Abstract
Growing tumor cells possess a distinct metabolic phenomenon that allows them to preferentially utilize glucose through aerobic glycolysis, which is referred to as the “Warburg effect.” Accumulating evidence suggests that microRNAs (miRNAs) could regulate such metabolic reprogramming. Our microarray analysis and quantitative real-time PCR validation revealed that miR-885-5p was strongly downregulated in hepatocellular carcinoma (HCC) tissues and cell lines. To investigate miR-885-5p’s biological functions in HCC progression, malignant phenotypes were analyzed in different types of hypoxic model and indicated that overexpression of miR-885-5p significantly inhibited HCC cell proliferation and migration and induced apoptosis in vitro and tumor growth in vivo. Subsequent investigations of whether miR-885-5p regulated the glycometabolic activity of cancer cells demonstrated that forced expression of miR-885-5p in SMMC-7721 cells significantly reduced glucose uptake and lactate production by repressing several key enzymes related to glycolysis. Particularly, miR-885-5p directly targets the 3′ UTR of hexokinase 2 (HK2), which is a key enzyme that catalyzes the irreversible first step of glycolysis and associates with poor patient outcomes. The miR-885-5p/HK2 axis strongly links aerobic glycolysis to carcinogenesis and may become a promising therapeutic target and prognostic predictor for HCC patients.
Keywords: Warburg effect, HCC, miRNAs, HK2, hypoxia, prognosis
Introduction
Altered energy metabolism is one of the typical hallmarks of cancer cells.1 Most cancer cells, including hepatocellular carcinoma (HCC) cells, have evolved several alterations in their metabolism to survive in unfavorable microenvironments while retaining their rapid cell growth ability.2 A classical metabolic adaptation of tumor cells is a shift to aerobic glycolysis as a main source of ATP rather than oxidative phosphorylation (OXPHOS), regardless of oxygen availability, a phenomenon referred to as the “Warburg effect,” which features high rates of glucose uptake and lactate production even in the presence of oxygen.3, 4 The dependency of cancer cell proliferation on accelerated glucose metabolism distinguishes cancer cells from their normal counterparts and could render them more vulnerable to the disruption of glucose metabolism. This glycolytic switch in cancer cells may provide an explanation for their extraordinary tolerance to extreme local hypoxia, or anoxia, and their capacity to compete with normal untransformed cells under normoxia. This metabolic switch may be reversible and partly result from aberrant regulation of glycolytic enzymes.5 However, the mechanisms driving aerobic glycolysis need further investigation. Therefore, understanding how the Warburg effect is regulated in cancer is particularly relevant for identifying new therapeutic interventions.
Multiple studies have revealed that altered metabolic pathways in cancers are tightly regulated by microRNAs (miRNAs).6, 7, 8 The downstream targets of a number of miRNAs are directly or indirectly connected to the metabolic regulation. Given the crucial role of miRNAs in cell growth and development processes, it is not surprising that alterations in the activity of these non-coding small RNA could lead to changes in metabolic pathways.
Hypoxia is a common situation in many types of solid tumors, where tumor cells proliferate rapidly and form large solid tumor masses, leading to obstruction and compression of surrounding blood vessels surrounding these masses. These abnormal blood vessels often do not function properly and result in poor oxygen supply to the central tumor regions.9 Thus, hypoxia may cause the tumor cells to acquire a more aggressive malignant phenotype.10 Hypoxia is known to induce changes in the expression of numerous miRNAs in the tumor microenvironment,11 and some studies have shown that miRNAs are associated with several key hypoxia-activated survival pathways and play important roles in hypoxic adaptation.12
In this study, we delineated that the aberrant expression of miR-885-5p could reprogram HCC metabolism and progression by directly repressing hexokinase 2 (HK2) and explored the miR-885-5p/HK2 axis’s potential as a therapeutic target and prognostic predictor.
Results
miR-885-5p Is Downregulated in HCC
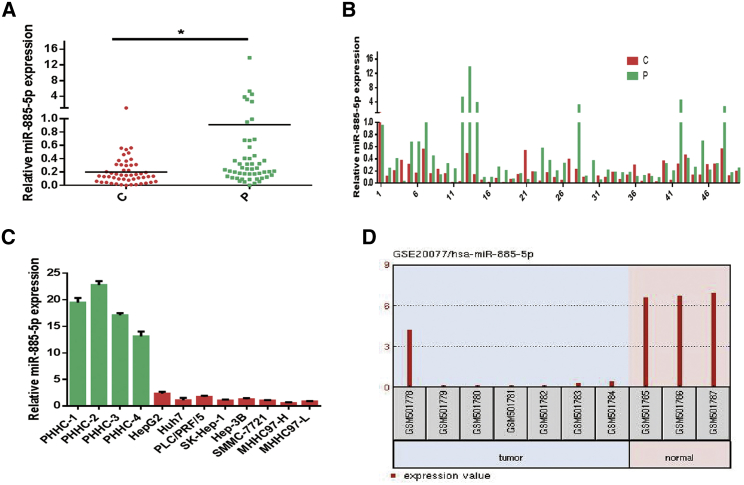
The expression level of miR-885-5p detected by quantitative real-time PCR showed that miR-885-5p was significantly downregulated in a majority of liver cancer tissues compared to the adjacent non-tumorous tissues (Figures 1A and 1B). Subsequently, we evaluated miR-885-5p in four normal primary human hepatocytes (PHHCs; PHHC-1, PHHC-2, PHHC-3, and PHHC-4) and eight liver cancer cell lines (HepG2, Huh7, PLC/PRF/5, SK-Hep-1, Hep-3B, SMMC-7721, MHHC97-H, and MHHC97-L). As shown in Figure 1C, the expression level of miR-885-5p in liver cancer cells was significantly downregulated compared to that in the four normal PHHCs. The results were consistent with our previous study,13 in which we analyzed the dataset GEO: GSE20077 and found that miR-885-5p was noted as one of the greatest changes of decreased expression (Figure 1D).
