Abstract
Background
The Salmonella enterica subsp. diarizonae serovar 61:k:1,5,(7) (SASd) has been found to be host-adapted to sheep, with a high prevalence in sheep herds worldwide. Infections are usually sub-clinical, however the serovar has the potential to cause diarrhea, abortions and chronic proliferative rhinitis. Although occurrence and significance of SASd infections in sheep have been extensively studied, the genetic mechanism underlying this unusual host-adaptation have remained unknown, due to a lack of (a) available high-quality genome sequence(s).
Results
We utilized Nanopore and Illumina sequencing technologies to generate a de novo assembly of the 4.88-Mbp complete genome sequence of the SASd strain 16-SA00356, isolated from the organs of a deceased sheep in 2016. We annotated and analyzed the genome sequence with the aim to gain a deeper understanding of the genome characteristics associated with its pathogenicity and host adaptation to sheep. Overall, we found a number of interesting genomic features such as several prophage regions, a VirB4/D4 plasmid and novel genomic islands. By comparing the genome of 16-SA00356 to other S. enterica serovars we found that SASd features an increased number of pseudogenes as well as a high level of genomic rearrangements, both known indicators of host-adaptation.
Conclusions
With this sequence, we provide the first complete and closed genome sequence of a SASd strain. With this study, we provide an important basis for an understanding of the genetic mechanism that underlie pathogenicity and host adaptation of SASd to sheep.
Keywords: Salmonella enterica subsp. diarizonae, Host-adaptation, Pseudogenes, Sheep
Background
Salmonella enterica subsp. diarizonae serovar 61:k:1,5,(7) (also designated as SASd) is a Gram-negative bacterium of the genus Salmonella. SASd is considered host-adapted to sheep, based on its wide distribution and high prevalence in sheep flocks worldwide [1–7]. SASd colonizes the intestines and tonsils of sheep and can be isolated from the faeces and nasal discharge of the animals [6]. Colonization might be chronic, with faecal shedding of the pathogen allowing transmission between individuals [8]. Although the serovar does not usually induce diseases [8–10], it has the potential to cause diarrhea [11], abortions [2] and chronic proliferative rhinitis [3, 6]. Over the last years, occurrence, distribution and impact of SASd infections in sheep have been extensively studied. However the potential genetic features underlying this unusual host-adaptation have remained unknown, due to a lack of available high-quality genome sequences. Here, we announce the first, complete and closed genome sequence of S. enterica subsp. diarizonae serovar 61:k:1,5,(7). Through genome analysis and a genome comparison study we identified numerous genetic features indicating host adaptation traits of SASd to sheep.
Methods
Strain isolation and characterization
Strain 16-SA00356 was isolated from an enriched pooled organ sample of an adult sheep that was found dead in Northern Germany in 2016. The sheep was postmortem diagnosed with liver cell necrosis, liver abscesses and serofibrinous peritonitis, most likely resulting from an infection with Fasciola hepatica. Detection of Salmonella spp. in the enriched culture was considered an incidental finding. Enrichment and isolation of Salmonella spp. was achieved by pooling small pieces of different organs (lung, liver, kidney, spleen and small intestine) in tetrathionate broth of Preuss, a selective medium for enrichment of Salmonella spp., followed by incubation for 12 h at 37 °C. After incubation, the broth was spread on three selective solid agar plates (Rambach agar, XLD agar, BSB agar), followed by another incubation cycle. Salmonella spp. colonies were confirmed through MALDI-TOF, before further subcultivation on Lysine Iron Agar (LIA) slants. The Salmonella isolate was serotyped by slide agglutination with the antigenic formula 61:k:1,5,(7). Antimicrobial susceptibility testing was performed by broth microdilution following CLSI guidelines (CLSI M07-A9) and EUCAST epidemiological cut-off values (ECOFFs; http://www.eucast.org/). The isolate was found to be sensitive to all tested antibiotics (ampicillin, chloramphenicol, ciprofloxacin, colistin, cefotaxime, gentamicin, nalidixic acid, sulfamethoxazole, ceftazidime, tetracycline and trimethoprim).
