Abstract
The Rag GTPases recruit the master kinase mTORC1 to lysosomes to regulate cell growth and proliferation in response to amino acid availability. The nucleotide state of Rag heterodimers is critical for their association with mTORC1. Our cryo-EM structure of RagA/RagC in complex with mTORC1 shows the details of RagA/C binding to the RAPTOR subunit of mTORC1 and explains why only the RagAGTP/RagCGDP nucleotide state binds mTORC1. Previous kinetic studies suggested that GTP binding to one Rag locks the heterodimer to prevent GTP binding to the other. Our crystal structures and dynamics show the mechanism for this locking, and explain how oncogenic hotspot mutations disrupt this process. In contrast to allosteric activation by RHEB, Rag heterodimer binding does not change mTORC1 conformation and activates mTORC1 by targeting it to lysosomes.
The mechanistic target of rapamycin complex 1 (mTORC1) is a Ser/Thr protein kinase complex that integrates signals from nutrient availability, energy, and growth factors to regulate cell growth, proliferation and metabolism (1). Upregulation of mTORC1 is associated with many diseases such as cancer, type-2 diabetes and defects in neurodevelopment(1, 2). The mTORC1 complex is a dimer of mTOR/RAPTOR/mLST8 heterotrimers (3). The mTORC1 kinase activity is tightly regulated by two classes of small GTPases, Rags and RHEB, both of which are necessary for activation (4, 5). In response to the abundance of nutrients, particularly amino acids, active Rag heterodimers bind the RAPTOR subunit of mTORC1 to recruit it to lysosomes (6–8), where mTORC1 can be allosterically stimulated by growth-factor activated RHEB (3, 9–12). Unlike RHEB, which carries a C-terminal farnesylation that weakly associates it to a variety of membranes (13, 14), Rags have no lipid modification. Instead, they associate with the heteropentameric Ragulator complex that has myristoyl and palmitoyl modifications at the N-terminus of its LAMTOR1 subunit that localize it to lysosomes (15, 16). Recurrent oncogenic mutations in RagC enhance its association with mTORC1, leading to increased mTORC1 signalling (17–19).
Both Rags and RHEB are members of the Ras-like superfamily of GTPases. However, unlike most members, Rags are obligate heterodimers, with RagA or RagB pairing with RagC or RagD (20). Analysis of the composite genome of Lokiarchaeum revealed that Rag GTPases have an archaeal origin closely related to the Arf family of Ras-like GTPases that are involved in vesicular sorting (21), but the mechanistic implication of this similarity was not clear.
Rag heterodimers have four possible nucleotide-binding states but are only active for mTORC1 binding when RagA or RagB is GTP-bound and RagC or RagD is GDP-bound (6, 7, 22). The GTPase domains communicate so that binding of GTP by one subunit inhibits GTP binding and induces the GTP hydrolysis by the other subunit (22). The nucleotide states of Rags are regulated by GTPase-activating proteins (GAPs), such as GATOR1 and folliculin (23–25) and guanine nucleotide exchange factors (GEFs) such as SLC38A9 and Ragulator (26, 27).
To elucidate how human Rags interact with mTORC1 and how RagC mutations activate mTORC1, we determined structures and dynamics of RagA/C complexes in isolation and bound to mTORC1. The structures revealed nucleotide-dependent conformational changes in Rags that are required for mTORC1 binding, enabling us to understand the mechanism by which oncogenic Rag mutations facilitate association with mTORC1.
HDX-MS shows that RagA/C protects the α-solenoid of RAPTOR
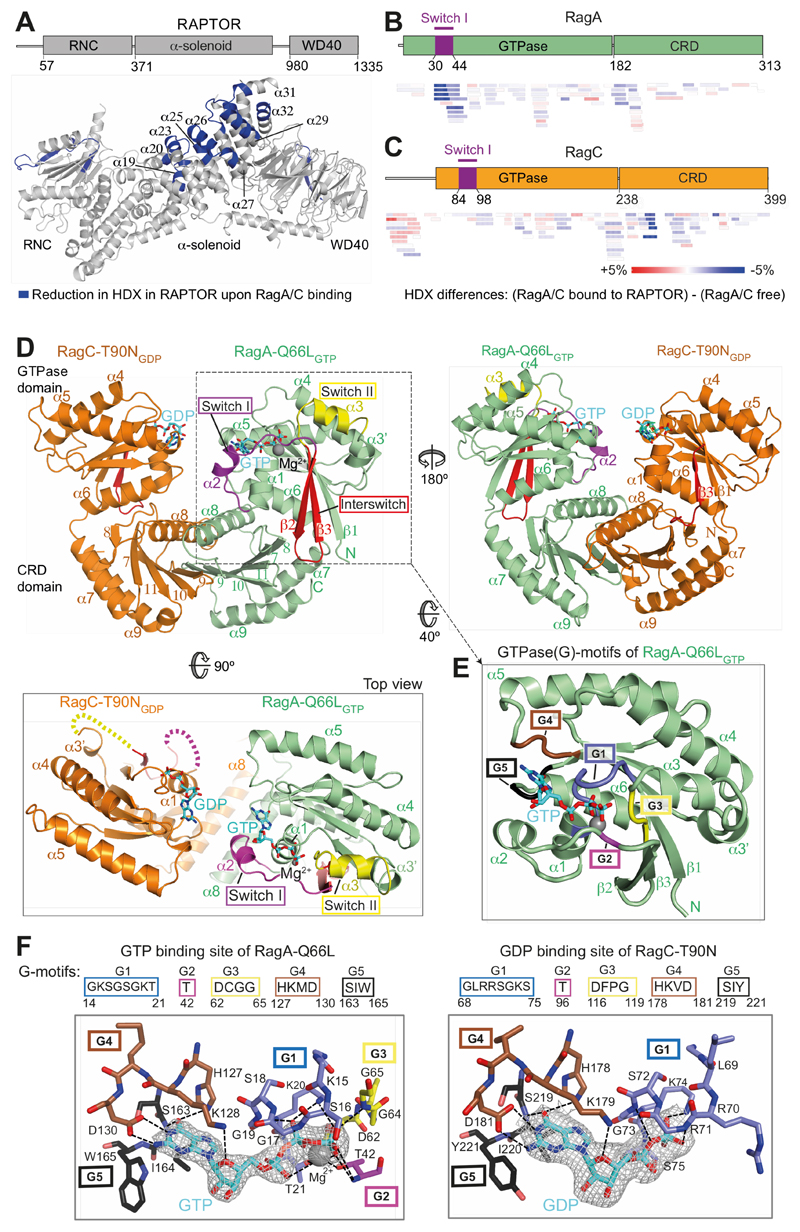
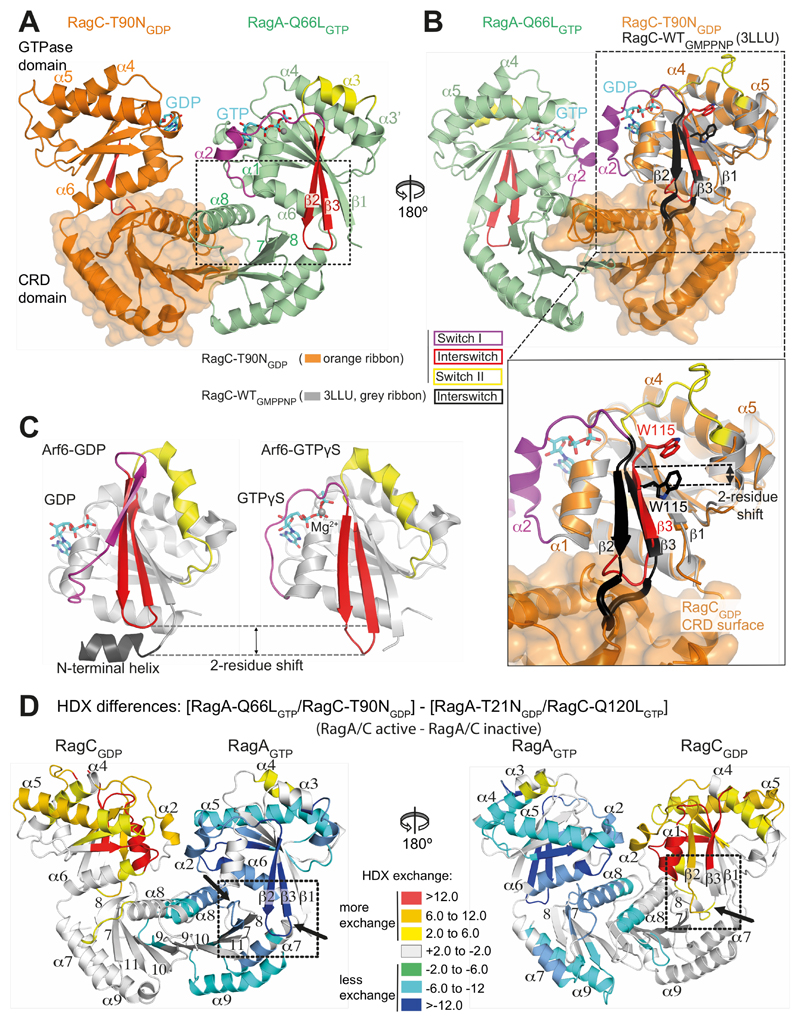
