Abstract
Influenza A viruses (IAV) are responsible for seasonal epidemics, and pandemics can arise from novel zoonotic influenza A viruses transmitting to humans1,2. IAV contain a segmented negative sense RNA genome that is transcribed and replicated by the viral RNA-dependent RNA polymerase, composed of the PB1, PB2, and PA subunits3–5. Although the high-resolution crystal structure of bat IAV polymerase (FluPolA) has been reported6, there are no complete structures available for human and avian FluPolA. Furthermore, the molecular mechanisms of viral RNA (vRNA) replication, which proceeds through a complementary RNA (cRNA) replicative intermediate and requires polymerase oligomerisation7–10, remain largely unknown. Here we report 3.0 – 4.3 Å resolution structures of polymerases from human A/NT/60/1968 (H3N2) and avian A/duck/Fujian/01/2002 (H5N1) IAVs, obtained by crystallography and cryo-electron microscopy (cryo-EM), in the presence or absence of cRNA or vRNA template. In solution, FluPolA forms dimers of heterotrimers through the PA C-terminal domain and the PB1 thumb and PB2 N1 subdomains. A cryo-EM structure of a monomeric FluPolA, bound to cRNA template, reveals a binding site for the 3′ cRNA at the dimer interface. Using a combination of cell-based and in vitro assays we show that the FluPolA dimer interface is required for initiation of vRNA synthesis during viral genome replication. Furthermore, we show that a nanobody, a single-domain antibody, which interferes with FluPolA dimerisation, inhibits vRNA synthesis and consequently virus replication in infected cells. Our study provides the first high-resolution structures of medically relevant FluPolA and offers novel insights into the replication mechanisms of the viral RNA genome. Furthermore, it identifies novel sites of FluPolA that could be targeted for antiviral drug development.
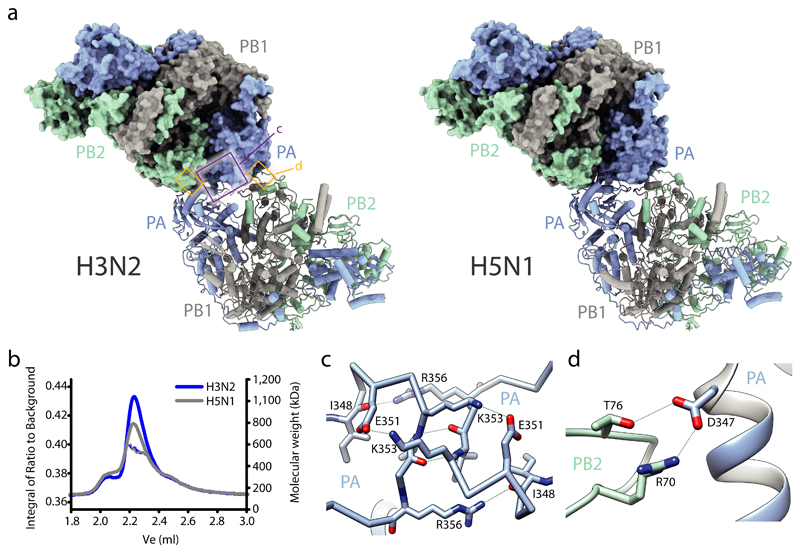
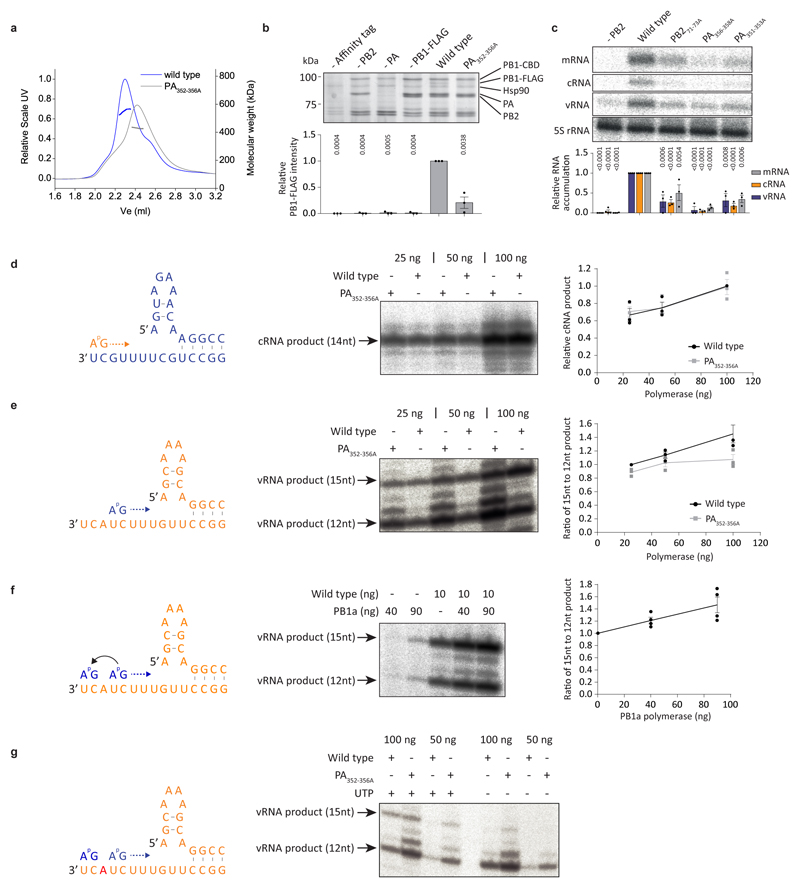
We used x-ray crystallography to solve the complete apo structures of human and avian FluPolA at 3.32 Å and 3.63 Å resolution, respectively (Extended Data Fig. 1a-f, Extended Data Table 1, Supplementary Fig. 1). Human and avian FluPolA are essentially identical structures forming dimers of heterotrimers with dimerisation mediated by the PA C-terminal domain, the PB1 thumb and PB2 N1 subdomains (Fig. 1a, b, Extended Data Fig. 1g-i). A key feature of this dimer interface is loop 352-356 in the PA C-terminal domain that interacts with the same loop of the second polymerase (Fig. 1c, Extended Data Fig. 1g). The dimer interface also involves hydrogen bonds between the PA C-terminal domain and PB2 N1 (Fig. 1d, Extended Data Fig. 1h). Mutation of PA residues 352-356 to alanines (PA352-356A) resulted in a shift towards a monomeric FluPolA heterotrimer (Extended Data Fig. 2a). FluPolA dimerisation through the same interface was also observed in mammalian cells (Extended Data Fig. 2b) and in previous studies of a truncated avian FluPolA 11.
Fig. 1. Structures of human H3N2 and avian H5N1 FluPolA.
a, Crystal structures of dimers of FluPolA heterotrimers from human H3N2 (left) and avian H5N1 (right) influenza A viruses. Regions at the dimer interface (shown in close-up in panels c and d) are boxed. b, SEC-SAXS analysis of human H3N2 and avian H5N1 FluPolA (n=3 independent experiments for H3N2 with similar results and n=1 for H5N1). Smooth lines reflect the relative UV signal of SEC and dotted lines indicate estimated molecular weight for each frame. Note that monomeric FluPolA heterotrimer has an approximate molecular weight of 255 kDa. c, d, Interactions between loops 352-356 of the PA C-terminal domains (c) and the PA C-terminal domain and PB2 N1 subdomain (d) at the FluPolA dimer interface. Dashed lines indicate hydrogen bonds.
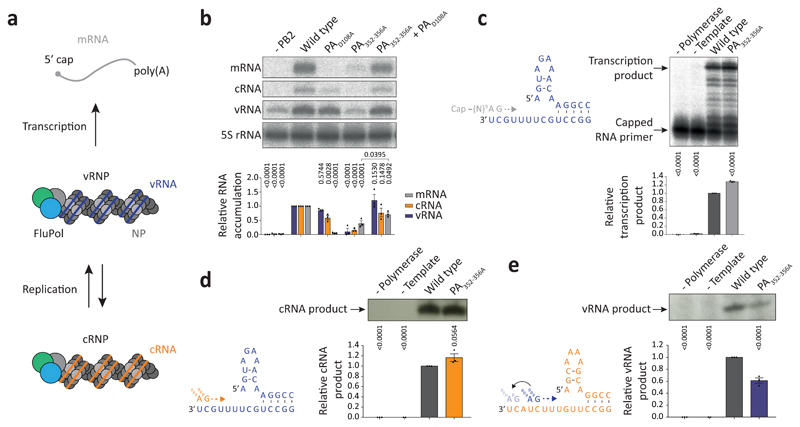
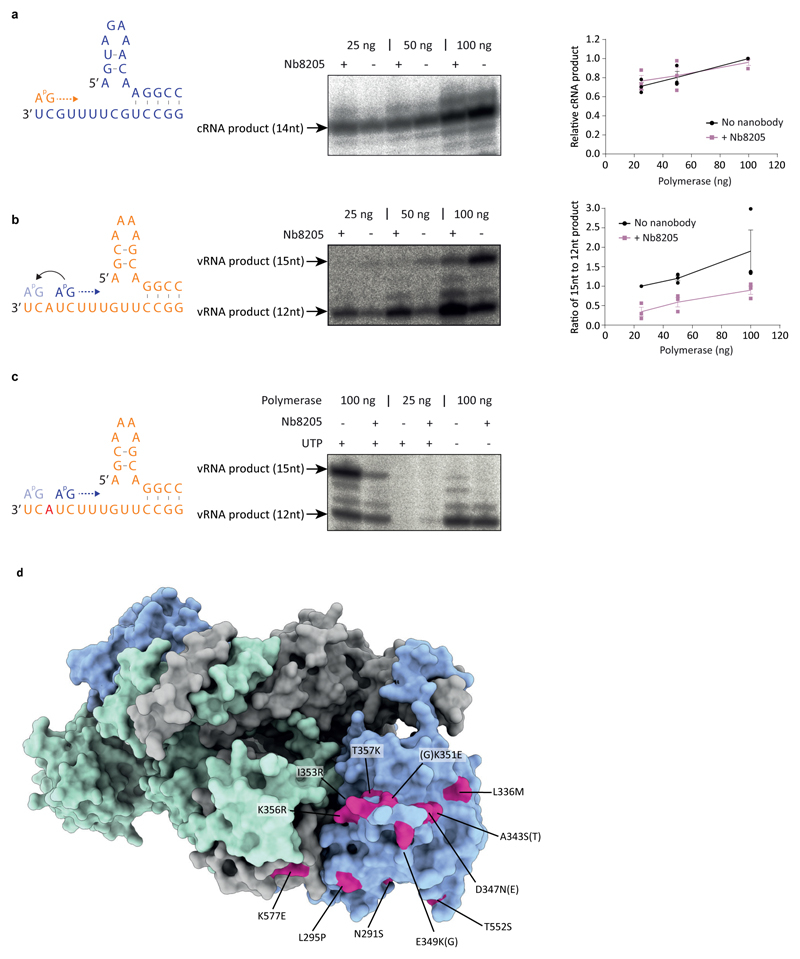
To assess the importance of dimerisation for FluPolA function we used a minireplicon assay measuring viral transcription and replication in the context of viral ribonucleoprotein complexes (vRNPs) (Fig. 2a). The PA352-356A dimer interface mutation and mutations designed to destabilise PB2 loop 71-76 and PA loop 352-356 at the dimer interface significantly decreased the synthesis of all viral RNAs (Fig. 2b, Extended Data Fig. 2c). Co-expression of PA352-356A with PAD108A, a transcription-deficient but replication-competent polymerase12,13, lead to a significant increase in mRNA signal, indicating that the PA352-356A mutant is specifically deficient in viral genome replication (Fig. 2b). In agreement, the PA352-356A mutation did not inhibit transcription in vitro using a vRNA template and capped RNA primer (Fig. 2c). It did not affect primer-independent cRNA synthesis on a vRNA template either but was deficient in vRNA synthesis on a cRNA template (Fig. 2d, e) that involves pppApG synthesis at positions 4 and 5 and subsequent realignment of the template14. In ApG dinucleotide-primed assays, the PA352-356A mutant showed activity on both vRNA and cRNA templates but on the cRNA template promoted the formation of a short 12 nucleotide and other incorrectly initiated products (Extended Fig. 2d, e). The ratio of full-length 15 to the short 12 nucleotide product was dependent on FluPolA concentration, and could be increased by adding a polymerase active site mutant (PB1a), suggesting the involvement of an intermolecular interaction between polymerases (Extended Data Fig. 2f, g). These data show that the FluPolA dimer interface is important for initiation of vRNA synthesis on the cRNA template and suggest that dimerisation promotes stabilisation of the replication complex and the correct positioning of the cRNA template to allow terminal pppApG-primed initiation during vRNA synthesis.
