Abstract
Worldwide, neoplasms of the gastrointestinal tract have a very high incidence and mortality. Among these, colorectal cancer, which includes colon and rectum malignancies, representing both highest incidence and mortality. While gallbladder cancer, another neoplasm associated to gastrointestinal tract occurs less frequently. Genetic factors, inflammation and nutrition are important risk factors associated with colorectal cancer development. Likewise, pathogenic microorganisms inducing intestinal dysbiosis have become an important scope to determine the role of bacterial infection on tumorigenesis. Interestingly, in human biopsies of different types of gastrointestinal tract cancer, the presence of different bacterial strains, such as Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica have been detected, and it has been considered as a high-risk factor to cancer development. Therefore, pathogens infection could contribute to neoplastic development through different mechanisms; including intestinal dysbiosis, inflammation, evasion of tumoral immune response and activation of pro-tumoral signaling pathways, such as β catenin. Here, we have reviewed the suggested bacterial molecular mechanisms and their possible role on development and progression of gastrointestinal neoplasms, focusing mainly on colon neoplasms, where the bacteria Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica infect.
1. Introduction
Worldwide, neoplasms affecting gastrointestinal tract are among the most frequent in incidence and mortality [1]. Gastrointestinal tract neoplasms are including: colon, rectum, stomach, pancreas, biliary tract and esophagus [2]. The main factors associated with development of gastrointestinal tract malignancies are alcohol consumption and smoking [3–5], high fat diets [6–9]; as well as, ageing, gender and race [10–13]. In addition, pathogenic microorganisms such as viruses and bacteria infecting the gastrointestinal tract, are being studied as possible triggers for development of neoplasms. In this regard, the role of Helicobacter pylori in the development of gastric cancer has been extensively studied [14]. However, other bacteria have also been associated with development of gastrointestinal neoplasms, especially in colon, rectum and gallbladder. This review describes the possible roles of Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica on cancer development. These bacteria have been considered as emerging pathogenic bacteria associated with development of colorectal cancer, which includes colon and rectum neoplasms, [15]. Here, we have focused on colon cancer, a neoplasia with a very high incidence on worldwide population, registering in 2018; 850,000 new cases and a mortality rate of 550,000 individuals [1].
2. Fusobacterium nucleatum
Fusobacterium nucleatum (F. nucleatum) is an adherent and invasive Gram-negative anaerobic bacterium. F. nucleatum resides mainly in oral cavity and is usually associated with periodontal disease [16]. Nevertheless, in last years, this bacterium has been detected in primary lesions [17], biopsies [18, 19], and stools [20] of patients with colon cancer, so bacterium has also been linked to development and progression of this neoplasia. In addition, different regions of human colon are colonized by F. nucleatum [21]. However, in patients with colon cancer, F. nucleatum has been located mainly on cecum and rectum [22, 23], where it is preferentially localized into tumor tissue [24, 25]. An important factor associated with F. nucleatum recruitment into tumor is over-expression of Gal-GalNAc molecules by tumor cells, which promote bacterial adhesion via Fap2 protein [26]. Likewise, high levels of anti-Fusobacterium IgA and IgG antibodies have been detected in sera of colon cancer patients [27], which could be used as biomarkers in early diagnosis of this neoplasia. Additionally, infection by F. nucleatum has been associated with a low survival of colon cancer patients [28], as well as increased resistance to chemotherapy treatment [29].
Previous studies have reported the association of F. nucleatum and colon cancer, although the presence of this bacterium in infected people is highly variable and inconsistent. In this regard, infection with F. nucleatum has been detected in 15% of North American population with colon cancer, while more than 60% of infected patients have been found in Chinese population [25, 28, 30, 31]. Interestingly, common characteristics found in all colon cancer patients with F. nucleatum infection were microsatellite instability (MSI), methylation phenotype of CpG island (CIMP), as well as BRAF and KRAS genes mutations [23, 25, 32].
On the other hand, infection with F. nucleatum in C57BL/6 APCMin/+ mice induced tumorigenesis regardless of colitis development [20], unlike the infection by enterotoxigenic Bacteroides fragilis, which initially produces colitis and subsequently tumors [33]. Therefore, several mechanisms inducing tumor by F. nucleatum have been proposed, including β catenin signaling pathway activation, which is upregulated in colon cancer [34]. In this pathway, β catenin is phosphorylated by PAK-1 through F. nucleatum-TLR4 interaction [35]. Likewise, binding of F. nucleatum FadA adhesin to E-cadherin expressed on host cells activates the Wnt/β catenin pathway promoting cell proliferation [36]. Additionally, a significant decrease on expression of TOX family proteins (thymocyte selection-associated high-mobility group box) after F. nucleatum infection has been shown [37]. These proteins regulate important cellular functions such as growth, apoptosis, DNA repair and metastatic processes [38]. Interestingly, an important decrease on TOX family proteins expression has been associated with advanced tumors.
Another mechanism associated with development and progression of colon cancer induced by F. nucleatum have been linked to inflammation. Thus, in colon cancer patients infected with F. nucleatum, an important increase on TNF-α and IL-10 expression levels have been shown in adenomas, a precursor lesion of colon cancer [17]; while into tumor, IL-6 and IL-8 increased levels were induced by F. nucleatum. Both IL-6 and IL-8 are proinflammatory cytokines regulated by NF-κB transcription factor, a link between inflammation and cancer; and NF-κB activation has been also shown in colon cancer [18, 36]. Additionally, F. nucleatum infection increased the chemokine CCL20 expression [39], a chemokine related with both colon cancer progression [40], and Th17 + lymphocytes mediated inflammatory response [41]. Likewise, F. nucleatum induced inflammation could be regulated by microRNAs, such as miR-135b; because a correlation between F. nucleatum and miR-135b overexpression in colon cancer patients has been found [42]. So it has suggested that miR-135b could also be used as a biomarker in early detection of colon cancer [43]. However, the role of F. nucleatum in development and progression of colon cancer remains to be understood.
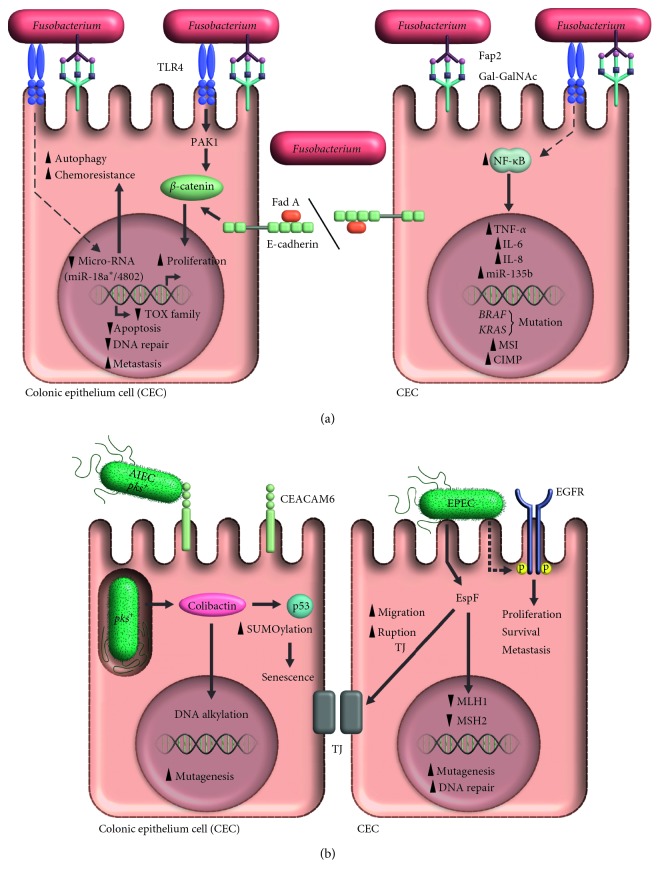
Finally, microsatellite instability (MSI) in colon cancer has been linked to capability to evade immune response by F. nucleatum infected tumor cells [31]. In this fact, CD3+ [32], and T CD4+ lymphocytes subsets were decreased into the tumor after F. nucleatum infection [37], but proportions of T CD8+, CD45RO+, or FOXP3+ lymphocytes subsets were not modified [32]. In addition, the binding of F. nucleatum Fap2 protein with TIGIT [44], a receptor with tyrosine-based inhibitory motif (ITIM) expressed on NK cells [45], leads to an important decreased on lymphocytes infiltration into tumor. This way, tumor is protected from an effective immune cells attack [44]. The proposed mechanisms are summarized in Figure 1(a).
