Abstract
The collapse of the polypeptide backbone is an integral part of protein folding. Using polyglycine as a probe, we explore the nonequilibrium pathways of protein collapse in water. We find that the collapse depends on the competition between hydration effects and intrapeptide interactions. Once intrapeptide van der Waal interactions dominate, the chain collapses along a nonequilibrium pathway characterized by formation of pearl-necklace-like local clusters as intermediates that eventually coagulate into a single globule. By describing this coarsening through the contact probability as a function of distance along the chain, we extract a time-dependent length scale that grows in a linear fashion. The collapse dynamics is characterized by a dynamical critical exponent z ≈ 0.5 that is much smaller than the values of z = 1–2 reported for nonbiological polymers. This difference in the exponents is explained by the instantaneous formation of intrachain hydrogen bonds and local ordering that may be correlated with the observed fast folding times of proteins.
Introduction
Changing the solvent condition from good to poor renders an extended polymer to undergo a collapse transition by forming a compact globule.1,2 Both experiments3,4 and simulations5,6 indicate that a protein also experiences such a collapse transition while folding into its native state. However, the nonequilibrium dynamics of the collapse of proteins is only poorly understood and an active research topic.7 Most previous studies consider only the hydrophobicity of apolar side chains of amino acids in a protein as the driving force for its collapse.8,9 In the present paper, we focus instead on the contributions by intrapeptide interactions, present even for residues with no or only weakly hydrophobic side chains10−13 where the collapse-driving forces are not necessarily proportional to the exposed surface. Our test system is polyglycine and has been chosen to connect our work with recent studies of homopolymer collapse dynamics14−17 that found nonequilibrium scaling laws as known for generic coarsening phenomena.18 Our hope is to establish such scaling laws also for the collapse of proteins. As a first stride toward this goal, here, we explore the kinetics of the collapse of polyglycine.
The collapse of homopolymers was first described by de Gennes’ seminal “sausage” model,19 but today, the phenomenological “pearl-necklace” picture by Halperin and Goldbart20 is more commonly used, both for flexible14,16,17,21−25 and semiflexible polymer models.26,27 In this picture, the collapse begins with nucleation of small local clusters (of monomers) leading to formation of an interconnected chain of (pseudo-) stable clusters, that is, the pearl-necklace intermediates. These clusters grow by eating up the unclustered monomers from the chain and subsequently coalesce, leading eventually to a single cluster. Finally, monomers within this final cluster rearrange to form a compact globule.
Of central interest in this context is the scaling of the collapse time τc with the degree of polymerization N (the number of monomers). While power-law scaling of the form
| 1 |
where z is the equilibrium dynamic critical exponent, has been firmly established, there is no consensus on the value of z. Molecular dynamics (MD) simulations provide much smaller values (z ≈ 1) than Monte Carlo (MC) simulations (z ≈ 2). This difference is often explained with the presence of hydrodynamics in the MD simulations, but a z value of 1 has been reported recently also for MC simulations.16 The pearl-necklace stage or the cluster growth kinetics can be understood by monitoring the time (t) dependence of the mean cluster size Cs(t), the relevant length scale. By drawing analogy with coarsening ferromagnets, it has been shown that scaling of the form
| 2 |
with growth exponent αc = 1 holds for flexible homopolymers.14,16
Protein collapse is much less understood. While it has been shown by modeling a protein as a semiflexible heteropolymer that the equilibrium scaling of the radius of gyration Rg with N is random-coil-like (Rg ∼ N3/5) in a good solvent and globule-like (Rg ∼ N1/3) in a poor solvent,28,29 there have been few attempts to explore nonequilibrium collapse pathways,30,31 and the corresponding scaling laws are not known. In order to probe the existence of such nonequilibrium scaling laws in protein collapse, we have simulated polyglycine chains (Gly)N of various numbers N of residues. This choice allows us to probe in a systematic way the collapse of the polypeptide chain, considering only homopolymers built from the simplest amino acid, namely, glycine. Our results show that, in water, there is a tug of war between collapse-disfavoring hydration effects and collapse-favoring intrapeptide interactions. For longer chains (N ≥ 15), the intrapeptide interactions win over the hydration effect leading to a collapse, making water in practice a poor solvent. We use these longer polyglycine chains to shed light on the collapse kinetics, with an emphasis on the presence of nonequilibrium scaling laws. Our results from all-atom MD simulations in the NVT ensemble using a hydrodynamics preserving thermostat suggest a collapse mechanism that relies on fast local ordering by formation of pearl-necklace structures, which eventually merge into a single globule. This process is characterized by a dynamic critical exponent z ≈ 0.5 much smaller than the exponents z = 1–2 observed for nonbiological polymers, and we speculate that this quicker local ordering during collapse enables the fast folding times seen in proteins.
Model and Methods
We construct (Gly)N molecules with hydrogenated N-terminus (−NH2) and C-terminus (−COOH). All-atom MD simulations are performed using standard GROMACS 5.0.2 tools, while CHARMM22 with CMAP corrections32,33 is used for interactions between the atoms. For studying the collapse dynamics, we first prepare an extended chain in the random-coil phase at 1500 K. This follows solvation of this extended chain in a simple cubic box with water (modeled by the TIP3P model34). The final MD run is performed at the desired quench temperature Tq = 290 K, which is lower than 310 K, roughly the collapse transition temperature of (Gly)N in water. The size of the box and the number of water molecules, of course, are dependent on N and are so chosen that the number density of water molecules is the same for all N. For the smallest N, that is, for N = 20, the default box size is 4.2 nm. Subsequently, the box sizes for longer chains are determined using the scaling relation Rg ∼ N3/5 for the radius of gyration discussed below. The size of the boxes should not have much role in the collapse provided that the two ends of the chain do not interact while using the periodic boundary condition. However, the number density of water molecules is supposed to play a role, which we keep the same for all N. For N = 20, the total number of water molecules used is 2000 giving a number density of 32 per nm–3, which is maintained for all N. After the solvation, we run our MD simulations using the velocity Verlet integration scheme with a time step δt of 2 fs in the NVT ensemble using the Nosé–Hoover thermostat that conserves linear momentum and thus is believed to be sufficient for preserving hydrodynamic effects.35 Here, we use chains of lengths N ∈ [20,50,75,100,150,200], and for each N, we generate 50 different initial configurations in the random-coil-like phase, except for N = 200 where this number is 15. Independent simulations starting from these different initial conditions are performed for every N. All these simulations are run up to time tf, which is 10 ns for N = 20, 20 ns for N ∈ [50,150], and 25 ns for N = 200. Unless otherwise mentioned, the results presented subsequently are all averaged over the aforesaid number of independent simulations for respective N.
