Abstract
Genomic imprinting, a phenomenon by which genes are expressed in a monoallelic, parent-of-origin-dependent fashion, is critical for normal brain development. Expression of imprinted genes is regulated via epigenetic mechanisms, including DNA methylation (5-methylcytosine, 5mC), and disruptions in imprinting can lead to disease. Early-life exposure to the endocrine disrupting chemical bisphenol A (BPA) is associated with abnormalities in brain development and behavior, as well as with disruptions in epigenetic patterning, including 5mC and DNA hydroxymethylation (5-hydroxymethylcytosine, 5hmC). Using an established mouse model of perinatal environmental exposure, the objective of this study was to examine the effects of perinatal BPA exposure on epigenetic regulation of imprinted gene expression in adult mice. Two weeks prior to mating, dams were assigned to control chow or chow containing an environmentally relevant dose (50 µg/kg) of BPA. Exposure continued until offspring were weaned at post-natal day 21, and animals were followed until 10 months of age. Expression of three imprinted genes—Pde10a, Ppp1r9a, and Kcnq1, as well as three genes encoding proteins critical for regulation of 5mC and 5hmC—Dnmt1, Tet1, and Tet2, were evaluated in the right cortex and midbrain using qRT-PCR. Perinatal BPA exposure was associated with a significant increase in adult Kcnq1 (p = 0.04) and Dnmt1 (p = 0.02) expression in the right cortex, as well as increased expression of Tet2 in the midbrain (p = 0.03). Expression of Tet2 and Kcnq1 were positively correlated in the midbrain. Analysis of 5mC and 5hmC at the Kcnq1 locus was conducted in parallel samples using standard and oxidative bisulfite conversion followed by pyrosequencing. This analysis revealed enrichment of both 5mC and 5hmC at this locus in both brain regions. No significant changes in 5mC and 5hmC at Kcnq1 were observed with perinatal BPA exposure. Together, these data suggest that perinatal BPA exposure results in altered expression of Kcnq1, Dnmt1, and Tet2 in the adult mouse brain. Further studies with larger sample sizes are necessary to understand the mechanistic basis for these changes, as well as to determine the implications they have for brain development and function.
Keywords: bisphenol A, imprinted genes, DNA methylation, DNA hydroxymethylation, brain
Introduction
The Developmental Origins of Health and Disease (DOHaD) hypothesis states that environmental exposures during critical windows of development influence the risk of diseases later in life (Barker, 2007). An important mechanism by which developmental exposures can affect long-term disease risk is via disruption of the normal epigenetic processes regulating gene expression (Dolinoy and Jirtle, 2008). One critical epigenetic mechanism of gene regulation is DNA methylation, the addition of a methyl group to the fifth position of cytosines in cytosine-phosphate-guanine (CpG) dinucleotides (5-methylcytosine, 5mC) (Jones, 2012). DNA methylation is critical for establishment of tissue-specific gene expression patterns, regulation of imprinted genes, maintenance of genome stability, and silencing of transposable elements (Jones, 2012). Because it is stable, mitotically heritable, and undergoes rapid reprogramming during early development, DNA methylation is particularly vulnerable to the effects of toxicant exposures (Dolinoy and Jirtle, 2008). 5mC can be further oxidized to 5-hydroxymethylcytosine (5hmC) (Lopez et al., 2017). 5hmC was initially proposed to be an intermediate in the DNA demethylation pathway; however, recent studies suggest that the genomic distribution, downstream targets, and functional roles of 5hmC are distinct from that of 5mC (Lopez et al., 2017). 5mC and 5hmC undergo dynamic changes during brain development, and both marks play a critical role in neuronal function, including learning and memory, during development and adulthood (Spiers et al., 2015; Jobe and Zhao, 2017; Spiers et al., 2017). Alterations in these marks are associated with neurodegenerative and psychiatric illnesses, suggesting a role for 5mC and 5hmC in disease pathogenesis (Zhao et al., 2017; Cui and Xu, 2018).
Genomic imprinting is an epigenetic mechanism resulting in mono-allelic, parent-of-origin expression of a subset of genes. Genomic imprinting is mediated in large part by DNA methylation, in cooperation with histone marks and long non-coding RNAs (Reik and Walter, 2001; Barlow and Bartolomei, 2014). During primordial germ cell development, the parental imprinting marks are erased, and new imprinting marks are then re-established, in a pattern consistent with the sex of the developing embryo (Reik and Walter, 2001). Because imprinted genes are functionally haploid, environmental exposures may be more likely to disrupt their expression and function, with potentially serious health consequences (Dolinoy et al., 2007). We and others have recently reported that perinatal environmental exposures disrupt programming of 5mC and 5hmC at imprinted loci in pancreatic islets (Susiarjo et al., 2013), blood (Li et al., 2016; Kochmanski et al., 2018), and mouse tail samples (Faulk et al., 2014), and lead to alterations in imprinted gene expression (Susiarjo et al., 2013; Kochmanski et al., 2018). Proper expression of imprinted genes is critical for normal neurodevelopment, and disruptions in imprinting are associated with several human brain disorders (Perez et al., 2016).
Bisphenol A (BPA) is a widely used, endocrine disrupting chemical found in a variety of consumer products, including food containers, receipt paper, and medical equipment. Widespread use of BPA has resulted in ubiquitous human exposure (Vandenberg et al., 2007; Vandenberg et al., 2010). Perinatal exposure to BPA is associated with a number of adverse neurodevelopmental, behavioral, and psychiatric outcomes in human and animal models, including attention deficit/hyperactivity behaviors, learning deficits, anxiety, and depression (Matsuda et al., 2012; Harley et al., 2013; Johnson et al., 2016; Xin et al., 2018). Furthermore, recent work suggests that perinatal BPA exposure may lead to altered expression of imprinted genes in the offspring brain (Drobna et al., 2018), and that BPA-induced alterations in DNA methylation may play a role in BPA-mediated adverse effects on brain development (Kundakovic et al., 2013; Kumar and Thakur, 2017; Drobna et al., 2018). Based on this evidence, we hypothesized that perinatal BPA exposure would result in altered epigenetic programming of imprinted gene expression in the brain, long after cessation of exposure. In particular, given our recent finding that perinatal BPA exposure alters DNA methylation and hydroxymethylation at imprinted loci in the blood (Kochmanski et al., 2018), we hypothesized that these changes in imprinted gene expression would be mediated by programming of 5hmC and/or 5mC. To address these questions, we utilized an established mouse model of perinatal BPA exposure to investigate whether BPA alters programming of imprinted gene expression at 10 months of age. For this study, we investigated three imprinted genes: Kcnq1, Ppp1r9a, and Pde10a. We previously found that these genes were differentially hydroxymethylated in the blood of mice after perinatal exposure to BPA (Kochmanski et al., 2018). Moreover, mutations and/or disruption in expression of these genes are associated with neurological and psychiatric illnesses (Goldman et al., 2009; Konopaske et al., 2015; Kelly, 2018).