Figure 1.
Oncogenic activity of Fusobacterium nucleatum and Escherichia coli. (a) Gal-GalNAc overexpression in colon cells promotes the recruitment of Fusobacterium nucleatum via the Fap2 protein. After interacting with TLR4, the bacterium activates the protein PAK 1 and in turn, β catenin; the latter can also be activated through the effect of FadA on E-Cadherin. Activation of these signaling pathways promotes cellular proliferation and decreases proteins of the TOX family, which are associated with decreased apoptosis, failures in DNA repair and increased metastases. Likewise, bacterial interaction with TLR4 and its signaling via MYD88, modulates specific microRNAs that activate the autophagy associated with chemotherapy resistance. Also, Fusobacterium nucleatum increases the inflammatory process characterized by the presence of cytokines such as TNF-α, IL-6 and IL-8, that are regulated by the transcription factor NF-κB, whose increased activation has also been documented in colon cancer. Fusobacterium nucleatum has also been shown to be associated with the development of mutations in the genes BRAF and KRAS, microsatellite instability (MSI) and the methylation phenotype in CpG islands (CIMP). (b). The Adherent Invasive Escherichia coli strain (AIEC) colonizes the intestinal epithelium and uses CEACAM6 to invade the cells of the colonic epithelium; once internalized, it produces colibactin, a cyclomodulin encoded by the pks island, that damages DNA by alkylation and promotes the development of mutations. Colibactin also fosters cellular senescence by favoring SUMOylation of p53. Infection with the Enteropathogenic Escherichia coli (EPEC) strain, promotes the autophosphorylation of EGFR, a protein associated with an increase in proliferation, survival and metastases; it also decreases the expression of the DNA repair proteins, MLH1 and MSH2, and favors the rupture of tight junctions, a process involved in the development of metastases. All these EPEC-dependent mechanisms have been associated with the EspF protein.
3. Escherichia coli
Escherichia coli (E. coli) is a Gram-negative bacterium widely distributed in nature, including human intestinal microbiome. The E. coli strains are classified into 5 phylogenetic groups: A, B1, B2, D, and E [46]. The main E. coli strains associated with human disease belong to B2 group and are also related to colon cancer [47, 48]. To date, the role of pathogenic E. coli strains in carcinogenesis is not completely known; however, chronic inflammation in gastrointestinal tract that they promote has been suggested as the trigger mechanism [49]. Because, this chronic inflammation induces pathologies such as Crohn's disease [50], an important risk factor to develop colon cancer [51]. Alternatively, molecular mechanisms induced directly by bacteria have been described. In vitro studies have shown that pathogenic strains such as Adherent-Invasive Escherichia coli (AIEC) and Enteropathogenic Escherichia coli (EPEC), secrete cyclomodulin colibactin [52] and effector protein EspF [53], respectively, which are involved in development and progression of colon cancer. Although the specific mechanisms associated to colon cancer induced by pathogenic E. coli have started to become elucidated recently. The molecular mechanisms associated to colon cancer and pathogenic E. coli are described in Figure 1(b).
3.1. Adherent-Invasive Escherichia coli
The main pathogenic E. coli strain found in tumor tissue from colon cancer patients is Adherent-Invasive Escherichia coli or AIEC [54]. On infection, AIEC binds to CEACAM6 (cellular adhesion receptor associated to carcinoembryonic antigen) [55], which is overexpressed on intestinal epithelial cells of both Crohn's disease and colon cancer patients [56]. To date, it is still unknown what induces overexpression of CEACAM6 on the intestinal epithelium in these patients, although it has been shown that IL-6 is related to induction of CEACAM6 expression [57]. Additionally, it well is known that infection with AIEC stimulates IL-6 production [58]. Taking all these finding together, it is suggested that AIEC could regulate its own infective capacity on intestinal epithelium by both increasing IL-6 production and CEACAM6 expression, and when bacterium has penetrated and invaded the intestinal epithelium, carcinogenesis could be induced through secretion of colibactin, although the true mechanism is not completely known.
3.2. Colibactin and the pks Island
Colibactin is a cyclomodulin encoded in the genotoxic pks island (polyketide island). The pks island has been found in different E. coli strains [59, 60]. Colibactin is a secondary metabolite produced by non-ribosomal peptide synthetase (NRPS)–polyketide synthase (PKS) (NRPS-PKS). Although the synthesis of colibactin is not completely known, it has been shown that a multi-enzymatic complex is required in which several genes of pks island participate [61, 62]. The main role of colibactin in carcinogenesis has been associated with DNA damage [63], by acting as an alkylating agent [64, 65], inducing DNA mutations and promoting tumor development.
On the other hand, because of the synthesis of colibactin has not yet been achieved, which has prevented the understanding of the molecular mechanism of this cyclomodulin, most studies designed to evaluate the role of colibactin in carcinogenesis have been limited to study the pks island function. In vitro infection of cell lines with E. coli pks + strains induced a cell cycle arrest, aneuploidy and tetraploidy [66, 67]; as well as, cell senescence via miR-20a-5P, which inhibits the expression of SUMO-specific protease 1 (SENP-1) [52]. SENP-1 is a protein that induce deSUMOylation of p53 [68], an important transcription factor involved in regulation of cellular senescence and development of colon cancer [69]. On the other hand, the role of the pks island has been evaluated in experimental murine models. The inflammatory environment in mice intestinal epithelium induced upon infection, both spreading of E. coli pks+ and increased risk of colon cancer were produced [49, 70]. In a xenotransplant murine model, infection with E. coli pks + strains lead to a significant increase in tumor size, while infection with E. coli pks–strains do not [52].
3.3. Enteropathogenic Escherichia coli
Enteropathogenic Escherichia coli or EPEC, is the second pathogenic strain of E. coli associated to colon cancer [71, 72], and it has been suggested that EPEC infection might be involved in some molecular pathways involved in colorectal tumorigenesis [72]. In vitro studies have shown that infection with EPEC stimulates macrophage-inhibitory cytokine-1 (MIC-1) production, a cytokine related to metastasis by inducing both, increasing survival and spreading of tumor cells through a GTPase Rho A-dependent pathway [73]. Likewise, autophosphorylation of EGFR receptor, was induced upon EPEC infection [74]; this is a upstream activator of both prosurvival phosphoinositide 3-kinase/Akt and proinflammatory mitogen-activated protein (MAP) kinase pathways. These molecular mechanisms have been associated with colon cancer [75], and poor prognosis in patients [76].