Genome sequencing and de novo assembly
Genomic DNA for both sequencing techniques was isolated from an overnight liquid culture using the PureLink® Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). Sequencing libraries for Illumina sequencing were prepared with the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Sequencing was performed in 2 × 251 cycles on the Illumina MiSeq benchtop using the MiSeq Reagent v3 600-cycle Kit (Illumina). A total number of 1,433,866 reads was generated, with 86% of bases above a quality score of 30 (> Q30) and an overall coverage of 60×. The paired Illumina reads were trimmed using Trimmomatic v0.36 [12] with option sliding window 4:20 and minlen 50 yielding 1,295,014 read pairs. To generate long reads for scaffolding, Oxford Nanopore MinION technology (ONT) was applied. MinION libraries were prepared using the Rapid barcoding kit (Oxford Nanopore Technologies, Oxford, UK), following the manufacturer’s instructions, and sequenced for approximately 16 hours using a FLO-MIN106 R9 flow cell generating 82,147 reads. The genome was assembled with Unicycler v0.4.4 [13], including Pilon v1.23 [14], providing the trimmed Illumina reads as paired short reads and the ONT reads as long reads with default parameters.
Genome annotation
Antibiotic resistance genes were identified with ResFinder v3.1 [15]. Salmonella pathogenicity islands (SPI) were detected with SPIFinder v1.0 with default parameters [16] and by BLAST against known Salmonella pathogenicity islands. Prophage regions were identified with PHASTER [15]. Pseudogenes were determined with Pseudofinder v0.10 [17] with standard parameters and length 0.8. Genomic rearrangements were detected with progressive Mauve v2.4.0, with standard parameters [18].
Comparative genomic analysis
The genome of the SASd isolate 16-SA00356 was compared to a set of well annotated S. enterica serovars which were chosen to represent different host ranges. All serovars together with their NCBI accession numbers and information regarding the size of their genomes, number of ORFs and GC content, are listed in Additional file 1: Table S1. Plasmid sequences were excluded from the comparative analysis.
Phylogenetic analysis
Phylogeny was inferred through alignment free genome comparison with feature frequency profiles (FFP v3.19) and through comparison of 107 essential single-copy core genes following the bcgTree pipeline (v1.1.0). The bcgTree pipeline was applied with default parameters as as described by Ankenbrand and Keller [19]. FPP was performed with default parameters and l-mer length 24 as described by Wang and Ash [20].