To map RagA/C interactions with the RAPTOR subunit of mTORC1, we carried out hydrogen/deuterium exchange mass spectrometry (HDX-MS). For this we used RagA-Q66LGTP/RagC-T90NGDP containing mutations that increase mTORC1 association (6, 7, 17, 19). The RagA-Q66L switch II mutation (Fig. S1 (28)) impairs GTP hydrolysis and is a potent activator of mTOR signalling, with mice bearing this mutation dying within one day of postnatal life (29). The RagC-T90N mutation binds only GDP and is the most frequent and potent oncogenic mutation in RagC (17, 19).
RagA/C heterodimers were monodisperse (Fig. S2A), and formed a 1:1:1 complex with RAPTOR (Fig. S2B,C). Most RAPTOR peptides showing decreased HDX upon RagA/C binding map to a contiguous surface in the region 541-678, encompassing three adjacent helical repeats of the RAPTOR α-solenoid (Fig. 1A, S3, Table S1). There was also reduced HDX in the insertion before the last two helices of the RAPTOR α–solenoid (peptides 760-780 and 805-812) and in the WD40 domain itself. On the Rag side of the interface, RagA switch I was protected from HDX by RAPTOR (Fig. 1B), while RagC switches were not (Fig. 1C, Table S1). To understand the context of the HDX-MS measured dynamics, we determined structures of RagA/C heterodimers both free and bound to mTORC1.
Fig. 1. The crystal structure of a RagA/C heterodimer and HDX-MS analysis of its interaction with RAPTOR.
(A) HDX-MS identified regions protected from HDX in the RAPTOR/RagA-Q66LGTP/RagCT90NGDP complex. Decreases in HDX (blue) of RAPTOR upon RagA/C binding are depicted on the RAPTOR structure (from PDB ID: 6BCX).
(B, C) Differences in HDX for RagA-Q66LGTP (B) and RagC-T90NGDP (C) upon RAPTOR binding for all the peptides at 0.3 s in D2O. Decreases in HDX are depicted in shades of blue, and increases are in shades of red.
(D) The crystal structure of RagA-Q66LGTP/RagC(34-399)-T90NGDP, highlighting the ordered switches in RagAGTP compared with disordered switches in RagC-T90NGDP oncogenic mutant.
(E) The conserved G-motifs of RagA-Q66LGTP that make up the nucleotide binding pocket.
(F) The 2mFo-DFc map (contoured at 1.2σ) for the GTP of RagA and the GDP of RagC together with the putative H-bonds that they make to the G-motifs. These are much more extensive for GTP than for GDP.
High-resolution crystal structures of active RagA/C heterodimers
HDX-MS identified RagC residues 1-34 as highly flexible (Fig. S2D). For crystallization, we truncated this region, producing a variant that bound RAPTOR the same as full-length RagA/C (Fig. S2E), enabling us to determine high-resolution crystal structures of RagA/C.
The 2.6 Å resolution crystal structure of RagA-Q66L/RagC(34-399)-T90N showed a compact arrangement of C-terminal CRD domains that mediate heterodimerization and N-terminal GTPase domains (Fig. 1D and Table S2). The overall fold of the GTPase domains is similar to other Ras-like GTPases, with conserved loops (G1-G5 motifs) that engage bound nucleotides, and regions known as switches that change conformation depending on whether GTP or GDP is bound (30). RagA is bound to GTP and has Mg2+ associated with the GTP γ-phosphate, while RagC is bound to GDP and has no bound Mg2+ (Fig. 1E,F). While RagA has switch I, interswitch and switch II ordered (Fig. 1D, S1), the RagC-T90NGDP GTPase domain has no density for all of switch I and interswitch strand β2 (residues 84-105) and all of switch II (116-130). The C-terminal CRDs have a roadblock fold consisting of a central five-stranded antiparallel β-sheet sandwiched between two α-helical layers (31). The whole complex has a pseudo-two-fold symmetry, with the two GTPase domains close to each other and their switches on opposite faces of the complex (Fig. 1D).