However, it has been shown that EPEC can degrade EGFR receptor via EspF protein [77]; this effector protein is internalized to epithelial cells through the E. coli type III secretion system [78]. Interestingly, this process can be inhibited by EspZ, another protein that is also internalized into epithelial cell through the same secretion system [77]. On the other hand, EspF has also been associated with other mechanism inducing cancer, such as decreasing levels of DNA repair proteins MLH1 and MSH2 (mismatch repair MMR) [53, 71], which are widely related to colon cancer [79]. Further, EspF could also contribute to colon cancer metastasis by promoting detachment and dissemination of tumor cells through rupturing tight junction proteins such as Occludin and Claudin on intestinal epithelium [80].
Finally, other proteins produced by pathogenic E. coli strains and related to carcinogenesis have been studied. These proteins include: (1) Cytolethal distending toxin (CDT), which blocks cell cycle [81], and induces malignant transformation of epithelial cells [82], (2) Cycle inhibiting factor (Cif), which induces nuclear DNA elongation on cells and stimulates DNA synthesis even when infected cells are not actively dividing [83] and (3) Cytotoxic Necrotizing Factor 1 (CNF1), which induces gene transcription and cellular proliferation by GTPases activation [84].
4. Bacteroides fragilis
The bacteroides is a normal inhabitant of human intestine and represent about 30% of intestinal microbiota [85]. These bacteria have a very important role on mucosal immune system development [86], and intestinal homeostasis [87]. Bacteroides fragilis (B. fragilis) is classified within bacteroides species and is an anaerobic Gram-negative bacterium colonizing about 0.5% to 2% of whole human intestine [86, 88, 89]. Two Bacteroides fragilis strain has been described: (a) non-toxigenic B. fragilis or NTBF and (b) toxigenic B. fragilis or ETBF, which is characterized by a 6 kb pathogenicity island encoding to a metalloproteinase, also known as B. fragilis toxin (BFT) or fragilysin [90], of which 3 isoforms have been identified [91].
It has been shown that while NTBF has a protective effect against the development of colitis and colon cancer [92], ETBF has been associated with a wide variety of clinical manifestations ranging from a simple diarrhea to inflammatory bowel disease and colitis [93], both considered as high-risk factors to develop colon cancer. ETBF has already been associated to colon cancer [88], because bacteria has been detected in stool and biopsies obtained from colon cancer patients [94], particularly in early cancer stages [95]. However, a very low proportion of ETBF has been detected in stools from healthy individuals [96].
Although role of enterotoxigenic B. fragilis in development of colon cancer has not been completely described; different studies have shown that carcinogenesis induced by ETBF is through BFT toxin, which is present in ETBF but not in NTBF bacteria strains. BTF toxin is a multifunctional protein; thus, it could induce to tumorigenesis through several mechanisms including activation of c-Myc [97], and consequently an increase on spermine oxidase (SMO) expression [98], an enzyme increasing reactive oxygen species (ROS), which favors cellular injury and carcinogenesis.
Another possible mechanism of ETBF toxin-mediated carcinogenesis, could be through host immune system dysregulation, inducing the recruitment and accumulation of Treg lymphocytes in intestinal lamina in response to bacteria [99], which subsequently suppress the mucosal Th1 response and polarizing to Th17 lymphocytes response [100] by increasing IL-17 secretion [33]. Interestingly, increased levels of IL-17 have been detected on early weeks post-infection, after that; its expression was decreased. However, in APCMin/+ mice, the early and temporary increased on IL-17 was enough to trigger tumorigenesis [101]. On this regards, it has been suggested that activation of Stat3 [102] and NF-κB [103] pathways by immune responding cells and colonic epithelial cells (CECs) may be involved [104]. Furthermore, ETBF also polarizes IL-17-secreting TCRγδ+ T lymphocytes [105], promoting the differentiation and recruitment of myeloid-derived suppressor cells (MDSC) into the tumor [106, 107], which has been associated with a poor prognosis of colon cancer patients [108]. Because IL-17 up regulates CXCL1, CXCL2 and CXCL5 chemokines expression, also has been involved on MDSC recruitment [104]. Additionally, T lymphocyte proliferation is inhibited by high levels of Nitric Oxide (NO), and arginase 1 (Arg1) a potent metabolic enzyme induced and produced by an increase on MDSC population [107], this way several mechanisms of evasion of anti-tumor immune response by tumor cells are generated.
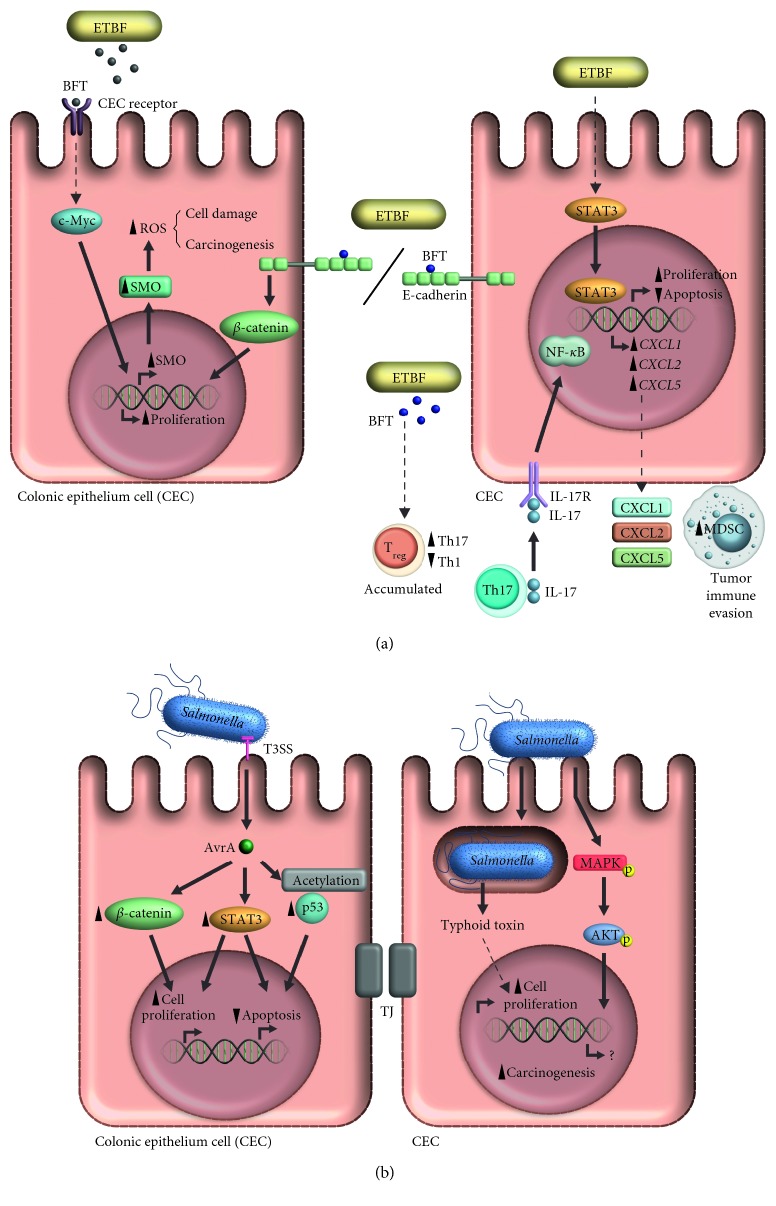
Finally, ETBF could trigger carcinogenesis through β catenin pathway activation, by disrupting the adherent E-cadherin gap junctions, similar than F. nucleatum, [109, 110]. The molecular carcinogenic mechanisms of ETBF are summarized in Figure 2(a).