Figure 1.
miR-885-5p Is Often Downregulated in HCC
(A and B) Relative miR-885-5p expressions measured by quantitative real-time PCR in liver cancer tissues and paired pericarcinomatous tissues (C and P, respectively) of 50 HCC patients were shown in (A) plot and (B) bar diagram, respectively. (C) Relative miR-885-5p expression in indicated hepatoma cell lines and PHHCs. (D) Expression levels of miR-885-5p in the GEO datasets. Data are presented as mean ± SEM of three independent experiments. *p < 0.05.
miR-885-5p Is Downregulated in Different Types of Hypoxic Model In Vitro
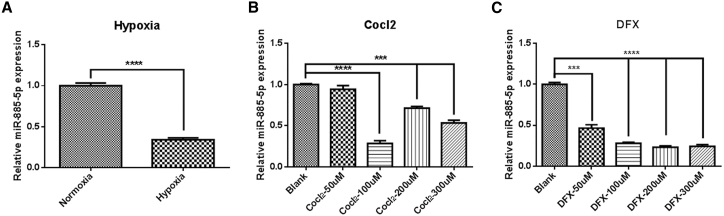
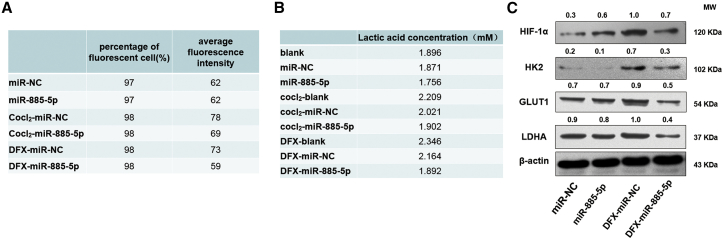
Hypoxia is a prominent micro-environmental feature in many types of solid tumors due to inadequate vascularization. Hypoxia is known to induce changes in the expression of a number of miRNAs and is considered to be an important proximal regulator of miRNA biogenesis and function. In order to simulate the microenvironment of tumor growth in vitro and further explore whether miR-885-5p is downregulated in a hypoxic microenvironment, three hypoxia models were constructed. As shown in Figure 2A, miR-885-5p showed a significant decrease under hypoxic conditions compared with normoxic conditions. In particular, the expression of miR-885-5p in the chemical-induced hypoxia models constructed by cobalt chloride (CoCl2) and deferoxamine (DFX) was decreased in a concentration-dependent manner (Figures 2B and 2C). Considering the reproducibility and stability of the hypoxic models, we selected the CoCl2 and DFX models and identified 100 μM as the optimal concentration.
Figure 2.
miR-885-5p Is Downregulated in Different Types of Hypoxic Model
SMMC-7721 cells were seeded in a hypoxia incubator or treated with different concentrations of chemical inducers (CoCl2 and DFX). (A) The expression of miR-885-5p under normoxia and hypoxia conditions. (B and C) Relative miR-885-5p expression in different concentrations of (B) CoCl2-induced and (C) DFX-induced hypoxia models. All data are shown as mean ± SEM of 3 independent experiments. ***p < 0.001, ****p < 0.0001.
Cationic Liposomes Can Deliver miR-885-5p Efficiently in HCC Cells In Vitro
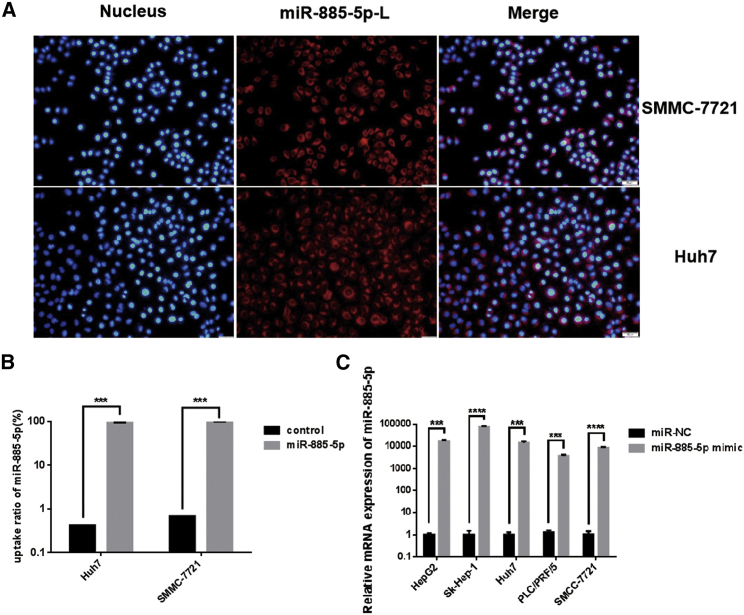
Nanocarriers were frequently selected for miRNA delivery due to their low toxicity, clinical potential, and the ease of production, so we constructed cationic liposomes to deliver miR-885-5p. First, we checked the delivery as well as the localization of miR-885-5p delivered by cationic liposomes with fluorescence. The liposomes were synthesized, and their characteristics were evaluated as previously described.14 miR-885-5p was labeled with cy3 (red), and the nucleus was counterstained with DAPI (blue) (Figure 3). After a 1.5-h incubation, SMMC-7721 and Huh7 cells treated with miR-885-5p mimic showed apparent red fluorescence in almost all cells, indicating an efficient and rapid uptake of miR-885-5p by hepatoma cells (Figure 3A). Meanwhile, we found that miR-885-5p located in the cytoplasm. Flow cytometry analysis also proved that miR-885-5p was absorbed by Huh7 and SMMC-7721 cells efficiently. The uptake ratio was 94.3% in Huh7 cells and 99.28% in SMMC-7721 after incubation with miR-885-5p mimic for 1.5 h (Figure 3B). Quantitative real-time PCR showed that miR-885-5p mimic enormously upregulated the expression of miR-885-5p in different HCC cell lines after treatment for 48 h (Figure 3C). All these results suggest an efficient uptake of miR-885-5p in HCC cells and the intact release of mature miR-885-5p.
Figure 3.