Fig. 2. Mutations at the FluPolA dimer interface inhibit cRNA to vRNA replication.
a, Scheme of transcription and replication by FluPolA in the context of viral ribonucleoproteins (vRNPs). b, vRNP reconstitution assay with the PA352-356A dimer mutant and complementation with the transcription-deficient PAD108A mutant. Data are mean ± s.e.m., n=3 independent transfections. Two-way ANOVA. P < 0.05 is considered significant. mRNA signals for PA352-356A with and without PAD108A were compared by two-tailed unpaired t-test. P < 0.05 is considered significant. c, Effect of the PA352-356A mutation on in vitro transcription by FluPolA primed with a capped RNA primer. Data are mean ± s.e.m., n=3 independent reactions. One-way ANOVA. P<0.05 is considered significant. d, e, Effect of the PA352-356A mutation on in vitro primer-independent replication by FluPolA on a vRNA (d) and cRNA (e) template. Data are mean ± s.e.m., n=3 independent reactions. One-way ANOVA. P < 0.05 is considered significant. For gel source data, see Supplementary Fig. 2.
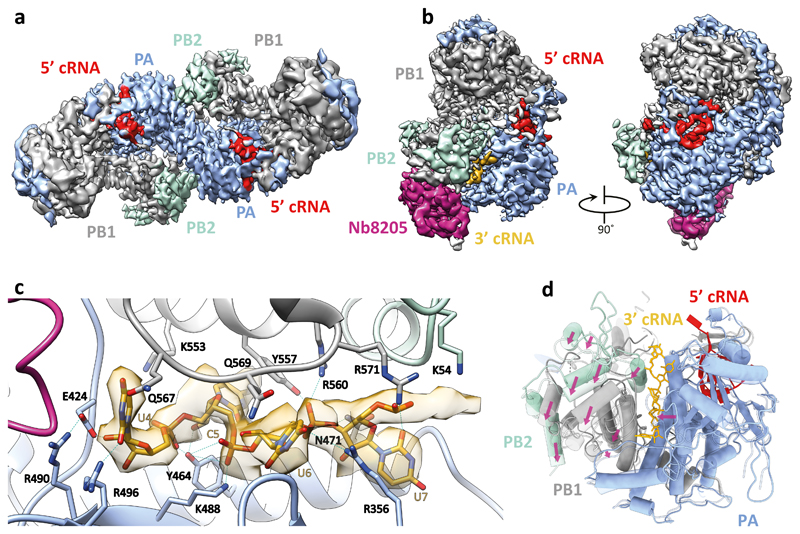
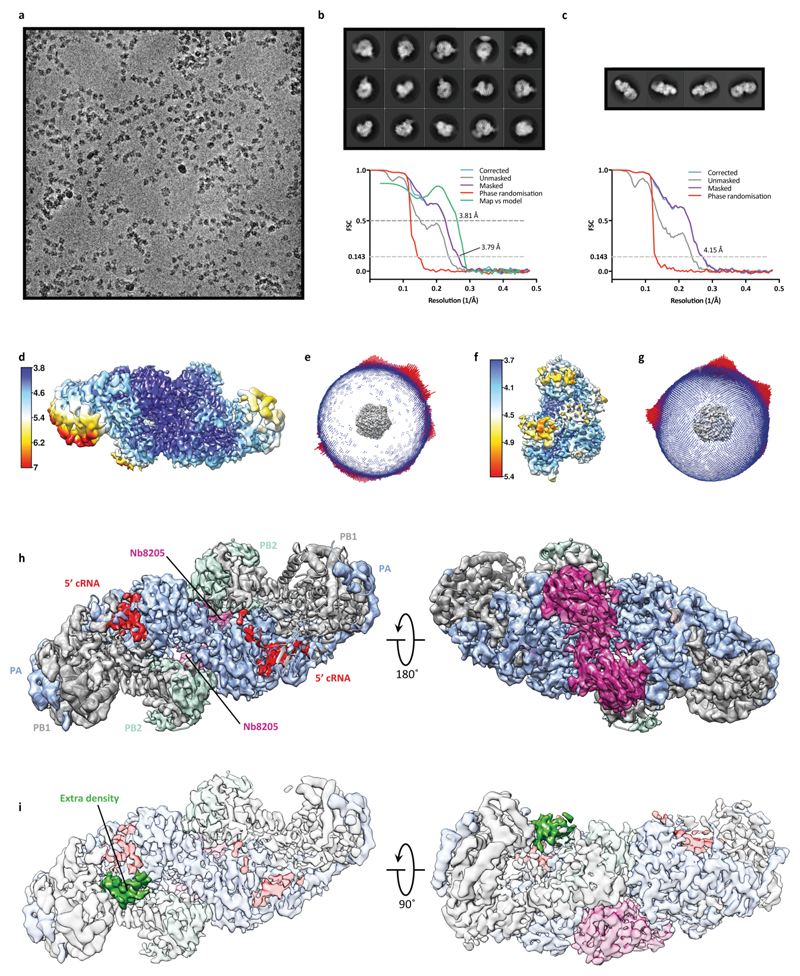
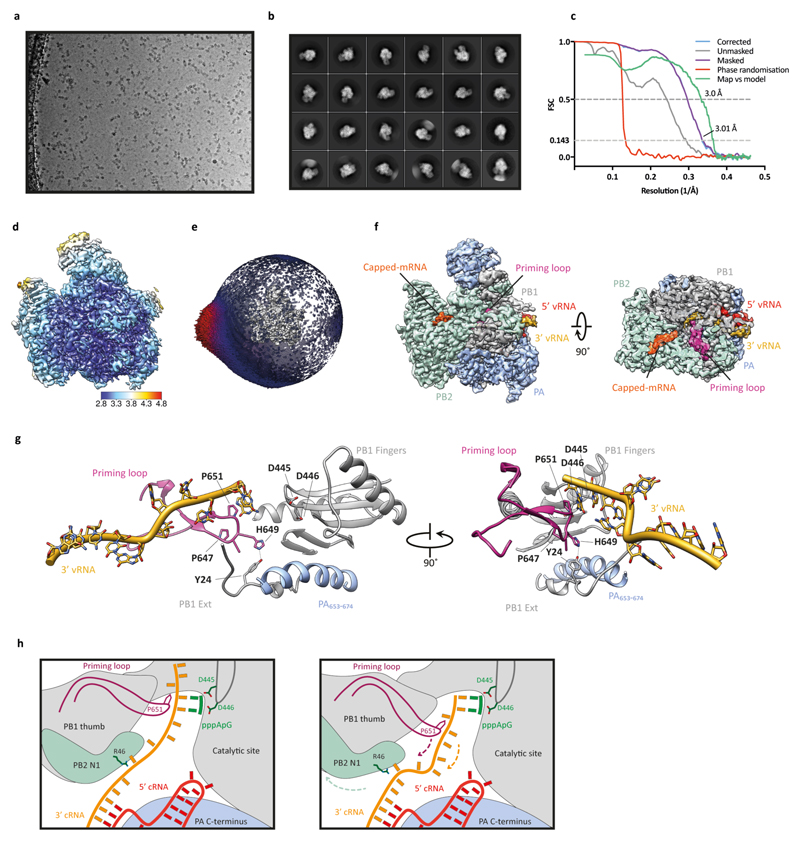
To investigate the binding of FluPolA to the cRNA template we determined the structure of dimeric FluPolA in the presence of cRNA promoter (comprised of 5′ and 3′ cRNA termini)3,4 at 4.07 Å resolution using cryo-EM (Fig. 3a, Extended Data Fig. 3a-f, Extended Data Table 2). Although the PA endonuclease and PB2 C-terminal domains could not be resolved, in agreement with previous observations that these domains are flexible15–17, unambiguous density was observed for the dimer interface, revealing a dimer interface essentially identical to that in the FluPolA crystal structures (Fig. 1a). Only the 5′ cRNA was clearly resolved in the density maps, showing a hook structure bound in a pocket formed by PA and PB1, as observed in previous FluPol structures with 5′ vRNA and cRNA6,16,18. Unresolved density around the template entry channel was observed suggesting that the 3′ cRNA has entered the active site but is highly dynamic (Extended Data Fig. 3g, h). These results confirm that cRNA-bound FluPolA can form dimers in solution.