Figure 2.
Oncogenic activity of Bacteroides fragilis and Salmonella enterica. (a) Enterotoxigenic Bacteroides fragilis (ETBF) stimulates carcinogenesis in colonic epithelium through the BFT toxin. This toxin leads to an increase in reactive oxygen species (ROS) by inducing spermine oxidase expression via c-Myc. Likewise, BFT cuts E-cadherin, thus activating β catenin which stimulates cellular proliferation. BFT also modulates the host's immune response by promoting Treg lymphocytes to polarize the response to Th17 lymphocytes, thus increasing IL-17 secretion which in turn, activates NF-κB in the colonic epithelium; this results in the secretion of the chemokines CXCL1, CXCL2 and CXCL5 that recruit MDSC, thus favoring evasion from the immune response. The presence of ETBF has also been associated with STAT3 activation. (b). Salmonella enterica releases two proteins that promote carcinogenesis: the typhoid toxin that induces cellular proliferation, and the AvrA protein that is internalized via the Type 3 Secretion System(T3SS). AvrA activates the β catenin and STAT3 pathways, and also causes the acetylation of p53. Additionally, Salmonella enterica leads to the activation of the MAPK/AKT pathway. The activation of these pathways promotes an increase in proliferation and cellular differentiation, and decreases apoptosis.
5. Salmonella enterica
Salmonella enterica represents a broad range of bacteria, including serotypes such as Salmonella Typhi (S. Typhi), Salmonella Paratyphi (S. Paratyphi), Salmonella Enteritidis (S. Enteritidis) and Salmonella Typhimurium (S. Typhimurium) [111]. In recent years, development of colon cancer [112], gallbladder cancer [113], and other gastrointestinal tract neoplasms have been associated with Salmonella enterica infection. Also, It has been found that bacteria may modulate host immune response [114], promoting carcinogenesis by both DNA damage and increasing proliferation, as well as cell migration through induction of chronic inflammation [115]. At least, two proteins of Salmonella enterica have been associated with an increased risk of developing colon cancer. The former is typhoid toxin; a cyclomodulin sharing features with the E. coli CDT [116], increasing cellular survival and promoting intestinal dysbiosis [117]. Both mechanisms are involved with development of inflammatory bowel disease and colon cancer [118]. The second protein is AvrA, an effector protein secreted by bacteria through type III Secretion System [119], and it has been detected in stool samples from colon cancer patients [120].
Thus, the main protein of Salmonella enterica associated with carcinogenesis is AvrA. It has been suggested that most important role of AvrA in colon cancer may be related to inflammatory and immune response dysregulation, through several mechanisms such as: inhibition of NF-ΚB signaling pathway [121], inhibition of IL-12, INF-γ and TNF-α secretion [122], inhibition of IL-6 transcription and increasing on IL-10 transcription [123]. On the other hand, AvrA has been associated to tumors on intestinal epithelium through activation of Wnt/β catenin, inducing cellular proliferation [124], by both β catenin phosphorylation (activation) and deubiquitination (decreased degradation) [125]. These mechanism are important in signaling pathway associated with colon cancer development [126]. Likewise, JAK/STAT signaling pathway is activated by AvrA [127], which regulates several mechanisms such as: apoptosis, cellular proliferation and differentiation, as well as inflammatory response, all these important events involved in carcinogenesis [128]. Additionally, the function of p53 transcription factor is affected by AvrA acetyl transferase activity [129], leading to cell cycle arrest and inhibition of apoptosis by decreasing pro-apoptotic proteins (such as Bax), dependent of p53 acetylation [130]. The carcinogenic mechanisms associated to Salmonella enterica are summarized in Figure 2(b).
5.1. Salmonella enterica and Gallbladder Cancer
Gallbladder cancer is the main type of neoplasia affecting the biliary tract. Worldwide, the incidence of this neoplasia is low. Interestingly, it has been shown that gallbladder cancer occurs more frequently in geographic regions with a high incidence of Salmonella infection [113, 131–134]. Therefore, a greater interest has been generated in searching for a possible association between Salmonella infection and development of gallbladder cancer. On this respect, Typhoidal Salmonella serotypes as S. Typhi and S. Paratyphi have been detected in most of the biopsies from patients with gallbladder cancer [113, 135–137], however, DNA traces of Non-typhoidal Salmonella serotypes as S. Typhimurium and S. Choleraesuis have also been found in gallbladder cancer biopsies [135]. These findings have suggested that Salmonella (which may be undetected for years, because it can produce biofilm on cholesterol biliary stones [138]), could represent an important risk factor in development of gallbladder cancer [132], because inflammation and epithelial injury associated to cholelithiasis is induced by Salmonella [139] and cholelithiasis is a common clinical manifestation in most patients with gallbladder cancer [137]. However, the mechanism triggering carcinogenesis by Salmonella enterica in gallbladder is not completely known, but it has been suggested that a chronic inflammation of gallbladder is induced [140], after bacteria arrival to gallbladder from either blood circulation or bile [141].
Additionally, recruitment of some immune cells, including activated macrophages expressing COX-2 is increased upon Salmonella enterica infection [142]. COX-2 is an important enzyme that promotes the development of gastrointestinal tract tumors [143, 144]. Also, bacteria induced inflammation leads to mutations of TP53 gene, increasing the risk of developing gallbladder cancer [145]. Finally, in vitro infection of cell lines and gallbladder organoids with S. Typhimurium, led to malignant transformation though MAPK and AKT signaling pathways activation. Similarly, in vivo activation of these signaling pathways resulted in tumor development in mice [134].
6. Conclusions
Recently, the number of publications referring an association between pathogenic bacteria and development of gastrointestinal tumors, has increased exponentially. The best example and widely reported is Helicobacter pylori and gastric cancer. However, emerging bacteria such as Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica have also been involved in development of cancer, particularly colon cancer.
In this review, it is suggested that infection by pathogenic bacteria may be a high-risk factor associated with the development of neoplasms in gastrointestinal tract. Mechanisms such as, inflammation, modulation and evasion of immune response and activation of signaling pathways, such as the β-catenin pathway; all are potential triggers of carcinogenesis.
The inducing tumor mechanisms can be evaluated in murine models, such as APCMin/+, a specific mice model to study intestinal tumorigenesis [146]. In this experimental model, developing colon cancer mechanisms by Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica have been identified. However, effects of coinfection with these bacteria and tumor development remains to be analyzed, because ETBF and E. coli pks + strains have been found simultaneously in patients with adenomatous polyps, a precursor lesion of colon cancer [147]. Nevertheless, ETBF is a very common bacterium in colon cancer patients but also in healthy individuals [96], so it remains to be elucidated whether ETBF has a role on induction of carcinogenesis. Another possible mechanism through bacteria may trigger cancer is by biofilm. This structure produced by a community of bacteria, more common in ascending colon [148], could increases carcinogenic metabolites concentration, such as polyamines [149], which are related to an important increase on reactive oxygen species. In addition, biofilm has been associated with decreased expression of E-cadherin on colonic epithelial cell, an over activation of IL-6 and Stat3 in epithelial cell [148], all these mechanisms are involved in colon cancer. The mechanisms above described, are used by Fusobacterium nucleatum, Escherichia coli, Bacteroides fragilis and Salmonella enterica. Therefore, further studies are required to understand the specific roles of these four bacteria in development of neoplasms on gastrointestinal tract, specifically in colon cancer.