Quality assurance
A single colony of 16-SA00356 was transferred to fresh LB medium to obtain a pure culture for genomic DNA extraction. After the genome sequence was obtained subspecies and serovar assignments were confirmed by the in silico typing tool SISTR v1.0.2 [21].
Results and discussion
General features
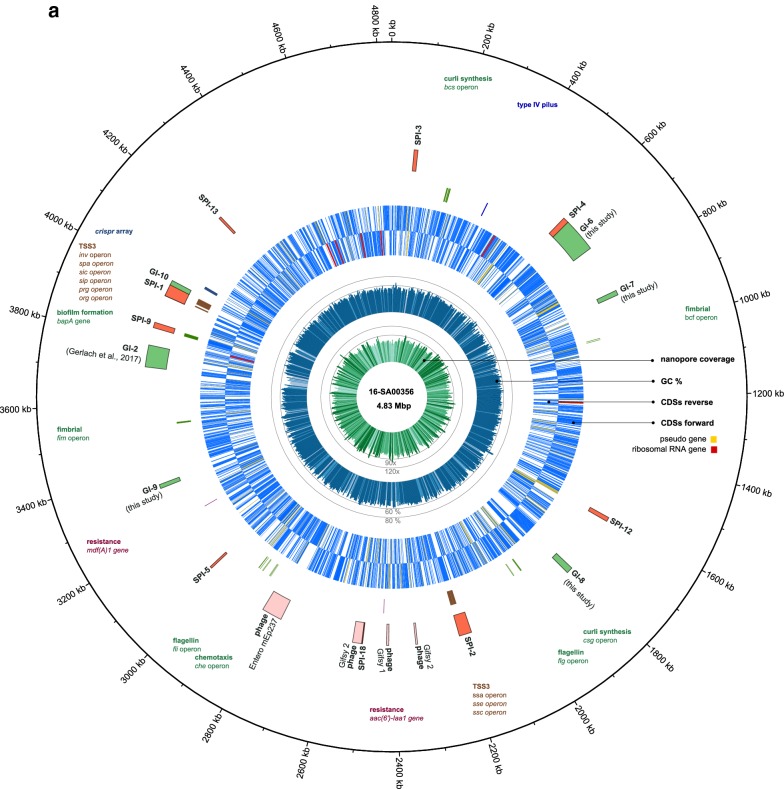
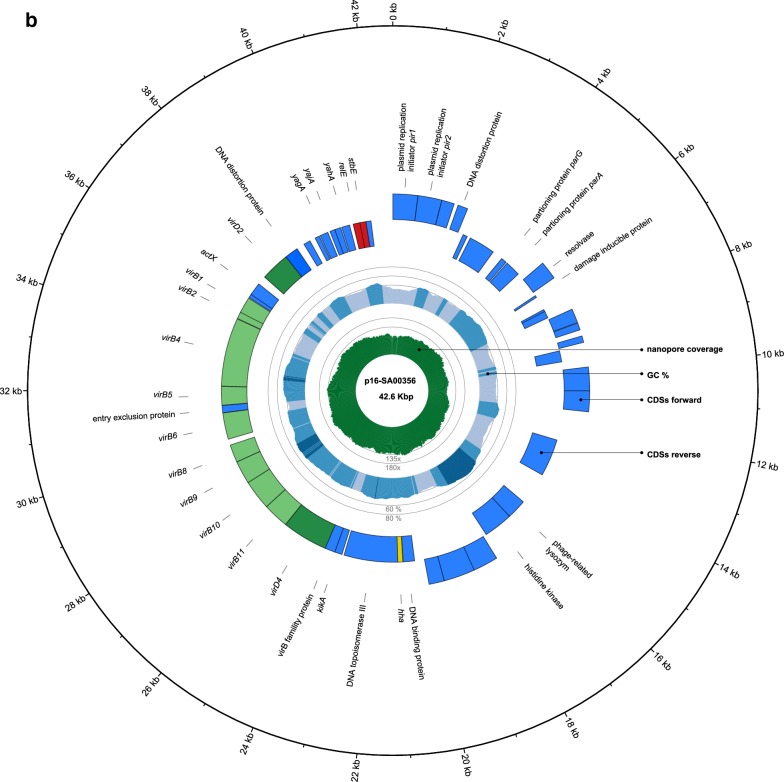
The genome of SASd isolate 16-SA00356 is composed of a circular chromosome of 4,832,672 bp (GC 51.49%) and a circular plasmid of 42,663 bp (GC 41.34%). A graphical representation of the annotated chromosome and plasmid is shown in Fig. 1. A total of 4687 CDSs, 80 tRNAs, 1 tmRNA and 22 rRNAs regions were predicted within the chromosome by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
Fig. 1.
Genome (a) and plasmid (b) map of S. enterica subsp. diarizonae serovar 61:k:1,5,(7), isolate 16-SA00356, displayed in Circos. The tracks from inside to outside represent the Nanopore sequencing coverage, GC content, reverse-strand CDSs, forward-strand CDSs and labeled genetic regions of interest such as important operons, Salmonella pathogenicity islands (SPI), genomic islands (GI) and prophage regions
Antibiotic resistance and pathogenicity
Although the strain was susceptible to eleven tested antibiotics, two antibiotic resistance genes homologous to aac(6′)-Iaa (Accession: NC_003197), an aminoglycoside acetyltransferase, and mdfA (Accession: Y08743), a macrolide–lincosamide–streptogramin B (MLS) resistance gene were identified. We found that the strain possesses the major pathogenicity islands SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-9, SPI-12, SPI-13, SPI-18 and GI-2 [22]. Furthermore, by comparing the sequence based similarity of the 16-SA00356 genome to five other S. enterica subsp. diarizonae serovars (see Additional file 2: Figure S1) we identified five novel genomic islands: GI-6 - GI-10. These novel genomic islands contain mainly proteins of unknown function and no major virulence genes could be attributed to them. In addition, we identified four incomplete prophage regions, with high similarity to the prophages Entero mEp237, Gifsy 1 and Gifsy 2. Overall, the resistance and virulence potential of SASd appears to be low. The absence of major antibiotic resistance genes can be attributed to the generally low antibiotic usage/low intensity farming practice of sheep.