Methods
Exposure Paradigm and Brain Collection
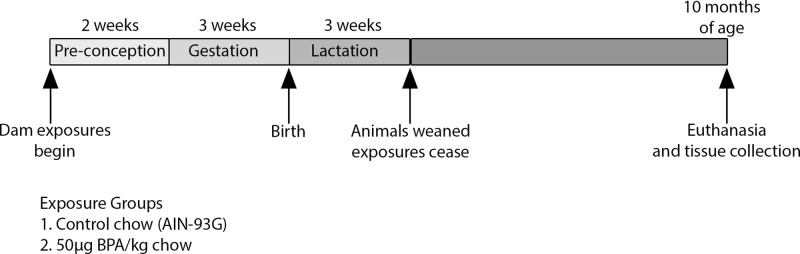
Mice utilized for breeding and exposure were obtained from a colony maintained for over 230 generations with the Avy (viable yellow agouti) allele passed through the male line, resulting in forced heterozygosity on a genetically invariant background with 93% identity to C57BL/6J (Waterland and Jirtle, 2003; Weinhouse et al., 2014). Two weeks prior to mating with Avy/a males, eight to ten-week-old virgin a/a dams were assigned to experimental diets consisting of control AIN-93G chow or chow containing 50 µg BPA/kg. Chow was supplied by Harlan Teklad, and BPA was supplied by the NTP (National Toxicology Program, Durham NC). The dose of BPA was chosen based on published data demonstrating that this results in human-relevant exposure levels (Anderson et al., 2012). Dams were kept on experimental diets ad libitum beginning 2 weeks prior to mating through pregnancy and lactation. On postnatal day 21, exposures ceased and offspring were weaned to control AIN-93G diet and followed until 10 months of age. Only wild-type, a/a offspring were followed for this study. At 10 months of age, mice were euthanized and the right cortex, left cortex, and midbrain of each animal were dissected out, flash-frozen and stored at -80°C for later use. This study utilized right cortex and midbrain samples (n = 6 control and 6 BPA-exposed) from the same a/a offspring in which BPA-induced changes in 5mC and 5hmC were identified in the blood (Kochmanski et al., 2018). Each exposure group had three males and three females. Figure 1 provides a summary of the exposure paradigm and experimental design. All animal studies were carried out in an AAALAC accredited facility, under approval by the University of Michigan Institutional Animal Care & Use Committee (IACUC) and in compliance with the Guide for the Care and Use of Laboratory Animals.
Figure 1.
Exposure paradigm and experimental timeline. Dams were exposed to control or BPA-containing chow starting two weeks prior to mating until weaning. Exposures ceased on offspring postnatal day 21, and animals were administered control chow until 10 months of age. Offspring brains were collected and sectioned at 10 months of age. Six mice (three males and three females) per group were analyzed in this study.
DNA and RNA Isolation
DNA and RNA were isolated from brain samples using the Qiagen Allprep DNA/RNA/miRNA Universal Kit (Qiagen, Cat. #80224). Briefly, frozen samples from the right cortex and midbrain of each mouse were thawed to the point where 30 mg could be excised from each brain section. Tissue samples were then homogenized using a TissueLyser (Qiagen) and treated according to the Qiagen Allprep DNA/RNA/miRNA Universal Kit Protocol. All isolated RNA and DNA samples were assessed for yield and purity using a NanoDrop spectrophotometer.
Analysis of Gene Expression Using Quantitative Real-Time PCR
Expression of three imprinted genes previously identified to be differentially hydroxymethylated with perinatal BPA exposure (Kochmanski et al., 2018)—Kcnq1, Pde10a, and Ppp1r9a and three genes encoding enzymes that catalyze the formation of 5mC and 5hmC—Dnmt1, Tet1 and Tet2 were quantified separately by brain region for each of the twelve mice using quantitative real-time PCR (qRT-PCR). qRT-PCR primers were designed using the online Genscript Real-time PCR Primer Design software, and specificity for all designed primers was checked using the NCBI Primer-BLAST online tool. See Supplementary Table 1 for primer sequences; 1000 ng of RNA from each sample was reverse transcribed into complementary DNA (cDNA) using the Bio-Rad iScript cDNA Synthesis Kit (Cat. #1708890). cDNA was diluted 1:5 in RNase-free water, mixed with 10 µM forward/reverse primers, nuclease-free water, and Bio-Rad iQ SYBR Green Supermix (Cat. #1708880). qRT-PCR was performed using the pre-programmed 2-step PCR+ melt curve protocol on a Bio-rad CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad; Hercules, CA). All target genes and three housekeeping genes (β-actin, 18S, GAPDH) for each brain sample were run in triplicate on the same plate. Samples in which the coefficient of variation between the triplicates was greater than 10% were discarded and rerun. Data were normalized to the average Ct value for all three housekeeping genes. Relative gene expression was calculated separately by brain region using the ΔΔCt method (Yuan et al., 2006; Rao et al., 2013). Statistical analysis was performed using a two-sided non-parametric Wilcoxon test using the stats package in R. Summary statistics were obtained using the EpiDisplay package in R. Graphs were generated using the ggplot2 package in R.
DNA Methylation and Hydroxymethylation Quantification
Pyrosequencing primers for Kcnq1 were designed using the PyroMark Assay Design Software 2.0 Methylation Analysis (CpG) Assay and the July 2007 NCBI37/mm9 mouse genome ( Supplementary Table 2 ). Primers were designed to target a CpG within an intronic region reported to be differentially hydroxymethylated with perinatal BPA exposure in the blood (Kochmanski et al., 2018); see Figure 4 for location. Primers were assessed for mispriming using the BiSearch: Primer Design and Search Tool. DNA samples and low and high mouse DNA methylation standards from EpigenDX were bisulfite and oxidative bisulfite treated according to the NuGEN TrueMethyl oxBS Module protocol (Cat. #0414-32). Briefly, 1 µg of input genomic DNA was adjusted with nuclease-free water to 50 µl and each genomic DNA sample was divided into two aliquots. Each aliquot underwent independent, parallel treatments and were either oxidative bisulfite converted or mock oxidative bisulfite converted according to the TrueMethyl protocol. The yield and purity of treated samples were quantified using a NanoDrop spectrophotometer. The target locus within Kcnq1 was PCR amplified in both the bisulfite and oxidative bisulfite converted samples. PCR products were verified using the QIAxcel automated DNA electrophoresis. DNA methylation levels were quantified using the PyroMark Q96 ID instrument (Qiagen). All plates were run with bisulfite and oxidative bisulfite converted low and high DNA methylation standards from EpigenDX. Samples that failed the PyroMark internal quality control assessment were rerun and only methylation percentages that passed were used for analysis. Bisulfite converted samples reveal the total level of 5mC + 5hmC, while oxidative bisulfite treated samples show total levels of 5mC. Thus, 5hmC levels were quantified by subtracting the result obtained from the oxidative bisulfite converted sample (5mC) from the bisulfite converted sample (5mC + 5hmC). For statistical analysis, Mann-Whitney and mixed linear effects models were used to asses differences in methylation and hydroxymethylation by exposure group. Analyses in the right cortex and midbrain were conducted separately. Exposure group, sex and methylation category (5mC or 5hmC) were included in all models, and a random factor for sample ID was included in all models to account for sample autocorrelation from matched 5mC and 5hmC percentages for each individual. The lme4 package within the statistical program R was used for these analyses. Raw pyrosequencing data can be found in Supplementary Table 4 . Correlation analyses were assessed with a Spearman Correlation Test using the ggpubr package in R. Correlation plots depict Ct values that have been normalized to the average Ct value of the three housekeeping genes.