Fig. 3. Structures of H3N2 FluPolA bound to cRNA promoter.
a, Cryo-EM map of dimer of FluPolA heterotrimers bound to cRNA promoter. b, Cryo-EM map of cRNA-bound FluPolA heterotrimer in complex with Nb8205. c, Close-up view of 3ʹ cRNA binding site. d, Comparison between monomeric (full colour) and dimeric (transparency) FluPolA polymerase reveals movement of the PB1 thumb/PB2 N1 subdomains (indicated by purple arrows) triggered by FluPolA dimerisation, resulting in the opening of the 3ʹ cRNA binding site.
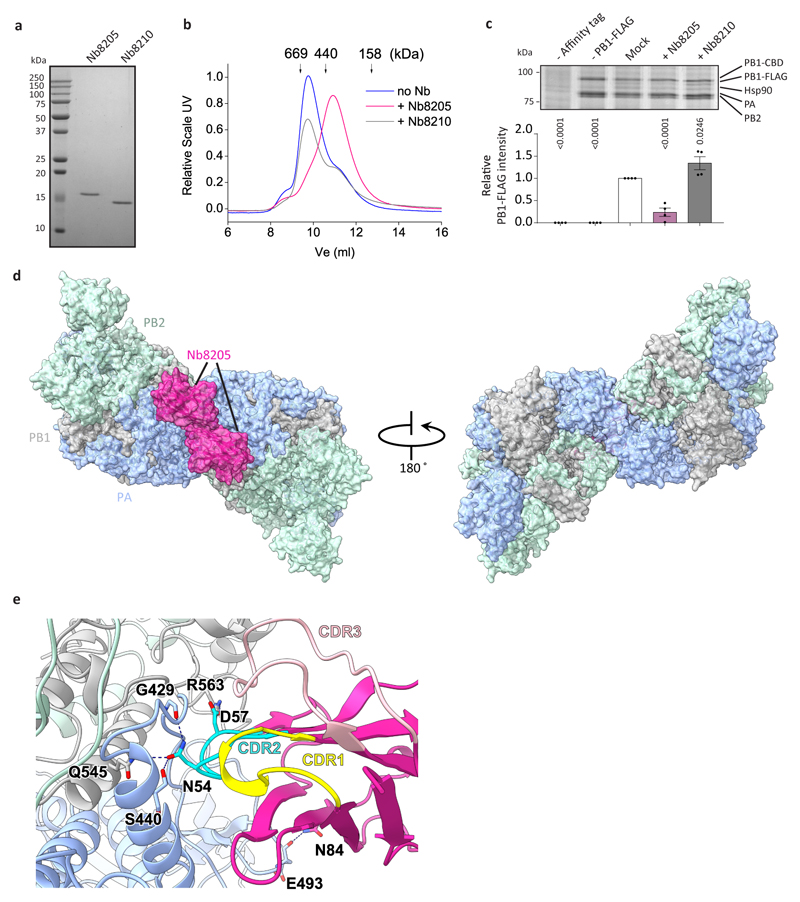
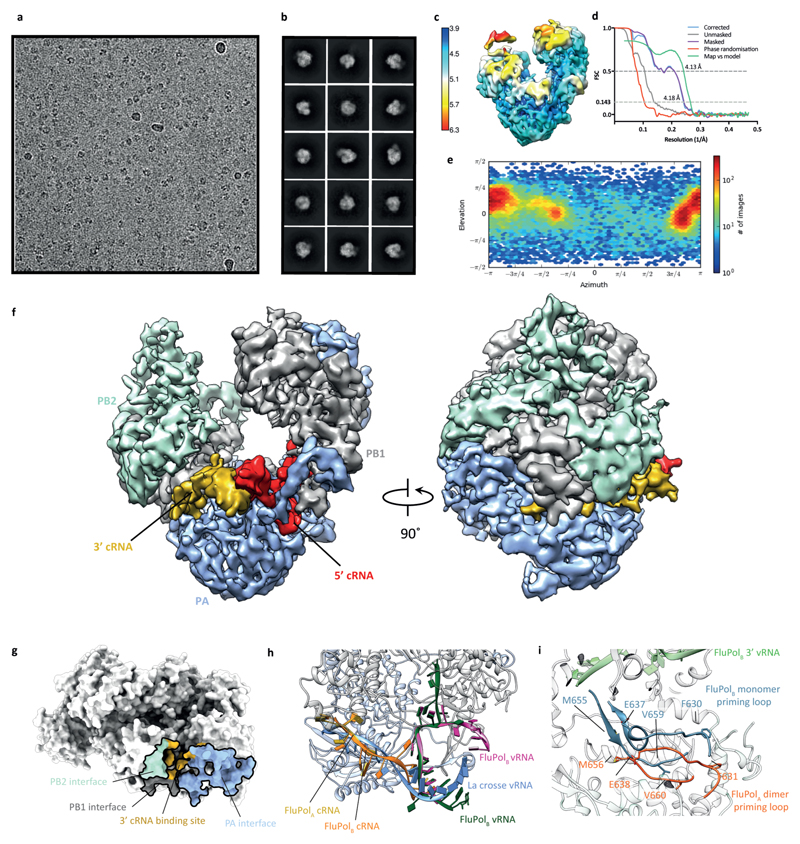
To gain further insight into FluPolA dimerisation and cRNA binding we used a nanobody (Nb8205) raised against FluPolA that reduces FluPolA dimerisation (Extended Data Fig. 4a-c). A crystal structure of the apo FluPolA-Nb8205 complex solved at 3.34 Å resolution revealed that the complementarity determining regions (CDRs) of Nb8205 interact with FluPolA at the PA C-terminal domain and PB1 thumb, a site close to the dimer interface (Extended Data Fig. 4d, e, Extended Data Table 1). We also solved the structures of monomeric and dimeric FluPolA bound to Nb8205 and cRNA promoter at 3.79 and 4.15 Å resolution, respectively, using cryo-EM (Fig. 3b, Extended Data Fig. 5a-h, Extended Data Table 2). The structure of the Nb8205-bound FluPolA dimer is essentially identical to the dimer observed in the absence of Nb8205, showing the same binding mode for the 5′ cRNA terminus with unresolved density around the template entry channel suggesting the presence of a dynamic 3′ cRNA terminus in the active site (Extended Data Fig. 5i). In contrast, the structure of the Nb8205-bound FluPolA monomer revealed the binding of both the 5′ and 3′ cRNA termini (Fig. 3b, Supplementary Video 1). The 5′ cRNA terminus is bound in the hook conformation as observed in the FluPolA dimer, while the 3′ cRNA terminus occupies a binding site formed between the PA C-terminal domain and the PB1 thumb and PB2 N1 subdomains. Only bases 4 to 8 of the 15 nucleotide 3′ cRNA could be resolved which are coordinated by residues of PB1 loop 553-571 and a series of charged residues of PA (E300, Y464, K488, R496) (Fig. 3c, Supplementary Video 1). We observed the same binding site for 3′ cRNA in a cryo-EM structure of the monomeric form of influenza B virus polymerase (FluPolB) (Extended Data Fig. 6a-f). This 3′ cRNA binding site lies in close vicinity to the FluPolA dimer interface (Extended Data Fig. 6g) and could represent a docking site for the 3′ cRNA in the replication pre-initiation state of FluPolA and could also accommodate the 3′ cRNA during replication elongation, after it is copied and extruded through the template exit channel. This site is distinct from the previously observed 3′ vRNA binding site in structures of bat FluPolA and human FluPolB but lies in a similar position as the 3′ vRNA of the La Crosse orthobunyavirus RNA polymerase (Extended Data Fig. 6h)6,18,19.
A comparison of the monomeric and dimeric FluPolA structures in complex with Nb8205 revealed that dimerisation induces a movement of a helical bundle formed by the PB1 thumb and PB2 N1 subdomains (Fig. 3d, Supplementary Video 2). This movement results in an opening of the 3′ cRNA binding site explaining the absence of 3′ cRNA at this site in the dimeric structure. Furthermore, dimerisation leads to rearrangements in the polymerase active site that could destabilise the 3′ cRNA binding, in agreement with the lack of density for the 3′ extremity of the 3′ cRNA in the dimer structure (Extended Data Fig. 5i). Specifically, the visible residues closest to the tip of the priming loop (PB1 residues E638 and M656) are moved away from the active site by approximately 7 Å, presumably pulling the tip out by a similar distance (Extended Data Fig. 6i, Supplementary Video 3). This movement of the priming loop and the destabilised binding of the 3′ cRNA could facilitate backtracking of the cRNA template during the initiation of vRNA synthesis, in agreement with previous data that the priming loop is required during the realignment of the pppApG initiating dinucleotide to the cRNA terminus20,21. In further support of this model we have solved the structure of FluPolA bound to the vRNA promoter and capped RNA by cryo-EM at 3.0 Å resolution (Extended Data Fig. 7a-f). This structure, with the flexible PA endonuclease and PB2 C-terminal domains fully resolved, revealed the interaction of a fully resolved priming loop with the 3′ vRNA in the active site (Extended Data Fig. 7g). The priming loop acts to buttress the template RNA positioning it next the catalytic aspartates (PB1 amino acid residues D445 and D446) to allow terminal initiation. This interaction is mediated by PB1 amino acid P651 at the tip of the priming loop that was found to be critical for priming loop function in our previous study21. The observed buttressing of the template by the priming loop is consistent with our proposed model for template realignment for vRNA synthesis triggered by polymerase dimerisation (Extended Data Fig. 7h, Supplementary Video 4).
