-MRSA mechanisms of squalene@CPC nanoemulsions is required for further elucidation.
Conclusions
The antimicrobial nanoparticles have attracted great interest as one of the suitable candidates to substitute for conventional antibiotics. In this study, we examined how the size difference in CPC-decorated nanoemulsions influences bacterial eradication. The nanoemulsions with small, medium, and large droplet sizes had high positive zeta potential, presenting great colloidal stability. The squalene@CPC nanosystems demonstrated substantial activity against bacteria and fungi, with the greatest potential to eliminate Gram-positive bacteria. Although the self-assembled micelles also showed broad and strong antibacterial activity, the poor stability led to the limited application. The killing of MRSA by the nanoemulsions was caused by membrane disruption initiated by electrostatic force. The degradation of DNA and interference in protein synthesis were also the possible mechanisms. The enhancement of anti-MRSA impact of the nanoemulsions with size reduction was observed in biofilm form but not in planktonic form. Compared to the larger ones, the small-sized nanoemulsions could facilely enter into the biofilm matrix to exert the stronger MRSA inhibition. The total DNA concentration in MRSA was also decreased following the reduction of droplet size. A slight difference in the bactericidal activity of different nanoformulations was detected in the evaluation platforms of MIC/MBC, agar diffusion assay, and SEM. The experimental results of this study can lead to the rational design of antibacterial nanoemulsions based on the desired application in antimicrobial coatings, foods, cosmetics, and mouth rinses. The nanoemulsions also have therapeutic significance to treat MRSA infection and act as the carriers of antibiotics.
Acknowledgments
The authors are grateful to the financial support from Ministry of Science and Technology of Taiwan (MOST-105-2320-B-182-010-MY3) and Chang Gung Memorial Hospital (CMRPG2F0491-3).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Dye C. After 2015: infectious diseases in a new era of health and development. Phil Trans R Soc B. 2014;369:20130426. doi: 10.1098/rstb.2013.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghanbar S, Fumakia M, Ho EA, Liu S. A new strategy for battling bacterial resistance: turning potent, non-selective and potentially non-resistance-inducing biocides into selective ones. Nanomed Nanotechnol Biol Med. 2018;14:471–481. doi: 10.1016/j.nano.2017.11.014 [DOI] [PubMed] [Google Scholar]
- 3.Hemeg HA. Nanomaterials for alternative antibacterial therapy. Int J Nanomed. 2017;12:8211–8225. doi: 10.2147/IJN.S132163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Z, Spilker T, Fan Y, et al. Nanoemulsion is an effective antimicrobial for methicillin-resistant Staphylococcus aureus in infected wounds. Nanomedicine. 2017;12:1177–1185. doi: 10.2217/nnm-2017-0025 [DOI] [PubMed] [Google Scholar]
- 5.Shao Y, Wu C, Wu T, et al. Eugenol-chitosan nanoemulsions by ultrasound-mediated emulsification: formulation, characterization and antimicrobial activity. Carbohydr Polym. 2018;193:144–152. doi: 10.1016/j.carbpol.2018.03.101 [DOI] [PubMed] [Google Scholar]
- 6.Park JB, Kang JH, Song KB. Antibacterial activities of a cinnamon essential oil with cetylpyridinium chloride emulsion against Escherichia coli O157: h7and Salmonella typhimurium in basil leaves. Food Sci Biotechnol. 2018;27:47–55. doi: 10.1007/s10068-017-0241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromm-Dornieden C, Rembe JD, Schäfer N, Böhm J, Stuermer EK. Cetylpyridinium chloride and miramistin as antiseptic substances in chronic wound management‒prospects and limitations. J Med Microbial. 2015;64:407–414. doi: 10.1099/jmm.0.000034 [DOI] [PubMed] [Google Scholar]
- 8.Latimer J, Munday JL, Buzza KM, Forbes S, Sreenivasan PK, McBain AJ. Antimicrobial and anti-biofilm activity of mouthrinses containing cetylpyridinium chloride and sodium fluoride. BMC Microbiol. 2015;15:169. doi: 10.1186/s12866-015-0501-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams CP, Walker KA, Obare SO, Docherty KM. Size-dependent antimicrobial effects of novel palladium nanoparticles. PLoS One. 2014;9:e85981. doi: 10.1371/journal.pone.0085981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravichandran AT, Pushpa KCS, Ravichandran K, Arun T, Ravidhas C, Muralidharan B. Effect of size reduction on the magnetic and antibacterial properties of ZnO:zr: mnnanoparticles synthesized by a cost-effective chemical method. J Mater Sci-Mater Electron. 2016;27:5825–5832. doi: 10.1007/s10854-016-4498-1 [DOI] [Google Scholar]