Plasmid
SASd isolate16-SA00356 was found to carry an IncX1/ColRNAI type plasmid of 42,663 bp which we named pSE16-SA00356. We found pSE16-SA00356 to harbour an almost complete conjugative Type IV secretion system (missing virB7), which has been linked to persistent infections in numerous pathogens [23]. The plasmid furthermore carries the RelE/StbE toxin/antitoxin system and a small Haemolysin expression-modulating protein Hha, although the complementary tomB antitoxin gene was not detected in our analysis. The addiction module RelE/StbE probably increases the stability and therefore the persistence of the plasmid in the microbial population.
Phylogenetic analysis
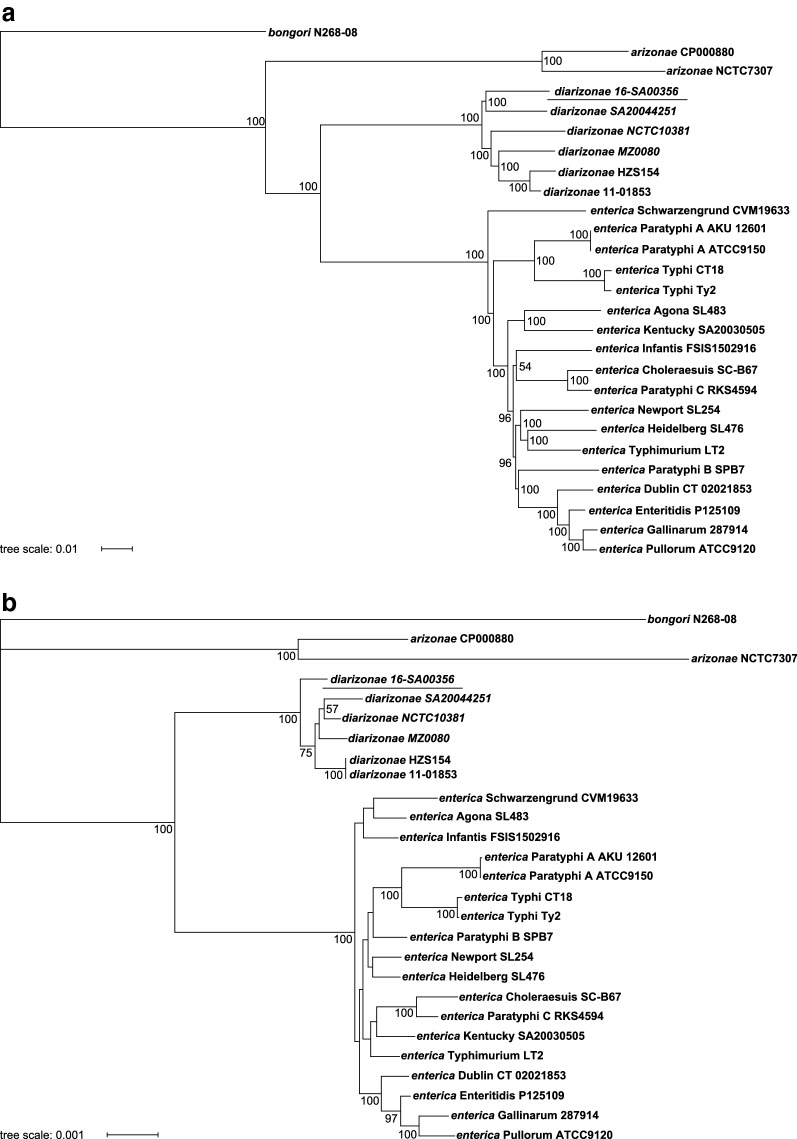
Phylogeny of the different Salmonella species was inferred through alignment-free genome comparison with feature frequency profiles (FFP) and through comparison of 107 essential single-copy core genes with bcgTree. The resulting phylogenetic trees are shown in Fig. 2 and both indicate that 16-SA00356 clusters within the group of the S. enterica subsp. diarizonae serovars. Bootstrap values attribute greater certainty to the pyhlogenetic tree obtained through FFP.