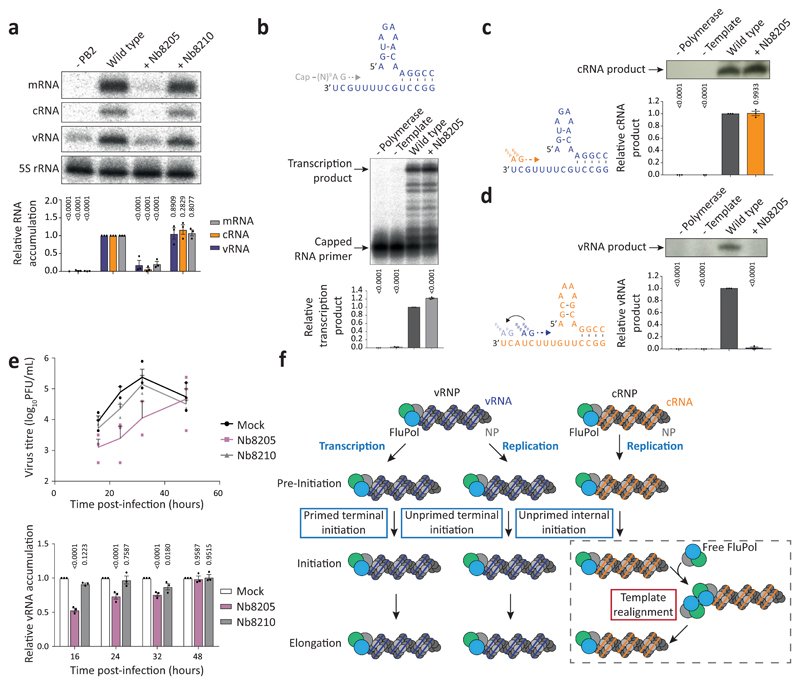
To address the effect of Nb8205 on FluPolA function, co-expression of Nb8205 in a minireplicon assay severely inhibited the accumulation of all viral RNAs, while another nanobody (Nb8210), also raised against FluPolA but not affecting FluPolA dimerisation (Extended Data Fig. 4a-c), had no significant effect (Fig. 4a). Nb8205 had no effect on capped RNA-primed transcription or cRNA synthesis on a vRNA template, but strongly inhibited vRNA synthesis on a cRNA template in vitro (Fig. 4b-d). Addition of Nb8205 to ApG dinucleotide-primed assays did not reduce activity but resulted in incorrectly initiated vRNA products on the cRNA template (Extended Data Fig. 8a-c). Nb8205 inhibited vRNA accumulation at 16 to 32 hours post-infection in cells infected with influenza A/WSN/33 virus and caused a significant reduction in virus titre, while Nb8210 had no or a much smaller effect (Fig. 4e).
Fig. 4. Nanobody Nb8205 that binds FluPolA at the dimer interface inhibits cRNA to vRNA replication and virus growth.
a, Effect of nanobodies on FluPolA activity in a vRNP reconstitution assay. Data are mean ± s.e.m., n=3 independent transfections. Two-way ANOVA. P < 0.05 is considered significant. b, Effect of nanobody on in vitro transcription by FluPolA primed with a capped RNA primer. Data are mean ± s.e.m., n=3 independent reactions. One-way ANOVA. P < 0.05 is considered significant. c, d, Effect of nanobody on in vitro primer-independent replication by FluPolA on a vRNA (c) and cRNA (d) template. Data are mean ± s.e.m., n=3 independent reactions. One-way ANOVA. P < 0.05 is considered significant. e, Effect of nanobodies on the growth of influenza A/WSN/33 virus and vRNA levels in infected HEK-293T cells. Data are mean ± s.e.m., n=3 independent transfections and infections. Two-way ANOVA (Nb8210: P = 0.8126; 0.4390; 0.8496; 0.8489, Nb8205: P = 0.1075; 0.0096; 0.0217; 0.9828, for 16, 24, 32, 48 hours post-infection). P < 0.05 is considered significant. For gel source data, see Supplementary Fig. 2. f, Model for the role of polymerase dimerisation in influenza virus genome replication.
Collectively, these data show that FluPolA dimerisation is required for the initiation of vRNA synthesis on the cRNA template during viral genome replication (Fig. 4f). Replication initiation on the cRNA template being dependent on dimerisation is consistent with our previous observations that vRNA synthesis requires a trans-activating polymerase8. A requirement for trans-activation through polymerase dimerisation provides an elegant mechanism for tuning the amount of vRNA synthesised - only once there is a sufficient level of newly-made free polymerase available in the cell is vRNA production initiated. This could help ensure that the virus does not produce vRNA that cannot be assembled into vRNPs and therefore could trigger an antiviral response through recognition by pathogen recognition receptors, e.g. RIG-I22,23. It is interesting to note that numerous avian to mammalian adaptive mutations have been observed at the dimer interface suggesting that dimerisation of FluPolA may be regulated in a host-specific manner (Extended Data Fig. 8d). In conclusion, the complete high-resolution structures of medically relevant human and avian FluPolA and the identification of novel sites involved in polymerase dimerisation and cRNA promoter binding will provide guidance for the development of influenza antivirals.
Methods
Cells
Human embryonic kidney 293T (HEK-293T) and Sf9 insect cells were sourced from the Cell Bank of the Sir William Dunn School of Pathology, University of Oxford. HEK-293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) and Sf9 cells were maintained in Sf-900 II serum free medium (Gibco). Cell lines have not been authenticated but tested negative for mycoplasma contamination.
Protein expression and purification
The three subunits of human influenza A/NT/60/1968 (H3N2) and avian influenza A/duck/Fujian/01/2002 (H5N1) virus polymerases were co-expressed in Sf9 cells from codon-optimized genes (GeneArt) cloned into a single baculovirus using the MultiBac system24. Mutagenesis of H3N2 FluPolA was carried out using the QuickChange Primer Design Program (Agilent). The influenza B/Panama/45/90 polymerase subunits were expressed as for FluPolA, however they were cloned into the biGBac system25 prior to bacmid generation. Expression and purification of wild-type and mutant FluPolA and FluPolB were performed as described8 with minor modifications. Size exclusion chromatography (SEC) was performed using 25 mM HEPES-NaOH, pH 7.5, 500 mM NaCl, and 5% (v/v) glycerol (Buffer A) on a Superdex 200 Increase 10/300 GL column (GE Healthcare). Pooled fractions from SEC were supplemented with 1 mM TCEP and the final product was concentrated to 3-5 mg ml−1 and used for crystallisation or flash-frozen in liquid nitrogen and stored at -80 °C until further use. All purification steps were performed at 4 ˚C.
Generation, expression and purification of nanobodies
Nanobodies targeting H5N1 FluPolA were generated following established protocols26. Plasmids pMESy4 encoding C-terminal His6-tagged nanobodies were transformed into E. coli strain WK6, grown in 2x YT medium containing 0.1% glucose, 2 mM MgCl2 and 100 mg ml−1 ampicillin at 37 °C until the A600 of the sample reached 0.7, and then induced with 1 mM IPTG and incubated overnight at 28 °C. Cells were collected and the periplasmic fraction was extracted using the modified osmotic shock protocol. The periplasmic extract containing nanobody was incubated with Ni-NTA agarose (Qiagen) for 1 h at room temperature. The beads were washed with 20 volumes of 50 mM K2HPO4:NaH2PO4, pH 7.0, 1 M NaCl, followed by 30 volumes of 50 mM K2HPO4:NaH2PO4, pH 6.0, 1 M NaCl. Nanobodies were eluted by the addition of 15 volumes of 50 mM Na-acetate, pH 4.6, 1 M NaCl. The eluate was neutralised by the addition of 5 volumes of 1 M Tris-HCl, pH 7.5 and concentrated using an Amicon Ultra centrifugal filter unit (Merck Millipore). Nanobodies were further purified on a Superdex 200 Increase 10/300 gel filtration column (GE Healthcare) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl. Nanobodies were concentrated to 4 mg ml−1, flash-frozen and stored at -20 °C.
Crystallisation, data collection and structure determination
Initial hits were found in sitting-drop vapour-diffusion experiments27 at 20 °C in conditions with 0.8-1.2 M phosphate buffer. After optimisation, H3N2 FluPolA protein was crystallised in the condition of 0.9-1.2 M K2HPO4/NaH2PO4, pH 6.7, and H5N1 FluPolA in the condition of 1 M K2HPO4/NaH2PO4, pH 6.9, 0.07% dichloromethane. Crystals appeared within 1-2 hours and grew to full size after 3-4 days. Crystals were cryo-protected by soaking for 10-30 seconds in 25% (v/v) glycerol or 20% (v/v) ethylene glycol in crystallisation buffer before flash-freezing in liquid nitrogen. H3N2 FluPolA was co-crystallised with nanobody Nb8205, added to FluPolA in a 1.5 fold molar excess, using the same conditions as for apo FluPolA. Additional nanobody was added during crystal stabilisation and freezing. Diffraction data were collected at -173 °C on beamlines I03 (wavelength 0.9159 Å) and I24 (wavelength 0.9686 Å) (for H3N2 FluPolA) and I04 (wavelength 0.9795 Å) (for H5N1 FluPolA and H3N2 FluPolA+Nb8205), at the Diamond Light Source, Didcot, UK. Data were processed using XDS28 and STARANISO29, using an anisotropic cut-off and the mean I/σ(I) value of 1.20 to determine the diffraction-limit surface. The structure of H5N1 FluPolA was solved by molecular replacement using Phaser-MR30 as implemented in PHENIX31 and a search model of the FluPolC structure15 (PDB ID: 5D98) with the PB2 627 domain deleted from the model. Phases of individual 627 domains were obtained by a second round of molecular replacement. After rigid-body refinement, the structure was rebuilt with COOT32 and refined with PHENIX31 and autoBUSTER33 until R-factor values converged. Translation-libration-screw (TLS) parameters and 4-fold torsion-angle non-crystallographic symmetry (NCS) were also applied in refinement. The structure of H3N2 FluPolA was solved by molecular replacement using the structure of H5N1 FluPolA (this study) as the search model and refined in the same way. To determine the structure of human FluPolA in complex with nanobody Nb8205, chain N of PDB ID: 3SN634 was used as a search model for the nanobody. In the final H3N2 FluPolA model 93.31 % residues are in the most favoured regions of the Ramachandran plot and 0.09 % are in the disallowed regions (90.93 % and 0.08 % for H5N1 FluPolA model, and 92.65 % and 0 % for H3N2 FluPolA+Nb8205 model, respectively).