- 11.Helmlinger J, Sengstock C, Groβ-Heitfeld C, et al. Silver nanoparticles with different size and shape: equal cytotoxicity, but different antibacterial effects. RSC Adv. 2016;6:18490. doi: 10.1039/C5RA27836H [DOI] [Google Scholar]
- 12.McCarthy H, Rudkin JK, Black NS, Gallagher L, O’Neill E, O’Gara JP. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol. 2015;5:1. doi: 10.3389/fcimb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan TL, Wang PW, Hung CF, Aljuffali IA, Dai YS, Fang JY. The impact of retinol loading and surface charge on the hepatic delivery of lipid nanoparticles. Colloids Surf B Biointerfaces. 2016;141:584–594. doi: 10.1016/j.colsurfb.2016.02.029 [DOI] [PubMed] [Google Scholar]
- 14.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5 [DOI] [PubMed] [Google Scholar]
- 15.Alalaiwe A, Wang PW, Lu PL, Chen YP, Fang JY, Yang SC. Synergistic anti-MRSA activity of cationic nanostructured lipid carriers in combination with oxacillin for cutaneous application. Front Microbiol. 2018;9:1493. doi: 10.3389/fmicb.2018.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoopman TC, Liu W, Joslin SN, Pybus C, Brautigam CA, Hansen EJ. Identification of gene products involved in the oxidative stress response of Moraxella catarrhalis. Infect Immun. 2011;79:745–755. doi: 10.1128/IAI.01060-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manteca A, Fernández M, Sánchez J. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology. 2005;151:3689–3697. doi: 10.1099/mic.0.28045-0 [DOI] [PubMed] [Google Scholar]
- 18.Beaudoin T, Yau YCW, Stapleton PJ, et al. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes. 2017;3:25. doi: 10.1038/s41522-017-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan TL, Wang PW, Al-Suwayeh SA, Chen CC, Fang JY. Skin toxicology of lead species evaluated by their permeability and proteomic profiles: a comparison of organic and inorganic lead. Toxicol Lett. 2010;197:19–28. doi: 10.1016/j.toxlet.2010.04.019 [DOI] [PubMed] [Google Scholar]
- 20.Franklyne JS, Mukherjee A, Chandrasekaran N. Essential oil micro- and nanoemulsions: promising roles in antimicrobial therapy tergeting human pathogens. Lett Appl Microbiol. 2016;63:322–334. doi: 10.1111/lam.12631 [DOI] [PubMed] [Google Scholar]
- 21.Doktorovova S, Shegokar R, Rakovsky E, et al. Cationic solid lipid nanoparticles (cSLN): structure, stability and DNA binding capacity correlation studies. Int J Pharm. 2011;420:341–349. doi: 10.1016/j.ijpharm.2011.08.042 [DOI] [PubMed] [Google Scholar]
- 22.Tavano L, Pinazo A, Abo-Riya M, et al. Cationic vesicles based on biocompatible diacyl glycerol-arginine surfactants: physicochemical properties, antimicrobial activity, encapsulation efficiency and drug release. Colloids Surf B Biointerfaces. 2014;120:160–167. doi: 10.1016/j.colsurfb.2014.04.009 [DOI] [PubMed] [Google Scholar]
- 23.Sharif HR, Williams PA, Sharif MK, et al. Influence of OSA-starch on the physico chemical characteristics of flax seed oil-eugenol nanoemulsions. Food Hydrocoll. 2017;66:365–377. doi: 10.1016/j.foodhyd.2016.12.002 [DOI] [Google Scholar]
- 24.Sasaki S. Transformation relationships among monomers, micelles, metastable solid, and stable solid in aqueous cetylpyridinium chloride solution. J Phys Chem B. 2010;114:11039–11045. doi: 10.1021/jp103293x [DOI] [PubMed] [Google Scholar]
- 25.Cui X, Qiao C, Wang S, Ding Y, Hao C, Li J. Synthesis, surface properties, and antibacterial activity of polysiloxane quaternary ammonium salts containing epoxy group. Colloid Polym Sci. 2015;293:1971–1981. doi: 10.1007/s00396-015-3588-6 [DOI] [Google Scholar]
- 26.van Hal SJ, Paterson DL. Systematic review and meta-analysis of the significance of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2011;55:405–410. doi: 10.1128/AAC.01133-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martín VI, de la Haba RR, Ventosa A, Congiu E, Ortega-Calvo JJ, Moyá ML. Colloidal and biological properties of cationic single-chain and dimeric surfactants. Colloids Surf B Biointerfaces. 2014;114:247–254. doi: 10.1016/j.colsurfb.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 28.Donsì F, Annunziata M, Vincensi M, Ferrari G. Design of nanoemulsion-based delivery systems of natural antimicrobials: effect of the emulsifier. J Biotechnol. 2012;159:342–350. doi: 10.1016/j.jbiotec.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 29.Sarwar A, Katas H, Zin NM. Antibacterial effects of chitosan-tripolyphosphate nanoparticles: impact of particle size molecular weight. J Nanopart Res. 2014;16:2517. doi: 10.1007/s11051-014-2517-9 [DOI] [Google Scholar]