Cellular Uptake of miR-885-5p in Hepatoma Cells and Release of miR-885-5p
(A) Cellular uptake of miR-885-5p-L (liposome) in SMMC-7721 and Huh7 cells visualized by fluorescence microscope. Hepatoma cells were incubated with cy3-labeled miR-885-5p-L (100 nM miR-885-5p) for 1.5 h and then stained with DAPI and observed under a fluorescence microscope (200×). (B) Cellular uptake of miR-885-5p in SMMC-7721 and Huh7 cells determined by flow cytometry analysis. SMMC-7721 and Huh7 cells were treated with miR-885-5p-L (100 nM miR-885-5p). (C) The expression levels of mature miR-885-5p in hepatoma cells were detected by quantitative real-time PCR. All data are presented as mean ± SEM of 3 independent experiments. ***p < 0.001; ****p < 0.0001.
miR-885-5p Is Negatively Associated with the Malignant Phenotypes of Liver Cancer and Exerts an Inhibitory Effect on Liver Cancer Cell Growth and Tumorigenicity In Vitro and In Vivo
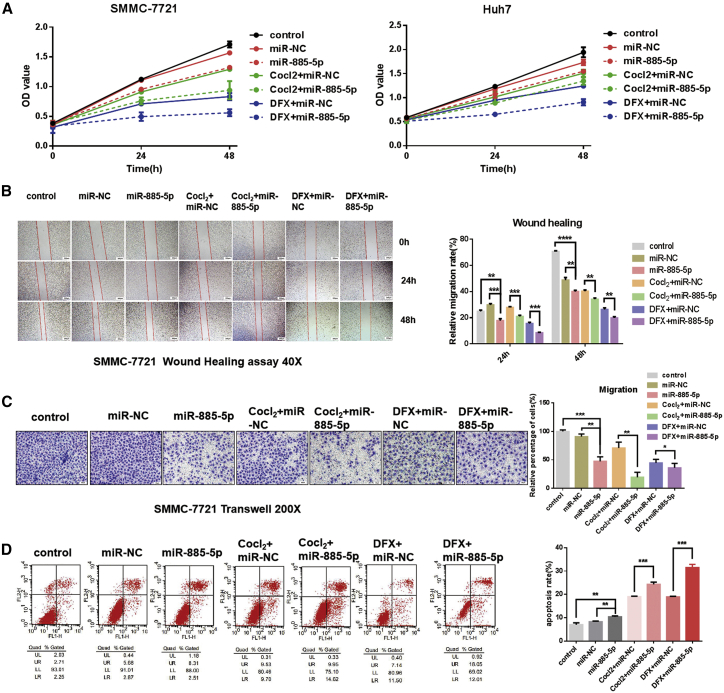
With subsequent functional assays, the CCK8 cell proliferation assay revealed compromised cell growth ability in SMMC-7721 cells transfected with miR-885-5p mimics (Figure 4A), and the inhibition effect was more obvious in an anoxic environment. Similar trends were observed in a wound healing assay (Figure 4B) and a Transwell cell migration assay (Figure 4C). In addition, an apoptotic assay showed apoptosis promoting the effect in miR-885-5p groups of different models (Figure 4D).
Figure 4.
miR-885-5p Negatively Regulated the Malignant Phenotypes of HCC In Vitro
The cells transfected with miR-NC and miR-885-5p under normoxia or hypoxia conditions induced by CoCl2 and DFX were subjected to CCK8 analysis, wound healing assay, cell migration, and apoptosis assay. (A) Effects of miR-885-5p on proliferation over different time periods of Huh7 and SMMC-7721 cells. (B) Images of cell migration from wound healing assay. Scratch wounds were made on confluent monolayer cultures 48 h post-transfection. Images of wound repair were taken at 0, 24, and 48 h after wound (40×). The percentage of wound closure was normalized to the wound area at hour 0. (C) Transwell assay of SMMC-7721 cells treated with miR-NC or miR-885-5p (200×). Histograms indicate the relative percentage of cells across the membrane. The relative percentage of migrating cells from control group is designated as 100%. (D) miR-885-5p induced cell apoptosis. Cell apoptosis was detected by annexin-V/propidium iodide combined-labeling flow cytometry in SMMC-7721 cells 48 h after transfection. Quadrants from lower left to upper left (counterclockwise) represent healthy, early apoptotic, late apoptotic, and necrotic cells, respectively. Apoptotic evaluation was determined by the percentage of apoptotic cell number in total cell number. All data are presented as mean ± SEM of 3 independent experiments. The comparisons were evaluated using t test. **p < 0.01; ***p < 0.001; ****p < 0.0001.
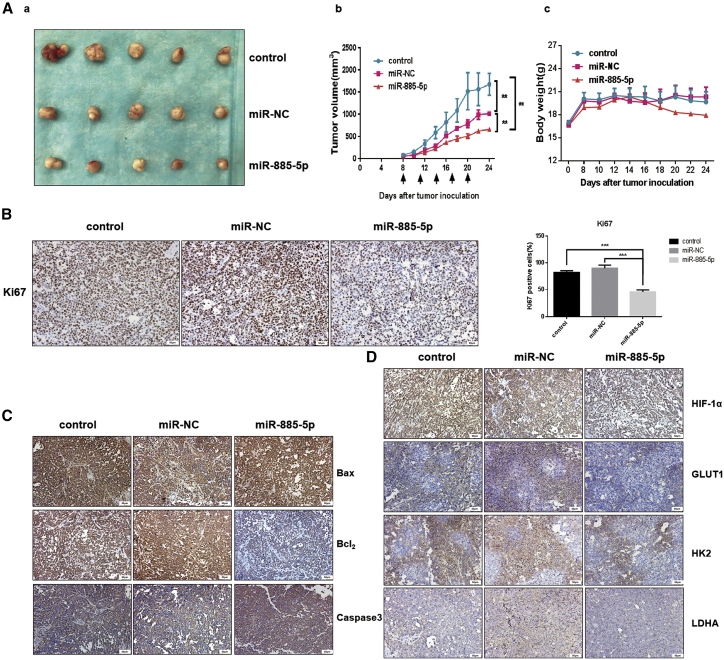
Further, we selected the Hep-3B cell line, which has a relatively high expression of miR-885-5p to perform cell function analysis with miR-885-5p inhibitor. The results were shown in Figure S1, which also was designed to verify the function of miR-885-5p under hypoxic conditions. Next, we studied the effects of miR-885-5p in vivo. Approximately 5 × 106 SMMC-7721 cells were injected subcutaneously into the right dorsal tissues of nude mice. Compared with the normal saline and microRNA negative control (miR-NC) groups, the miR-885-5p-treated group showed obvious suppressing effects of tumor growth, which could be presented by gross morphology (Figure 5Aa). As shown in Figure 5Ab, the tumor growth curves indicated that the miR-885-5p-treated group had the smallest volume and maintained persistent tumor suppression. Furthermore, no distinct body weight decline was shown among these groups (Figure 5Ac). As shown in Figure 5C, the expressions of Bax and caspase-3 analyzed by immunohistochemistry techniques were significantly increased, while the expression of Bcl-2 was downregulated evidently. As a marker of proliferation, the expression of Ki-67 was found to have decreased remarkably in the miR-885-5p-treated group (Figure 5B). Collectively, all of these data fully demonstrated that miR-885-5p could suppress the malignant phenotypes and tumorigenicity of liver cancer cells both in vitro and in vivo.