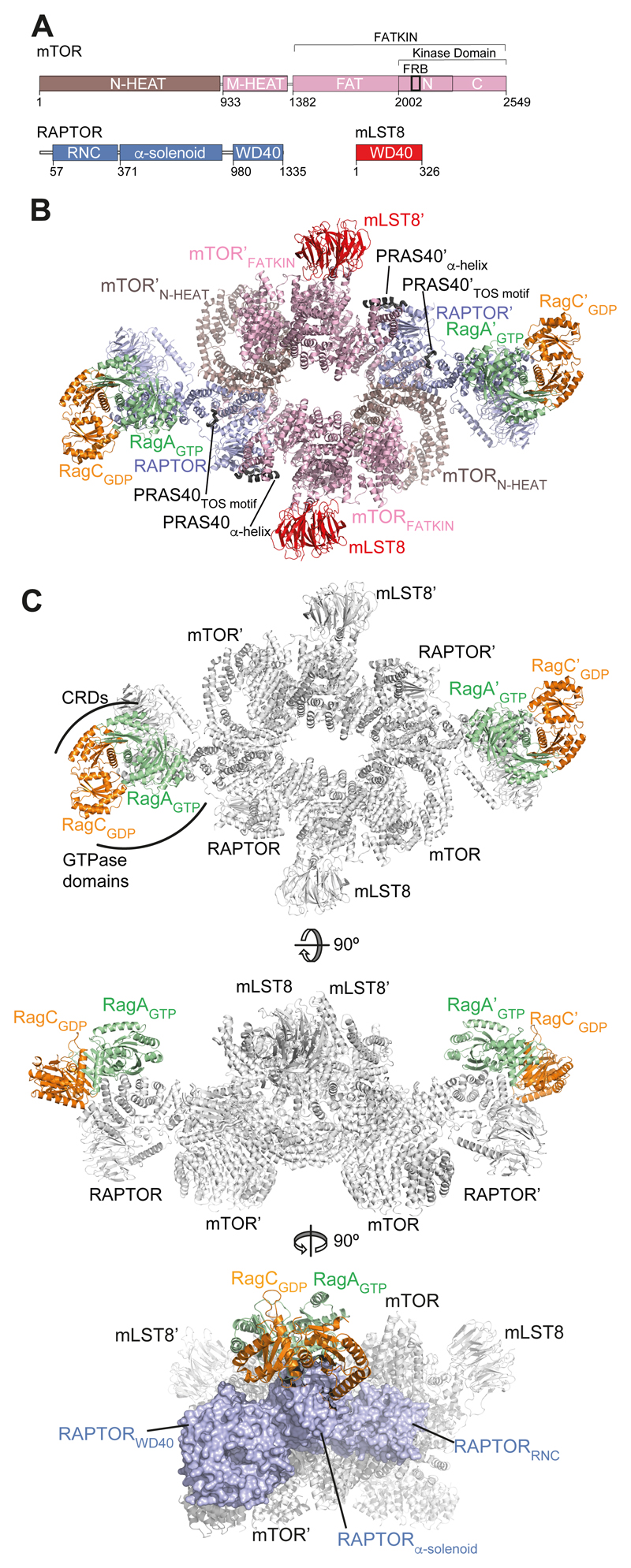
The cryo-EM structure of mTORC1 bound to RagA/C
To understand how active RagA/C interacts with intact mTORC1, we used electron cryo-microscopy (cryo-EM). In order to stabilize mTORC1 bound to RagA-Q66LGTP/RagC-T90NGDP (RagA/C), we used chemical crosslinking and expressed the RagA/C heterodimer with RagC fused to another mTORC1-binding protein, PRAS40, which is largely disordered (Fig. S4, Table S3). Using this mTORC1-RagA/C complex (Fig. S5A), we generated a final 4.1 Å resolution reconstruction of mTORC1 (Fig. S5, S6A, Table S4). This reconstruction showed extra density adjacent to the α-solenoid region of RAPTOR (Fig. S6A) that HDX-MS identified as the RagA/C binding site. The TOS motif from PRAS40 (fused to RagC) contacts a groove between the RNC and the α-solenoid of RAPTOR, as also observed by HDX-MS (Fig. S4C-E).
Focused classification with signal subtraction (32) showed that about 9.5% of particles were bound to RagA/C, corresponding to 90,809 particles, and reconstruction of the mTORC1-RagA-Q66LGTP/RagC-T90NGDP complex at 5.5 Å resolution revealed density for the RagA/C into which we could readily fit our high-resolution RagA/C crystal structure (Figs. 2, S6B-D). RagA/C interacts with the convex surface of the RAPTOR α-solenoid (Fig. 3A). The GTPase-containing ends of the horseshoe-shaped RagA/C heterodimer are closest to the RAPTOR α-solenoid, with the CRDs pointing away from RAPTOR (Fig. 2B,C). The RagA GTPase domain makes much more extensive RAPTOR contacts than RagC, and the interface agrees with our HDX-MS analysis (Fig. 3A,B). The overall conformation of mTORC1 bound to RagA/C heterodimers is nearly identical to the conformation of apo-mTORC1 (3).
Fig. 2. Architecture of the mTORC1-RagA/C complex.
(A) Schematic representation of mTORC1 components (mTOR, RAPTOR and mLST8).
(B) Overall cryo-EM-based model of the mTORC1-RagA/C complex. The two ordered regions of the PRAS40 moiety of the fusion construct are also shown.
(C) Three views of mTORC1-RagA/C complex show RagA/C sitting on top of the RAPTOR α-solenoid, with the GTPase domains making most of the interactions.