7. Future Perspectives
Worldwide, colon cancer has very high incidence and mortality. Here we have described that infection with either bacteria such as F. nucleatum, E. coli, B. fragilis or S. enterica represent an important risk factor that promote cell transformation (carcinogenesis). In this regards, detection of promoting carcinogenesis bacterial proteins, such as cyclomodulin, colibactin, BFT, AvrA or EspF may be used as a biomarker for early detection of colon cancer, as it has been proposed for Fap2 [150]. Because early detection of tumor can increase both healing and survival. Moreover, it would generate new and appropriate strategies to block bacterial proteins activity, thus complementing the traditional treatment to neoplasms of gastrointestinal tract.
Acknowledgments
R. L. P. Acknowledges support from CONACYT (CB-2013-01-222446, INFR-2015-01-255341, PN-2015-01-1537) and Federal Funds (HIM-2016-114 SSA. 1333, HIM-2017-041 SSA. 1325).
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bijlsma M. F., Sadanandam A., Tan P., Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nature Reviews Gastroenterology & Hepatology. 2017;14(6):333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 3.Park S.-Y., Wilkens L. R., Setiawan V. W., Monroe K. R., Haiman C. A., Le Marchand L. Alcohol intake and colorectal cancer risk in the multiethnic cohort study. American Journal of Epidemiology. 2019;188(1):67–76. doi: 10.1093/aje/kwy208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanella G., Archibugi L., Stigliano S., Capurso G. Alcohol and gastrointestinal cancers. Current Opinion in Gastroenterology. 2019;35(2):107–113. doi: 10.1097/mog.0000000000000502. [DOI] [PubMed] [Google Scholar]
- 5.Fagunwa I. O., Loughrey M. B., Coleman H. G. Alcohol, smoking and the risk of premalignant and malignant colorectal neoplasms. Best Practice & Research Clinical Gastroenterology. 2017;31(5):561–568. doi: 10.1016/j.bpg.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Triff K., McLean M. W., Callaway E., Goldsby J., Ivanov I., Chapkin R. S. Dietary fat and fiber interact to uniquely modify global histone post-translational epigenetic programming in a rat colon cancer progression model. International Journal of Cancer. 2018;143(6):1402–1415. doi: 10.1002/ijc.31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadella S., Burks J., Al-Sabban A., et al. Dietary fat stimulates pancreatic cancer growth and promotes fibrosis of the tumor microenvironment through the cholecystokinin receptor. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2018;315(5):G699–G712. doi: 10.1152/ajpgi.00123.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arita S., Kinoshita Y., Ushida K., Enomoto A., Inagaki-Ohara K. High-fat diet feeding promotes stemness and precancerous changes in murine gastric mucosa mediated by leptin receptor signaling pathway. Archives of Biochemistry and Biophysics. 2016;610:16–24. doi: 10.1016/j.abb.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Fowler A. J., Richer A. L., Bremner R. M., Inge L. J. A high-fat diet is associated with altered adipokine production and a more aggressive esophageal adenocarcinoma phenotype in vivo. The Journal of Thoracic and Cardiovascular Surgery. 2015;149(4):1185–1191. doi: 10.1016/j.jtcvs.2014.11.076. [DOI] [PubMed] [Google Scholar]
- 10.Tramontano A. C., Nipp R., Mercaldo N. D., Kong C. Y., Schrag D., Hur C. Survival disparities by race and ethnicity in early esophageal cancer. Digestive Diseases and Sciences. 2018;63(11):2880–2888. doi: 10.1007/s10620-018-5238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashktorab H., Kupfer S. S., Brim H., Carethers J. M. Racial disparity in gastrointestinal cancer risk. Gastroenterology. 2017;153(4):910–923. doi: 10.1053/j.gastro.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo G., Zhang Y., Guo P., Wang L., Huang Y., Li K. Global patterns and trends in stomach cancer incidence: age, period and birth cohort analysis. International Journal of Cancer. 2017;141(7):1333–1344. doi: 10.1002/ijc.30835. [DOI] [PubMed] [Google Scholar]
- 13.Øines M., Helsingen L. M., Bretthauer M., Emilsson L. Epidemiology and risk factors of colorectal polyps. Best Practice & Research Clinical Gastroenterology. 2017;31(4):419–424. doi: 10.1016/j.bpg.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Maleki Kakelar H., Barzegari A., Dehghani J., et al. Pathogenicity of Helicobacter pylori in cancer development and impacts of vaccination. Gastric Cancer. 2019;22(1):23–36. doi: 10.1007/s10120-018-0867-1. [DOI] [PubMed] [Google Scholar]
- 15.Tamas K., Walenkamp A. M. E., de Vries E. G. E., et al. Rectal and colon cancer: not just a different anatomic site. Cancer Treatment Reviews. 2015;41(8):671–679. doi: 10.1016/j.ctrv.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Han Y. W. Fusobacterium nucleatum: a commensal-turned pathogen. Current Opinion in Microbiology. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy A. N., Araujo-Perez F., Azcarate-Peril A., Yeh J. J., Sandler R. S., Keku T. O. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053653.e53653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellarin M., Warren R. L., Freeman J. D., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt A. D., Gevers D., Pedamallu C. S., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baselga A. D., Chun E., Robertson L., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Omar J., White A., Ambrose C., McDonald J., Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14(6):301–309. doi: 10.1016/j.anaerobe.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Mima K., Cao Y., Chan A. T., et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clinical and Translational Gastroenterology. 2016;7(11) doi: 10.1038/ctg.2016.53.e200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito M., Kanno S., Nosho K., et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. International Journal of Cancer. 2015;137(6):1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 24.Yoshii H. J., Kim J. H., Bae J. M., Kim H. J., Cho N.-Y., Kang G. H. Prognostic impact of Fusobacterium nucleatum depends on combined tumor location and microsatellite instability status in stage II/III colorectal cancers treated with adjuvant chemotherapy. Journal of Pathology and Translational Medicine. 2019;53(1):40–49. doi: 10.4132/jptm.2018.11.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahara T., Yamamoto E., Suzuki H., et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Research. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.can-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shureiqi J., Emgård J. E. M., Zamir G., et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host & Microbe. 2016;20(2):215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellul H. F., Li L. F., Guo S. H, et al. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Scientific Reports. 2016;6 doi: 10.1038/srep33440.33440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mima K., Nishihara R., Qian Z. R., et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostic T., Guo F., Yu Y., et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y.-Y., Ge Q.-X., Cao J., et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World Journal of Gastroenterology. 2016;22(11):3227–3233. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada T., Zhang X., Mima K., et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunology Research. 2018;6(11):1327–1336. doi: 10.1158/2326-6066.cir-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song K., Sukawa Y., Nishihara R., et al. Fusobacterium nucleatum and T Cells in colorectal carcinoma. JAMA Oncology. 2015;1(5):653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostic S., Rhee K.-J., Albesiano E., et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Medicine. 2009;15(9):1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Housseau A., Amerizadeh F., ShahidSales S., et al. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: rational and progress. Journal of Cellular Biochemistry. 2017;118(8):1979–1983. doi: 10.1002/jcb.25903. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., Peng Y., Yu J., et al. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8(19):31802–31814. doi: 10.18632/oncotarget.15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen T., Li Q., Zhang X., et al. TOX expression decreases with progression of colorectal cancers and is associated with CD4 T-cell density and Fusobacterium nucleatum infection. Human Pathology. 2018;79:93–101. doi: 10.1016/j.humpath.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Yu X., Li Z. TOX gene: a novel target for human cancer gene therapy. American Journal of Cancer Research. 2015;5(12):3516–3524. [PMC free article] [PubMed] [Google Scholar]