Figure 5.
miR-885-5p Inhibited Tumorigenicity of HCC In Vivo
(A) Xenograft tumors in nude mice. (a) Morphology of tumors excised from mice at the end of experiment. (b) Growth curves of xenograft tumors. Overexpression of miR-885-5p reduced the volume of the xenograft tumors. (c) The body weight of mice after sacrifice. The results are displayed as mean ± SEM of 5 animals per group. (B) Ki-67 staining in biopsies (200×). For each treatment, 3 independent fields from different biopsies were counted. (C) Immunohistochemical analysis of apoptosis proteins Bcl-2, Bax, and Caspase-3 (200×). (D) miR-885-5p suppressed the expression of HIF-1ɑ, GLUT1, HK2, and LDHA in xenograft microenvironments. The immunohistochemical analysis from excised tumors of Warburg-effect-related enzymes was evaluated. Protein signal was stained by DAB (brown), and cell nucleus was stained by DAPI (blue). **p < 0.01; ***p < 0.001.
miR-885-5p Suppresses Warburg Effect by Targeting Several Crucial Enzymes in Liver Cancer Cells
Proliferating cancer cells preferentially use aerobic glycolysis to support growth, which is a metabolic alteration commonly referred to as the “Warburg effect.” In recent research, miRNAs have emerged as very significant regulators of glucose metabolism.15 We further tested whether miR-885-5p modulates the glycolytic phenotype in cultured cells and found that forced expression of miR-885-5p in SMMC-7721 cells significantly reduced glucose uptake and lactate production (Figures 6A and 6B).
Figure 6.
miR-885-5p Suppresses the Warburg Effect by Regulating Several Glucose Metabolism-Related Enzymes in HCC
(A and B) Glucose uptake activity (A) and lactate production (B) in SMMC-7721 cells with ectopic expression of miR-885-5p in different hypoxic models. (C) miR-885-5p suppressed the expression of HIF-1ɑ, GLUT1, HK2, and LDHA at protein level in the DFX-induced hypoxia model.
In order to support proliferation and to satisfy the increased biosynthetic demands, the rapidly growing cancer cells make use of the hypoxic reaction and develop a great appetite for glucose. Transcription factor hypoxia-inducible factor-1α (HIF-1α) mediates the expression of many enzymes in the glycolytic pathway, which allows tumors to survive under hypoxic conditions by metabolizing glucose to lactate through anaerobic glycolysis.16, 17, 18, 19 Hexokinases catalyze the first committed step of glucose metabolism. HK2 expresses at a high level in cancer cells and plays a role in glucose uptake together with GLUT1 (glucose transporter 1).20, 21, 22, 23, 24, 25 LDHA is a key enzyme responsible for lactate production from pyruvate, the final step in aerobic glycolysis.26, 27 The protein levels of GLUT1, LDHA, and HIF-1α in different HCC cell lines by western blot are presented in Figure S2.
Since hypoxia is a key phenomenon in cancers, to elucidate the mechanisms for how miR-885-5p suppressed the “Warburg effect,” we identified its potential target in the regulation of glycolysis under hypoxia in liver cancer. We detected the protein expression of critical enzymes involved in glycolysis metabolism in SMMC-7721 cells with miR-885-5p mimics transfected. As shown in Figure 6C, western blot analysis revealed that overexpression of miR-885-5p resulted in a significant decrease in the expression of HIF-1α, GLUT1, HK2, and LDHA at the protein level in DFX-induced hypoxia models. Similar results were also verified in Huh7 cells in Figure S3. In xenograft microenvironments, an overexpression of miR-885-5p remarkably inhibited the expression of the enzymes related to the “Warburg effect” in vivo, as described earlier, compared with the controls (Figure 5D).
Taken together, these data indicate that the “Warburg effect” is inhibited by miR-885-5p via its suppression of HIF-1α, GLUT1, HK2, and LDHA under hypoxia and suggest the therapeutic potential of miR-885-5p in liver cancer treatment.
HK2 Is a Direct Target of miR-885-5p and Prognostic Biomarker in Liver Cancer
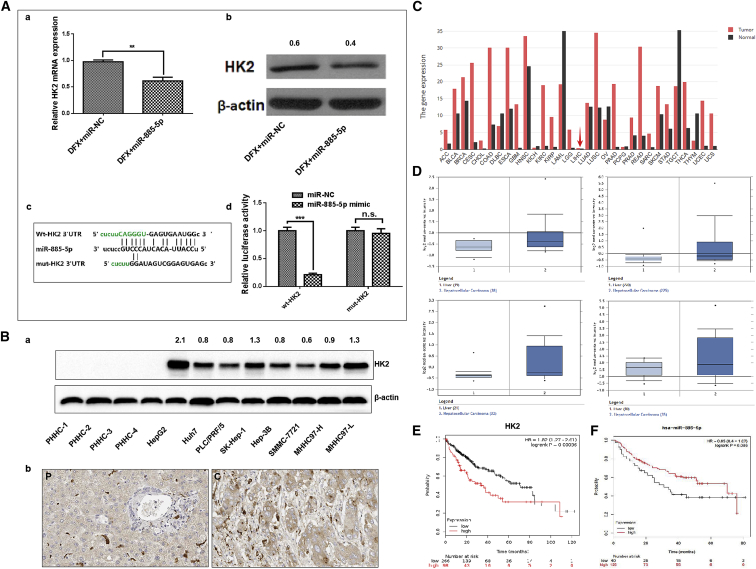
As shown in Figure 7Ab, western blot analysis revealed that the overexpression of miR-885-5p resulted in a significant decrease in HK2 protein expression in DFX-induced hypoxia models. Further experiments indicated that miR-885-5p had similar inhibitory effects on HK2 mRNA levels (Figure 7Aa). Then, we used the online tool TargetScan (http://www.targetscan.org) to predict the miR-885-5p-binding region of HK2. Accordingly, there was a potential miR-885-5p-binding fragment in the 3′ UTR of HK2 (Figure 7Ac). We found that forced expression of miR-885-5p significantly reduced the activity of the luciferase reporter with the wild-type 3′ UTR of HK2, while no effects were found with the mutated form (Figure 7Ad). Collectively, these data suggested that miR-885-5p directly repressed HK2 expression by targeting its 3′ UTR.