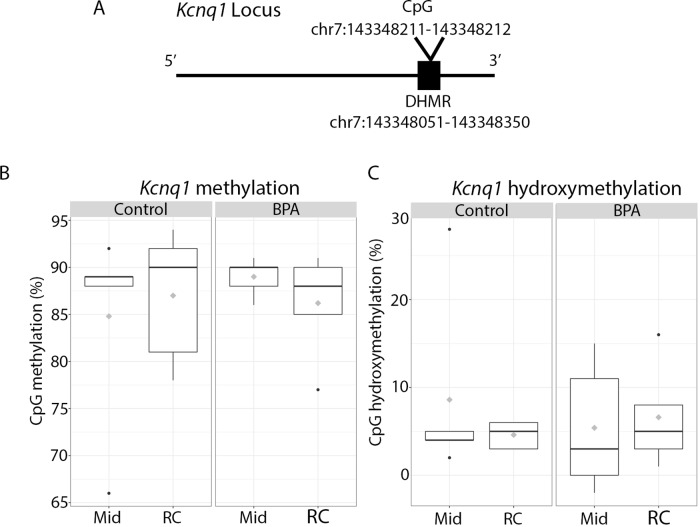
Figure 4.
Enrichment of 5mC and 5hmC at the Kcnq1 locus in 10 month old animals exposed to control vs. BPA. (A) Schematic of the Kcnq1 locus, depicting the location of the CpG analyzed in this study. This CpG falls within an intronic region that was previously identified to be differentially hydroxymethylated with BPA exposure (DHMR) in the blood (J Kochmanski et al., 2018). (B, C) Box plots depicting CpG methylation (B) and hydroxymethylation (C) in the midbrain (Mid) and right cortex (RC) in control vs. exposed offspring. Raw pyrosequencing data can be found in Supplementary Table 4 . Five samples from each exposure group/brain region passed the PyroMark quality control assessment and were used in the analysis. n = 5 mice per exposure/brain region.
Results
Litter Parameters and Phenotypic Data
To investigate the effects of BPA exposure on epigenetic regulation of imprinted gene expression, we utilized brain tissue from a previous study in an established mouse model of perinatal BPA exposure (Kochmanski et al., 2018) (see Methods). Dams were exposed to BPA or control chow beginning two weeks prior to mating until weaning at post-natal day (PND) 21 ( Figure 1 ). Thus, offspring were exposed to BPA during pregnancy and lactation. After PND21, exposures ceased and all animals received control chow until euthanasia at 10 months of age. Developmental BPA exposure had no effect on litter size, ratio of males to females, or the a/a to Avy/a genotypic ratio (Kochmanski et al., 2018). As noted previously, BPA exposure resulted in a significant reduction in the number of pups per litter that survived at weaning (Kochmanski et al., 2017).
Perinatal BPA Exposure Alters Imprinted Gene Expression in the Mouse Brain
To assess the effects of perinatal BPA exposure on expression of imprinted genes, we used qRT-PCR to measure expression of Kcnq1, Pde10a, and Ppp1r9a in the brains of 10-month-old mice. Notably, although BPA exposure was discontinued at three weeks of age, Kcnq1 expression in the right cortex was significantly higher in BPA-exposed animals compared to controls at 10 months of age (p = 0.04) ( Figure 2A and Table 1 ). In the midbrain, there was a trend toward increased expression of Kcnq1 in BPA-exposed animals that did not reach statistical significance (p = 0.13) ( Figure 2B and Table 1 ). Pde10a expression was higher in the BPA exposed group compared to the control group in both the right cortex and midbrain, although the differences did not reach statistical significance (p = 0.18 and 0.13 for right cortex and midbrain respectively, Figures 2C, D and Table 1 ). Ppp1r9a expression in the right cortex and midbrain did not differ between control and BPA-exposed mice ( Figures 2E, F and Table 1 ). Collectively, these data demonstrate that perinatal BPA exposure altered Kcnq1 expression in the brains of adult mice, months after discontinuation of exposure.
Figure 2.