Fig. 2.
Phylogenetic analysis of Salmonella species with Salmonella bongori N268-08 (CP006608) as outgroup. a NJ tree based on the comparison of complete genome sequences with the alignment-free feature frequency profiles (FFP) method. Numbers at nodes designate bootstrap support values generated using 100 permutations. b Best-scoring maximum-likelihood tree based on the comparison of the amino acid sequences of 107 essential single-copy core genes with bcgTree. Numbers at nodes designate bootstrap support values resulting from 100 bootstrap replicates. Only bootstrapping values greater than 50 are displayed
Pseudogenes
Recent intracellular pathogens have a higher number of pseudogenes that results from the fact that adaptation to an intracellularly lifestyle causes bacteria to gradually loses genes no longer needed in their new environment. Nuccio and Bäumler [24] propose that Salmonella serovars could be divided into a group with a low number of pseudogenes and those with a high number of pseudogenes, with the later group referred as the extraintestinal pathovars. When comparing the percentage of pseudogenes normalized to the total number of ORFs among different Salmonella serovars we found SASd isolate16-SA00356 to possess a medium number of pseudogenes. An overview of the results of our analysis is shown in Table 1. Overall, host-adapted and host-restricted serovars such as S. enterica subsp. enterica serovar Choleraesuis (pigs), S. enterica subsp. enterica serovar Typhi (humans), S. enterica subsp. enterica serovars Gallinarum and Pullorum (birds) feature a higher percentage of pseudogenes (6.5–7.6%), than those reported to have a broad host range i.e. S. enterica subsp. enterica serovar Typhimurium (4.9%). Interestingly, the genome of the SASd isolate 16-SA00356 features a comparable number of pseudogenes (6.0%), to the cattle-adapted S. enterica subsp. enterica serovars Dublin (5.7%) and Kentucky (5.5%). Together with the fact that among the investigated S. enterica subsp. diarizonae serovars, SASd possesses the highest number of pseudogenes, these results further indicate a host-adaptation to sheep.
Table 1.
Correlation between number of pseudogenes and host range of the respective organism
| Serovar | Host range | ORFs | Pseudogenes | % |
|---|---|---|---|---|
| S. enterica subsp. enterica serovar Infantis FSIS1502916 | Broad [26] | 4407 | 206 | 4.67 |
| S. enterica subsp. enterica serovar Agona SL483 | Broad [27] | 4444 | 213 | 4.79 |
| S. enterica subsp. enterica serovar Schwarzengrund CVM19633 | Broad [27] | 4410 | 212 | 4.81 |
| S. enterica subsp. enterica serovar Paratyphi B SPB7 | Broad [27] | 4549 | 220 | 4.84 |
| S. enterica subsp. diarizonae serovar 60:r:z HZS154 | Unknown | 4689 | 228 | 4.86 |
| S. enterica subsp. enterica serovar Typhimurium LT2 | Broad [27, 28] | 4504 | 220 | 4.88 |
| S. enterica subsp. enterica serovar Heidelberg SL476 | Broad [27] | 4565 | 223 | 4.88 |
| S. enterica subsp. diarizonae serovar 65:c:z SA20044251 | Unknown | 4461 | 218 | 4.89 |
| S. enterica subsp. enterica serovar Newport SL254 | Broad [27] | 4489 | 220 | 4.90 |
| S. enterica subsp. enterica serovar Enteritidis P125109 | Broad [27] | 4352 | 223 | 5.12 |