Figure 7.
HK2 Is a Direct Target of miR-885-5p, and Both Are Prognostic Biomarkers in HCC
(A) The mRNA and protein levels were measured by (a) qRT-PCR and (b) western blot in SMMC-7721 cells transfected with miR-885-5p mimics. (c) Alignment of potential miR-885-5p binding site in the 3′ UTR of HK2. (d) Luciferase assay. SMMC-7721 cells were transfected with a dual-luciferase reporter vector containing the 3′ UTR of HK2 or a mutated 3′ UTR. At 24 h after the transfection, cells were further transfected with miR-885-5p or miR-NC, and dual-luciferase activity was measured. (B) The expression of HK2 in liver and prognostic analysis. (a) The protein expression levels of HK2 in indicated hepatoma cell lines and PHHCs. (b) The expression level of HK2 in human liver specimens of cancer tissues and paired pericarcinomatous tissues (C and P, respectively) (200×). (c) Bar plot of the gene expression profile across all tumor samples and paired normal tissues from TCGA datasets. The height of the bars represents the median expression of a certain tumor type or normal tissue. The red arrow marks the expression of the HK2 gene in liver cancer tissue and its matched paracancerous tissues. (d) The expression level of HK2 in normal liver and hepatocellular carcinoma from 4 different Human Genome Arrays in TCGA datasets. (e) KM analysis of the correlation between HK2 expression and OS in the 364-patient TCGA cohort. (f) KM analysis of the correlation between miR-885-5p expression and overall survival in the 165-patient cohort. Log-rank tests were used to determine statistical significance. Data were presented as mean ± SEM of three independent experiments. **p < 0.01; ***p < 0.001; n.s., not significant.
The bar plot of the HK2 gene expression profile across all tumor samples and paired normal tissues from The Cancer Genome Atlas (TCGA) datasets is shown in Figure 7Bc; as the red arrow indicates, the median expressions of liver cancer and corresponding normal tissue were 0.44 and 0.29, respectively. Data of liver and hepatocellular carcinoma from different Human Genome Arrays (Human Genome U133A 2.0 Array, Affiymetrix Human Genome HT U133A Array, and Human Genome U133A Plus 2.0 Array) in TCGA datasets (Figure 7Bd) showed that the gene expression was higher in liver cancer than in normal liver tissue. Immunohistochemistry of HK2 gene in human tissue samples (Figure 7Bb) presented stronger expression signals in tumor tissues. Furthermore, the protein expression levels of HK2 were highly expressed in eight liver cancer cell lines, whereas they were hardly detectable in PHHC cells (Figure 7Ba). In addition, the protein-protein interaction network of HK2-related genes based on the STRING database in Figure S4 revealed 10 predicted functional partners, most of which focused on the glucose metabolic process, especially gluconeogenesis.
Importantly, Kaplan-Meier (KM) survival analyses revealed that patients with high HK2 expression had poorer survival in the TCGA cohort (Figure 7Be). Cox proportional hazards regression analyses indicated that HK2 expression is an independent risk factor in the TCGA cohort (hazard rate = 1.82, 95% confidence interval 1.27–2.61, p = 0.00086). Meanwhile, liver cancer patients from TCGA datasets with decreased miR-885-5p expression had poorer overall survival (Figure 7Bf; hazard rate = 0.65, 95% confidence interval 0.4–1.07, p = 0.086).
Taken together, these data suggest that HK2 is a direct target of miR-885-5p and a potential prognostic biomarker in liver cancer.
Clinical Survival Period of the miR-885-HK2 Axis in Liver Cancer
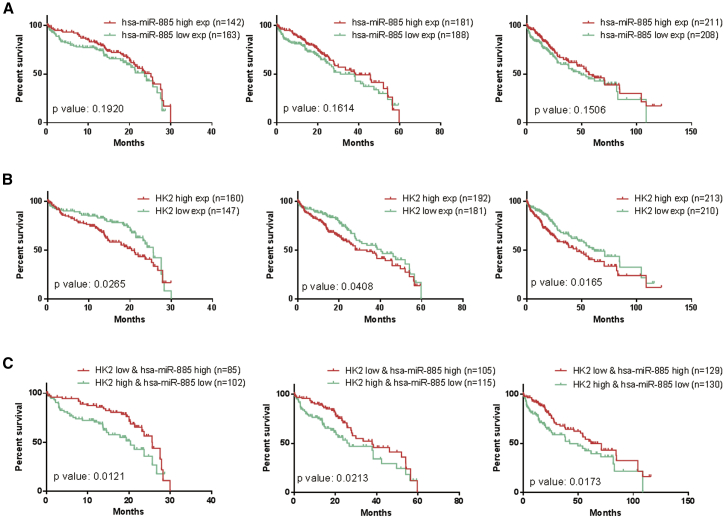
To understand their clinical significance in human HCC, KM survival analysis was used to analyze the relationship between their expressions and the overall survival (OS; in months) in TCGA HCC samples. Our analysis showed that miR-885 and HK2 both significantly affected the OS time of liver cancer patients (Figures 8A and 8B). High expression of miR-885 can serve as an indicator for better prognosis, while high expression of HK2 is an indicator for worse prognosis.
Figure 8.