In the following, we briefly discuss three standard observables that we use for the analyses of our simulation data. (i) The squared radius of gyration for a polymer of length N (the number of monomers) is calculated as
| 3 |
For (Gly)N, the chain length is determined from N, the number of residues or repeating units that contain a fixed set of atoms. Thus, Rg2 for (Gly)N is calculated considering all the atoms present in all the residues. However, the scaling can still be checked in terms N, as is done here. (ii) The static structure factor is calculated as
| 4 |
where  is the scattering wave vector. In general, S(
is the scattering wave vector. In general, S( ) is the Fourier transform of the radial
distribution function and is directly comparable with the experimentally
observed X-ray scattering. For an isotropic system, as in the case
here, everything depends on q = |
) is the Fourier transform of the radial
distribution function and is directly comparable with the experimentally
observed X-ray scattering. For an isotropic system, as in the case
here, everything depends on q = |  |. As explained above, in the case for measuring Rg2, for S(
|. As explained above, in the case for measuring Rg2, for S( ), too, we use all the atoms in all the
residues. (iii) Hydrogen bonds are calculated using the standard GROMACS
tool gmx hbond. It considers all possible donors and acceptors and
decides for the existence of a hydrogen bond if the distance between
them is less than 0.35 nm and the hydrogen-donor-acceptor angle is
less than 30°.
), too, we use all the atoms in all the
residues. (iii) Hydrogen bonds are calculated using the standard GROMACS
tool gmx hbond. It considers all possible donors and acceptors and
decides for the existence of a hydrogen bond if the distance between
them is less than 0.35 nm and the hydrogen-donor-acceptor angle is
less than 30°.
Results
Evolution of Short Chains
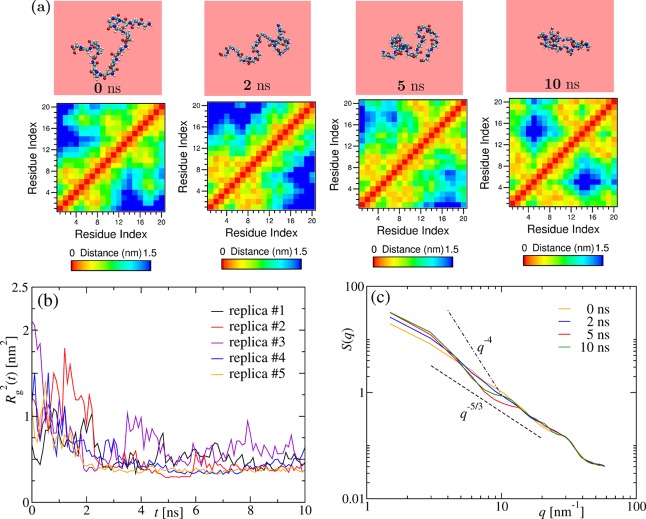
We begin our analysis with a rather short chain, that is, (Gly)20. The time evolution snapshots during the collapse in water at a temperature Tq = 290 K, well below the corresponding collapse transition temperature, are shown in Figure 1a. In a protein, collapse leads eventually to folding characterized by formation of distinct native contacts among the residues. We show for this reason in the lower panel the residue contact maps where we define two residues as being in contact if they are within a distance of rc = 1.5 nm. The red stripe along the diagonals depicts the self-contacts. The size of the extended (Gly)20 chain is 2.0 nm; thus, almost all the mutual distances between the residues fall under rc. This makes it difficult to capture segregation or formation of any local structures on length scales comparable to rc. Only late in the trajectories do we find a signature for loop formation, which is also apparent in the snapshot at t = 10 ns. The emergence of such a loop is due to a competition between the hydration effects and the intrapeptide interactions leading to residue–residue contacts along the chain, although there are trapped water molecules. The interplay can be deduced from the nonmonotonous behavior of the squared radius of gyration Rg2 as a function of time in Figure 1b, obtained from five different replicas chosen randomly out of the total 50 independent simulations. Note that, for all the cases, Rg2 decays eventually to the equilibrium value.
Figure 1.
Time evolution of a short polypeptide. (a) The
upper row shows
typical representative snapshots from the time evolution [of replica
#1 shown in panel (b)] for the collapse of the (Gly)20 chain
in water at Tq = 290 K, starting from
an extended state at t = 0 ns. The lower row shows
the corresponding residue contact maps where two residues along the
chain are in contact if the distance between them is less than 1.5
nm. (b) Time dependence of the squared radius of gyration Rg2(t) shown for
five different replicas chosen randomly out of the total 50 independent
simulations for (Gly)20. (c) Illustration of the structural
evolution of the chain during the collapse shown via structure factors S(q) averaged over 50 independent simulations
for (Gly)20, as a function of the modulus q = | | of the wave vector
| of the wave vector  , at four different times
as indicated.
The dashed lines with power-law decay exponents 5/3 and 4 correspond
to the expected behavior for an extended chain and crumpled globule,
respectively.
, at four different times
as indicated.
The dashed lines with power-law decay exponents 5/3 and 4 correspond
to the expected behavior for an extended chain and crumpled globule,
respectively.
In order to probe further the structural evolution of the chain along the collapse of (Gly)20, we calculate the static structure factor S(q) at different times. Figure 1c shows S(q) for the times corresponding to the snapshots. At t = 0 ns, within the range q ∈ [3,30] nm–1, the chain can be described as an extended coil with S(q) ∼ q–1/ν,36 where ν = 3/5 is the critical (Flory) exponent describing the scaling of Rg ∼ Nν for a self-avoiding polymer. With time, the decay exponent should increase from −5/3 and is expected to approach −4 in order to be consistent with the globule-like behavior of S(q) ∼ q–4.36 Although the slope in our data in Figure 2c gradually increases with time, it does not appear to approach −4. This again could be due to the still ongoing interplay between the hydration effect and the intrapeptide interactions that hinders the chain to form a compact globule; however, extending the simulations up to 20 ns does not change the overall behavior. Similar observations are made for all systems (Gly)N having a chain length of N < 50 residue units.