- 30.Loo CY, Rohanizadeh R, Young PM, et al. Combination of silver nanoparticles and curcumin nanoparticles for enhanced anti-biofilm activities. J Agric Food Chem. 2016;64:2513–2522. doi: 10.1021/acs.jafc.5b04559 [DOI] [PubMed] [Google Scholar]
- 31.Chatterjee T, Chatterjee BK, Chakrabarti P. Modeling of growth kinetics of Vibrio cholera in presence of gold nanoparticles: effect of size and morphology. Sci Rep. 2017;7:9671. doi: 10.1038/s41598-017-09357-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chihalova K, Chudobova D, Michalek P, et al. Staphylococcus aureus and MRSA growth and biofilm formation after treatment with antibiotics and SeNPs. Int J Mol Sci. 2015;16:24656–24672. doi: 10.3390/ijms161024656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, Radovic-Moreno AF, Wu J, Langer R, Shi J. Nanomedicine in the management of microbial infection‒overview and perspectives. Nano Today. 2014;9:478–498. doi: 10.1016/j.nantod.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng F, He T, Huang S, et al. Cetylpyridinium chloride mouth rinses alleviate experimental gingivitis by inhibiting dental plaque maturation. Int J Oral Sci. 2016;8:182–190. doi: 10.1038/ijos.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baelo A, Levato R, Julián E, et al. Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J Control Release. 2015;209:150–158. doi: 10.1016/j.jconrel.2015.04.028 [DOI] [PubMed] [Google Scholar]
- 36.Cheow WS, Chang MW, Hadinoto K. The roles of lipid in anti-biofilm efficacy of lipid-polymer hybrid nanoparticles encapsulating antibiotics. Colloids Surf A Physicochem Eng Asp. 2011;389:158–165. doi: 10.1016/j.colsurfa.2011.08.035 [DOI] [Google Scholar]
- 37.Zhou C, Wang H, Bai H, et al. Tuning antibacterial activity of cyclodextrin-attached cationic ammonium surfactants by a supramolecular approach. ACS Appl Mater Interf. 2017;9:31657–31666. doi: 10.1021/acsami.7b11528 [DOI] [PubMed] [Google Scholar]
- 38.Nance WC, Dowd SE, Samarian D, et al. A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials. J Antimicrob Chemother. 2013;68:2550–2560. doi: 10.1093/jac/dkt211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C, Hu J, Zhang S, et al. Molecular mechanisms of antibacterial and antitumor actions of designed surfactant-like peptides. Biomaterials. 2012;33:592–603. doi: 10.1016/j.biomaterials.2011.09.059 [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Kumar KV, Rasheed S, Geng RX, Zhou CH. Design, synthesis, and antimicrobial evaluation of novel quinolone imidazoles and interactions with MRSA DNA. Chem Biol Drug Des. 2015;86:648–655. doi: 10.1111/cbdd.12532 [DOI] [PubMed] [Google Scholar]
- 41.Ganeshnarayan K, Shah SM, Libera MR, Santostefano A, Kaplan JB. Poly-N-acetylglucosamine matrix polysaccharide impedes fluid convection and transport of the cationic surfactant cetylpyridinium chloride through bacterial biofilms. Appl Environ Microbiol. 2009;75:1308–1314. doi: 10.1128/AEM.01900-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss A, Moore BD, Tremblay MHJ, Chaput D, Kremer A, Shaw LN. The ω subunit governs RNA polymerase stability and transcriptional specificity in Staphylococcus aureus. J Bacteriol. 2017;199:e00459–16. doi: 10.1128/JB.00459-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekine S, Tagami S, Yokoyama S. Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Curr Opin Struct Biol. 2012;22:110–118. doi: 10.1016/j.sbi.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 44.Murakami KS. Structural biology of bacterial RNA polymerase. Biomolecules. 2015;5:848–864. doi: 10.3390/biom5020848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Czerwonka D, Domagalska J, Pyta K, et al. Structure-activity relationship studies of new rifamycins containing l-amino acid esters as inhibitors of bacterial RNA polymerases. Eur J Med Chem. 2016;116:216–221. doi: 10.1016/j.ejmech.2016.03.061 [DOI] [PubMed] [Google Scholar]