OS Analysis of the miR-885-HK2 Axis in HCC
The x axis represents the OS month, and the y axis represents the survival rate. (A) KM survival analysis of miR-885 at 30, 60, and 120 months. (B) KM survival analysis of miR-885 target gene HK2 at 30, 60, and 120 months. (C) KM survival analysis of miR-885-HK2 combinations at 30, 60, and 120 months. The data for prognostic analysis of OS come from TCGA cohort; log-rank tests were used to determine statistical significance.
Interestingly, KM survival analysis and log-rank test suggested that the group with low expression of HK2 and high expression of miR-885 had a significantly longer OS rate than that with high expression of HK2 and low expression of miR-885 in liver cancer patients (Figure 8C).
Discussion
Most cancer cells reprogram cellular glucose metabolism to fulfill their anabolic demands. Among cancer cells, HCC cells probably display the most comprehensive reprogramming of glucose metabolism.28 The liver is the primary organ site for the regulation of whole-body metabolism and energy homeostasis; thus, aberrant energy metabolism is closely related with the pathogenesis of many liver diseases as well as the progression of liver disease to carcinomas.29 HCC is a complex multi-step process arising from a combination of genetic and epigenetic alterations, somatic mutations, genomic instability, and environmental factors.30 Recent studies have demonstrated that miRNAs play critical roles in energy metabolism.8 Although some dysregulated miRNAs have been reported to be able to alter glucose metabolism in liver cancer cells8 and play important roles in the tumor metabolic regulatory network,7 the underlying mechanism by which miRNAs modulate cancer metabolism is still largely unclear.
Noticeably, miRNAs are both upregulated (miR-21, miR-221, miR-222, miR-224, and miR-17-92) and downregulated (miR-29, miR-122, miR-200, miR-123, miR-199a, miR-199b, and let-7) in association with HCC and have been extensively reviewed.31 In this study, we found that miR-885-5p was downregulated in liver tumor tissues and eight liver cancer cell lines. Our previous study13 of microarray data from the GEO database also supported the result that miR-885-5p was noted as one of the greatest changes of decreased expression in liver cancer. Hypoxia is a prominent micro-environmental feature in many types of solid tumors due to inadequate vascularization. We found that hypoxia further downregulated miR-885-5p expression and thus induced cancer cells to acquire relatively less aggressive phenotypes in vitro, such as lower abilities of proliferation, migration, and apoptosis promotion. Under a hypoxic microenvironment in vivo, overexpression of miR-885-5p has significantly inhibited tumor growth and led to tumor cell apoptosis. Although some dysregulated miRNAs have been reported to be responsible for altering glucose metabolism in liver cancer cells,32 a metabolomic approach would be ideal to obtain a global perspective on such regulations. Interestingly, we found that miR-885-5p suppressed lactate production and glucose consumption of liver cancer cells, and the suppression was especially significant especially under hypoxia.
Hypoxic cells tend to consume more glucose in order to meet their energy needs. In tumor microenvironments, the HIF-1α (one oxygen-insensitive subunit of HIF-1) oxygen-dependent degradation pathway is halted, resulting in elevated levels of HIF-1α. HIF-1α upregulates many enzymes of the glycolytic pathway, including HK2, by binding to hypoxia-responsive elements (HREs) in the HK2 promoter.33 In human glioblastoma multiforme (GBM), GBM cells depleted of HK2 showed a significant compensatory increase in glucose uptake. An enhanced glucose uptake was associated with the increased GLUT1 expression at the transcript level in cells depleted of HK2 and HK1. Furthermore, HK2-depleted cells showed significantly reduced LDHA transcription, correlating with the reduction in lactate. In our research, overexpression of miR-885-5p resulted in a significant decrease in the expression of HIF-1α, GLUT1, HK2, and LDHA at the protein level in both DFX-induced hypoxia models and xenograft microenvironments.
Hexokinases catalyze the first irreversible step in the glycolytic pathway by phosphorylating glucose to G6P in an ATP-dependent manner. There are four major isozymes encoded by separate genes: HK1, HK2, HK3, and HK4.34 The major isozyme is HK2, and it is exclusively upregulated in many types of cancers, although it is not expressed in most adult tissues, including adult hepatocytes. It contributes to aerobic glycolysis, which distinguishes cancer cells from the normal cells and is, at least in part, responsible for the accelerated glucose flux. Therefore, it is regarded as a key player in the Warburg effect and has been proposed as a therapeutic target for cancers.23 Furthermore, high levels of HK2 expression and activity in glycolytic cancers are manifested by the use of positron emission tomography (PET) to visualize tumors. Previous studies have shown that HK2 is directly regulated by miR-199a-5p in liver cancer cells,35 revealing a new mechanism for HK2 regulation by miRNAs in human cancers. Our study verified that HK2 was also a direct target and negatively regulated by miR-885-5p in HCC. Overexpression of HK2 might be the reason for poor prognosis in downregulated miR-885 expression cases, but further prospective multicenter studies with more adequate design and larger sample size are needed to verify it, whereas our findings provide a direction for future research.
In conclusion, our observations found that miR-885-5p could regulate the glycometabolism of HCC cells in the hypoxic tumor microenvironment and demonstrated that miR-885-5p is a suppressor for HCC partially by inhibiting several enzymes involved in the glycolysis pathway and sequentially restrained malignancies of HCC in vitro and in vivo. Importantly, we further indicated that HK2 was a direct target for miR-885-5p. Since the miR-885-5p/HK2 axis is abnormally regulated in cancer, which correlates with prognosis and controls the Warburg effect, this axis is worthy of further study for HCC therapy.
Materials and Methods
Clinical Specimens
A total of 50 liver cancer specimens and matched adjacent non-tumorous tissues were collected from surgical specimens of HCC patients undergoing partial hepatectomy and immediately stored in liquid nitrogen. All human materials were obtained with informed consent, and our study was approved by the local Research Ethics Committee at the Tongji Hospital of Huazhong University of Science and Technology. Normal PHHCs were isolated using a modified four-step retrograde perfusion technique as reported previously.36, 37
Expression Datasets
The microarray dataset of GEO: GSE20077 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo). The dataset was used to analyze deregulated miRNAs in liver cancer, and the expression levels of precursor miR-885-5p were also retrieved from this dataset.