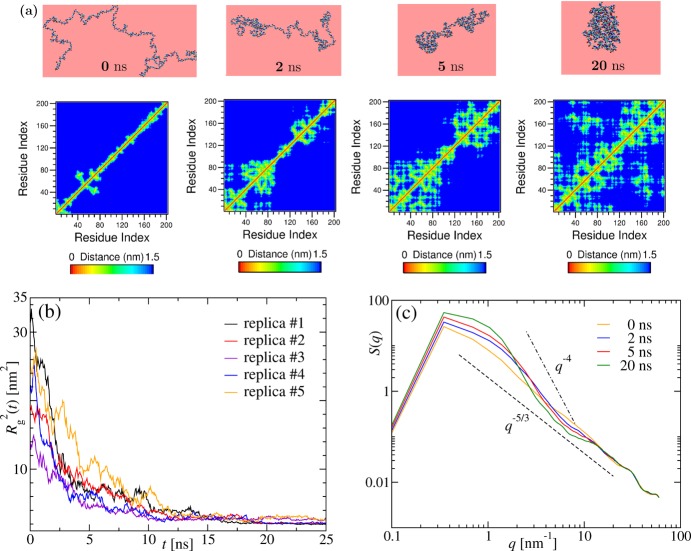
Figure 2.
Pearl-necklace formation during collapse of a long chain. (a) Same as in Figure 1a but for (Gly)200 and correspondingly at different times, as mentioned. (b) Time dependence of the squared radius of gyration Rg2(t) obtained from five different replicas chosen randomly out of the total 15 independent simulations for (Gly)200. (c) Structure factors S(q) averaged over the total 15 independent simulations for (Gly)200 at times indicated in the figure. The dashed lines have the same meaning as in Figure 1c but are plotted with different prefactors.
Evolution of Long Chains
For longer chains, the collapse is more pronounced, and we finally encounter characteristic features reminiscent of the homopolymer collapse. For instance, in the upper row of Figure 2a, we present snapshots of the collapse of (Gly)200 at Tq = 290 K. The sequence of these snapshots demonstrates a process that starts with local ordering of the residues along the chain. These local structures later merge with each other before finally forming a single globule at t = 20 ns. The emergence of these local arrangements is similar to the formation of local clusters in the pearl-necklace picture of homopolymer collapse.14,16,17,20,21 The resemblance becomes even more obvious when looking at the corresponding contact maps in the lower row of Figure 2a. The box-like clustering along the diagonal indicates formation of pearls along the chain (see particularly at t = 2 and 5 ns) that are reminiscent of the ones observed during the collapse of a semiflexible homopolymer in ref (27). However, we do not see the antiparallel hairpins that are associated with the diamond-shaped internal order within these boxes. An idea about the variation of these contact maps for different independent simulations and other chain lengths can be obtained from the Supporting Information.
In order to check for the presence of a competition between hydration effects and the intrapeptide interactions, we probe again the time dependence of Rg2 as measured in five independent simulations. Data are presented in Figure 2b. Unlike for the shorter (Gly)20 chain, the radius of gyration is now monotonically decreasing. This can be explained by the assumption that, for longer chains, the intrachain interactions overcome the hydration effects. A similar picture emerges from Figure 2c. The plots of the structure factor S(q) as a function of time demonstrate how the extended coil behavior of S(q) ∼ q–5/3 at t = 0 ns gradually changes to a globule-like behavior of S(q) ∼ q–4 at t = 20 ns.
Relaxation Dynamics
Next, we analyze the number of intramolecular (protein–protein npp) and intermolecular (protein–water npw) hydrogen (H) bonds. The time dependence of npp measured for different N and normalized by the respective values at tf (the maximum time up to which the simulations are run; for details see the Model and Methods section) is plotted in the main frame of Figure 3a. Data for all N in Figure 3a attain a saturation value of 1 at the same time, demonstrating a reasonable overlap of the data. Similar observations can be made in panel (b), which shows that the decay of npw(t)/npw(0) to the saturation value happens at almost the same time for different N, leading again to nicely overlapping curves. In the inset of Figure 3a, the time dependence of npp for (Gly)20 is nonmonotonous, whereas the npw data in the inset of Figure 3b exhibit a jump at an early time before reaching saturation. This again confirms the hydration effects for smaller chains. The overlap of the hydrogen-bond kinetics for large N (>20) implies that the collapse dynamics, that is, the presence of any scaling of the collapse time with respect to the chain length, shall not depend on the intrapeptide hydrogen bonds. Equilibrium studies, too, suggest that collapse is driven by the intrapeptide van der Waals interactions rather than the hydrogen-bond formation.7
Figure 3.
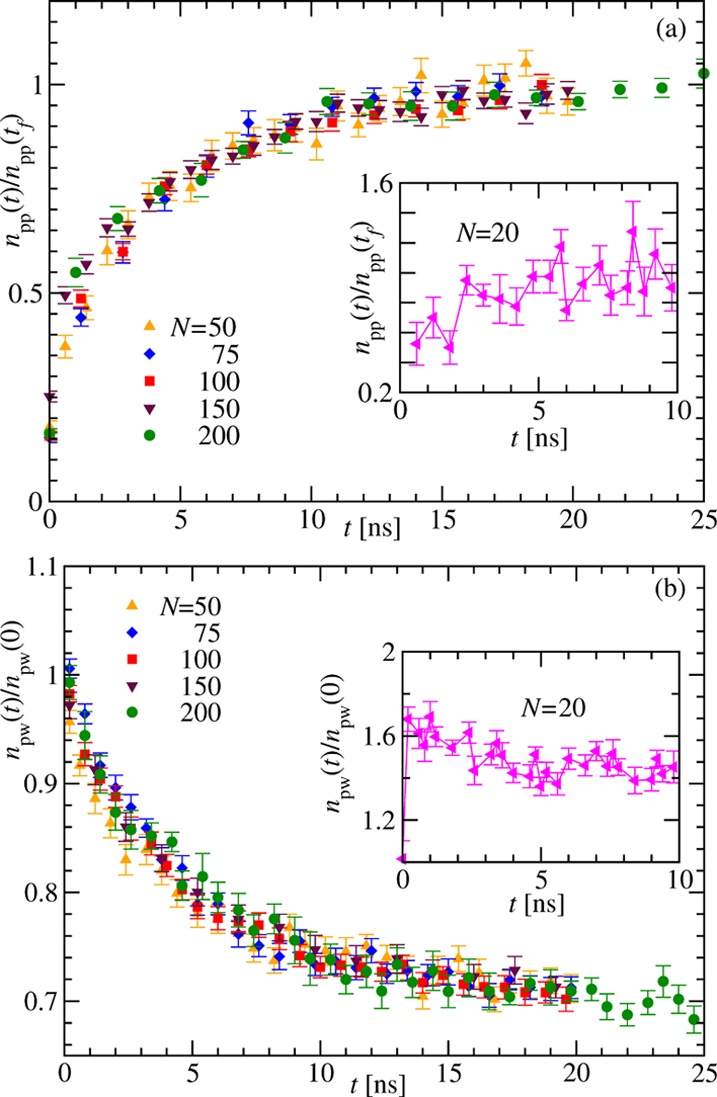
Kinetics of H bonding. (a) Time dependence of the number of protein–protein hydrogen bonds npp(t) during the collapse of (Gly)N for different N. To make the curves fall within the same scale, the data is normalized with npp(tf); tf is the maximum run time the simulations are done. The inset shows the time dependence of npp(t) for (Gly)20. (b) Same as in panel (a) but for the number of protein–water hydrogen bonds npw(t). Here, the normalization is done with npw(0). The inset shows the variation of npw(t) with time for (Gly)20. The error bars in all the plots here correspond to the standard error of the mean calculated while averaging the data obtained from a number (mentioned in the Model and Methods section) of independent simulations.