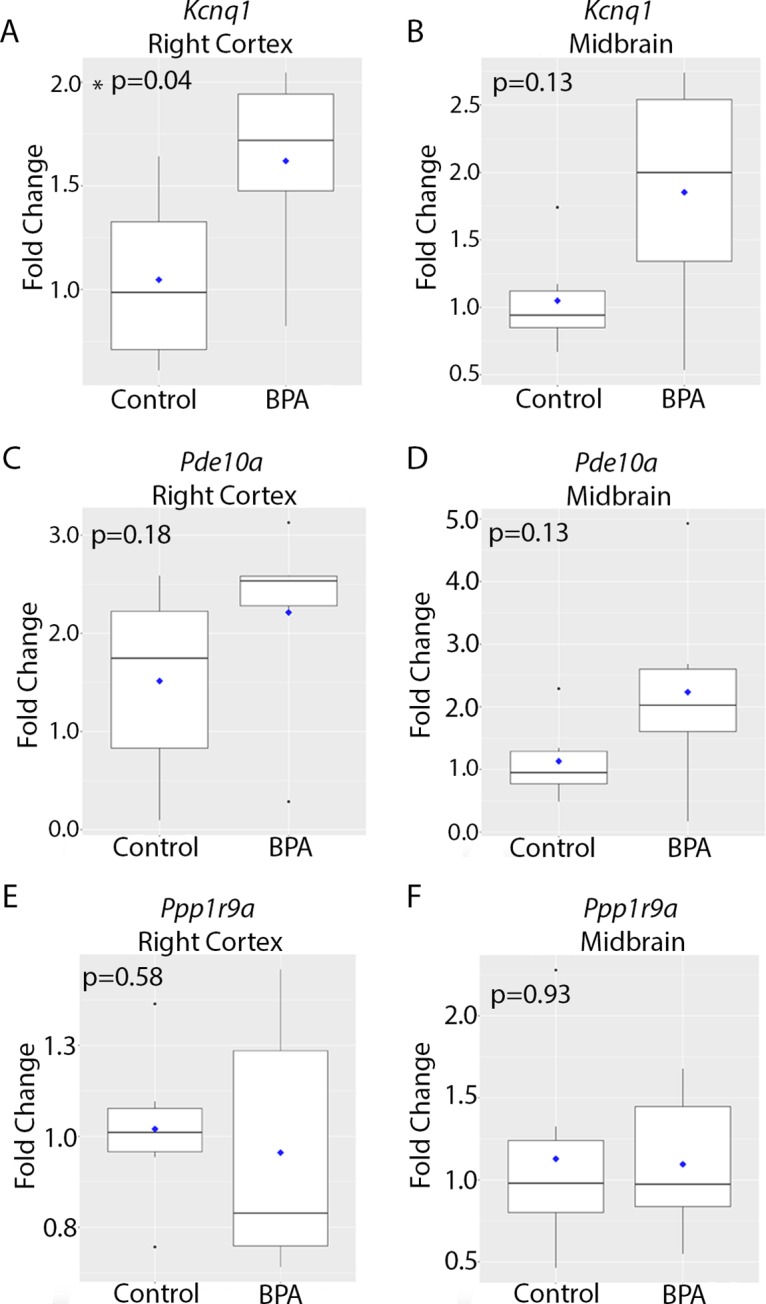
Imprinted gene expression in the brain at 10 months of age in animals exposed to control vs. BPA. Box plots depicting qRT-PCR data for Kcnq1 (A, B), Pde10a (C, D), and Pp1r9a (E, F) in the right cortex (A, C, E) and midbrain (B, D, F). Data were analyzed using a two-sided non-parametric Wilcoxon test. Graphs depict the fold change in gene expression relative to the average of the control samples. n = 6 mice per exposure.
Table 1.
Kcnq1 qRT-PCR Summary Statistics.
| Gene | Region | Exposure | N | Meana (SD) | Mediana (Range) | Mann-Whitney test (α = 0.05) |
|---|---|---|---|---|---|---|
| Kcnq1 | Right Cortex | Control | 6 | 16.18 (0.60) | 16.13 (15.46-16.89) | 0.04 |
| BPA | 6 | 15.54 (0.49) | 15.40 (15.14-16.46) | |||
| Midbrain | Control | 6 | 16.43 (0.47) | 16.51 (15.63-17.01) | 0.13 | |
| BPA | 6 | 15.73 (0.90) | 15.44 (14.97-17.33) | |||
| Pde10a | Right Cortex | Control | 6 | 10.20 (1.84) | 9.40 (8.83-13.60) | 0.18 |
| BPA | 9.36 (1.31) | 8.86 (8.55-12.01) | ||||
| Midbrain | Control | 6 | 12.56 (0.78) | 12.66 (11.36-13.60) | 0.13 | |
| BPA | 11.91 (1.66) | 11.56 (10.26-15.08) | ||||
| Ppp1r9a | Right Cortex | Control | 6 | 7.601 (0.32) | 7.587 (7.15-8.13) | 0.58 |
| BPA | 7.749 (0.52) | 7.944 (7.06-8.24) | ||||
| Midbrain | Control | 6 | 8.753 (0.77) | 9.783 (7.57-9.86) | 0.93 | |
| BPA | 8.726 (0.61) | 8.794 (8.01-9.62) | ||||
| Dnmt1 | Right Cortex | Control | 6 | 10.374 (0.29) | 10.442 (9.88-10.63) | 0.01 |
| BPA | 9.471 (0.65) | 9.454 (8.65-10.46) | ||||
| Midbrain | Control | 6 | 10.334 (0.42) | 10.299 (9.66-10.78) | 0.39 | |
| BPA | 9.895 (0.60) | 10.008 (8.84-10.53) | ||||
| Tet2 | Right Cortex | Control | 6 | 13.867 (0.39) | 14.055 (13.32-14.19) | 0.13 |
| BPA | 13.111 (1.42) | 12.888 (11.46-15.54) | ||||
| Midbrain | Control | 6 | 14.195 (0.63) | 14.427 (13.08-14.77) | 0.03 | |
| BPA | 12.912 (0.65) | 13.166 (12.09-13.53) |
Mean and median values are the Ct values for each gene after normalization to housekeeping gene expression (delta Ct).
Effect of BPA Exposure on Expression of Epigenetic Writers
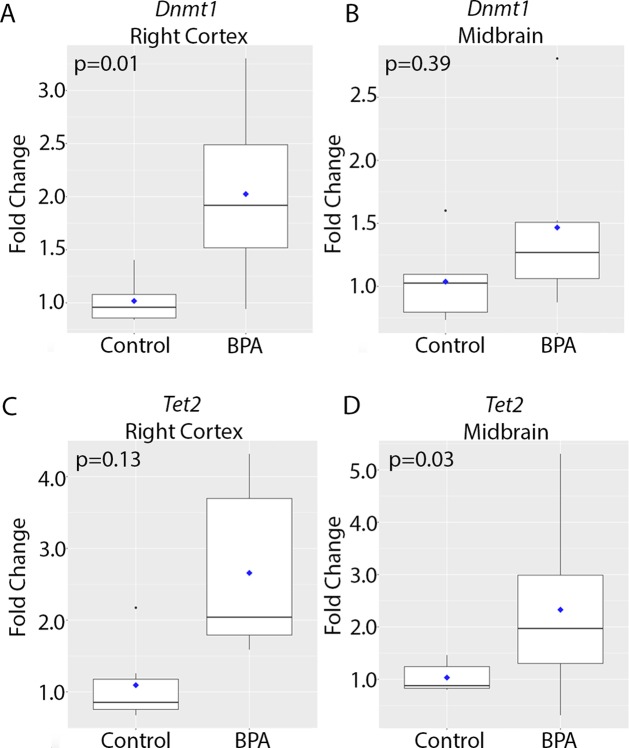
We next investigated whether epigenetic mechanisms contributed to the observed increase in Kcnq1 expression in the brains of BPA-exposed animals. The conversion of cytosine to 5mC is catalyzed by DNA methyltransferases, including Dnmt1, and further oxidation of 5mC to 5hmC is mediated by Tet methylcytosine dioxygenases, including Tet1 and Tet2 (Jones, 2012; Antunes et al., 2019). Therefore, we investigated expression of Dnmt1, Tet1, and Tet2 using qRT-PCR. Dnmt1 expression was significantly higher in the right cortex of BPA-exposed mice compared to the control group (p = 0.01, Figure 3A and Table 1 ). In the midbrain, Dnmt1 expression did not significantly differ between control and BPA-exposed groups (p = 0.4, Figure 3B and Table 1 ). Developmental BPA exposure had no significant effect on Tet1 expression in either the right cortex or midbrain ( Supplementary Figure 1 and Supplementary Table 3 ). However, Tet2 exhibited a trend toward increased expression in the right cortex (p = 0.13, Figure 3C and Table 1 ), and was significantly higher in the midbrain of BPA-exposed mice compared to controls (p = 0.03, Figure 3D and Table 1 ). Given the increased expression of Dnmt1 and Tet2 with BPA exposure, and their roles in epigenetic regulation of gene expression, we next evaluated whether expression of Dnmt1 and Tet2 were correlated with Kcnq1 expression. Concomitant with the observed increase in Kcnq1 expression with BPA exposure, expression of Tet2 exhibited a trend toward significant correlation in the right cortex (p = 0.08), and was significantly positively correlated with Kcnq1 expression in the midbrain ( Supplementary Figures 2A, B ). Dnmt1 and Kcnq1 expression exhibited a trend toward significant correlation in the right cortex (p = 0.06), and were not significantly correlated in the midbrain ( Supplementary Figures 2C, D ). Together, these data suggest that developmental BPA exposure leads to reprogramming of Dnmt1 and Tet2 expression in a brain region-dependent manner, concomitant with increased expression of Kcnq1.