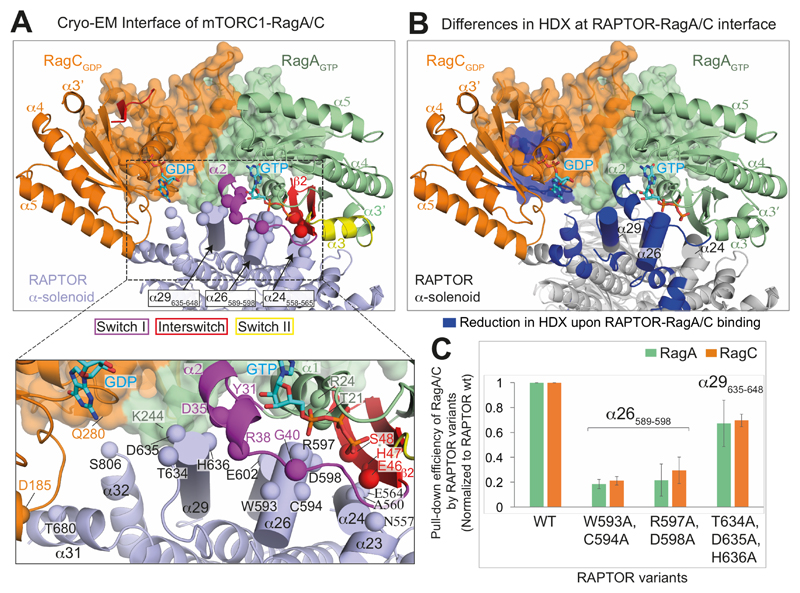
Fig. 3. Interface between RAPTOR and the RagA/C complex.
(A) Close-up views of RagA/C binding to the RAPTOR subunit of mTORC1. The CRDs are shown as transparent surfaces. RAPTOR helices contacting switch I and interswitch of RagA are shown as cylinders. Spheres mark RAPTOR/RagA interface residues.
(B) View of the interface, illustrating regions with a decrease in HDX (blue) upon formation of the RagA/C/RAPTOR complex.
(C) Mutational analysis of the binding interface. Strep-tagged wild-type RAPTOR (WT) and three different RAPTOR mutants (WC(593,594)AA, RD(597,598)AA and TDH(634-636)AAA) were assayed for their ability to pull-down RagA-Q66LGTP/RagC-T90NGDP in vitro. The pull-down efficiencies of RAPTOR mutants were normalized to WT RAPTOR. Values are means from three independent experiments, and error bars show standard deviations.
Three helices from RAPTOR (α24, α26 and α29) in the region 546-650 of the RAPTOR α-solenoid make an extensive network of interactions with switch I and interswitch strand β2 of the RagA-Q66LGTP (Fig. 3A). The GTPase domain of RagCGDP forms limited interactions with RAPTOR, including a contact of RagC-D185 at the N-terminus of helix α5 with T680 in RAPTOR helix α31. Although the GTPase domains form most of the interface with RAPTOR, there are some contacts with the CRD domains. These involve the C-terminus of RagA helix α8 (S243 and K244) and the N-terminus of RagC helix α8 (Q280), which come close together and engage two adjacent structural elements of RAPTOR, helix α29 (T639, N643, M646) and the end of the long, mostly disordered insertion after α31.
Mutations of RAPTOR residues in helices α26 (W593/C594 or R597/D598) and α29 (T634/D635/H636) that contact the RagA-Q66LGTP switch I/interswitch greatly reduce binding to RagA/C (Fig. 3C). Previously, RagA mutations were identified that prevent RAPTOR interaction (33), and they map to the interface with RAPTOR in our structure.
We also determined the cryo-EM structure of mTORC1-RagA-Q66LGTP/RagC-T90NGDP where RagA/C was not covalently fused to PRAS40. Importantly, in both cryo-EM structures, the RagA/C heterodimer interacts with RAPTOR in the same manner (Figs. S7 and S8).
Cancer-associated mutations in RagA/C affect communication between GTPase domains
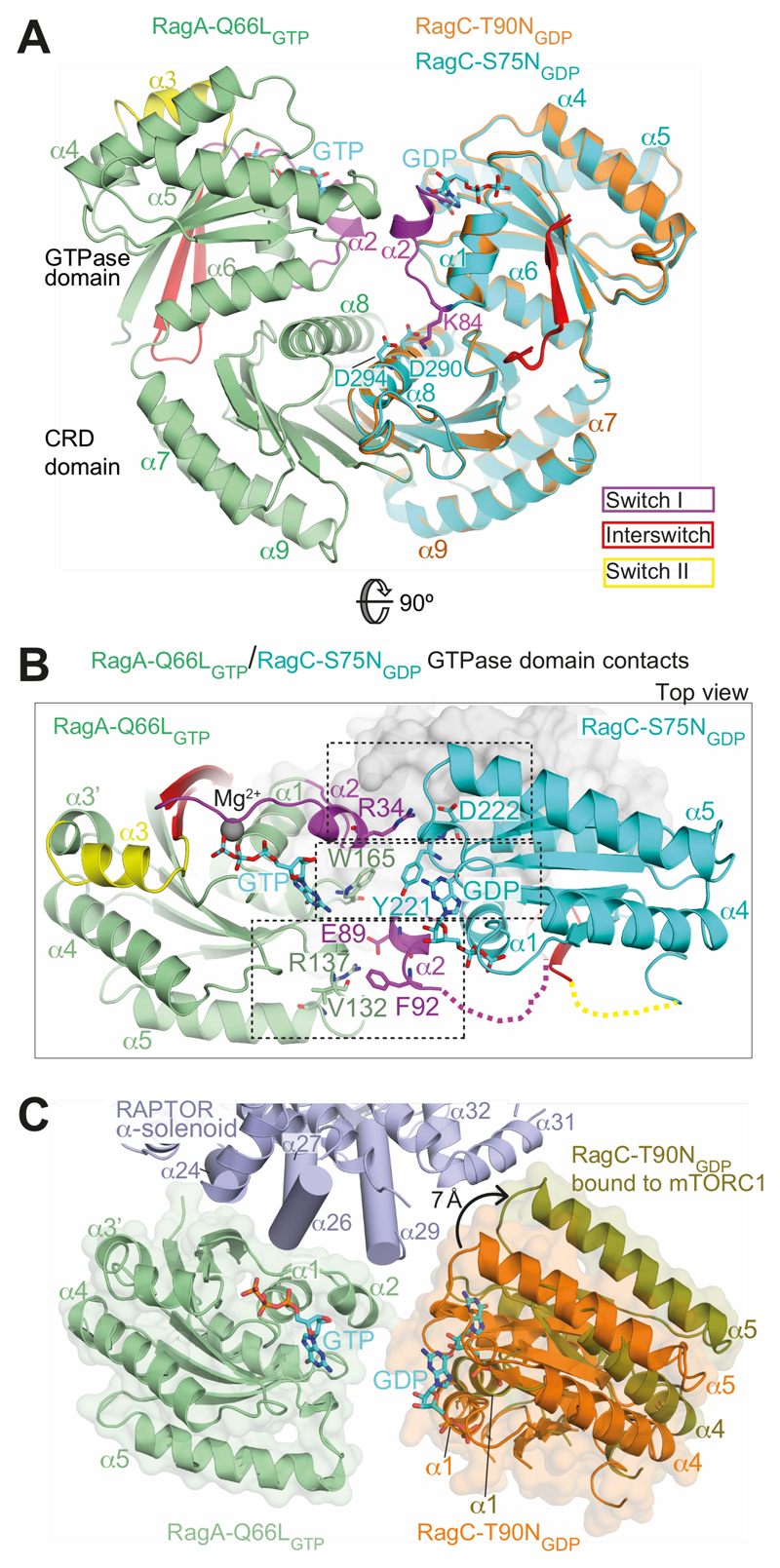
Cancer-associated mutations in RagC increase mTORC1 binding (17–19), and we wanted to gain insights into the structural basis for this effect. The mutations cluster in various nucleotidesensing elements of RagC: the P-loop (e.g., S75), switch I (e.g., T90), interswitch (e.g., W115, D116) and switch II (e.g., P118) (Fig. S1). The RagC-T90N mutant had switch regions disordered (Fig. 1D). To see whether this disorder is specific for the T90N mutation, we also determined a 2.5 Å resolution crystal structure of RagA-Q66LGTP/RagC-S75NGDP (Table S2). The RagC-S75N mutation in the P-loop impairs GTP binding by eliminating the interaction of S75 with Mg2+. The structures of RagC-S75NGDP and the RagC-T90NGDP are very similar, except that RagC-S75NGDP has helix α2 of switch I (residues 86-93) ordered in one of the two heterodimers in the crystal asymmetric unit (Fig. 4A), suggesting that S75N destabilizes, but does not completely disorder switch I. This shows that T90N causes a greater perturbation in switch I than S75N, consistent with the more potent phenotype of the T90N mutation in cells (17). The structure of the isolated wild-type RagC GTPase domain bound to a GTP analog (PDB ID 3LLU (34)) shows a completely ordered switch I and helix α2 that closely superimposes with this helix in RagC-S75NGDP. In the RagC-S75NGDP structure, OG1 of the T90 side chain is close to the O2’ of the bound nucleotide (3.7 Å), so it is likely that the larger T90N substitution leads to disorder of helix α2.
Fig. 4. Interactions between GTPase domains in the RagA/C heterodimer.
(A) Comparison of the RagA-Q66LGTP/RagC-T90NGDP with RagA-Q66LGTP/RagC-S75NGDP, illustrating ordering of helix α2 in switch I of RagC-S75N. Superposition was on the RagA subunit.
(B) Three sets of interactions between RagAGTP and RagCGDP GTPase domains.
(C) A change in the orientation of RagA/C GTPase domains in the free RagA/C relative to RagA/C bound to mTORC1. Superposition was on the RagA subunit. The view is similar to (B).