Cryo-EM sample preparation
Purified dimer fraction of H3N2 FluPolA was mixed with a 1.2 fold molar excess of 5′ and 3′ cRNA promoters (5′ cRNA: 5′-pAGCAAAAGCAGGCC-3′; 3′ cRNA: 5′-GGCCUUGUUUCUACU-3′) or vRNA promoters (5′ vRNA: 5′-pAGUAGAAACAAGGCC-3′; 3′ vRNA: 5′-GGCCUGCUUUUGCU-3′) and, if stated, a 2.5 fold molar excess of purified nanobody Nb8205 to prepare cRNA-bound FluPolA-nanobody complex. The samples were injected on a Superdex 200 Increase 10/300 GL column (GE Healthcare) running in 25 mM HEPES-NaOH, pH 7.5, 500 mM NaCl. The fractions of interest were concentrated, and protein surface charges were neutralized by adding 0.001% glutaraldehyde for 20 min on ice in order to minimize preferential orientation of particles. After quenching the reaction by adding Tris-HCl, pH 8.0 to a final concentration of 100 mM, the sample was re-injected on a Superdex 200 Increase 10/300 GL column (GE Healthcare) running in 25 mM HEPES-NaOH, pH 7.5, 500 mM NaCl. The fractions of interest were concentrated to 1 mg ml−1 and diluted three fold into 25 mM HEPES-NaOH, pH 7.5, 37.5 mM NaSCN, 0.0075% (v/v) Tween 20 prior to grid preparation. A volume of 3.5 μl of cRNA-bound FluPolA at a concentration of 0.35 mg ml−1 was placed on glow discharged carbon-coated (40 nm film) copper C-flat grids (Protochips) with 2 μm holes and 1.0 μm spacing before blotting for 3.5 s and flash-freezing in liquid ethane. Purified dimer fraction of FluPolB was mixed with a 1.2 fold molar excess of 5′ and 3′ cRNA promoters (5′ cRNA: 5′-pAGCAAAAGCAGGCC-3′; 3′ cRNA: 5′-GGCCUUGUUUCUACU-3′) and the sample was processed as described for FluPolA and concentrated to 0.35 mg ml−1. A volume of 3.5 μl of cRNA-bound FluPolB was used to prepare grids as described above. All grids were prepared using a Vitrobot mark IV (FEI) at 95-100% humidity.
Cryo-EM image collection and processing
Cryo-EM data were collected on a 300 kV Titan Krios microscope (Thermo Fisher Scientific) fitted with a GIF Quantum energy filter (Gatan) at either the Division of Structural Biology (Strubi) or Electron Bio-Imaging Centre (eBIC). For the FluPolA data sets a Volta Phase Plate (Thermo Fisher Scientific) was used. Micrographs were recorded in counting mode using a K2 Summit (Gatan) direct electron detector (or K3 for the FluPolA-vRNA dataset). For sample-specific data collection parameters, see Extended Data Table 2. Movie data were processed using MotionCor2-1.1.035, with a 5 by 5 patch-based alignment, keeping all the frames and dose weighting up to the total exposure. The contrast transfer function and additional phase-shift of full dose non-weighted micrographs was estimated using Gctf-v1.18 or Gctf-v1.0636 for the FluPolB dataset. Poor-quality images were discarded after manual inspection. For cRNA-bound dimeric FluPolA, 56,070 particles were manually picked from the dose-weighted micrographs using the RELION 3.037 manual picking tool, then extracted in a 250 pixel box and subjected to one round of 2D classification resulting in 16 classes with 43,316 selected particles. 3D classification with alignment into three different classes was performed. Two of the classes containing a total number of 36,913 particles were selected and refined to 4.2 Å with C2 symmetry. Bayesian polishing and per particle CTF refinement were performed in RELION 3.0 improving map resolution up to 4.07 Å. The same particles were also refined without symmetry resulting in a map with a resolution of 4.34 Å. Local resolution estimation and sharpening was performed by the RELION sharpening tool using a -100 Å2 and -80 Å2 B-factor, respectively. For the cRNA-bound FluPolA-Nb8205 complex, a first of set of 406,945 particles has been automatically picked with the template picker implemented in cryoSPARC v2.538 using 2D classes from cRNA-bound FluPolA as a template and then 2D classified. A final set of 34,162 particles containing only the dimeric form of the complex has been exported to RELION 3.0 in a 250 pixel box while the monomeric classes have been used as template for another round of automatic picking. From an initial set of 505,860 particles, 216,066 particles containing only the monomeric form were selected after 2D classification and exported into RELION 3.0 in a 200 pixel box. Both dimer and monomer data sets were refined individually to a resolution at 4.38 Å and 4.1 Å using C1 symmetry, respectively. Bayesian polishing and per particle CTF refinement were performed in RELION 3.0 improving map resolution up to 4.15 Å and 3.79 Å, respectively. For the vRNA-bound FluPolA, a first of set of 2,210,168 particles has been automatically picked with the template picker implemented in cryoSPARC v2.538 using 2D classes from cRNA-bound FluPolA as a template and then 2D classified. A final set of 432,160 particles containing only the monomeric form of the complex has been exported to RELION 3.0 in a 250 pixel box. The data were refined to a resolution at 3.3 Å using C1 symmetry. Bayesian polishing and per particle CTF refinement were performed in RELION 3.0 improving map resolution up to 2.9 Å. A final iteration of 3D classification was performed in RELION 3.0, giving a final map at a resolution of 3.01 Å with a final set of 170,144 particles. For cRNA-bound FluPolB, a first of set of 1,012,085 particles was automatically picked with the template picker implemented in cryoSPARC v2.538 using 2D classes of the cRNA-bound FluPolA-Nb8205 complex as a template. After 2D classification, the best views were selected for another round of automatic picking. From an initial set of 324,395 particles, 41,549 particles were selected after 3D classification and exported to RELION 3.0 in a 200 pixel box. The data were refined to a resolution at 4.18 Å using C1 symmetry. The structures were modelled by first fitting an initial model into the locally sharpened map using UCSF Chimera39. One cycle of rigid body real space refinement followed by manual adjustment in Coot40 was performed to correctly position the Cα chain into the density. Finally, cycles of PHENIX31 real space refinement and manual building in Coot40 were used to improve model geometry. Map-to-model comparison in PHENIX mtriage validated that no over-fitting was present in the structures. Model geometry was validated for all models using MolProbity41. All map and model statistics are detailed in Extended Data Table S2.
Analytical size-exclusion chromatography (SEC)
Analytical SEC experiments were performed on a Superdex 200 Increase 10/300 GL column (GE Healthcare) using Buffer A. Wild-type or mutant FluPolA were loaded via a 100 μl sample loop at a concentration of 10 μM. For investigating the effect of nanobodies, the nanobody was added to FluPolA in a 1.5 fold molar excess and incubated on ice for one hour before injection.
Size-exclusion chromatography coupled small-angle X-ray scattering (SEC-SAXS)
SEC-SAXS experiments were performed on beamline B21 at the Diamond Light Source, Didcot, UK. 45 μl purified FluPolA at a concentration of 12 μM was injected on a Shodex KW-403 size exclusion column under a flow rate of 0.16 ml min−1 at 20˚C in Buffer A. Data were collected using continuous 3 second exposures. The data were buffer-subtracted, scaled, merged, and analysed using the ScÅtter 3.0 software (http://www.bioisis.net/scatter). Molecular weight of each individual frame was estimated from DATASW42.
Size-exclusion chromatography coupled multi-angle light scattering (SEC-MALS)
SEC-MALS experiments were performed on beamline B21 at the Diamond Light Source, Didcot, UK. 45 μl purified FluPolA at a concentration of 4 μM was injected on a Shodex KW-403 size exclusion column under a flow rate of 0.16 ml min−1 at 20˚C in Buffer A in the absence of glycerol. An 18-angle multi-angle light scattering instrument (DAWN HELEOS, WYATT) was used to collect light scattering data and the data were processed with ASTRA (WYATT).
Plasmids
Plasmids pcDNA-PB1, pcDNA-PB1a, pcDNA-PB1-FLAG, pcDNA-PB1-TAP (PB1 fused to a C-terminal tandem affinity purification [TAP] tag that consists of a calmodulin binding domain [CBD], a tobacco etch virus [TEV] protease cleavage site, and two copies of protein A), pcDNA-PB2, pcDNA-PB2-TAP, pcDNA-PA, pcDNA-PAD108A, pcDNA-NP and pPOLI-NA have been described12,43–47. Plasmids pcDNA-PB271-73A, pcDNA-PA351-353A, pcDNA-PA356-358A, and pcDNA-PA352-356A, encoding mutant PB2 and PA polymerase subunits, were generated from pcDNA-PB2 and pcDNA-PA using site-directed PCR mutagenesis. Plasmids pcDNA-Nb8205 and pcDNA-Nb8210, to express nanobodies Nb8205 and Nb8210 in mammalian cells, were generated by PCR amplification using pMESy4 plasmids as templates and cloning into pcDNA3A.
FluPolA dimerisation assay
Dimerisation of FluPolA in human embryonic kidney 293T (HEK-293T) cells was assessed as described48. Protein complexes were analysed by SDS-PAGE and silver staining using SilverXpress (Invitrogen) and bands of PB1-FLAG and PB1-CBD quantitated in ImageJ49.
RNP reconstitution assay and primer extension analysis
Approximately 0.2x106 HEK-293T cells were transfected with 0.2 μg of each pcDNA plasmid encoding PB1, PB2, PA, NP and pPOLI plasmid encoding neuraminidase (NA) vRNA segment, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plasmids encoding mutant proteins or nanobodies were included as indicated. For the complementation assay with a transcription-deficient but replication-competent PAD108A polymerase equal amounts of pcDNA-PAD108A and pcDNA-PA356-358A were co-transfected as described previously13,48. Cells were harvested 20 hours post-transfection and total cellular RNA was extracted using TRI reagent (Sigma) according to the manufacturer’s instructions. Viral RNA levels were analysed using primer extension as described50. Briefly, RNA was reverse transcribed using 32P-labelled primers specific to positive and negative sense viral RNAs, with a primer specific to cellular 5S rRNA as a loading control. Transcripts were separated by 6% denaturing PAGE and visualised by phosphorimaging on an FLA-5000 scanner (Fuji). Analysis was carried out using ImageJ49 and Prism 7 (GraphPad). Viral RNA levels were normalised to the 5S rRNA loading control.