| S. enterica subsp. diarizonae serovar 61:i:z NCTC10381 | Unknown | 4828 | 249 | 5.16 |
| S. enterica subsp. diarizonae serovar 50:k:z MZ0080 | Unknown | 4684 | 245 | 5.23 |
| S. enterica subsp. diarizonae serovar 60:r:z 11-01853 | Unknown | 4326 | 227 | 5.25 |
| S. enterica subsp. enterica serovar Kentucky SA20030505 | Bovine-adapted [28] | 4427 | 242 | 5.47 |
| S. enterica subsp. enterica serovar Dublin CT_02021853 | Bovine-adapted [27, 28] | 4580 | 261 | 5.70 |
| S. enterica subsp. diarizonae serovar 61:k:1,5,(7) 16-SA00356 | Sheep-adapted [5] | 4461 | 269 | 6.03 |
| S. enterica subsp. enterica serovar Paratyphi C RKS4594 | Human-restricted [29] | 4615 | 285 | 6.18 |
| S. enterica subsp. enterica serovar Paratyphi A AKU_12601 | Human-restricted [27, 28] | 4340 | 281 | 6.47 |
| S. enterica subsp. enterica serovar Paratyphi A ATCC 9150 | Human-restricted [27, 28] | 4337 | 281 | 6.48 |
| S. enterica subsp. enterica serovar Choleraesuis SC-B67 | Porcine-adapted [27] | 4566 | 296 | 6.48 |
| S. enterica subsp. enterica serovar Typhi CT18 | Human-restricted [27, 28] | 4665 | 307 | 6.58 |
| S. enterica subsp. enterica serovar Typhi Ty2 | Human-restricted [27, 28] | 4621 | 314 | 6.80 |
| S. enterica subsp. enterica serovar Pullorum ATCC 9120 | Avian-restricted [29, 30] | 4474 | 334 | 7.47 |
| S. enterica subsp. enterica serovar Gallinarum 287/91 | Avian-restricted [27, 28] | 4453 | 336 | 7.55 |
Host ranges were inferred from the literature (when available). The number of ORFs and pseudogenes was determined with Pseudofinder [17]. Only complete genome sequences were considered. Plasmid sequences were excluded from the analysis
Genome rearrangements
Host restricted pathogens often exhibit genomic rearrangements [25] and by comparing the genome of SASd strain16-SA00356 to other S. enterica subsp. diarizonae serovars, we were able to detect large scale genome rearrangements with many inversions as shown in Additional file 3: Figure S2.
Conclusion
Overall, this study found a number of interesting genomic features linked to pathogenicity and host specificity of SASd to sheep. Among these, we detected increased pseudogene formation, large scale genomic rearrangements, a VirB4/D4 plasmid and novel genomic islands. The complete genome sequence generated in this study forms an important basis for further understanding of the pathogenicity and host adaptation of SASd, as well as a high-quality reference for future genome comparison studies.
Supplementary information
Additional file 1: Table S1. List of S. enterica serovars analyzed in this study.
Additional file 2: Figure S1. Sequence based similarity of five S. enterica subsp. diarizonae serovars to S. enterica subsp. diarizonae serovar 61:k:1,5,(7), isolate 16-SA00356. The sequence similarity is shown by color-coded tracks which from inside to outside represent (i) S. enterica subsp. diarizonae SA20044251, (ii) S. enterica subsp. diarizonae NCTC10381, (iii) S. enterica subsp. diarizonae MZ0080 and (iv) S. enterica subsp. diarizonae HZS154 and (v) S. enterica subsp. diarizonae 11-01853. The location of genetic regions of interest such as Salmonella pathogenicity islands (SPI), genomic islands (GI) and prophage regions are indicated.