RagA/RagC GTPase domain contacts can be grouped into three sets (Fig. 4B). One set is at the center of the interface where the G5-motifs of the two domains meet, with RagA-W165 near the equivalent of this residue in RagC, Y221 (Fig. 1E, Fig. S1). The second set involves interactions between RagA switch I helix α2 and RagC loop immediately following the G5-motif. In particular, there is a water-mediated interaction between RagA-R34 and the side chain of RagC-D222 (Fig. 4B). The third set consists of interactions between RagC switch I helix α2 and RagA G4/α5 loop. This set is present in the complex with RagC-S75NGDP where RagC-E89 makes a salt-link with RagA-R137, and RagC-F92 contacts the RagA loop 131-133 (Fig. 4B), but is absent in the complex with RagC-T90NGDP, where RagC switch I is completely disordered. This demonstrates that oncogenic mutations significantly perturb the interface between the GTPase domains.
Rearrangements within the RagA/C heterodimer when bound to mTORC1
Comparing the crystal structure of free RagA-Q66LGTP/RagC-T90NGDP and the cryo-EM structure of RagA/C bound to mTORC1 reveals a shift in the interface between the GTPase domains by ~7 Å (Fig. 4C, Movie S1). This creates a more open space between the two GTPase domains, which in the free RagA/C would be kept closer by interactions involving switch I helix α2. This might explain why the oncogenic RagC-T90N mutation, which has helix α2 disordered, binds more easily to RAPTOR, since the RagC helix α2/RagA interactions are already disrupted before the heterodimer binds to RAPTOR. A similar structural change could occur in the RagC-L91P mutant associated with follicular lymphomas (17). Residue K84 in the α1-α2 loop of RagC switch I forms salt-links with residues D290 and D294 of helix α8 in the CRD (Fig. 4A). The lymphoma-associated mutation RagC-K84T (17) would likely disrupt this interaction, which could facilitate RagC rearrangement relative to RagA as seen in the complex with mTORC1.
Comparison of the CRDs from the free RagA/C with the mTORC1-bound RagA/C shows a small shift in the orientation of the two CRDs so that in the mTORC1-bound form, the top surface of the CRDs that embraces the GTPase domains is more splayed (Fig. 4C, S9, Movie S1). Interestingly, comparing the CRD dimer from the free RagA/C with the CRD dimer bound to the Ragulator (35), also shows shifts between the two CRDs (Fig. S9). Together this indicates that the interface between the CRD domains has some flexibility. This could be exploited by interactors such as the Ragulator to exert changes on the GTPase domains via the CRDs, which could contribute to the established role of the Ragulator as a GEF for RagC (27).
Structural basis for relaying nucleotide binding to the CRDs
Although only the RagAGTP/RagCGDP state binds RAPTOR, the reverse, inactive state, RagAGDP/RagCGTP, is essential for terminating mTORC1 activation. Furthermore, some Rag interactors such as Galectin-8 preferentially associate with this state (36). Therefore, a structural understanding of both states is important. In the active heterodimer, nucleotide-sensitive elements in the GTPase domain of each Rag form contacts with its own CRD. In RagAGTP, switch I and the β2-β3 interswitch firmly engage the CRD. The switch I interaction is at the center of the heterodimer interface, primarily with the CRD helix α8, while the β2-β3 interaction is at the outer edge of the CRD (Fig. 5A, S10A). These two interactions flank a central contact involving both non-switch (α1 and α6) and switch (β2-β3) contacts with the β7-β8 hairpin of the CRD. In the GTP-loaded state of RagA, the tip of the interswitch protrudes beyond the GTPase domain and slips into a pocket on the surface of the CRD (Fig. 5A, S10B). The interswitch tip bound in the CRD pocket may be an element of the structural basis for the “locked” state for RagA that was proposed based on a kinetic study of the communication between GTPase domains in the Rag heterodimers (22). In contrast, the interswitch loop in RagCGDP is in a retracted position and partially disordered. Interestingly, in the structure of the isolated GTPases domain of GTP-bound RagC (PDB ID 3LLU (34)), the interswitch protrudes beyond the GTPase domain in a manner equivalent to the RagAGTP. This is accomplished by a two-residue shift in the register of strand β3 relative to β1 so that residues 109-115 in the RagC-T90NGDP are in the positions of residues 111-117 of the GTP-bound RagC (Fig. 5B). The extended interswitch in the RagCGTP would clash with its CRD, suggesting that a change in the relative orientations of the GTPase and CRD domains would be required to accommodate GTP binding by RagC (Fig. S10B). All of the contacts of the GTPase domain with the CRD constitute possible mechanisms for nucleotide binding to imprint onto the CRD.