Virus growth analysis
Approximately 106 HEK-293T cells were transfected with 5 μg of pcDNA plasmid encoding Nb8205, Nb8210, or an empty pcDNA3 vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 24 hours post-transfection, cells were infected with influenza A/WSN/33 virus at an MOI 0.1. Media was collected 12, 24, 36 and 48 hours post-infection, and virus titres were determined by plaque assay. At each time point, total cellular RNA was also extracted using TRI reagent (Sigma) and vRNA levels were analysed by primer extension50.
In vitro transcription assays
In vitro transcription assays were carried out as described21. Briefly, a cap-1 structure was added to a synthetic 11-nucleotide RNA (5′-ppGAAUACUCAAG-3′) (ChemGenes) by mixing 1 μM of RNA with 0.25 μM [α-32P]GTP (3000 Ci mmol−1; Perkin-Elmer), 0.8 mM S-adenosylmethionine, 0.5 U μl−1 vaccinia virus capping enzyme (NEB) and 2.5 U μl−1 2′-O-methyltransferase (NEB) in a 20 μl reaction at 37 °C for 1 hour. The product was isolated by 16% denaturing PAGE, excised, eluted overnight in dH2O, and desalted using NAP-10 columns (GE Healthcare). Transcription reactions were performed using ∼1,500 c.p.m. capped RNA primer in a 3 μl reaction mixture containing 1mM ATP, 0.5 mM CTP, 0.5 mM UTP, 0.1 μM GTP, 5 mM MgCl2, 1 mM DTT, 2 U μl−1 RNasin, 0.5 μM 5′ vRNA promoter, 0.5 μM 3′ vRNA promoter, 70 ng Nb8205 (if indicated) and 100 ng H3N2 FluPolA. Reactions were incubated for 10 min at 30 °C and stopped by the addition of an equal volume of 80% formamide, 1 mM EDTA, and bromophenol blue and xylene cyanol dyes, followed by incubation at 95 °C for 3 min. Products were resolved by 20% denaturing PAGE and visualised by phosphorimaging on an FLA-5000 scanner (Fuji). Analysis was carried out using ImageJ49 and Prism 7 (GraphPad).
In vitro replication assays
Primer-independent and ApG extension assays were carried out as described21. Briefly, 3 μl reaction mixtures containing 1 mM ATP, 0.5 mM CTP, 0.5 mM UTP (if indicated), 0.1 μM GTP, 0.05 μM [α-32P]GTP (3000 Ci mmol−1; Perkin-Elmer), 0.25 mM ApG (if indicated), 5 mM MgCl2, 1 mM DTT, 2 U μl−1 RNasin, 0.5 μM 5′ vRNA or cRNA promoter, 0.5 μM 3′ vRNA or cRNA promoter, 70 ng Nb8205 (if indicated) and 10-100 ng H3N2 FluPolA were incubated at 37 °C for 4 hours. Where PB1a active site mutant polymerase was added, the concentrations of 5′ and 3′ cRNA promoters were adjusted accordingly to maintain a constant molar ratio of promoter to polymerase. Reactions were stopped by addition of an equal volume of 80% formamide, 1 mM EDTA, and bromophenol blue and xylene cyanol dyes, followed by incubation at 95 °C for 3 min. Products were resolved by 20% denaturing PAGE and visualised by phosphorimaging on an FLA-5000 scanner (Fuji). Analysis was carried out using imageJ49 and Prism 7 (GraphPad).
Extended Data
Extended Data Fig. 1. Subunit organisation of FluPolA heterotrimers.
a, b, Views of the structure of human H3N2 (a) and avian H5N1 (b) FluPolA heterotrimers, coloured according to subunit. c–e, Structures of human H3N2 FluPolA subunits PA (c), PB1 (d) and PB2 (e), coloured and labelled by domain. f, Domain maps of each H3N2 FluPolA subunit. g-i, The 2D Fo – mFc electron density maps of FluPolA dimer interface as shown in Fig. 1c (g, stereo view), and 1d (h, stereo view), as well as of the complete FluPolA dimer (i), are shown in gray mesh (contoured at 1.5 σ, all from the H3N2 FluPolA structure model).
Extended Data Fig. 2. Effect of mutations at the dimer interface on FluPolA dimerisation and activity.
a, SEC-MALS analysis of wild type and PA352-356A mutant H3N2 FluPolA (n=1 independent experiment). Smooth lines reflect the relative UV signal of SEC and dotted lines indicate estimated molecular weight for each frame. Note that monomeric FluPolA heterotrimer has an approximate molecular weight of 255 kDa. b, Effect of PA352-356A mutation on FluPolA dimerisation in HEK-293T cells. Data are mean ± s.e.m., n=3 independent transfections. One-way ANOVA. P < 0.05 is considered significant. c, Effect of mutations designed to destabilise PB2 and PA loops at the FluPolA dimer interface on FluPolA activity in a vRNP reconstitution assay. Data are mean ± s.e.m., n=3 independent transfections. Two-way ANOVA. P < 0.05 is considered significant. d, e, Effect of PA352-356A mutation on in vitro ApG-primer replication by FluPolA on a vRNA (d) and cRNA (e) template. f, Effect of an active site polymerase mutant (PB1a) on in vitro ApG-primer replication by FluPolA on a cRNA template. Data are mean ± s.e.m., n=4 independent reactions. g, Omitting UTP from in vitro ApG-primer replication by FluPolA on a cRNA template affects the synthesis of the 15 nucleotide full-length vRNA but not of the 12 nucleotide short vRNA indicating that the 12 nucleotide product is derived from internal initiation by the ApG dinucleotide at positions 4 and 5 of the cRNA template. The position in the template at which UTP is required is indicated in red. Representative data from n=2 independent reactions. For gel source data, see Supplementary Fig. 2.
Extended Data Fig. 3. Single-particle cryo-EM analysis of human H3N2 FluPolA bound to cRNA promoter.
a, Representative micrograph of cRNA-bound FluPolA heterotrimer particles embedded in vitreous ice. b, Representative 2D class averages. c, FSC curves for 3D reconstruction using gold-standard refinement in RELION, indicating overall map resolution of 4.07 Å and the model-to-map FSC. Curves are shown for phase randomisation, unmasked, masked and phase-randomisation-corrected masked maps. d, 3D reconstruction locally filtered and coloured according to RELION local resolution. e, Angular distribution of particle projections with the cryo-EM map shown in grey. f, Cryo-EM density of the PA loop 352-356 at the dimer interface. g, Cryo-EM map of cRNA-bound FluPolA dimer refined without symmetry imposed (C1), revealing an extra density (green) located next to the 3ʹ end of the 5ʹ cRNA close to the template entry channel. h, Close-up views highlighting cryo-EM extra density (dark green) with the 3′ vRNA strand from the superimposed FluPolB structure51 (PDB: 5MSG, light green) inserting into the polymerase active site. Localisation of the 3ʹ vRNA shows that bases are positioned in the extra density facing the density corresponding to the 3ʹ end of the 5ʹ cRNA, suggesting the presence of a promoter RNA duplex region as observed in vRNA-bound FluPolB 51. The extra density is consistent with the presence of a 3ʹ cRNA in one of the heterotrimers of the cRNA-bound FluPolA dimer, oriented towards the polymerase active site.
Extended Data Fig. 4. The effect of Nb8205 on FluPolA dimerisation.
a, SDS-PAGE of purified nanobodies (n=1 independent experiment). B, Analytical SEC of FluPolA in complex with nanobodies (n=4 for Nb8205 and n=2 for Nb8210, with similar results). c, Effect of nanobodies on FluPolA dimerisation in HEK-293T cells. Data are mean ± s.e.m., n=4 independent transfections. One-way ANOVA. P < 0.05 is considered significant. For gel source data, see Supplementary Fig. 2. d, Crystal structure of H3N2 FluPolA in complex with Nb8205. e, Close-up view of FluPolA-Nb8205 interactions. Residues involved in hydrogen bonding interactions are labelled and hydrogen bonds are indicated with dashed lines. The complementarity determining regions (CDRs) are coloured individually and labelled.
Extended Data Fig. 5. Single-particle cryo-EM analysis of monomeric and dimeric cRNA-bound human H3N2 FluPolA heterotrimer in complex with Nb8205.
a, Representative micrograph of cRNA-bound FluPolA in complex with Nb8205 embedded in vitreous ice. b, Representative 2D class averages. c, FSC curves for the 3D reconstruction using gold-standard refinement in RELION, indicating overall map resolution of 3.79 Å and 4.15 Å for the monomeric and dimeric FluPolA form, respectively, and the model-to-map FSC. Curves are shown for phase randomisation, unmasked, masked and phase-randomisation-corrected masked maps. d, f, 3D reconstruction locally filtered and coloured according to RELION local resolution for the dimeric (d) and monomeric (f) form. e, g, Angular distribution of particle projections for the dimeric (e) and monomeric (g) form with the cryo-EM map shown in grey. h, Dimer of FluPolA heterotrimers bound to cRNA promoter and Nb8205 rigid body fitted into the cryo-EM map of dimeric cRNA-bound FluPolA heterotrimer in complex with Nb8205. i, Cryo-EM map of the dimeric cRNA-bound FluPolA heterotrimer in complex with Nb8205 revealing an extra density (green) located next to the 3ʹ end of the 5ʹ cRNA, as observed for the cRNA-bound FluPolA dimer (Extended Data Fig. 3g, h).
Extended Data Fig. 6. Single-particle cryo-EM analysis of cRNA-bound FluPolB.
a, Representative micrograph of cRNA-bound FluPolB heterotrimer particles embedded in vitreous ice b, Representative 2D class averages. c, 3D reconstruction locally filtered and coloured according to RELION local resolution. d, FSC curves for the 3D reconstruction using gold-standard refinement in RELION, indicating overall map resolution of 4.18 Å and the model-to-map FSC. Curves are shown for the phase randomisation, unmasked, masked, phase-randomisation-corrected masked maps. e, Angular distribution of particle projections according to cryoSPARC v2.5 non-uniform refinement. f, Cryo-EM map of cRNA-bound FluPolB. g, Comparison of the dimerisation interface and the 3ʹ cRNA binding site in H3N2 FluPolA (PDB: 6QNW and 6QPG). h, 3ʹ cRNA binding site in FluPolA and FluPolB overlaps with the previously identified 3′ vRNA binding site in the La Crosse orthobunyavirus polymerase19 (PDB: 5AMQ). Sites of 3ʹ vRNA binding at surface of the polymerase in FluPolB (PDB: 4WRT) and in the polymerase active site for FluPolB (PDB: 5MSG) are shown for comparison6,51. i, Comparison of the structure of dimeric FluPolA to monomeric FluPolB 51 (PDB: 5MSG) reveals a movement of the priming loop that protrudes from the PB1 thumb subdomain into the polymerase active site. Resolved PB1 residues closest to the tip of the priming loop, E638 and M656, move away from the corresponding E637 and M655 residues in FluPolB and the polymerase active site, indicated by the end of the 3ʹ vRNA, by approximately 7 Å.