Additional file 3: Figure S2. Mauve alignment of 16-SA00356, SA20044251, NCTC10381, MZ0080, HZS154 and 11-01853. Colored blocks indicate individual locally collinear blocks (LCB). Homologous LCBs are connected with lines. 16-SA00356 is set as the reference genome.
Acknowledgements
We thank Ernst Junker for his excellent laboratory assistance.
Authors’ contributions
LU and BM designed the study. CJ and IS provided the isolate and information to the isolate and host organism. MB performed the sequencing. LU, CD and SHT conducted the bioinformatic analysis. BM supervised the project. LU wrote the manuscript and created the figures. All authors read and approved the final manuscript.
Funding
This work was supported by the German Federal Institute for Risk Assessment (BfR) (Research Project Number 1322-716).
Availability of data and materials
Nucleotide sequences were deposited in GenBank under the accession numbers CP034074 (chromosome) and CP034075 (plasmid). The datasets supporting the conclusions of this article are included within the article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13099-019-0330-9.
References
- 1.Amagliani G, Petruzzelli A, Carloni E, Tonucci F, Foglini M, Micci E, et al. Presence of Escherichia coli O157, Salmonella spp., and Listeria monocytogenes in raw ovine milk destined for cheese production and evaluation of the equivalence between the analytical methods applied. Foodborne Pathog Dis. 2016;13:626–632. doi: 10.1089/fpd.2016.2159. [DOI] [PubMed] [Google Scholar]
- 2.Davies RH, Evans SJ, Chappell S, Kidd S, Jones YE, Preece BE. Increase in Salmonella enterica subspecies diarizonae serovar 61:k:l,5,(7) in sheep. Vet Rec. 2001;149:555–557. doi: 10.1136/vr.149.18.555. [DOI] [PubMed] [Google Scholar]
- 3.Lacasta D, Ferrer LM, Ramos JJ, Bueso JP, Borobia M, Ruiz de Arcaute M, et al. Chronic proliferative rhinitis associated with Salmonella enterica subspecies diarizonae serovar 61:k:1,5,(7) in sheep in Spain. J Comp Pathol. 2012;147:406–409. doi: 10.1016/j.jcpa.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Methner U, Moog U. Occurrence and characterisation of Salmonella enterica subspecies diarizonae serovar 61:k:1,5,(7) in sheep in the federal state of Thuringia, Germany. BMC Vet Res. 2018;14:401. doi: 10.1186/s12917-018-1741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sörén K, Lindblad M, Jernberg C, Eriksson E, Melin L, Wahlström H, et al. Changes in the risk management of Salmonella enterica subspecies diarizonae serovar 61:(k):1,5,(7) in Swedish sheep herds and sheep meat due to the results of a prevalence study 2012. Acta Vet Scand. 2015;57:6. doi: 10.1186/s13028-015-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokar-Regenscheit N, Overesch G, Giezendanner R, Roos S, Gurtner C. Salmonella enterica subspecies diarizonae serotype 61:k:1,5,(7) associated with chronic proliferative rhinitis and high nasal colonization rates in a flock of Texel sheep in Switzerland. Prev Vet Med. 2017;145:78–82. doi: 10.1016/j.prevetmed.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Dargatz DA, Marshall KL, Fedorka-Cray PJ, Erdman MM, Kopral CA. Salmonella prevalence and antimicrobial susceptibility from the National Animal Health Monitoring System Sheep 2011 study. Foodborne Pathog Dis. 2015;12:953–957. doi: 10.1089/fpd.2015.2016. [DOI] [PubMed] [Google Scholar]
- 8.Lacasta D, Figueras L, Bueso JP, De las Heras M, Ramos JJ, Ferrer LM, et al. Experimental infection with Salmonella enterica subspecies diarizonae serotype 61:k:1,5,(7) in sheep: study of cell mediated immune response. Small Rumin Res. 2017;149:28–33. doi: 10.1016/j.smallrumres.2017.01.011. [DOI] [Google Scholar]
- 9.Bonke R, Wacheck S, Bumann C, Thum C, Stüber E, König M, et al. High prevalence of Salmonella enterica subsp. diarizonae in tonsils of sheep at slaughter. Food Res Int. 2012;45:880–884. doi: 10.1016/j.foodres.2011.01.050. [DOI] [Google Scholar]
- 10.Sandberg M, Alvseike O, Skjerve E. The prevalence and dynamics of Salmonella enterica IIIb 61:k:1,5,(7) in sheep flocks in Norway. Prev Vet Med. 2002;52:267–275. doi: 10.1016/S0167-5877(01)00250-1. [DOI] [PubMed] [Google Scholar]
- 11.Wray C, Wray A. Salmonella in domestic animals. Wallingford: CABI Pub; 2000. [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SPIFinder 1.0. https://cge.cbs.dtu.dk/services/SPIFinder/. Accessed 23 Apr 2019.