- 39.Ye X., Wang R., Bhattacharya R., Boulbes D. R., et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prevention Research. 2017;10(7):398–409. doi: 10.1158/1940-6207.capr-16-0178. [DOI] [PubMed] [Google Scholar]
- 40.Petrosino V. O., Rubie C., Kölsch K., et al. CCR6/CCL20 chemokine expression profile in distinct colorectal malignancies. Scandinavian Journal of Immunology. 2013;78(3):298–305. doi: 10.1111/sji.12087. [DOI] [PubMed] [Google Scholar]
- 41.Chin C.-C., Chen C.-N., Kuo H.-C., et al. Interleukin-17 induces CC chemokine receptor 6 expression and cell migration in colorectal cancer cells. Journal of Cellular Physiology. 2015;230(7):1430–1437. doi: 10.1002/jcp.24796. [DOI] [PubMed] [Google Scholar]
- 42.Proença M. A., Biselli J. M., Succi M., et al. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World Journal of Gastroenterology. 2018;24(47):5351–5365. doi: 10.3748/wjg.v24.i47.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee K., Ferguson L. R. MicroRNA biomarkers predicting risk, initiation and progression of colorectal cancer. World Journal of Gastroenterology. 2016;22(33):7389–7401. doi: 10.3748/wjg.v22.i33.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gur C., Ibrahim Y., Isaacson B., et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shussman Y., Peng H., Sun R., et al. Contribution of inhibitory receptor TIGIT to NK cell education. Journal of Autoimmunity. 2017;81:1–12. doi: 10.1016/j.jaut.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhuri R. R., Henderson I. R. The evolution of the Escherichia coli phylogeny. Infection, Genetics and Evolution. 2012;12(2):214–226. doi: 10.1016/j.meegid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Deshpande N. P., Wilkins M. R., Mitchell H. M., Kaakoush N. O. Novel genetic markers define a subgroup of pathogenic Escherichia coli strains belonging to the B2 phylogenetic group. FEMS Microbiology Letters. 2015;362(22) doi: 10.1093/femsle/fnv193. [DOI] [PubMed] [Google Scholar]
- 48.Swidsinski A., Khilkin M., Kerjaschki D., et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115(2):281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 49.Arthur J. C., Perez-Chanona E., Mühlbauer M., et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes A., Boudeau J., Bulois P., et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 51.Mattar M. C., Lough D., Pishvaian M. J., Charabaty A. Current management of inflammatory bowel disease and colorectal cancer. Gastrointestinal Cancer Research. 2011;4(2):53–61. [PMC free article] [PubMed] [Google Scholar]
- 52.Cougnoux A., Dalmasso G., Martinez R., et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63(12):1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 53.Pezet O. D., Scanlon K. M., Donnenberg M. S. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. MBio. 2013;4(3) doi: 10.1128/mbio.00152-13.e00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buc E., Dubois D., Sauvanet P., et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056964.e56964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barnich N., Carvalho F. A., Glasser A.-L., et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. Journal of Clinical Investigation. 2007;117(6):1566–1574. doi: 10.1172/jci30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darfeuille-Michaud K. S., Kim J.-T., Lee S.-J., et al. Overexpression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in colorectal cancer. Clinica Chimica Acta. 2013;415:12–19. doi: 10.1016/j.cca.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Kim R., Watzig G. H., Tiwari S., Rose-John S., Kalthoff H. Interleukin-6 trans-signaling increases the expression of carcinoembryonic antigen-related cell adhesion molecules 5 and 6 in colorectal cancer cells. BMC Cancer. 2015;15:p. 975. doi: 10.1186/s12885-015-1950-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lapaquette P., Bringer M.-A., Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cellular Microbiology. 2012;14(6):791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 59.Lee E., Lee Y. Prevalence of Escherichia coli carrying pks islands in bacteremia patients. Annals of Laboratory Medicine. 2018;38(3):271–273. doi: 10.3343/alm.2018.38.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson J. R., Johnston B., Kuskowski M. A., Nougayrede J.-P., Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. Journal of Clinical Microbiology. 2008;46(12):3906–3911. doi: 10.1128/jcm.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Lanen S. G. SAM cycles up for colibactin. Nature Chemical Biology. 2017;13(10):1059–1061. doi: 10.1038/nchembio.2479. [DOI] [PubMed] [Google Scholar]
- 62.Zha L., Jiang Y., Henke M. T., et al. Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nature Chemical Biology. 2017;13(10):1063–1065. doi: 10.1038/nchembio.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vizcaino M. I., Crawford J. M. The colibactin warhead crosslinks DNA. Nature Chemistry. 2015;7(5):411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balskus E. P. Colibactin: understanding an elusive gut bacterial genotoxin. Natural Product Reports. 2015;32(11):1534–1540. doi: 10.1039/c5np00091b. [DOI] [PubMed] [Google Scholar]
- 65.Wilson M. R., Jiang Y., Villalta P. W., et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363(6428) doi: 10.1126/science.aar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cuevas-Ramos G., Petit C. R., Marcq I., Boury M., Oswald E., Nougayrede J.-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences. 2010;107(25):11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nougayrede J.-P., Homburg S., Taieb F., et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 68.Yates K. E., Korbel G. A., Shtutman M., Roninson I. B., DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7(5):609–621. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandurangan A. K., Divya T., Kumar K., Dineshbabu V., Velavan B., Sudhandiran G. Colorectal carcinogenesis: insights into the cell death and signal transduction pathways: a review. World Journal of Gastrointestinal Oncology. 2018;10(9):244–259. doi: 10.4251/wjgo.v10.i9.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arthur J. C., Gharaibeh R. Z., Muhlbauer M., et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nature Communications. 2014;5:p. 4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maddocks O. D., Short A. J., Donnenberg M. S., Bader S., Harrison D. J. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005517.e5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magdy A., Elhadidy M., Abd Ellatif M. E., et al. Enteropathogenic Escherichia coli (EPEC): does it have a role in colorectal tumourigenesis? a prospective cohort study. International Journal of Surgery. 2015;18:169–173. doi: 10.1016/j.ijsu.2015.04.077. [DOI] [PubMed] [Google Scholar]
- 73.Choi H. J., Kim J., Do K. H., Park S.-H., Moon Y. Enteropathogenic Escherichia coli-induced macrophage inhibitory cytokine 1 mediates cancer cell survival: an in vitro implication of infection-linked tumor dissemination. Oncogene. 2013;32(41):4960–4969. doi: 10.1038/onc.2012.508. [DOI] [PubMed] [Google Scholar]
- 74.Roxas J. L., Koutsouris A., Viswanathan V. K. Enteropathogenic Escherichia coli-induced epidermal growth factor receptor activation contributes to physiological alterations in intestinal epithelial cells. Infection and Immunity. 2007;75(5):2316–2324. doi: 10.1128/iai.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z., Gao S., Wang D., Song D., Feng Y. Colorectal cancer cells are resistant to anti-EGFR monoclonal antibody through adapted autophagy. American Journal of Translational Research. 2016;8(2):1190–1196. [PMC free article] [PubMed] [Google Scholar]