Figure 3.
Expression of genes encoding epigenetic modifying enzymes in the brain at 10 months of age in animals exposed to control vs. BPA. Box plots depicting qRT-PCR data for Dnmt1 (A, B) and Tet2 (C, D) in the right cortex (A, C) and midbrain (B, D). Data were analyzed using a two-sided non-parametric Wilcoxon test. Graphs depict the fold change in gene expression relative to the average of the control samples. n = 6 mice per exposure.
Effect of BPA on 5mC and 5hmC at the Kcnq1 Locus
Given the roles for Dnmt1 and Tet2 in regulation of 5mC and 5hmC, respectively, we next investigated whether perinatal BPA exposure resulted in altered 5mC and/or 5hmC at the Kcnq1 locus. To address this question, we performed bisulfite conversion and oxidative bisulfite conversion of paired DNA samples from control and BPA-exposed right cortex and midbrain (see Methods). We chose a CpG site within an intronic region of the Kcnq1 locus in which differential 5hmC was observed in blood samples of the same BPA-exposed mice (Kochmanski et al., 2018) ( Figure 4A ). Importantly, we discovered both 5mC and 5hmC at this CpG in both regions of the brain. The mean levels of 5mC in the right cortex were 87% and 86.2% in the control and BPA-exposed groups, respectively, and mean 5mC levels in the midbrain were 84.8% and 89% with control and BPA exposure, respectively ( Figure 4B and Table 2 ). We observed no significant differences in 5mC with BPA exposure in either brain region ( Figure 4B and Table 2 ). Levels of 5hmC at this locus were much lower than 5mC, consistent with literature demonstrating a lower level of 5hmC genome-wide compared to 5mC (Lopez et al., 2017). In the right cortex, mean 5hmC levels were 5.4% and 6.6% in the control and BPA-exposed group, respectively. In the midbrain, mean 5hmC levels were 8.6% and 4.6% in the control and BPA group, respectively ( Figure 4C and Table 2 ). No significant differences in 5hmC levels were identified in either brain region with BPA exposure ( Figure 4C and Table 2 ). We next investigated whether expression of Dnmt1 and Tet2 were associated with levels of 5mC and 5hmC at the Kcnq1 locus. We observed no correlation between Dnmt1 expression and levels of 5mC and 5hmC in the right cortex ( Supplementary Figure 3A ). However, we observed modest positive and negative correlations between Dnmt1 expression and levels of 5mC and 5hmC, respectively, within the Kcnq1 locus in the midbrain ( Supplementary Figure 3B ). No significant correlations were observed between Tet1 or Tet2 expression and 5mC/5hmC in either brain region ( Supplementary Figures 3C–F ). Finally, we determined whether Kcnq1 expression correlated with levels of either 5mC or 5hmC at the Kcnq1 locus. This analysis did not show significant correlation between Kcnq1 expression and 5mC or 5hmC levels ( Supplementary Figures 4A, B ). These findings demonstrate enrichment with both 5mC and 5hmC at the Kcnq1 locus, but suggest that BPA-induced changes in Kcnq1, Dnmt1 and Tet2 expression are not associated with altered 5mC or 5hmC at this site.
Table 2.
DNA Methylation and Hydroxymethylation at Kcnq1.
| Sample | Exposure | Na | % DNA methylation (SD) |
P value (Mann-Whitney test) |
Beta valuec (P Value) |
|---|---|---|---|---|---|
| 5mC RCb | Control | 5 | 87.0 (7.0) | 0.6 | 1.2 (0.7) |
| BPA | 5 | 86.2 (5.6) | |||
| 5hmC RC | Control | 5 | 5.4 (7.3) | 0.6 | |
| BPA | 5 | 6.6 (5.9) | |||
| 5mC Midb | Control | 5 | 84.8 (10.6) | 0.8 | -4.0 (0.4) |
| BPA | 5 | 89.0 (2.0) | |||
| 5hmC Mid | Control | 5 | 8.6 (10.9) | 0.9 | |
| BPA | 5 | 4.6 (1.5) |
aNumber of samples that passed pyrosequencing quality control and were included in statistical analysis. bRC, Right Cortex, Mid, Midbrain. cBeta value is the interaction term calculated using a linear mixed effects model. Methylation category (5mC or 5hmC), exposure group, and sex were included as terms in all models.
Discussion
It is increasingly clear that exposures to endocrine disrupting chemicals, including BPA, during the perinatal period may interfere with normal programming of brain development and function (Patisaul, 2019). Normal development is orchestrated by epigenetic processes, and several studies have demonstrated that developmental BPA exposure results in disruption of the brain epigenome (Kundakovic et al., 2013; Kumar and Thakur, 2017; Drobna et al., 2018). In this work, we investigated the effects of perinatal BPA exposure on epigenetic regulation of imprinted genes in the brains of adult mice. We first examined expression of three imprinted genes, including Kcnq1, Pde10a, and Ppp1r9a. all of which have been reported to be expressed in the brain (Coskran et al., 2006; Goldman et al., 2009; Konopaske et al., 2015). We found that Kcnq1 expression was significantly upregulated in the right cortex of 10 month old mice. To our knowledge, this is the first report demonstrating modulation of Kcnq1 expression by developmental BPA exposure. Kcnq1 encodes a voltage-gated potassium channel that is critical for normal cardiac repolarization, and it is also expressed in neurons (Goldman et al., 2009). Mutations in this channel are associated with seizures and fatal cardiac arrhythmias (Goldman et al., 2009). Moreover, recent evidence suggests that a single nucleotide polymorphism affecting Kcnq1 expression in the brain is associated with schizophrenia spectrum disorder (Bruce et al., 2017). In light of this evidence and our findings, the functional consequences of increased Kcnq1 expression in the brain as a result of environmental exposures warrant further investigation.