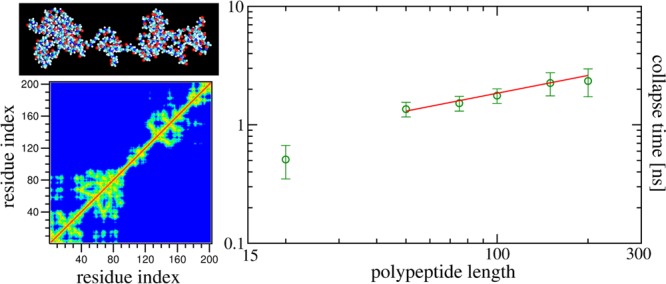
The overlap of the hydrogen-bond data does not allow one to calculate the collapse time τc from the time evolution of this quantity. More suitable for this purpose is the decay of the average squared radius of gyration Rg2 depicted in Figure 4a. The nonoverlapping data are consistent with the respective solid lines obtained from the previously proposed fit16,17
| 5 |
where b0 corresponds to the value of Rg2(t) in the collapsed
state and b1 and β are associated
nontrivial fitting
parameters. The obtained values of β (see the inset of Figure 4a) indicate a very
weak dependence on N, similar to the case of the
earlier studied collapse of synthetic homopolymers.16 Although the above fit yields a collapse time τc, more accurate estimates can be calculated from the time
when Rg2(t) has decayed to 50% of its total decay, that is, 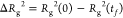 . We plot the measured values of τc for different chain length N (including N = 20) in Figure 4b to check for scaling of the form in eq 1. Due to the competition between hydration
effects and intrapeptide interactions that dominate for smaller N, one expects distinct scaling forms for small and large N. Our data indeed hint at the existence of two such scaling
regions. Especially interesting is the consistency of our data for
large N with the solid line having z = 0.5. This exponent suggests that the dynamics is faster than the
one observed in MC simulations of nonbiological homopolymers.16 Surprisingly, it is even faster than in the
case of homopolymer collapse in the presence of hydrodynamics.22,23 We conjecture that the more rapid collapse is due to the almost
instantaneous presence of intrachain hydrogen bonds that hasten local
ordering; that is, once the intrachain hydrogen bonds are formed,
the Cα atoms along the backbone come closer to form clusters,
which in turn coalesce to finally form a single globule. This latter
phase of the collapse is guided by the diffusive dynamics, which is
certainly N-dependent. However, in combination with
the initial N-independent phase of local pearl-necklace
formation, one observes an overall collapse time that is weakly dependent
on N. Simulations of longer chains would be desirable
to confirm the value of z = 0.5 and the super-fast
collapse mechanism in hydrogen-bonded polymers; however, such simulations
were computationally too costly to be considered in the present study.
. We plot the measured values of τc for different chain length N (including N = 20) in Figure 4b to check for scaling of the form in eq 1. Due to the competition between hydration
effects and intrapeptide interactions that dominate for smaller N, one expects distinct scaling forms for small and large N. Our data indeed hint at the existence of two such scaling
regions. Especially interesting is the consistency of our data for
large N with the solid line having z = 0.5. This exponent suggests that the dynamics is faster than the
one observed in MC simulations of nonbiological homopolymers.16 Surprisingly, it is even faster than in the
case of homopolymer collapse in the presence of hydrodynamics.22,23 We conjecture that the more rapid collapse is due to the almost
instantaneous presence of intrachain hydrogen bonds that hasten local
ordering; that is, once the intrachain hydrogen bonds are formed,
the Cα atoms along the backbone come closer to form clusters,
which in turn coalesce to finally form a single globule. This latter
phase of the collapse is guided by the diffusive dynamics, which is
certainly N-dependent. However, in combination with
the initial N-independent phase of local pearl-necklace
formation, one observes an overall collapse time that is weakly dependent
on N. Simulations of longer chains would be desirable
to confirm the value of z = 0.5 and the super-fast
collapse mechanism in hydrogen-bonded polymers; however, such simulations
were computationally too costly to be considered in the present study.
Figure 4.