Fig. 5. Structural basis for communicating nucleotide binding within the RagA/C heterodimer.
(A) Both switch I and the interswitch make nucleotide-state-dependent direct contacts with the CRD.
(B) Superposition of GTP-bound RagC (PDB ID 3LLU, in gray) on the GDP-bound RagC (from our RagA-Q66LGTP/RagC-T90NGDP complex). The interswitch of GTP-bound RagC is black and GDP-bound RagC is red. The W115 position illustrates a two-residue shift in strand β3 (relative to strand β1). The β2/β3 loop toggles between a retracted conformation in the GDP state and an extended conformation in the GTP state that would clash with the CRD, if there were no conformational changes.
(C) The structures of Arf6 bound to either GDP (PDB ID 1E0S (45)) or GTPγs (PDB ID 2J5X (46)), with switches colored as in (A). The interswitch toggle couples nucleotide binding with membrane binding by the N-terminal helix.
(D) Differences in HDX between the active (RagAGTP/RagCGDP) and inactive (RagAGDP/RagCGTP) states, illustrating changes in the CRDs, in addition to the expected changes in the GTPase domains. In RagCGDP, disordered regions in the switches have been modelled to illustrate all of the HDX changes.
Dynamics of the active and the reverse, inactive state of Rags
To gain a better understanding of the conformational changes that occur in the reverse state, we tried but did not succeed in obtaining diffracting crystals for this heterodimer. As an alternative strategy, we used HDX-MS to examine differences in conformation between the active (RagA-Q66LGTP/RagC-T90NGDP) and reverse (RagA-T21NGDP/RagC-Q120LGTP) states (Fig. 5D, Table S5). Overall, both Rags showed less HDX throughout the GTPase domains when GTP-bound compared with GDP-bound, indicating a more compact domain when bound to GTP. Furthermore, upon GTP binding to RagA, there is a distinct protection in its CRD domain (e.g., in CRD helix α8, β7/β8 and the hinge that are engaged with the GTPase domain), suggesting communication of the nucleotide state to the CRD (Fig. 5D, Movie S2). Residues making up a pocket on the surface of the RagA CRD that accommodate the interswitch in GTP-bound RagA have increased exchange in the reverse state, which we attribute to retraction of the RagA interswitch, exposing the RagA CRD pocket. Consistent with this, the GDP-bound RagA interswitch has increased HDX (displayed on the RagAGTP crystal structure in Fig. 5D and on a RagAGDP model in Movie S2). For RagC, there is less change in HDX in the CRD upon GTP binding. Although RagC interswitch shows GTP-dependent HDX protection, the pocket on the RagC CRD analogous to the RagA pocket has no significant concomitant changes in protection, so currently we do not know the position of the interswitch in the GTP-bound RagC.
Implications for yeast TORC1 signalling
Comparing yeast Gtr1GTP/Gtr2GDP structure (PDB ID 4ARZ (37)) with the human RagAGTP/RagCGDP indicates very large conformational differences, both in switches and in the relative orientations of GTPase domains, with Gtr2 GTPase domain rotated about 36° relative to the RagC GTPase domain (Fig. S11A). The arrangement of the Gtr1/2 GTPase domains is not compatible with binding to mTORC1 in the same manner as RagA/C (Fig. S11B). This suggests that the Gtr1/2 GTPase domains may reorient in order to bind Kog1, the yeast homologue of RAPTOR. The very different conformation of the Gtr2GDP switch I region and the extreme orientation of the Gtr2 GTPase domain relative to RagC may reflect a fundamental difference between RagC and Gtr2. This could explain why RagA-Q66L can complement a Gtr1 deficient strain, while neither wild-type nor a GDP-bound mutant RagC could complement Gtr2 deficiency in yeast (38). Despite these differences, the binding site of Gtrs on Kog1 maps to a similar region on the Kog1 α-solenoid (39).
Given the role of the Rag heterodimers in recruiting mTORC1 to lysosomes, the constitutive association of yeast TORC1 with the vacuole is surprising (5). A recent report elegantly showed that upon glucose starvation yeast TORC1 forms striking inactive, vacuole-associated helical tubes named TOROIDs and that the TOROID formation is antagonized by active Rags (Gtr1GTP/Gtr2GDP) in cells (40). Fitting our RagA/C heterodimers into the cryo-EM reconstruction of the tubes, in accordance with the arrangement present in our RagA/C/mTORC1 complex, suggests that the Gtr1/2 binding would not be compatible with the TORC1 arrangement in the TORIODs (Fig. S11C). This might mean that Gtr1/2 binding could directly regulate assembly/disassembly of the tubes to activate TORC1. Further work is needed to test this structure-based proposal.
Discussion
RagA/C binding causes no conformational change in mTORC1, suggesting that the role of the Ragulator/Rags complex is to localise mTORC1 to lysosomes where it can be allosterically activated by RHEB. Rag/RAPTOR interaction requires a GTP-loaded RagA, so that RagA switch I and interswitch are ordered, since they make most of the interactions with RAPTOR. A reverse state of Rags with GDP-loaded RagA and GTP-loaded RagC does not bind RAPTOR as well (6, 7), because RagAGDP would have the switch regions disordered, while RagCGTP could not interact with RAPTOR, since RagC residues analogous to RAPTOR-binding residues of RagA are not conserved (Fig. S12).