Programming of imprinted gene expression is governed by epigenetic mechanisms, particularly DNA methylation (Dolinoy et al., 2007). Therefore, we investigated the effects of perinatal BPA exposure on expression of Dnmt1, which encodes the DNA methyltransferase that is primarily responsible for maintenance of DNA methylation patterns across cell divisions (Jones, 2012). Recent evidence from our lab and others suggests a potential role for 5hmC in regulation of imprinted gene expression (Yamaguchi et al., 2013; Kochmanski et al., 2018). Therefore, we also investigated expression of the Tet methylcytosine dioxygenases Tet1 and Tet2, which catalyze oxidation of 5mC to 5hmC (Antunes et al., 2019). Dnmt1, Tet1, and Tet2 are expressed in neurons (Fan et al., 2001; Hsieh et al., 2016; Mi et al., 2015), and are also critical for differentiation of oligodendrocyte progenitors (Zhao et al., 2014; Moyon et al., 2017). Interestingly, expression of Dnmt1 and Tet2 were significantly increased in the right cortex and midbrain, respectively, of 10 month old animals, and exhibited trends toward increased expression in the other brain regions. These findings are consistent with research from others demonstrating changes in Dnmt1 expression in the brain with perinatal BPA exposure (Kundakovic et al., 2013; Zhou et al., 2013; Kumar and Thakur, 2017). In contrast with the time point in later adulthood investigated in this work, these studies identified BPA-induced changes in Dnmt1 expression at much earlier time points. The striking consistency of this finding by multiple research groups across several time points, and with varying experimental parameters (i.e. dose, method of administration of BPA), suggests that the effects of BPA on Dnmt1 expression in the brain are robust. Prior to this study, the effects of perinatal BPA exposure on Tet enzyme expression had not yet been examined. However, low dose BPA exposure results in altered subcellular localization of Tet2 in hypothalamic neurons (Kurian et al., 2016), suggesting that BPA may target this enzyme via multiple mechanisms. Mutations and altered expression of Dnmt1 and Tet2 are associated with age-related cognitive decline and neurological diseases (Baets et al., 2015; Cui and Xu, 2018; Gontier et al., 2018). The effects of BPA on brain Dnmt and Tet enzyme levels across the lifecourse, and the health-related implications these changes, represent an important area for future studies.
The relationships between levels of 5mC/5hmC and gene expression are nuanced and dependent upon the genomic context (Jones, 2012). Methylation of CpGs at gene promoters is associated with repression of gene expression (Jones, 2012). In contrast, enrichment with 5hmC is generally associated with gene activation during development and in the adult brain (Ficz et al., 2011; Lin et al., 2017). Given the role for Dnmt and Tet proteins in regulation of imprinted gene expression (Barlow and Bartolomei, 2014; Liu et al., 2015), we investigated whether the expression of Tet2 and Dnmt1 were correlated with Kcnq1 expression. In keeping with the function of Tet enzymes in gene activation, we observed a significant positive correlation between Tet2 and Kcnq1 expression in the midbrain. Indeed, we observed enrichment with 5hmC at the Kcnq1 locus in the brain, an observation that has not yet been reported. However, no significant correlations were observed between expression of Dnmt1 or Tet2 and 5mC/5hmC at the Kcnq1 locus. Moreover, there were no significant correlations between the levels of 5mC and 5hmC at the Kcnq1 locus and expression of Kcnq1. Thus, given the small sample size, we were unable to determine potential mechanistic links between BPA exposure, alterations in 5mC and 5hmC at the Kcnq1 locus, and increased Kcnq1 expression. Further studies with larger sample sizes and assessment of additional CpGs in the Kcnq1 locus are necessary to address this question. The observation of increased expression of Kcnq1, Dnmt1 and Tet2 with BPA exposure is notable, given that BPA exposure ceased at 3 weeks of age. The mechanism by which developmental BPA exposure mediates long-term programming of gene expression is unclear. However, recent evidence suggests that BPA can induce activation of the histone methyltransferase, MLL1, resulting in long-term programming of histone methylation (Wang et al., 2016). Whether a similar mechanism is responsible for our observations remains to be determined.
There are several limitations to this study. First, with six animals per exposure group, the sample size was small. Due to the variability inherent in in vivo studies, we were unable to ascertain statistically significant changes in 5mC and 5hmC with BPA exposure. In addition to increasing sample size, interrogation of additional CpGs in the Kcnq1 locus may reveal further mechanistic insight into BPA-induced changes in Kcnq1 expression. Moreover, although each exposure group contained three males and three females per group, the experiments were not adequately powered to look at sex-specific effects. Basal patterns of 5mC and 5hmC, and the effects of endocrine disruptors, including BPA, are highly sex-specific (Spiers et al., 2015; McCabe et al., 2017; Spiers et al., 2017). Indeed, sex-specific effects of BPA on Dnmt1 expression have been reported (Kundakovic et al., 2013; Zhou et al., 2013; Wright et al., 2017). Thus, future studies controlling for sex may yield important biological insight, as well as potentially reduce variability in the data. Finally, it is increasingly clear that there is significant tissue and cellular heterogeneity with respect to gene expression and patterns of epigenetic marks (Cheow et al., 2016; Cusanovich et al., 2018). As bulk right cortex and midbrain were used in this study, the effects of BPA exposure on specific cellular populations in the brain are unclear. Nevertheless, in spite of these limitations, this is the first report to demonstrate that perinatal BPA exposure alters expression of Kcnq1 and Tet2 in the adult mouse brain, long after cessation of exposure. Moreover, consistent with previous studies, we provide further evidence that Dnmt1 is a target of BPA exposure. Given the critical roles for Dnmt1 and Tet2 in programming 5mC/5hmC during development and in adulthood, alterations in the expression of these enzymes may result in genome-wide epigenetic changes. The potential health implications of this programming by BPA exposure should be further investigated.
Data Availability Statement
All datasets generated/analyzed for this study are included in the manuscript/ Supplementary files .
Ethics Statement
The animal study was reviewed and approved by University of Michigan Institutional Animal Care and Use Committee (IACUC).
Author Contributions
MM, JK, LS, JG, JC, and DD planned experiments and assisted with troubleshooting. MM, JK, and TJ performed experiments. MM, JC, JG, DD, and LS analyzed data. MM and LS wrote manuscript.