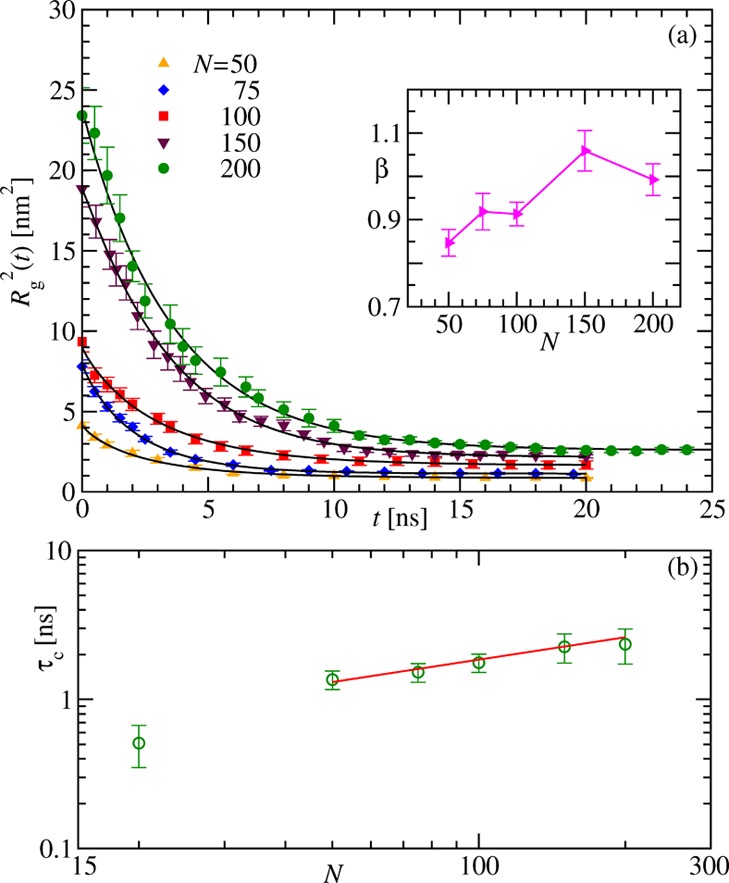
Scaling of the collapse time. (a) Variation of the average squared radius of gyration Rg2(t) with time for different chain lengths as indicated. The solid black lines are respective fits using eq 5, and the corresponding β obtained is shown as a function of N in the inset. (b) Dependence of the collapse times τc extracted from the time decay of Rg2 on the number of residues N. The solid line represents the behavior τc ∼ Nz with z = 0.5. For all the data shown in the main frames, the error bars correspond to the standard error of the mean, while in the inset, they result from the fitting exercise.
Cluster Growth Kinetics
In a final step, we want to
quantify the coarsening kinetics of the pearls observed in Figure 2a. A measure of the
relevant length scale, that is, the mean cluster or pearl size Cs(t), can be obtained from
a box plot analysis of the contact maps.27 Conjecturing that the collapse is driven by the intrapeptide van
der Waals attraction of the backbone, we extract Cs(t) from an analysis of the contact
probability P(cij) as
a function of the contour distance cij = | i – j| between any
two Cα atoms at the ith and jth positions along the chain.37 Two Cα
atoms are said to have contact if they are within a cutoff distance rc. Using rc = 2.5
nm, we show in Figure 5a values of P(cij) calculated
at different times during the collapse of (Gly)200. These
contact probabilities indicate indeed a growing length scale as their
decay slows with time. At the beginning, for t =
0 ns, the chain is in the extended state and P(cij) decays according to a power law 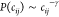 with an exponent γ = 1.5, as expected
in a good solvent.38 As time progresses,
this power-law behavior appears at larger cij after crossing over from a plateau-like behavior for small cij, which marks the local ordering along the
chain. For any reasonable choice of rc, the form of the curves stays unchanged as demonstrated in Figure 5b. Similarly, the
form of the curve also does not depend on the chain length N as illustrated in the inset of Figure 5b where we use rc = 2.5 nm and choose the point in time t = 2 ns.
Results analogous to Figure 5a for N = 100 and N = 150
are presented in the Supporting Information.
with an exponent γ = 1.5, as expected
in a good solvent.38 As time progresses,
this power-law behavior appears at larger cij after crossing over from a plateau-like behavior for small cij, which marks the local ordering along the
chain. For any reasonable choice of rc, the form of the curves stays unchanged as demonstrated in Figure 5b. Similarly, the
form of the curve also does not depend on the chain length N as illustrated in the inset of Figure 5b where we use rc = 2.5 nm and choose the point in time t = 2 ns.
Results analogous to Figure 5a for N = 100 and N = 150
are presented in the Supporting Information.
Figure 5.
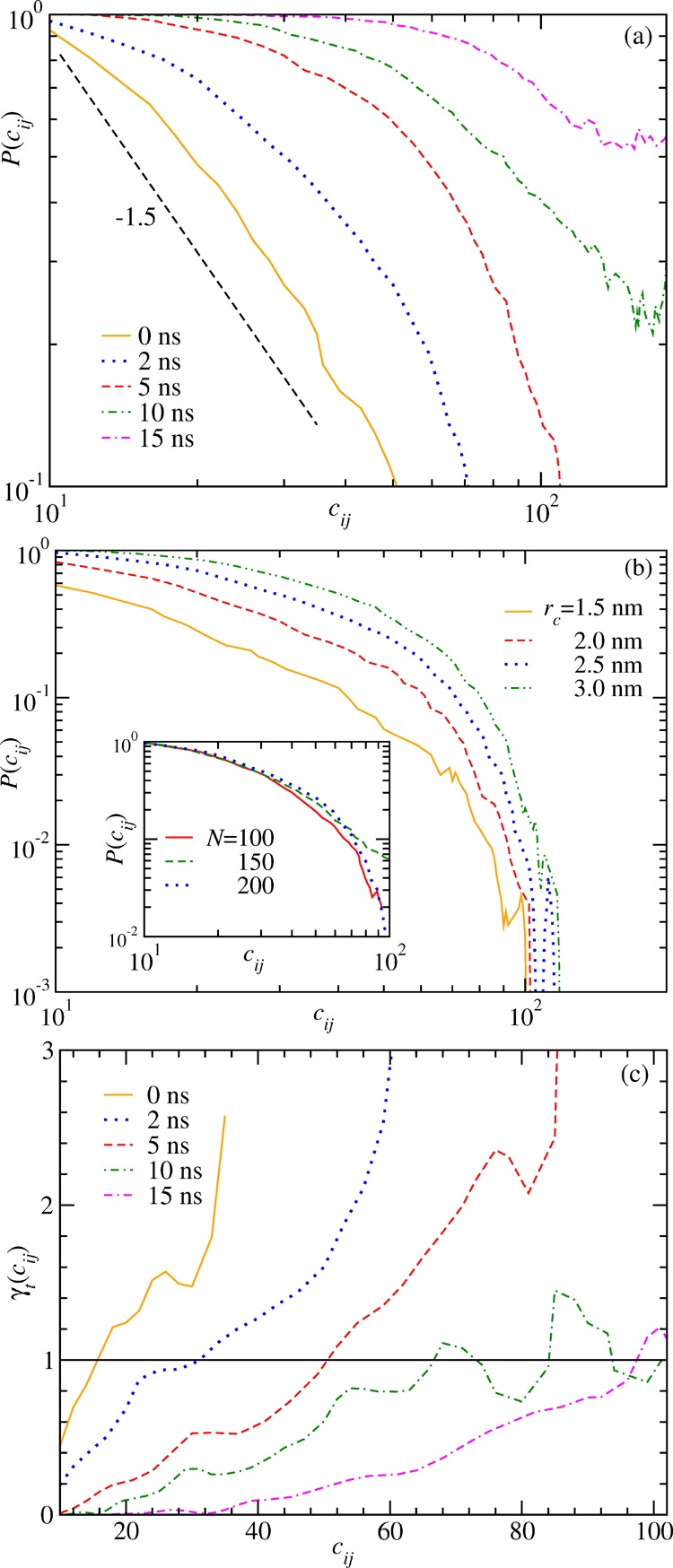
Contact probabilities along the contour of the chain. (a) Contact probability P(cij) calculated using the cutoff rc = 2.5 nm as a function of the distance cij along the chain at five different times during collapse of (Gly)200. The dashed line there represents a power-law decay with an exponent γ = 1.5 as expected in a good solvent.38 (b) P(cij) at a fixed time t = 2 ns using different rc as indicated, demonstrating the consistency of the proportionality behavior of the estimated contact probability. The inset shows P(cij) at t = 2 ns using rc = 2.5 nm for different N. (c) Discrete slope γt obtained from eq 6 as a function of cij for the times presented in panel (a). The solid line is for γt = 1, marking the crossover value that gives the measure of the length scale Cs(t).