- 76.De Robertis M., Loiacono L., Fusilli C., et al. Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancer. Clinical Cancer Research. 2017;23(1):159–170. doi: 10.1158/1078-0432.ccr-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia-Foncillas J. L., Ryan K., Vedantam G., Viswanathan V. K. Enteropathogenic Escherichia coli dynamically regulates EGFR signaling in intestinal epithelial cells. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2014;307(3):G374–G380. doi: 10.1152/ajpgi.00312.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elliott S. J., Sperandio V., Giron J. A., et al. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infection and Immunity. 2000;68(11):6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pino M. S., Chung D. C. Microsatellite instability in the management of colorectal cancer. Expert Review of Gastroenterology & Hepatology. 2011;5(3):385–399. doi: 10.1586/egh.11.25. [DOI] [PubMed] [Google Scholar]
- 80.Peralta-Ramirez J., Hernandez J. M., Manning-Cela R., et al. EspF Interacts with nucleation-promoting factors to recruit junctional proteins into pedestals for pedestal maturation and disruption of paracellular permeability. Infection and Immunity. 2008;76(9):3854–3868. doi: 10.1128/iai.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fais T., Delmas J., Serres A., Bonnet R., Dalmasso G. Impact of CDT toxin on human diseases. Toxins. 2016;8(7) doi: 10.3390/toxins8070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Graillot V., Dormoy I., Dupuy J., et al. Genotoxicity of cytolethal distending toxin (CDT) on isogenic human colorectal cell lines: potential promoting effects for colorectal carcinogenesis. Frontiers in Cellular and Infection Microbiology. 2016;6:p. 34. doi: 10.3389/fcimb.2016.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taieb F., Nougayrède J.-P., Watrin C., Samba-Louaka A., Oswald E. Escherichia coli cyclomodulin Cif induces G2arrest of the host cell cycle without activation of the DNA-damage checkpoint-signalling pathway. Cellular Microbiology. 2006;8(12):1910–1921. doi: 10.1111/j.1462-5822.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 84.Fabbri A., Travaglione S., Fiorentini C. Escherichia coli cytotoxic necrotizing factor 1 (CNF1): toxin biology, in vivo applications and therapeutic potential. Toxins. 2010;2(2):283–296. doi: 10.3390/toxins2020282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arumugam M., Raes J., Pelletier E., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wexler H. M. Bacteroides: the good, the bad, and the nitty-gritty. Clinical Microbiology Reviews. 2007;20(4):593–621. doi: 10.1128/cmr.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu J., Gordon J. I. Honor thy symbionts. Proceedings of the National Academy of Sciences. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sears C. L., Geis A. L., Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. Journal of Clinical Investigation. 2014;124(10):4166–4172. doi: 10.1172/jci72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holton J. Enterotoxigenic Bacteroides fragilis. Current Infectious Disease Reports. 2008;10(2):99–104. doi: 10.1007/s11908-008-0018-7. [DOI] [PubMed] [Google Scholar]
- 90.Pierce J. V., Bernstein H. D. Genomic diversity of enterotoxigenic strains of Bacteroides fragilis. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0158171.e0158171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sears C. L. The toxins of Bacteroides fragilis. Toxicon. 2001;39(11):1737–1746. doi: 10.1016/s0041-0101(01)00160-x. [DOI] [PubMed] [Google Scholar]
- 92.Lee Y. K., Mehrabian P., Boyajian S., et al. The protective role of Bacteroides fragilis in a murine model of colitis-associated colorectal cancer. mSphere. 2018;3(6) doi: 10.1128/msphere.00587-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rhee K.-J., Wu S., Wu X., et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infection and Immunity. 2009;77(4):1708–1718. doi: 10.1128/iai.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sartor J. I., Aitchison A., Purcell R. V., Greenlees R., Pearson J. F., Frizelle F. A. Screening for enterotoxigenic Bacteroides fragilis in stool samples. Anaerobe. 2016;40:50–53. doi: 10.1016/j.anaerobe.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 95.Purcell R. V., Pearson J., Aitchison A., Dixon L., Frizelle F. A., Keenan J. I. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171602.e0171602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boleij A., Hechenbleikner E. M., Goodwin A. C., et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clinical Infectious Diseases. 2015;60(2):208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Platz A. V., Krasnov G. S., Lipatova A. V., et al. The dysregulation of polyamine metabolism in colorectal cancer is associated with overexpression of c-Myc and C/EBPbeta rather than enterotoxigenic Bacteroides fragilis infection. Oxidative Medicine and Cellular Longevity. 2016;2016:11. doi: 10.1155/2016/2353560.2353560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goodwin A. C., Shields C. E. D., Wu S., et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proceedings of the National Academy of Sciences. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Casero A. L., Housseau F. Procarcinogenic regulatory T cells in microbial-induced colon cancer. OncoImmunology. 2016;5(4) doi: 10.1080/2162402x.2015.1118601.e1118601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geis A. L., Fan H., Wu X., et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discovery. 2015;5(10):1098–1109. doi: 10.1158/2159-8290.cd-15-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeStefano Shields C. E., Van Meerbeke S. W., Housseau F., et al. Reduction of murine colon tumorigenesis driven by enterotoxigenic Bacteroides fragilis using cefoxitin treatment. Journal of Infectious Diseases. 2016;214(1):122–129. doi: 10.1093/infdis/jiw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wick E. C., Rabizadeh S., Albesiano E., et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflammatory Bowel Diseases. 2014;20(5):821–834. doi: 10.1097/mib.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Housseau S., Powell J., Mathioudakis N., Kane S., Fernandez E., Sears C. L. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor- B pathway. Infection and Immunity. 2004;72(10):5832–5839. doi: 10.1128/iai.72.10.5832-5839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chung L., Thiele Orberg E., Geis A. L., et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host & Microbe. 2018;23(2):203–214. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tam F., Wu S., Wick E. C., et al. Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Research. 2016;76(8):2115–2124. doi: 10.1158/0008-5472.can-15-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iyadorai P., Wu D., Ni C., et al. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40(5):785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen E., Fan H., Tam A. J., et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunology. 2017;10(2):421–433. doi: 10.1038/mi.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ganguly E., Euvrard R., Thibaudin M., et al. Accumulation of MDSC and Th17 cells in patients with metastatic colorectal cancer predicts the efficacy of a FOLFOX-bevacizumab drug treatment regimen. Cancer Research. 2016;76(18):5241–5252. doi: 10.1158/0008-5472.can-15-3164. [DOI] [PubMed] [Google Scholar]
- 109.Bengrine-Lefevre A. G., Shiryaev S. A., Strongin A. Y. Distinct interactions with cellular E-cadherin of the two virulent metalloproteinases encoded by a Bacteroides fragilis pathogenicity island. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113896.e113896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu S., Rhee K. J., Zhang M., Franco A., Sears C. L. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. Journal of Cell Science. 2007;120(11):1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spanò S. Mechanisms of Salmonella Typhi host restriction. Biophysics of Infection. 2016;915:283–294. doi: 10.1007/978-3-319-32189-9_17. [DOI] [PubMed] [Google Scholar]