Extended Data Fig. 7. Single-particle cryo-EM analysis of human H3N2 FluPolA bound to vRNA promoter.
a, Representative micrograph of vRNA-bound FluPolA heterotrimer particles embedded in vitreous ice. b, Representative 2D class averages. c, FSC curves for 3D reconstruction using gold-standard refinement in RELION, indicating overall map resolution of 3.01 Å and the model-to-map FSC. Curves are shown for phase randomisation, unmasked, masked and phase-randomisation-corrected masked maps. d, 3D reconstruction locally filtered and coloured according to RELION local resolution. e, Angular distribution of particle projections with the cryo-EM map shown in grey. f, Cryo-EM map of vRNA-bound FluPolA heterotrimer revealing the presence of a fully resolved priming loop. g, Close-up views highlighting the stacking of the 3′ vRNA by the priming loop. h, Cartoon illustration of the role of polymerase dimerisation in template realignment during replication initiation on a cRNA template. Base-pairing between the 5′ and 3′ cRNA positions bases 4 and 5 of the 3′ cRNA next to the catalytic aspartates (PB1 amino acid residues D445 and D446) in the active site to allow internal replication initiation by the synthesis of a pppApG dinucleotide. The priming loop stacks the cRNA template through PB1 amino acid P651 (left panel). Rotation of the PB1 thumb/PB2-N1 domain triggered by polymerase dimerisation results in a movement of the priming loop and backtracking of the stacked template (arrows). Backtracking is also facilitated by an interaction of PB2 amino acid residue R46 with the 3′ cRNA introducing a ‘kink’ in the template. Backtracking positions bases 1 and 2 of the cRNA template opposite the pppApG dinucleotide that remains coordinated by the catalytic aspartates. The resulting replication complex is ready to extend the pppApG dinucleotide by incorporating the next incoming NTP (right panel).
Extended Data Fig. 8. Effect of Nb8205 on FluPolA activity and mapping of host adaptive mutations at the FluPolA dimer interface.
a, b, Effect of Nb8205 on in vitro ApG-primer replication by FluPolA on a vRNA (a) and cRNA (b) template. Data are mean ± s.e.m., n=3 independent reactions. c, Omitting UTP from in vitro ApG-primer replication by FluPolA on a cRNA template affects the synthesis of the 15 nucleotide full-length vRNA but not of the 12 nucleotide short vRNA. The position in the template at which UTP is required is indicated in red. Representative data from n=2 independent reactions. For gel source data, see Supplementary Fig. 2. d, Crystal structure of H3N2 FluPolA with amino acid residues implicated in avian to mammalian host adaptation of influenza A viruses indicated52–66.
Extended Table 1. Crystallographic data collection and refinement statistics.
| H3N2 FluPolA
PDB: 6QNW |
HSNl FluPolA
PDB: 6QPF |
H3N2 FluPolA-Nb8205 PDB: 6QPG |
|
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 336.5, 191.9, 235.7 | 337.1, 192.9, 235.7 | 335.2, 192.9, 235.1 |
| α, β, γ (°) | 90.0, 91.5, 90.0 | 90.0, 91.5, 90.0 | 90.0, 91.8, 90.0 |
| Resolution* (Å) | 235.6-3.3 (3.92-3.32) | 235.6-3.6 (4.27-3.63) | 235.0-3.3 (3.87-3.34) |
| R merge * (ellipsoidal) | 0.46 (2.24) | 0.18 (0.61) | 0.34 (1.24) |
| I / σI* | 4.8 (2.0) | 2.9 (1.6) | 4.8 (1.8) |
| Completeness* (%, ellipsoidal) | 95.3 (80.7) | 92.7 (72.4) | 91.9 (74.5) |
| Redundancy* | 14.4 (13.7) | 3.4 (3.4) | 7.0 (6.9) |
| Refinement | |||
| Resolution (Å) | 135.34-3.32 | 235.61-3.63 | 234.98-3.34 |
| No. reflections | 123,065 | 81,274 | 89,539 |
| R work / R free (%) | 23.5/27.9 | 27.7/32.3 | 25.6/30.1 |
| No. atoms | |||
| Protein | 67,232 | 67,760 | 70,549 |
| Ligand/ion | 0 | 0 | 0 |
| Water | 0 | 0 | 0 |
| B-factors (Å2) | |||
| Protein | 117.0 | 101.0 | 78.0 |
| Ligand/ion | - | - | - |
| Water | - | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.003 | 0.002 |
| Bond anales (°) | 0.62 | 0.71 | 0.62 |
Values in parentheses are for highest-resolution shell.
Number of crystals used for each dataset: 4 (6QNW), 1 (6QPF), and 2 (6QPG).
Extended Table 2. Cryo-EM data collection, refinement and validation statistics.
| FluPolA-cRNA | FluPolA-cRNA-Nb8205 | FluPolB-cRNA | Flu PolA-vRNA | |||
|---|---|---|---|---|---|---|
| Dimer | Monomer | Dimer | Monomer | Monomer | ||
| EMD-4664 | EMD-4663 | EMD-4661 | EMD-4666 | EMD-4660 | EMD-4986 | |
| PDB 6QX8 | PDB 6QX3 | PDB 6QXE | PDB 6QWL | PDB 6RR7 | ||
| Data collection and processing | ||||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 | ||
| Voltage (kV) | 300 | 300 | 300 | 300 | ||
| Electron exposure (e–/Å2) | 32.92 | 30.00 | 31.10 | 36.60 | ||
| Defocus range (μm) | -0.5 to -0.7 | -0.5 to -0.7 | -1.3 to -2.5 | -0.5 | ||
| Pixel size (Å) | 1.043 | 1.080 | 1.080 | 1.085 | ||
| Symmetry imposed | Cl | C2 | Cl | Cl | Cl | Cl |
| Initial particle images (no.) | 56,070 | 505,860 | 406,945 | 1,012,085 | 2,210,168 | |
| Final particle images (no.) | 36,913 | 52,932 | 27,861 | 41,549 | 170,144 | |
| Map resolution (Å) | 4.34 | 4.07 | 3.79 | 4.15 | 4.18 | 3.01 |
| (FSC threshold 0.143) | ||||||
| Map resolution range (Å) | 3.97-7.82 | 3.84-6.35 | 3.70-5.41 | 3.85-7.43 | 3.90-6.30 | 2.82-5.03 |
| 3DFSC spherity* | 0.758 | 0.787 | 0.851 | 0.772 | 0.781 | 0.909 |
| CryoEF score** | 0.41 | 0.53 | 0.72 | 0.50 | 0.68 | 0.50 |
| Refinement | ||||||
| Initial model used (PDB code) | - | 6QNW | 6QPG | 6QX8/6QPG | 5EPI | 6QNW |
| Model resolution (Å) | 4.34 | 4.07 | 3.79 | 4.15 | 4.18 | 3.01 |
| (FSC threshold 0.143) | ||||||
| Model resolution range (Å) | 3.97-7.82 | 3.84-6.35 | 3.70-5.41 | 3.85-7.43 | 3.90-6.30 | 2.82-5.03 |
| Map sharpening B factor (Å2) | -80 | -100 | -110 | -95 | -57 | -65 |
| Model composition | ||||||
| Non-hydrogen atoms | - | 38,596 | 21,589 | - | 10,409 | 36,306 |
| Protein residues | - | 2,384 | 1,337 | - | 1,244 | 2,228 |
| Nucleotide (RNA) | - | 24 | 14 | - | 26 | 30 |
| B factors (Å2) | ||||||
| Protein | - | 68.60 | 71.24 | - | 151.24 | 22.39 |
| Nucleotide (RNA) | - | 64.94 | 86.46 | - | 207.12 | 24.68 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | - | 0.007 | 0.004 | - | 0.008 | 0.007 |
| Bond angles (°) | - | 1.158 | 0.750 | - | 1.006 | 0.820 |
| Validation | ||||||
| MolProbity score | - | 1.85 | 1.66 | - | 2.1 0 | 1.31 |
| Clashscore | - | 6.48 | 3.24 | - | 10.07 | 1.90 |
| Poor rotamers (%) | - | 0.14 | 0 | - | 0.93 | 0 |
| Ramachandran plot | ||||||
| Favored (%) | - | 91.86 | 90.34 | - | 89.07 | 94.88 |
| Allowed (%) | - | 7.97 | 9.66 | - | 10.85 | 5.12 |
| Disallowed (%) | - | 0.17 | 0 | - | 0.08 | 0 |
Supplementary Material
Acknowledgements
We thank G.G. Brownlee and F. Vreede for plasmids, I. Berger for the MultiBac system and S. Cusack for sharing and discussing unpublished data on the 3′ promoter binding site. We thank K. Harlos and T. Walter for assistance with crystallisation and K. Dent and D. Clare for cryo-EM assistance. We thank G.G. Brownlee, D. Stuart, and A. te Velthuis, as well as members of the Fodor and Grimes laboratories, for helpful comments and discussions. We thank Instruct-ERIC, part of the European Strategy Forum on Research Infrastructures (ESFRI), Instruct-ULTRA (EU H2020 Grant 731005), and the Research Foundation - Flanders (FWO) for support with nanobody discovery. This work was supported by Medical Research Council (MRC) programme grants MR/K000241/1 and MR/R009945/1 (to E.F.), Wellcome Investigator Award 200835/Z/16/Z (to J.M.G.), MRC Studentships (to A.P.W. and I.S.M.), Wellcome Studentship 092931/Z/10/Z (to N.H.). The authors would like to thank Diamond Light source for beamtime (proposals MX10627, MX14744, and MX19946), and for access and support of the cryo-EM facilities at the UK national Electron Bio-Imaging Centre (eBIC) (proposal EM14856), funded by the Wellcome, MRC and BBSRC. Further EM provision was provided through the OPIC electron microscopy facility which was founded by a Wellcome JIF award (060208/Z/00/Z) and is supported by a Wellcome equipment grant (093305/Z/10/Z). Computation used the Oxford Biomedical Research Computing (BMRC) facility, a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre. Financial support was provided by a Wellcome Trust Core Award (203141/Z/16/Z). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Part of this work was supported by Wellcome administrative support grant 203141/Z/16/Z.