- 17.Pseudofinder. https://github.com/filip-husnik/pseudo-finder. Accessed 15 May 2019.
- 18.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ankenbrand MJ, Keller A. bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome. 2016;59:783–791. doi: 10.1139/gen-2015-0175. [DOI] [PubMed] [Google Scholar]
- 20.Wang A, Ash GJ. Whole genome phylogeny of Bacillus by feature frequency profiles (FFP) Sci Rep. 2015;5:13644. doi: 10.1038/srep13644. [DOI] [Google Scholar]
- 21.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach RG, Walter S, McClelland M, Schmidt C, Steglich M, Prager R, et al. Comparative whole genome analysis of three consecutive Salmonella diarizonae isolates. Int J Med Microbiol IJMM. 2017;307:542–551. doi: 10.1016/j.ijmm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Voth DE, Broederdorf LJ, Graham JG. Bacterial Type IV secretion systems: versatile virulence machines. Fut Microbiol. 2012;7:241–257. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuccio S-P, Bäumler AJ. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio. 2014;5:e00929-14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews TD, Schmieder R, Silva GGZ, Busch J, Cassman N, Dutilh BE, et al. Genomic comparison of the closely-related Salmonella enterica serovars Enteritidis, Dublin and Gallinarum. PLoS ONE. 2015;10:e0126883. doi: 10.1371/journal.pone.0126883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, Zurfluh K, et al. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol. 2017;8:1322. doi: 10.3389/fmicb.2017.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. The Salmonella enterica pan-genome. Microb Ecol. 2011;62:487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner JR, Kingsley RA. Evolution of Salmonella within hosts. Trends Microbiol. 2018;26:986–998. doi: 10.1016/j.tim.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–255. doi: 10.1017/S0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Hu Y, Chen J, Liu Z, Han J, Sun L, et al. Identification of Salmonella enterica serovar pullorum antigenic determinants expressed in vivo. Infect Immun. 2013;81:3119–3127. doi: 10.1128/IAI.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of S. enterica serovars analyzed in this study.
Additional file 2: Figure S1. Sequence based similarity of five S. enterica subsp. diarizonae serovars to S. enterica subsp. diarizonae serovar 61:k:1,5,(7), isolate 16-SA00356. The sequence similarity is shown by color-coded tracks which from inside to outside represent (i) S. enterica subsp. diarizonae SA20044251, (ii) S. enterica subsp. diarizonae NCTC10381, (iii) S. enterica subsp. diarizonae MZ0080 and (iv) S. enterica subsp. diarizonae HZS154 and (v) S. enterica subsp. diarizonae 11-01853. The location of genetic regions of interest such as Salmonella pathogenicity islands (SPI), genomic islands (GI) and prophage regions are indicated.
Additional file 3: Figure S2. Mauve alignment of 16-SA00356, SA20044251, NCTC10381, MZ0080, HZS154 and 11-01853. Colored blocks indicate individual locally collinear blocks (LCB). Homologous LCBs are connected with lines. 16-SA00356 is set as the reference genome.
Data Availability Statement
Nucleotide sequences were deposited in GenBank under the accession numbers CP034074 (chromosome) and CP034075 (plasmid). The datasets supporting the conclusions of this article are included within the article and its additional files.