Funding
This work was supported by the Rackham Graduate Student Research Grant, University of Michigan, the UM NIEHS/EPA Children’s Environmental Health and Disease Prevention Center P01 ES022844/RD83543601, the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center (P30 ES017885), and the UM NIEHS Institutional Training Grant T32 ES007062.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of Dolinoy Lab for their advice and input on this work, especially Dr. Kari Neier.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00951/full#supplementary-material
References
- Anderson O. S., Nahar M. S., Faulk C., Jones T. R., Liao C., Kannan K., et al. (2012). Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A’. Environ. Mol. Mutagen. 53, 334–342. 10.1002/em.21692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes C., Sousa N., Pinto L., Marques C. J. (2019). TET enzymes in neurophysiology and brain function. Neurosci. Biobehav. Rev. 102, 337–344. 10.1016/j.neubiorev.2019.05.006 [DOI] [PubMed] [Google Scholar]
- Baets J., Duan X., Wu Y., Smith G., Seeley W. W., Mademan I., et al. (2015). Defects of mutant DNMT1 are linked to a spectrum of neurological disorders’. Brain 138, 845–861. 10.1093/brain/awv010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. J. (2007). The origins of the developmental origins theory. J. Intern. Med. 261, 412–417. 10.1111/j.1365-2796.2007.01809.x [DOI] [PubMed] [Google Scholar]
- Barlow D. P., Bartolomei M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, 1–20. 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce H. A., Kochunov P., Paciga S. A., Hyde C. L., Chen X., Xie Z., et al. (2017). Potassium channel gene associations with joint processing speed and white matter impairments in schizophrenia. Genes Brain. Behav. 16, 515–521. 10.1111/gbb.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow L. F., Courtois E. T., Tan Y., Viswanathan R., Xing Q., Tan R. Z., et al. (2016). Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13, 833–836. 10.1038/nmeth.3961 [DOI] [PubMed] [Google Scholar]
- Coskran T. M., Morton D., Menniti F. S., Adamowicz W. O., Kleiman R. J., Ryan A. M., et al. (2006). Immunohistochemical localization of phosphodiesterase 10A in multiple mammalian species. J. Histochem. Cytochem. 54, 1205–1213. 10.1369/jhc.6A6930.2006 [DOI] [PubMed] [Google Scholar]
- Cui D., Xu X. (2018). DNA methyltransferases, DNA methylation, and age-associated cognitive function. Int. J. Mol. Sci. 19, 1315–1331. 10.3390/ijms19051315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D. A., Hill A. J., Aghamirzaie D., Daza R. M., Pliner H. A., Berletch J. B., et al. (2018). A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–24 e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy D. C., Jirtle R. L. (2008). Environmental epigenomics in human health and disease. Environ. Mol. Mutagen. 49, 4–8. 10.1002/em.20366 [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C., Weidman J. R., Jirtle R. L. (2007). Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod. Toxicol. 23, 297–307. 10.1016/j.reprotox.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Drobna Z., Henriksen A. D., Wolstenholme J. T., Montiel C., Lambeth P. S., Shang S., et al. (2018). Transgenerational Effects of Bisphenol A on gene expression and DNA methylation of imprinted genes in brain. Endocrinology 159, 132–144. 10.1210/en.2017-00730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Beard C., Chen R. Z., Csankovszki G., Sun Y., Siniaia M., et al. (2001). DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797. 10.1523/JNEUROSCI.21-03-00788.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C., Liu K., Barks A., Goodrich J. M., Dolinoy D. C. (2014). Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9, 934–941. 10.4161/epi.29024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Branco M. R., Seisenberger S., Santos F., Krueger F., Hore T. A., et al. (2011). Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. 10.1038/nature10008 [DOI] [PubMed] [Google Scholar]
- Goldman A. M., Glasscock E., Yoo J., Chen T. T., Klassen T. L., Noebels J. L. (2009). Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Sci. Transl. Med. 1, 2ra6. 10.1126/scitranslmed.3000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontier G., Iyer M., Shea J. M., Bieri G., Wheatley E. G., Ramalho-Santos M., et al. (2018). Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981. 10.1016/j.celrep.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley K. G., Gunier R. B., Kogut K., Johnson C., Bradman A., Calafat A. M., et al. (2013). Prenatal and early childhood bisphenol A concentrations and behavior in school-aged children. Environ. Res. 126, 43–50. 10.1016/j.envres.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M. C., Lai C. Y., Ho Y. C., Wang H. H., Cheng J. K., Chau Y. P., et al. (2016). Tet1-dependent epigenetic modification of BDNF expression in dorsal horn neurons mediates neuropathic pain in rats. Sci. Rep. 6, 37411. 10.1038/srep37411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe E. M., Zhao X. (2017). DNA methylation and adult neurogenesis. Brain Plast. 3, 5–26. 10.3233/BPL-160034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. A., Javurek A. B., Painter M. S., Ellersieck M. R., Welsh T. H., Jr., Camacho L., et al. (2016). Effects of developmental exposure to bisphenol A on spatial navigational learning and memory in rats: a CLARITY-BPA study. Horm. Behav. 80, 139–148. 10.1016/j.yhbeh.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- Kelly M. P. (2018). Cyclic nucleotide signaling changes associated with normal aging and age-related diseases of the brain. Cell Signal. 42, 281–291. 10.1016/j.cellsig.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J. J., Marchlewicz E. H., Cavalcante R. G., Perera B. P. U., Sartor M. A., Dolinoy D. C. (2018). Longitudinal Effects of developmental bisphenol A exposure on epigenome-wide DNA hydroxymethylation at imprinted loci in mouse blood. Environ. Health Perspect. 126, 077006. 10.1289/EHP3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J., Marchlewicz E. H., Dolinoy D. C. (2018). Longitudinal effects of developmental bisphenol A, variable diet, and physical activity on age-related methylation in blood. Environ. Epigenet. 4, dvy017. 10.1093/eep/dvy017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochmanski J., Marchlewicz E. H., Savidge M., Montrose L., Faulk C., Dolinoy D. C. (2017). Longitudinal effects of developmental bisphenol A and variable diet exposures on epigenetic drift in mice. Reprod. Toxicol. 68, 154–163. 10.1016/j.reprotox.2016.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopaske G. T., Subburaju S., Coyle J. T., Benes F. M. (2015). Altered prefrontal cortical MARCKS and PPP1R9A mRNA expression in schizophrenia and bipolar disorder. Schizophr. Res. 164, 100–108. 10.1016/j.schres.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Thakur M. K. (2017). Effect of perinatal exposure to bisphenol-A on DNA methylation and histone acetylation in cerebral cortex and hippocampus of postnatal male mice. J. Toxicol. Sci. 42, 281–289. 10.2131/jts.42.281 [DOI] [PubMed] [Google Scholar]