The structures suggest how the nucleotide-bound state of one GTPase domain is communicated both between subunits to the paired GTPase domain and within a subunit to its CRD (Figs. 4, 5, Movie S2). First, consistent with communication between GTPase domains (22), both RagA and RagC have helix α2 (in switch I) contacting the paired GTPase domain, and filling the space between them (Fig. 4A,B). Second, there are several sets of interactions between the GTPase and CRD domains within a subunit that HDX suggests are dynamic and nucleotide dependent, including switch I and the interswitch. The interswitch of Rag GTPases apparently undergoes a nucleotide-dependent register shift of strand β3 relative to β1 that could be part of a mechanism to transmit nucleotide binding information from the GTPase domain to the CRD (Fig. 5). This is analogous to conformational changes that accompany transition from the GDP to GTP bound states of Arf family GTPases (Fig. 5C)(41, 42) and is consistent with the evolutionary relationship of the Rags to the Arf family (21). In Arfs, this interswitch toggle between retracted and protruded conformations coordinates membrane binding with GTP loading (Fig. 5B). In Rags, the interswitch toggle could be part of a mechanism that rotates one GTPase domain via a CRD fulcrum relative to the other GTPase domain. This would change RagA/RagC GTPase domain contacts, making it less favourable for the heterodimer to accommodate GTP in both GTPase domains at the same time as kinetically observed (22).
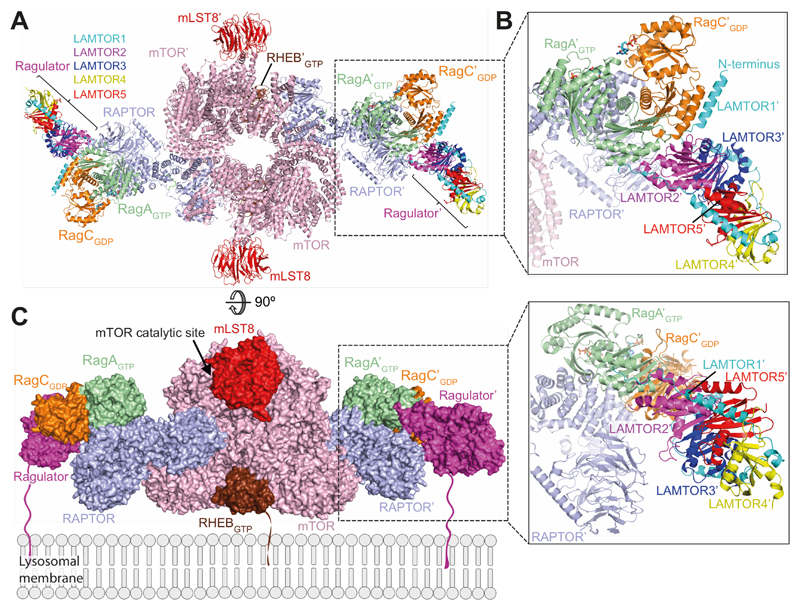
mTORC1 activity is intricately regulated in a signal and location-specific manner. Membrane compartments act as signaling platforms that serve to co-localize mTORC1 with its activating G-protein RHEB, which is targeted transiently to most endomembranes by farnesylation (13). The lysosomal activity of mTORC1 in amino-acid signalling is achieved through its dynamic interface with the Rags-Ragulator lysosomal scaffold (8, 43). Rags couple mTORC1 to lysosomes by binding to RAPTOR with their GTPase domains and to the Ragulator with their CRDs. Two RHEB molecules bind mTORC1 cooperatively, due to large allosteric changes in mTOR that are incompatible with a mixed mTOR dimer (3). In contrast, two soluble RagA/C heterodimers bind independently to mTORC1, because RagA/C binding does not introduce conformational changes in mTORC1. We propose an organization of active mTORC1 on membranes based on our structure of mTORC1-RagA/C complex, the previously published structure of mTORC1-RHEB complex (3) and the crystal structure of Ragulator bound to RagA/C CRDs (35, 44) (Fig. 6). In this model, RagA/C associated with membranes through the Ragulator (via lipidated N-terminus of LAMTOR1) and RHEB associated through Cterminal farnesylation, can be bound at the same time to mTORC1, yet still allow the mTOR active sites to face the cytosol. The RHEB-binding surface of mTORC1 would be near a RHEB-containing membrane, and the first ordered residue of the LAMTOR1 subunit of Ragulator (residue 96) would be about 105 Å from the membrane surface, suggesting that the 95 flexible N-terminal LAMTOR1 residues could easily reach the membrane. Further structural and kinetic analysis of mTORC1 complexes on membranes will be essential to fully appreciate the roles of structural dynamics of mTORC1 with its regulators and the roles of membranes in regulation of mTORC1.
Fig. 6. Model of mTORC1-RHEB-RagA/C-Ragulator complex.
(A) A ribbon diagram of the mTORC1-RHEB-RagA/C-Ragulator complex. RAPTOR-RagA/C was superimposed on RAPTOR in mTORC1-RHEB complex (PDB ID 6BCU (3)). The CRD of the mTORC1-RagA/C-RHEB complex was superimposed onto the CRD of the crystal structure of Ragulator-CRD domain complex (PDB ID 6EHR (35)).
(B) Expanded view of the Ragulator/Rags/RAPTOR interface.
(C) A model of the complex on a lysosomal membrane. The lipid-modified regions of LAMTOR1 and RHEB that anchor them to membranes are depicted in arbitrary conformations.
Supplementary Material
One Sentence Summary.
Regulatory mechanisms embodied in the architecture of a RagA/C complex with mTORC1
Acknowledgments
We thank Giuseppe Cannone, Andreas Boland, Sjors Scheres, Garib Murshudov, Takanori Nakane, Rangana Warshamanage, Wenjuan Zhang, Paul Emsley, the MRC-LMB EM facility and the Scientific Computing (Jake Grimmett and Toby Darling) for assistance and advice. We thank Stephen McLaughlin, Sarah Maslen, Fabrice Gorrec and Minmin Yu from the LMB Facilities, Alistair Siebert for help with eBIC microscopes and Stephanie Monaco and Christoph Mueller-Dieckmann for assistance with ESRF beamlines ID30B and ID30-A3. We acknowledge Diamond Light Source for access to eBIC funded by the Wellcome Trust, MRC, and BBSRC.
Funding:
MA was supported by a FEBS fellowship and an EMBO fellowship (ALTF 1171-2016) and GRM by St. Catharine’s College fellowship. KBR was supported by a Junior Fellowship from Tuberous Sclerosis Association, a Researcher Mobility Grant from the Royal Society of Chemistry and a Travelling Fellowship from the Company of Biologists. DMS was supported by grants from the NIH (R01 CA103866, R01 CA129105 and R37 AI047389), Department of Defense (W81XWH15-1-0230), and Lustgarten Foundation to DMS. DMS is an HHMI investigator and an ACS Research Professor. The work was supported by the Medical Research Council [MC_U105184308 to RLW] and Cancer Research UK (grant C14801/A21211 to RLW).