Moreover, we also acquired the expression data of HK2 and miR-885-5p in TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), as well as related clinical information. Then, patients were divided into two groups according to the median expression of HK2 or miR-885-5p, and KM survival analysis was then conducted.
Chemicals for Preparation of Liposomes
Dicyclohexylcarbodiimide (DCC), N-hydroxysuccinimide (NHS), cholesterol, DMSO, (NH4)2SO4, CHCl3, and Sepharose CL-4B chromatography media were purchased from Sigma-Aldrich Chemical (St. Louis, MO, USA). 1, 2-Dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP), monomethoxy polyethylene glycol 2000-distearoyl phosphatidylethanolamine (mPEG-DSPE), and PEG-bis-amine (3,350 Da) were purchased from Avanti Polar Lipid (Alabaster, AL, USA). PD-10 desalting columns were purchased from GE Healthcare Biosciences (Pittsburgh, PA, USA). All reagents and solvents were of analytical or high-performance liquid chromatography (HPLC) grade and were used without further purification. The lipid compositions of liposomes were DOTAP/Chol/mPEG-DSPE at a molar ratio of 40:55:5, respectively.
Cell Culture, Treatment, and Transfection
HepG2, Huh7, PLC/PRF/5, SK-Hep-1, Hep-3B, SMMC-7721, MHHC97-H, MHHC97-L, and four PHHC cells were cultured in DMEM (GIBCO), supplemented with 10% fetal bovine serum (FBS; GIBCO) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere with 5% CO2. The hypoxic condition was created by culturing cells in a hypoxia chamber flushed with 1% O2, 5% CO2, and 94% N2 mixture gas. CoCl2 and DFX were obtained from Sigma Chemical (St. Louis, MO, USA); dissolved in distilled water at high concentrations; and then filtered through a sterile, pathogen-free nylon filter (pore size: 0.22 mm; MSI, Westborough, MA, USA). Cells were treated with CoCl2 and DFX, which were adopted to simulate a low-oxygen environment.
The miR-885-5p mimics (sense, 5′-uccauuacacuacccugccucu-3′) and NC (sense, 5′-uucuccgaacgugucacgutt-3′) were synthesized by RiboBio (Guangzhou, China). Cells were transfected with miRNAs using cationic liposomes as described earlier.14 Cell transfections were performed using liposomes at the proper time point.
Quantitative Real-Time PCR
Total RNA was extracted from clinical specimens or cells using TRIzol reagent (Invitrogen) according to the protocol. The first-strand cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara). Quantitative real-time PCR was performed in the ABI7500 system using SYBR Green methods. The relative gene expression was measured and normalized to β-actin by the 2−ΔΔCT method. The primers used in the study were as follows: HK2, forward, 5′- AACAGCCTGGACGAGAGCATC-3′, and reverse, 5′-AGGTCAAACTCCTCTCGCCG-3′; β-actin, forward, 5′-TGATGATATCGCCGCGCTC-3′, ανδ reverse, 5′-CCATCACGCCCTGGTGC-3′. Mature miRNA was quantified using miRNA assays. The relative miRNA expression was measured and normalized to U6.
Western Blot
Total cellular protein was extracted using RIPA buffer in the presence of a proteinase and phosphatase inhibitor cocktail. An amount of 30 μg protein was loaded to 10% SDS-PAGE and then transferred onto the polyvinylidene fluoride (PVDF) membranes. Antibodies used for blotting were as followed: HK2, HIF-1α, GLUT1, and LDHA were purchased from abcam. β-actin was used as normalized control.
Cellular Uptake Assay
Cells grown in a monolayer were washed once with DMEM and then were incubated with miR-885-5p mimic (cy3 labeled) at 37°C. After treatment for 1.5 h, the cells were rinsed with PBS for 3 times and fixed with 4% paraformaldehyde solution for 15 min. The solution of DAPI was used for the staining of nucleus. Samples were observed with a fluorescence microscope (Olympus, Japan) and then measured using a flow cytometer (Becton Dickinson, San Jose, CA, USA).
Cell Proliferation Assay
Cell proliferation was determined using the CCK8 (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s protocol.
Cell Migration Assay
The migration assay was performed by using Transwell insert chambers (6.5 mm in diameter, 8-mm pore size; Corning, Corning, NY, USA). For migration assay, 3 × 104 SMMC-7721 transfected cells were placed into the upper chamber in serum-free medium in triplicate 24 h after transfection. The lower chamber was filled with 600 mL DMEM, with 10% FBS as chemoattractant. After incubation for 48 h at 37°C in a 5% CO2 humidified incubator, nonmigrating cells in the upper chambers were removed by using a cotton swab, and cells that migrated to the lower surface of membrane were fixed with methanol and stained with 0.2% crystal violet. Migrating cells were scored by counting at least three fields per membrane under a light microscope. A wound healing assay was carried out as previously described,38 and the relative cell motility rate was presented in a bar graph.
Flow-Cytometric Analysis of Apoptosis and Cell Cycle
Cells were seeded in 6-well plates and harvested 48 h following transfection with miR-885-5p mimic or NC. For apoptosis analysis, apoptotic cells were measured using an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit according to the manufacturer’s instructions (LiankeBio, Nanjing, China). For flow-cytometric analysis, 20,000 cells were examined for each sample and detected using a FACSAria flow cytometer and FACSDiva 4.1 software (BD Biosciences, Franklin Lakes, NJ, USA). For cell-cycle analysis, the cell samples were harvested 48 h after transfection and fixed with 75% ethanol at 4°C overnight according to the manufacturer’s instructions. Cell-cycle distributions were immediately analyzed on a flow cytometer (BD Biosciences). Each experiment was performed in triplicate.
Luciferase Assay
To verify the precise target of miR-885-5p, the pMIR-REPORT system (Applied Biosystems) was applied. SMMC-7721 cells were cultured in 96-well plates and cotransfected with pLUC-HK2 3′ UTR-wild or pLUC-HK2 3′ UTR-mut plasmid and miR-885-5p mimic or NC. pMIR-REPORT β-gal plasmid served as an internal transfection efficiency control. 48 h after transfection, luciferase and β-galactosidase activities were measured using the Dual Light System. For each transfection, luciferase activity was averaged from five replicates.