The crossover point in the decay of P(cij) as a function of cij is estimated from the discrete local slope calculated as37
| 6 |
Plots of γt(cij) as a function of cij are shown Figure 5c for the data presented in Figure 5a. The crossing of the data with the γt = 1 line happens at larger cij as t increases, and thus, this crossover point gives a measure of the pearl size Cs(t). The obtained Cs(t) values for three different N are shown as a function of t on a double-log scale in the main frame of Figure 6. The flattening of the data for very large t is due to finite-size effects when no more ordering is possible due to the eventual formation of a single globule. For large t, before hitting finite-size effects, the growth resembles a power law Cs(t) = ANtαc, where the amplitude AN depends still on the chain length N as the considered N is not large enough. Hence, P(cij) calculated using the same rc will overlap with each other, a fact that is demonstrated in the inset of Figure 5b. However, since their form stays invariant in the large t regime, they apparently follow the same power law. To estimate the exponent αc of the power-law growth (eq 2) on a double-log scale, one needs to have data ranging over several decades, which is not the case with our data for Cs(t). Thus, to distinguish the consistency of the data with either αc = 2/3 or αc = 1 behavior as shown in the main frame of Figure 6 by the dashed and the solid lines, respectively, is not so easy. In such cases, instead of eq 2, it is advantageous to describe the growth as
| 7 |
by considering a crossover time t0 and cluster size Cs(t0). This approach, originally developed for ferromagnets,39 was already necessary in our earlier work for describing the collapse of nonbiological homopolymers.14,16,17 Using the transformation
| 8 |
one finds 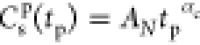 , with the shifted time tp = t – t0. If αc = 1 in eq 7, the transformation in eq 8 is invariant under any choice of t0 in the post-crossover regime. This is demonstrated
in the inset of Figure 4 where we plot Csp(tp) as a function
of tp on a log–log scale for two
different choices of t0 as mentioned.
The data for both the cases are consistent with a linear power-law
behavior
, with the shifted time tp = t – t0. If αc = 1 in eq 7, the transformation in eq 8 is invariant under any choice of t0 in the post-crossover regime. This is demonstrated
in the inset of Figure 4 where we plot Csp(tp) as a function
of tp on a log–log scale for two
different choices of t0 as mentioned.
The data for both the cases are consistent with a linear power-law
behavior 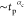 having αc = 1 as represented
by the solid line there. This further consolidates our finding of
a linear growth of clusters.
having αc = 1 as represented
by the solid line there. This further consolidates our finding of
a linear growth of clusters.
Figure 6.
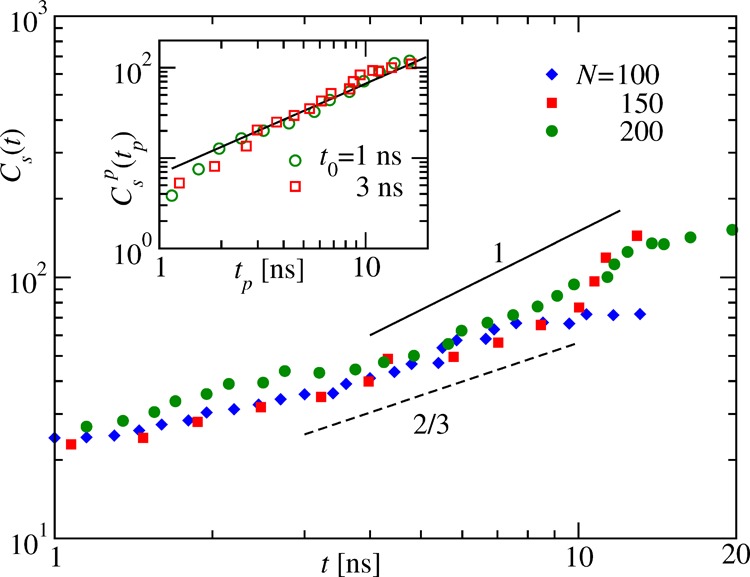
Cluster growth during the collapse. The main
frame shows the growth
of the mean cluster or pearl size Cs(t) with time for different N. The solid
and the dashed lines represent power-law behavior Cs(t) ∼ tα with α = 1 and 2/3, respectively. The inset shows
the plot of Csp(tp) as a function
of the shifted time tp = t – t0 for N =
200 with two different choices of t0.
The solid line there represents a linear power-law behavior 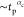 with αc = 1.
with αc = 1.