Footnotes
Author contributions: MA, GRM, OP, AB, JK, CMJ, and KBR conducted the research. MA, GRM, AB, BS and RLW analyzed data. MA, OP, DMS and RLW developed the experimental plan. MA and RLW wrote the draft. All authors reviewed and edited the drafts.
Competing interests: Authors declare no competing interests.
Data and materials availability: Materials supporting findings are available from RLW upon reasonable request. Coordinates for crystal structures of RagA/C GTPase heterodimers were deposited in the Protein Data Bank, accession codes 6S6A and 6S6D. The cryo-EM reconstruction of the mTORC1/Rag complex was deposited with EMDB (EMD-10132 and EMD-10133) and PDB (entries 6SB0 and 6SB2).
References
- 1.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–128. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicastro R, Sardu A, Panchaud N, De Virgilio C. The Architecture of the Rag GTPase Signaling Network. Biomolecules. 2017;7:48. doi: 10.3390/biom7030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera RM, Zoncu R. The Lysosome as a Regulatory Hub. Annu Rev Cell Dev Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & Development. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 12.Stocker H, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 13.Angarola B, Ferguson SM. Weak Membrane Interactions Allow Rheb to Activate mTORC1 Signaling Without Major Lysosome Enrichment. bioRxiv. 2019;29:2213. doi: 10.1091/mbc.E19-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacevic M, et al. A spatially regulated GTPase cycle of Rheb controls growth factor signaling to mTORC1. bioRxiv. 2018 472241. [Google Scholar]
- 15.Nada S, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28:477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y, et al. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okosun J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48:183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long PA, et al. De novo RRAGC mutation activates mTORC1 signaling in syndromic fetal dilated cardiomyopathy. Hum Genet. 2016;135:909–917. doi: 10.1007/s00439-016-1685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ying ZX, et al. Recurrent Mutations in the MTOR Regulator RRAGC in Follicular Lymphoma. Clin Cancer Res. 2016;22:5383–5393. doi: 10.1158/1078-0432.CCR-16-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 21.Klinger CM, Spang A, Dacks JB, Ettema TJG. Tracing the Archaeal Origins of Eukaryotic Membrane-Trafficking System Building Blocks. Mol Biol Evol. 2016;33:1528–1541. doi: 10.1093/molbev/msw034. [DOI] [PubMed] [Google Scholar]
- 22.Shen K, Choe A, Sabatini DM. Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Mol Cell. 2017;68:552–565.e8. doi: 10.1016/j.molcel.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsun Z-Y, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen K, Sabatini DM. Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci. 2018;115:9545–9550. doi: 10.1073/pnas.1811727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.see supplementary materials.
- 29.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 31.Levine TP, et al. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases. 2013;4:62–69. doi: 10.4161/sgtp.24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong R, et al. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011;25:1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nedyalkova L, et al. Crystal structure of the nucleotide-binding domain of Ras-related GTP-binding protein C. doi: 10.2210/pdb3llu/pdb. [DOI] [Google Scholar]
- 35.de Araujo MEG, et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. 2017;358:377–381. doi: 10.1126/science.aao1583. [DOI] [PubMed] [Google Scholar]
- 36.Jia J, et al. Galectins Control mTOR in Response to Endomembrane Damage. Mol Cell. 2018;70:120–135.e8. doi: 10.1016/j.molcel.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong J-H, et al. Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem. 2012;287:29648–29653. doi: 10.1074/jbc.C112.384420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 39.Sekiguchi T, Kamada Y, Furuno N, Funakoshi M, Kobayashi H. Amino acid residues required for Gtr1p-Gtr2p complex formation and its interactions with the Ego1p-Ego3p complex and TORC1 components in yeast. Genes Cells. 2014;19:449–463. doi: 10.1111/gtc.12145. [DOI] [PubMed] [Google Scholar]
- 40.Prouteau M, et al. TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature. 2017;550:265–269. doi: 10.1038/nature24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherfils J. Encoding Allostery in mTOR Signaling: The Structure of the Rag GTPase/Ragulator Complex. Mol Cell. 2017;68:823–824. doi: 10.1016/j.molcel.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence RE, et al. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol. 2018;20:1052–1063. doi: 10.1038/s41556-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yonehara R, et al. Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01762-3. 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ménétrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat Struct Biol. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- 46.Pasqualato S, Ménétrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teo H, Perisic O, Gonzalez B, Williams R. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Wallis NG, Allen MD, Broadhurst RW, Lessard IA, Perham RN. Recognition of a surface loop of the lipoyl domain underlies substrate channelling in the pyruvate dehydrogenase multienzyme complex. J Mol Biol. 1996;263:463–474. doi: 10.1006/jmbi.1996.0589. [DOI] [PubMed] [Google Scholar]
- 49.Burke JE, et al. Structures of PI4KIIIβ complexes show simultaneous recruitment of Rab11 and its effectors. Science. 2014;344:1035–1038. doi: 10.1126/science.1253397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masson GR, et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat Meth. 2019;16:595–602. doi: 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Gorrec F, Löwe J. Automated Protocols for Macromolecular Crystallization at the MRC Laboratory of Molecular Biology. J Vis Exp. 2018:e55790–e55790. doi: 10.3791/55790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorrec F. The MORPHEUS II protein crystallization screen. Acta Crystallogr F Struct Biol Commun. 2015;71:831–837. doi: 10.1107/S2053230X1500967X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy A. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov G, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Meth Enzymol. 2010;481:109–126. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- 60.Boland A, et al. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat Struct Biol. 2017;24:414–418. doi: 10.1038/nsmb.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Meth. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Meth. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Meth. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 69.Yang H, et al. 4.4 Å Resolution Cryo-EM structure of human mTOR Complex 1. Protein Cell. 2016;7:878–887. doi: 10.1007/s13238-016-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aylett CHS, et al. Architecture of human mTOR complex 1. Science. 2015;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 71.Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat Commun. 2016;7 doi: 10.1038/ncomms11016. 11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naydenova K, Russo CJ. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00782-3. 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Meth. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf*GTP. Nat Struct Biol. 2010;17:876–881. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen K, et al. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 2018;556:64–69. doi: 10.1038/nature26158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.