Footnotes
Author contributions
H.F., A.P.W., L.C., J.R.K., J.M.G. and E.F. conceived and designed the study. H.F., L.C., J.R.K., and N.H. carried out cloning of recombinant baculoviruses and protein purification. H.F. J.R.K. performed crystallisations, data collection and analysis, model building and refinement. L.C. and J.R.K. collected and processed electron microscopy data and built and refined models with assistance from D.K. and I.S.M. A.P.W. performed functional assays and analysed data. J.S. performed dimerisation assays in mammalian cells and E.F. analysed the data. E.P. and J.S. designed and generated Nb8205 and Nb8210 and N.H. performed nanobody expression and purification. J.M.G and E.F. supervised the structural and functional studies, respectively. H.F., A.P.W., L.C., J.R.K, J.M.G. and E.F. wrote the manuscript, with input from all co-authors.
Competing interests The authors declare no competing interests.
Data availability
All data are available from the corresponding authors and/or included in the manuscript or Supplementary Information. Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6QNW (H3N2 FluPolA), 6QPF (H5N1 FluPolA) and 6QPG (H3N2 FluPolA+Nb8205). Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-4661 (monomeric H3N2 FluPolA+cRNA+Nb8205), EMD-4663 and 4664 (dimeric H3N2 FluPolA+cRNA), EMD-4666 (dimeric H3N2 FluPolA+cRNA+Nb8205), EMD-4660 (monomeric FluPolB+cRNA), and EMD-4986 (monomeric H3N2 FluPolA+vRNA+capped RNA) with the corresponding atomic coordinates deposited in the Protein Data Bank with accession numbers 6QX3, 6QX8, 6QXE, 6QWL, 6RR7, respectively.
References
- 1.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–51. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostafa A, Abdelwhab EM, Mettenleiter TC, Pleschka S. Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses. 2018;10 doi: 10.3390/v10090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res. 2017;234:103–117. doi: 10.1016/j.virusres.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Te Velthuis AJ, Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol. 2016;14:479–93. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker AP, Fodor E. Interplay between Influenza Virus and the Host RNA Polymerase II Transcriptional Machinery. Trends Microbiol. 2019;27:398–407. doi: 10.1016/j.tim.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–60. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 7.Jorba N, Coloma R, Ortin J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 2009;5:e1000462. doi: 10.1371/journal.ppat.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc Natl Acad Sci U S A. 2013;110:E4238–E4245. doi: 10.1073/pnas.1315068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorba N, Area E, Ortin J. Oligomerization of the influenza virus polymerase complex in vivo. J Gen Virol. 2008;89:520–4. doi: 10.1099/vir.0.83387-0. [DOI] [PubMed] [Google Scholar]
- 10.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–4. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang S, et al. Cryo-EM structure of influenza virus RNA polymerase complex at 4.3 A resolution. Mol Cell. 2015;57:925–935. doi: 10.1016/j.molcel.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 12.Hara K, Schmidt FI, Crow M. Brownlee GG Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol. 2006;80:7789–98. doi: 10.1128/JVI.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manz B, Brunotte L, Reuther P, Schwemmle M. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat Commun. 2012;3:802. doi: 10.1038/ncomms1804. [DOI] [PubMed] [Google Scholar]
- 14.Deng T, Vreede FT. Brownlee GG Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol. 2006;80:2337–48. doi: 10.1128/JVI.80.5.2337-2348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengrung N, et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature. 2015;527:114–7. doi: 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thierry E, et al. Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol Cell. 2016;61:125–37. doi: 10.1016/j.molcel.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serna Martin I, et al. A Mechanism for the Activation of the Influenza Virus Transcriptase. Mol Cell. 2018;70:1101–1110 e4. doi: 10.1016/j.molcel.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich S, et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:3616. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 19.Gerlach P, Malet H, Cusack S, Reguera J. Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter. Cell. 2015;161:1267–79. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oymans J, Te Velthuis AJW. A Mechanism for Priming and Realignment during Influenza A Virus Replication. J Virol. 2018;92 doi: 10.1128/JVI.01773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Te Velthuis AJ, Robb NC, Kapanidis AN, Fodor E. The role of the priming loop in influenza A virus RNA synthesis. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.29. 16029. [DOI] [PubMed] [Google Scholar]
- 22.Killip MJ, Fodor E, Randall RE. Influenza virus activation of the interferon system. Virus Res. 2015;209:11–22. doi: 10.1016/j.virusres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Te Velthuis AJW, et al. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat Microbiol. 2018;3:1234–1242. doi: 10.1038/s41564-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends in Biochemical Sciences. 2012;37:4957. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weissmann F, et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci U S A. 2016;113:E2564–9. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9 doi: 10.1038/nprot.2014.039. 67493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallographica Section D-Biological Crystallography. 2005;61 doi: 10.1107/S0907444905007808. 651657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66 doi: 10.1107/S0907444909047337. 12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tickle IJ, et al. STARANISO. Cambridge, United Kingdom: Global Phasing Ltd; 2018. ( http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi) [Google Scholar]
- 30.Mccoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40 doi: 10.1107/S0021889807021206. 658674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–80. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shkumatov AV, Strelkov SV. DATASW, a tool for HPLC-SAXS data analysis. Acta Crystallogr D Biol Crystallogr. 2015;71:1347–50. doi: 10.1107/S1399004715007154. [DOI] [PubMed] [Google Scholar]
- 43.Deng T, Sharps J, Fodor E, Brownlee GG. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J Virol. 2005;79:8669–74. doi: 10.1128/JVI.79.13.8669-8674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fodor E, et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol. 2002;76:8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fodor E, et al. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–82. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J Virol. 2004;78:9144–53. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vreede FT, Jung TE, Brownlee GG. Brownlee GG Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol. 2004;78:9568–72. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsson-Payant BE, Sharps J, Hengrung N, Fodor E. The Surface-Exposed PA(51-72)-Loop of the Influenza A Virus Polymerase Is Required for Viral Genome Replication. J Virol. 2018;92 doi: 10.1128/JVI.00687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol. 2009;90:1398–407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- 51.Reich S, Guilligay D, Cusack S. An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase. Nucleic Acids Res. 2017;45:3353–3368. doi: 10.1093/nar/gkx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bussey KA, et al. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J Virol. 2011;85:7020–8. doi: 10.1128/JVI.00522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, et al. The PA-gene-mediated lethal dissemination and excessive innate immune response contribute to the high virulence of H5N1 avian influenza virus in mice. J Virol. 2013;87:2660–72. doi: 10.1128/JVI.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ilyushina NA, et al. Adaptation of pandemic H1N1 influenza viruses in mice. J Virol. 2010;84:8607–16. doi: 10.1128/JVI.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamiki H, et al. A PB1-K577E Mutation in H9N2 Influenza Virus Increases Polymerase Activity and Pathogenicity in Mice. Viruses. 2018;10 doi: 10.3390/v10110653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee CY, et al. Novel mutations in avian PA in combination with an adaptive mutation in PR8 NP exacerbate the virulence of PR8-derived recombinant influenza A viruses in mice. Vet Microbiol. 2018;221:114–121. doi: 10.1016/j.vetmic.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Liedmann S, et al. New virulence determinants contribute to the enhanced immune response and reduced virulence of an influenza A virus A/PR8/34 variant. J Infect Dis. 2014;209:532–41. doi: 10.1093/infdis/jit463. [DOI] [PubMed] [Google Scholar]
- 58.Mehle A, Dugan VG, Taubenberger JK, Doudna JA. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J Virol. 2012;86:1750–7. doi: 10.1128/JVI.06203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann G, Macken CA, Kawaoka Y. Identification of amino acid changes that may have been critical for the genesis of A(H7N9) influenza viruses. J Virol. 2014;88:4877–96. doi: 10.1128/JVI.00107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng X, et al. Amino Acid Substitutions HA A150V, PA A343T, and PB2 E627K Increase the Virulence of H5N6 Influenza Virus in Mice. Front Microbiol. 2018;9:453. doi: 10.3389/fmicb.2018.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slaine PD, et al. Adaptive Mutations in Influenza A/California/07/2009 Enhance Polymerase Activity and Infectious Virion Production. Viruses. 2018;10 doi: 10.3390/v10050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu R, et al. Multiple amino acid substitutions are involved in the adaptation of H9N2 avian influenza virus to mice. Vet Microbiol. 2009;138:85–91. doi: 10.1016/j.vetmic.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Xu G, et al. Prevailing PA Mutation K356R in Avian Influenza H9N2 Virus Increases Mammalian Replication and Pathogenicity. J Virol. 2016;90:8105–14. doi: 10.1128/JVI.00883-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamaji R, et al. Mammalian adaptive mutations of the PA protein of highly pathogenic avian H5N1 influenza virus. J Virol. 2015;89:4117–25. doi: 10.1128/JVI.03532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, et al. Multiple amino acid substitutions involved in enhanced pathogenicity of LPAI H9N2 in mice. Infect Genet Evol. 2011;11:1790–7. doi: 10.1016/j.meegid.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 66.Zhong G, et al. Mutations in the PA Protein of Avian H5N1 Influenza Viruses Affect Polymerase Activity and Mouse Virulence. J Virol. 2018;92 doi: 10.1128/JVI.01557-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naydenova K, Russo CJ. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun. 2017;8:629. doi: 10.1038/s41467-017-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding authors and/or included in the manuscript or Supplementary Information. Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6QNW (H3N2 FluPolA), 6QPF (H5N1 FluPolA) and 6QPG (H3N2 FluPolA+Nb8205). Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-4661 (monomeric H3N2 FluPolA+cRNA+Nb8205), EMD-4663 and 4664 (dimeric H3N2 FluPolA+cRNA), EMD-4666 (dimeric H3N2 FluPolA+cRNA+Nb8205), EMD-4660 (monomeric FluPolB+cRNA), and EMD-4986 (monomeric H3N2 FluPolA+vRNA+capped RNA) with the corresponding atomic coordinates deposited in the Protein Data Bank with accession numbers 6QX3, 6QX8, 6QXE, 6QWL, 6RR7, respectively.