Discussion
In summary, we have investigated the nonequilibrium pathways by which polyglycine [(Gly)N] collapses in water. For short chains, the pathway has few noticeable features and is driven by the competition between the hydration of the peptide, opposing the collapse, and the intrapeptide attractions, favoring the collapse.7 For chains with N > 20, the importance of hydration effects decreases, and the kinetics of hydrogen bonds indicates that van der Waals interactions of the backbone dominate7 and drive the collapse. The nonequilibrium intermediates seen during the collapse exhibit local ordering or clustering that is analogous to the phenomenological pearl-necklace picture known to be valid for the earlier studied coarse-grained homopolymer models.20 Using the contact probability of the Cα atoms in the backbone, we extract a relevant dynamic length scale, that is, cluster size Cs, that as in simple homopolymer models grows linearly with time (Cs ∼ t).16 We believe that this linear growth is a result of the Brownian motion of the clusters and subsequent coalescence as in the case of droplet growth in fluids.40
Especially intriguing is that the scaling of the collapse time with the length of the chain indicates a faster dynamics, with a critical exponent z ≈ 0.5 instead of z ≈ 1 that was seen in earlier homopolymer collapse studies,22,23 which considered simplified models describing non-hydrogen-bonded polymers such as polyethylene and polystyrene.41 The smaller exponent found in this study may be connected with a mechanism that allows a more rapid collapse in amino acid based polymers than seen in nonbiological homopolymers such as poly(N-isoporpylacrylamide) and polystyrene where collapse times of 300 ms up to 350 s have been reported,42,43 respectively. We conjecture that the smaller exponent z is characteristic of collapse transitions in amino acid based polymers where the presence of intrachain hydrogen bonding immediately seeds (transient) local ordering, while in non-hydrogen-bonded polymers such local ordering only happens as a consequence of diffusive motion.
This connection of rapid collapse with a quick appearance of local ordering would also have implications for possible folding mechanisms and may explain the fast folding times of proteins (typically in the μs–ms range for proteins with 100–200 residues). While the topology and roughness of protein folding funnels vary with sequence, making it difficult to establish the scaling laws observed for amino acid based homopolymers, we expect to see a comparable mechanism at work during protein folding. In this picture, the formation of the intermediate and transient secondary structures and other local ordering hasten the collapse of the protein chain, which in turn enables formation of the long-range contacts that stabilize the final fold. While the latter part would be modulated by the protein sequence, the fast collapse itself appears to be a polymer property involving only the backbone.
In order to test this conjecture, one would need to repeat first our above investigation for the other 19 amino acids verifying whether the scenario observed in this paper for polyglycine applies to all amino acids. Such studies and their extension to protein-like heteropolymers are beyond the scope of our current paper; however, the presented results demonstrate already that our approach provides a general platform to understand various conformational transitions that occur in biomolecules via local ordering. Another example would be, for instance, the helix–coil transition of polyalanine where the short-time dynamics has already been explored44,45 or the study of two-time properties such as aging and dynamical scaling in collapse and folding15,17 Hence, the main advantage of our approach is that it allows for a clearer separation between polymer properties and sequence-dependent factors in folding and structural changes of proteins.
Acknowledgments
This project was funded by the Deutsche Forschungsgemeinschaft
(DFG, German Research Foundation) under project nos. 189 853 844 –
SFB/TRR 102 (project B04) and JA 483/33-1 and the National Institutes
of Health (NIH) under grants GM120578 and GM120634. It was further
supported by the Deutsch-Französische Hochschule (DFH-UFA)
through the Doctoral College “ ” under grant no. CDFA-02-07 and
the Leipzig Graduate School of Natural Sciences “BuildMoNa”.
U.H.E.H. thanks the Institut für Theoretische Physik and especially
the Janke group for kind hospitality during his sabbatical stay at
Universität Leipzig.
” under grant no. CDFA-02-07 and
the Leipzig Graduate School of Natural Sciences “BuildMoNa”.
U.H.E.H. thanks the Institut für Theoretische Physik and especially
the Janke group for kind hospitality during his sabbatical stay at
Universität Leipzig.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.macromol.9b00562.
Contact maps from four further independent simulations for chain lengths 20 and 200; contact maps for chain lengths 50, 75, 100, and 150; contact probabilities and their time-dependent discrete local slopes for chain lengths 100 and 150 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Stockmayer W. H. Problems of the statistical thermodynamics of dilute polymer solutions. Macromol. Chem. Phys. 1960, 35, 54–74. 10.1002/macp.1960.020350103. [DOI] [Google Scholar]
- Nishio I.; Sun S.-T.; Swislow G.; Tanaka T. First observation of the coil-globule transition in a single polymer chain. Nature 1979, 281, 208–209. 10.1038/281208a0. [DOI] [Google Scholar]
- Pollack L.; Tate M. W.; Finnefrock A. C.; Kalidas C.; Trotter S.; Darnton N. C.; Lurio L.; Austin R. H.; Batt C. A.; Gruner S. M.; Mochrie S. G. J. Time resolved collapse of a folding protein observed with small angle x-ray scattering. Phys. Rev. Lett. 2001, 86, 4962–4965. 10.1103/PhysRevLett.86.4962. [DOI] [PubMed] [Google Scholar]
- Sadqi M.; Lapidus L. J.; Muñoz V. How fast is protein hydrophobic collapse?. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 12117–12122. 10.1073/pnas.2033863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. J.; Thirumalai D. Kinetics and thermodynamics of folding in model proteins. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 6369–6372. 10.1073/pnas.90.13.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G.; Thirumalai D. Collapse precedes folding in denaturant-dependent assembly of ubiquitin. J. Phys. Chem. B 2017, 121, 995–1009. 10.1021/acs.jpcb.6b13100. [DOI] [PubMed] [Google Scholar]