- Kundakovic M., Gudsnuk K., Franks B., Madrid J., Miller R. L., Perera F., et al. (2013). Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc. Natl. Acad. Sci. U.S.A. 110, 9956–9961. 10.1073/pnas.1214056110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian J. R., Louis S., Keen K. L., Wolfe A., Terasawa E., Levine J. E. (2016). The Methylcytosine dioxygenase ten-eleven translocase-2 (tet2) enables elevated GnRH gene expression and maintenance of male reproductive function. Endocrinology 157, 3588–3603. 10.1210/en.2016-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xie C., Murphy S. K., Skaar D., Nye M., Vidal A. C., et al. (2016). Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ. Health Perspect. 124, 666–673. 10.1289/ehp.1408577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin I. H., Chen Y. F., Hsu M. T. (2017). Correlated 5-hydroxymethylcytosine (5hmC) and gene expression profiles underpin gene and organ-specific epigenetic regulation in adult mouse brain and liver. PLoS One 12, e0170779. 10.1371/journal.pone.0170779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Mao S. Q., Ray C., Zhang Y., Bell F. T., Ng S. F., et al. (2015). Differential regulation of genomic imprinting by TET proteins in embryonic stem cells. Stem Cell Res. 15, 435–443. 10.1016/j.scr.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez V., Fernandez A. F., Fraga M. F. (2017). The role of 5-hydroxymethylcytosine in development, aging and age-related diseases. Ageing Res. Rev. 37, 28–38. 10.1016/j.arr.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Matsuda S., Matsuzawa D., Ishii D., Tomizawa H., Sutoh C., Nakazawa K., et al. (2012). Effects of perinatal exposure to low dose of bisphenol A on anxiety like behavior and dopamine metabolites in brain. Prog. Neuropsychopharmacol. Biol. Psychiatry 39, 273–279. 10.1016/j.pnpbp.2012.06.016 [DOI] [PubMed] [Google Scholar]
- McCabe C., Anderson O. S., Montrose L., Neier K., Dolinoy D. C. (2017). Sexually dimorphic effects of early-life exposures to endocrine disruptors: sex-specific epigenetic reprogramming as a potential mechanism. Curr. Environ. Health Rep. 4, 426–438. 10.1007/s40572-017-0170-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y., Gao X., Dai J., Ma Y., Xu L., Jin W. (2015). A novel function of TET2 in CNS: sustaining neuronal survival. Int. J. Mol. Sci. 16, 21846–21857. 10.3390/ijms160921846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyon S., Ma D., Huynh J. L., Coutts D. J. C., Zhao C., Casaccia P., et al. (2017). Efficient remyelination requires DNA methylation. eNeuro 4, e0336–16. 10.1523/ENEURO.0336-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patisaul H. B. (2019). Achieving CLARITY on bisphenol A, brain and behaviour. J. Neuroendocrinol. e12730. 10.1111/jne.12730 [DOI] [PMC free article] [PubMed]
- Perez J. D., Rubinstein N. D., Dulac C. (2016). New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu. Rev. Neurosci. 39, 347–384. 10.1146/annurev-neuro-061010-113708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X., Huang X., Zhou Z., Lin X. (2013). An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 3(3), 71–85 [PMC free article] [PubMed] [Google Scholar]
- Reik W., Walter J. (2001). Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32. 10.1038/35047554 [DOI] [PubMed] [Google Scholar]
- Spiers H., Hannon E., Schalkwyk L. C., Bray N. J., Mill J. (2017). 5-hydroxymethylcytosine is highly dynamic across human fetal brain development. BMC Genomics 18, 738. 10.1186/s12864-017-4091-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers H., Hannon E., Schalkwyk L. C., Smith R., Wong C. C., O’Donovan M., et al. (2015). Methylomic trajectories across human fetal brain development. Genome. Res. 25, 338–352. 10.1101/gr.180273.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M., Sasson I., Mesaros C., Bartolomei M. S. (2013). Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet. 9, e1003401. 10.1371/journal.pgen.1003401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg L. N., Chahoud I., Heindel J. J., Padmanabhan V., Paumgartten F. J., Schoenfelder G. (2010). ‘Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 118, 1055–1070. 10.1289/ehp.0901716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg L. N., Hauser R., Marcus M., Olea N., Welshons W. V. (2007). Human exposure to bisphenol A (BPA). Reprod. Toxicol. 24, 139–177. 10.1016/j.reprotox.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Wang Q., Trevino L. S., Wong R. L., Medvedovic M., Chen J., Ho S. M., et al. (2016). Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol. Endocrinol. 30, 856–871. 10.1210/me.2015-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R. A., Jirtle R. L. (2003). Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300. 10.1128/MCB.23.15.5293-5300.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse C., Anderson O. S., Bergin I. L., Vandenbergh D. J., Gyekis J. P., Dingman M. A., et al. (2014). Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ. Health Perspect. 122, 485–491. 10.1289/ehp.1307449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. C., Johnson S. A., Hao R., Kowalczyk A. S., Greenberg G. D., Ordones Sanchez E., et al. (2017). Exposure to extrinsic stressors, social defeat or bisphenol A, eliminates sex differences in DNA methyltransferase expression in the amygdala. J. Neuroendocrinol. 29, 1–9. 10.1111/jne.12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F., Fischer E., Krapp C., Krizman E. N., Lan Y., Mesaros C., et al. (2018). Mice exposed to bisphenol A exhibit depressive-like behavior with neurotransmitter and neuroactive steroid dysfunction. Horm. Behav. 102, 93–104. 10.1016/j.yhbeh.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Hong K., Liu R., Inoue A., Shen L., Zhang K., et al. (2013). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339. 10.1038/cr.2013.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. S., Reed A., Chen F., Stewart C. N. J. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7, 85. 10.1186/1471-2105-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhu Y., Yang J., Li L., Wu H., De Jager P. L., et al. (2017). A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimers Dement. 13, 674–688. 10.1016/j.jalz.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Dai J., Ma Y., Mi Y., Cui D., Ju G., et al. (2014). Dynamics of ten-eleven translocation hydroxylase family proteins and 5-hydroxymethylcytosine in oligodendrocyte differentiation. Glia 62, 914–926. 10.1002/glia.22649 [DOI] [PubMed] [Google Scholar]
- Zhou R., Chen F., Chang F., Bai Y., Chen L. (2013). Persistent overexpression of DNA methyltransferase 1 attenuating GABAergic inhibition in basolateral amygdala accounts for anxiety in rat offspring exposed perinatally to low-dose bisphenol A. J. Psychiatr. Res. 47, 1535–1544. 10.1016/j.jpsychires.2013.05.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated/analyzed for this study are included in the manuscript/ Supplementary files .