- 112.Mughini-Gras L., Schaapveld M., Kramers J., et al. Increased colon cancer risk after severe Salmonella infection. PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0189721.e0189721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koshiol J., Wozniak A., Cook P., et al. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Medicine. 2016;5(11):3310–3235. doi: 10.1002/cam4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levine D. L., Chaudhary A., Miller S. I. Salmonellae interactions with host processes. Nature Reviews Microbiology. 2015;13(4):191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kuper H., Adami H.-O., Trichopoulos D. Infections as a major preventable cause of human cancer. Journal of Internal Medicine. 2000;248(3):171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 116.Grasso F., Frisan T. Bacterial genotoxins: merging the DNA damage response into infection biology. Biomolecules. 2015;5(3):1762–1782. doi: 10.3390/biom5031762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Del Bel Belluz L., Guidi R., Pateras I. S., et al. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLOS Pathogens. 2016;12(4) doi: 10.1371/journal.ppat.1005528.e1005528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kang M., Martin A. Microbiome and colorectal cancer: unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Seminars in Immunology. 2017;32:3–13. doi: 10.1016/j.smim.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 119.Ye Z., Petrof E. O., Boone D., Claud E. C., Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. The American Journal of Pathology. 2007;171(3):882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu R., Bosland M., Xia Y., Zhang Y. G., Kato I., Sun J. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget. 2017;8(33):55104–55115. doi: 10.18632/oncotarget.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu X., Lu R., Xia Y., Wu S., Sun J. Eukaryotic signaling pathways targeted by Salmonella effector protein AvrA in intestinal infection in vivo. BMC Microbiology. 2010;10(1):p. 326. doi: 10.1186/1471-2180-10-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu R., Wu S., Liu X., Xia Y., Zhang Y. G., Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010505.e10505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu R., Liu X., Wu S., et al. Consistent activation of the β-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2012;303(10):G1113–G1125. doi: 10.1152/ajpgi.00453.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu X., Lu R., Wu S., Sun J. Salmonella regulation of intestinal stem cells through the Wnt/β-catenin pathway. FEBS Letters. 2010;584(5):911–916. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu R., Wu S., Zhang Y.-g., et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014;3(6):p. e105. doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li Z., Vogelstein B., Kinzler K. W. Phosphorylation of beta-catenin at S33, S37, or T41 can occur in the absence of phosphorylation at T45 in colon cancer cells. Cancer Research. 2003;63(17):5234–5235. [PubMed] [Google Scholar]
- 127.Lu R., Wu S., Zhang Y. G., et al. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer. Neoplasia. 2016;18(5):307–316. doi: 10.1016/j.neo.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu R., Zhang Y. G., Sun J. STAT3 activation in infection and infection-associated cancer. Molecular and Cellular Endocrinology. 2017;451:80–87. doi: 10.1016/j.mce.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu S., Ye Z., Liu X., et al. Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2010;298(5):G784–G794. doi: 10.1152/ajpgi.00526.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamaguchi H., Woods N. T., Piluso L. G., et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. Journal of Biological Chemistry. 2009;284(17):11171–11183. doi: 10.1074/jbc.m809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lazcano-Ponce E. C., Miquel J. F., Munoz N., et al. Epidemiology and molecular pathology of gallbladder cancer. CA: A Cancer Journal for Clinicians. 2001;51(6):349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 132.Nagaraja V., Eslick G. D. Systematic review with meta-analysis: the relationship between chronic Salmonella typhicarrier status and gall-bladder cancer. Alimentary Pharmacology & Therapeutics. 2014;39(8):745–750. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 133.Randi G., Franceschi S., La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. International Journal of Cancer. 2006;118(7):1591–1602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 134.Scanu T., Spaapen R. M., Bakker J. M., et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host & Microbe. 2015;17(6):763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 135.Song P., Barreto S. G., Sahoo B., et al. Non-typhoidal Salmonella DNA traces in gallbladder cancer. Infectious Agents and Cancer. 2016;11:p. 12. doi: 10.1186/s13027-016-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dongol S., Thompson C. N., Clare S., et al. The microbiological and clinical characteristics of invasive Salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047342.e47342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Walawalkar Y. D., Gaind R., Nayak V. Study on Salmonella Typhi occurrence in gallbladder of patients suffering from chronic cholelithiasis-a predisposing factor for carcinoma of gallbladder. Diagnostic Microbiology and Infectious Disease. 2013;77(1):69–73. doi: 10.1016/j.diagmicrobio.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 138.Crawford R. W., Rosales-Reyes R., Ramirez-Aguilar M. d. l. L., Chapa-Azuela O., Alpuche-Aranda C., Gunn J. S. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proceedings of the National Academy of Sciences. 2010;107(9):4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pilgrim C. H. C., Groeschl R. T., Christians K. K., Gamblin T. C. Modern perspectives on factors predisposing to the development of gallbladder cancer. HPB. 2013;15(11):839–844. doi: 10.1111/hpb.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Menendez A., Arena E. T., Guttman J. A., et al. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. The Journal of Infectious Diseases. 2009;200(11):1703–1713. doi: 10.1086/646608. [DOI] [PubMed] [Google Scholar]
- 141.Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. Typhoid fever. New England Journal of Medicine. 2002;347(22):1770–1782. doi: 10.1056/nejmra020201. [DOI] [PubMed] [Google Scholar]
- 142.Carotti S., Guarino M. P. L., Cicala M., et al. Effect of ursodeoxycholic acid on inflammatory infiltrate in gallbladder muscle of cholesterol gallstone patients. Neurogastroenterology & Motility. 2010;22(8):866–e232. doi: 10.1111/j.1365-2982.2010.01510.x. [DOI] [PubMed] [Google Scholar]
- 143.Ye Y., Liu M., Yuan H., et al. COX-2 regulates Snail expression in gastric cancer via the Notch1 signaling pathway. International Journal of Molecular Medicine. 2017;40(2):512–522. doi: 10.3892/ijmm.2017.3011. [DOI] [PubMed] [Google Scholar]
- 144.Sorski L., Melamed R., Matzner P., et al. Reducing liver metastases of colon cancer in the context of extensive and minor surgeries through β-adrenoceptors blockade and COX2 inhibition. Brain, Behavior, and Immunity. 2016;58:91–98. doi: 10.1016/j.bbi.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Espinoza J. A., Bizama C., García P., et al. The inflammatory inception of gallbladder cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2016;1865(2):245–254. doi: 10.1016/j.bbcan.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moser A. R., Luongo C., Gould K. A., McNeley M. K., Shoemaker A. R., Dove W. F. ApcMin: a mouse model for intestinal and mammary tumorigenesis. European Journal of Cancer. 1995;31(7-8):1061–1064. doi: 10.1016/0959-8049(95)00181-h. [DOI] [PubMed] [Google Scholar]
- 147.Dejea C. M., Fathi P., Craig J. M., et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Anders C. M., Wick E. C., Hechenbleikner E. M., et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the National Academy of Sciences. 2014;111(51):18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stein C. H., Dejea C. M., Edler D., et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metabolism. 2015;21(6):891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Uritboonthai L. A., Jr., Afable A. C. F., Belza P. J. O., et al. Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer. Infectious Agents and Cancer. 2018;13:p. 11. doi: 10.1186/s13027-018-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]