- Asthagiri D.; Karandur D.; Tomar D. S.; Pettitt B. M. Intramolecular interactions overcome hydration to drive the collapse transition of Gly15. J. Phys. Chem. B 2017, 121, 8078–8084. 10.1021/acs.jpcb.7b05469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959, 14, 1–63. 10.1016/S0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry 2002, 29, 7133–7155. 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Bolen D. W.; Rose G. D. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 2008, 77, 339–362. 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- Tran H. T.; Mao A.; Pappu R. V. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J. Am. Chem. Soc. 2008, 130, 7380–7392. 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- Holthauzen L. M. F.; Rösgen J.; Bolen D. W. Hydrogen bonding progressively strengthens upon transfer of the protein urea-denatured state to water and protecting osmolytes. Biochemistry 2010, 49, 1310–1318. 10.1021/bi9015499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel D. P.; Johnson C. M.; Lum J. K.; Neuweiler H. Backbone-driven collapse in unfolded protein chains. J. Mol. Biol. 2011, 409, 250–262. 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- Majumder S.; Janke W. Cluster coarsening during polymer collapse: Finite-size scaling analysis. Europhys. Lett. 2015, 110, 58001 10.1209/0295-5075/110/58001. [DOI] [Google Scholar]
- Majumder S.; Janke W. Evidence of aging and dynamic scaling in the collapse of a polymer. Phys. Rev. E 2016, 93, 032506 10.1103/PhysRevE.93.032506. [DOI] [PubMed] [Google Scholar]
- Majumder S.; Zierenberg J.; Janke W. Kinetics of polymer collapse: Effect of temperature on cluster growth and aging. Soft Matter 2017, 13, 1276–1290. 10.1039/C6SM02197B. [DOI] [PubMed] [Google Scholar]
- Christiansen H.; Majumder S.; Janke W. Coarsening and aging of lattice polymers: Influence of bond fluctuations. J. Chem. Phys. 2017, 147, 094902 10.1063/1.4991667. [DOI] [PubMed] [Google Scholar]
- Bray A. J. Theory of phase-ordering kinetics. Adv. Phys. 2002, 51, 481–587. 10.1080/00018730110117433. [DOI] [Google Scholar]
- de Gennes P.-G. Kinetics of collapse for a flexible coil. J. Phys., Lett. 1985, 46, 639–642. 10.1051/jphyslet:019850046014063900. [DOI] [Google Scholar]
- Halperin A.; Goldbart P. M. Early stages of homopolymer collapse. Phys. Rev. E 2000, 61, 565–573. 10.1103/PhysRevE.61.565. [DOI] [PubMed] [Google Scholar]
- Byrne A.; Kiernan P.; Green D.; Dawson K. A. Kinetics of homopolymer collapse. J. Chem. Phys. 1995, 102, 573–577. 10.1063/1.469437. [DOI] [Google Scholar]
- Abrams C. F.; Lee N.-K.; Obukhov S. P. Collapse dynamics of a polymer chain: Theory and simulation. Europhys. Lett. 2002, 59, 391–397. 10.1209/epl/i2002-00207-5. [DOI] [Google Scholar]
- Kikuchi N.; Ryder J. F.; Pooley C. M.; Yeomans J. M. Kinetics of the polymer collapse transition: The role of hydrodynamics. Phys. Rev. E 2005, 71, 061804 10.1103/PhysRevE.71.061804. [DOI] [PubMed] [Google Scholar]
- Reddy G.; Yethiraj A. Implicit and explicit solvent models for the simulation of dilute polymer solutions. Macromolecules 2006, 39, 8536–8542. 10.1021/ma061176+. [DOI] [Google Scholar]
- Guo J.; Liang H.; Wang Z.-G. Coil-to-globule transition by dissipative particle dynamics simulation. J. Chem. Phys. 2011, 134, 244904 10.1063/1.3604812. [DOI] [PubMed] [Google Scholar]
- Montesi A.; Pasquali M.; MacKintosh F. C. Collapse of a semiflexible polymer in poor solvent. Phys. Rev. E 2004, 69, 021916 10.1103/PhysRevE.69.021916. [DOI] [PubMed] [Google Scholar]
- Lappala A.; Terentjev E. M. “Raindrop” coalescence of polymer chains during coil-globule transition. Macromolecules 2013, 46, 1239–1247. 10.1021/ma302364f. [DOI] [Google Scholar]
- Wilkins D. K.; Grimshaw S. B.; Receveur V.; Dobson C. M.; Jones J. A.; Smith L. J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 1999, 38, 16424–16431. 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- Uversky V. N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke I. R.; Williams D. R. M. Collapse dynamics of block copolymers in selective solvents: Micelle formation and the effect of chain sequence. Macromolecules 2003, 36, 2149–2157. 10.1021/ma020741f. [DOI] [Google Scholar]
- Pham T. T.; Dünweg B.; Prakash J. R. Collapse dynamics of copolymers in a poor solvent: Influence of hydrodynamic interactions and chain sequence. Macromolecules 2010, 43, 10084–10095. 10.1021/ma101806n. [DOI] [Google Scholar]
- MacKerell A. D. Jr.; Bashford D.; Bellott M.; Dunbrack R. L. Jr.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell A. D. Jr.; Feig M.; Brooks C. L. III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25, 1400–1415. 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Frenkel D.; Smit B.. Understanding Molecular Simulations: From Algorithms to Applications; Academic Press: San Diego, 2002. [Google Scholar]
- Rubenstein M.; Colby R.. Polymer Physics (Chemistry); Oxford University Press: Oxford, 2003. [Google Scholar]
- Scolari V. F.; Mercy G.; Koszul R.; Lesne A.; Mozziconacci J. Kinetic signature of cooperativity in the irreversible collapse of a polymer. Phys. Rev. Lett. 2018, 121, 057801 10.1103/PhysRevLett.121.057801. [DOI] [PubMed] [Google Scholar]
- de Gennes P.-G.Scaling Concepts in Polymer Physics; AIP: Melville, New York, 1980. [Google Scholar]
- Majumder S.; Das S. K. Domain coarsening in two dimensions: Conserved dynamics and finite-size scaling. Phys. Rev. E 2010, 81, 050102 10.1103/PhysRevE.81.050102. [DOI] [PubMed] [Google Scholar]
- Binder K.; Stauffer D. Theory for the slowing down of the relaxation and spinodal decomposition of binary mixtures. Phys. Rev. Lett. 1974, 33, 1006–1009. 10.1103/PhysRevLett.33.1006. [DOI] [Google Scholar]
- Kremer K.; Grest G. S. Dynamics of entangled linear polymer melts: A molecular–dynamics simulation. J. Chem. Phys. 1990, 92, 5057–5086. 10.1063/1.458541. [DOI] [Google Scholar]
- Xu J.; Zhu Z.; Luo S.; Wu C.; Liu S. First observation of two-stage collapsing kinetics of a single synthetic polymer chain. Phys. Rev. Lett. 2006, 96, 027802 10.1103/PhysRevLett.96.027802. [DOI] [PubMed] [Google Scholar]
- Chu B.; Ying Q.; Grosberg A. Y. Two-stage kinetics of single-chain collapse: Polystyrene in cyclohexane. Macromolecules 1995, 28, 180–189. 10.1021/ma00105a024. [DOI] [Google Scholar]
- Arashiro E.; Drugowich de Felício J. R.; Hansmann U. H. E. Short-time dynamics of the helixcoil transition in polypeptides. Phys. Rev. E 2006, 73, 040902 10.1103/PhysRevE.73.040902. [DOI] [PubMed] [Google Scholar]
- Arashiro E.; Drugowich de Felício J. R.; Hansmann U. H. E. Short-time dynamics of polypeptides. J. Chem. Phys. 2007, 126, 045107 10.1063/1.2430709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.