Lactate Production and Glucose Uptake
Cells were cultured in a 6-well plate. After transfection with miR-885-5p mimics or miR-NC for 48 h, culture media was harvested for measuring the lactate. The extracellular lactate was measured using the Lactic Acid Assay Kit (Jianchengbio, Nanjing, China) according to the manufacturer’s instructions.
Cells were cultured in a 6-well plate. After transfection with miR-885-5p mimics or miR-NC for 48 h, cells were refreshed with serum-starved (0.1% FBS) and glucose-free DMEM. Sixteen hours later, cells were treated with 50 mM 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose (2-NBDG) (Invitrogen) for 1 h, and then glucose uptake was quantified using flow cytometry according to the manufacturer’s instructions as well as published reference.39
Animal Experiments
BALB/c athymic nude mice (male, 3–4 weeks old) were purchased from Beijing HFK Bioscience (Beijing, China) and bred in pathogen-free conditions. All animal experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of Tongji Medical College. SMMC-7721 cells were inoculated subcutaneously into the left flank of BALB/c nude mice (5 × 106 cells per mouse). 8 days later, tumors of comparable size were established. Mice with tumor formation were randomly divided into three groups of five mice each, and then the mice received intratumoral administration of normal saline, miR-885-5p mimic, or miR-NC (10 nmol miRNA per mouse each time), respectively, for five times (days 8, 11, 14, 17, and 20). Tumor dimension was monitored every other day, and their volumes were calculated by length and width by using the formula: volume = length × width × width/2. Tumor tissues were collected and processed for immunohistochemical analysis.
Immunohistochemistry (IHC)
Tumor tissues were collected, fixed in 4% paraformaldehyde, and embedded in paraffin after the mice were euthanized. The slides were sectioned and stained with H&E; tumor tissues were also used for IHC, and Ki-67 was used for histological evaluation. Ki-67 signal was stained by diaminobenzidine (DAB; brown), and cell nucleus was counterstained by hematoxylin (blue) (Figure 5D). The primary antibodies, anti-HK2 (1:100; Abcam), anti-GLUT1 (1:100; Abcam), anti-LDHA (1:100; Abcam), anti-HIF-1α (1:100; Abcam), anti-Ki-67 (1:200; Abcam), anti-Bax (1:100; Proteintech), anti-Bcl2 (1:100; Proteintech), and anti-Caspase-3 (1:100; Proteintech) were applied to the tissue sections and allowed to incubate overnight at 4°C. Then, samples were further processed as per manufacturer’s instructions and a series of procedures.15
Statistical Analysis
Representative data from series of at least three independent experiments carried out in triplicate are presented as mean ± SD, unless otherwise indicated. The statistical difference between each group was assessed by unpaired two-tailed Student t test using GraphPad 5.0 software. Two-tailed p values < 0.05 were considered statistically significant. Single-factor survival analysis was carried out using the KM method, and the log-rank method was used to identify the difference in survival curve. p values < 0.05 indicate that the difference has statistical significance.
Author Contributions
F.X. made substantial contributions to conception, designed and performed the experiments, interpreted the data, and wrote the manuscript. J.-J.Y. designed and performed the experiments. Y.G. performed the animal experiments. Q.Z. and X.-X.H. designed the experiments and revised the article critically for important intellectual content. Y.C. and H.-L.W. participated in revision. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (nos. 81772969, 81670554, and 81302112), the Natural Science Fund for Distinguished Young Scholars of Hubei Province (no. 2017CFA068), and the Applied Basic Research Program of Wuhan City (no. 2017060201010155). We thank Dr. Fan Wang for his contribution to the bioinformatic data process in this paper.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.002.
Contributor Information
Xing-Xing He, Email: xxhe@tjh.tjmu.edu.cn.
Qiu Zhao, Email: qiuzhao@whu.edu.cn.
Supplemental Information
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 4.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., Lee C.T., Lopaschuk G.D., Puttagunta L., Bonnet S. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Chan B., Manley J., Lee J., Singh S.R. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356(2 Pt A):301–308. doi: 10.1016/j.canlet.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Gao P., Sun L., He X., Cao Y., Zhang H. MicroRNAs and the Warburg effect: new players in an old arena. Curr. Gene Ther. 2012;12:285–291. doi: 10.2174/156652312802083620. [DOI] [PubMed] [Google Scholar]
- 8.Rottiers V., Näär A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powis G., Kirkpatrick L. Hypoxia inducible factor-1alpha as a cancer drug target. Mol. Cancer Ther. 2004;3:647–654. [PubMed] [Google Scholar]
- 10.Tan D.S., Agarwal R., Kaye S.B. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006;7:925–934. doi: 10.1016/S1470-2045(06)70939-1. [DOI] [PubMed] [Google Scholar]
- 11.Nallamshetty S., Chan S.Y., Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013;64:20–30. doi: 10.1016/j.freeradbiomed.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen G., Li X., Jia Y.F., Piazza G.A., Xi Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013;34:336–341. doi: 10.1038/aps.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X.X., Chang Y., Meng F.Y., Wang M.Y., Xie Q.H., Tang F., Li P.Y., Song Y.H., Lin J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 14.Xu F., Liao J.Z., Xiang G.Y., Zhao P.X., Ye F., Zhao Q., He X.X. MiR-101 and doxorubicin codelivered by liposomes suppressing malignant properties of hepatocellular carcinoma. Cancer Med. 2017;6:651–661. doi: 10.1002/cam4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh P.K., Mehla K., Hollingsworth M.A., Johnson K.R. Regulation of aerobic glycolysis by microRNAs in cancer. Mol. Cell. Pharmacol. 2011;3:125–134. [PMC free article] [PubMed] [Google Scholar]
- 16.Denko N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 17.Larbi A., Zelba H., Goldeck D., Pawelec G. Induction of HIF-1alpha and the glycolytic pathway alters apoptotic and differentiation profiles of activated human T cells. J. Leukoc. Biol. 2010;87:265–273. doi: 10.1189/jlb.0509304. [DOI] [PubMed] [Google Scholar]
- 18.Maynard M.A., Ohh M. The role of hypoxia-inducible factors in cancer. Cell. Mol. Life Sci. 2007;64:2170–2180. doi: 10.1007/s00018-007-7082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semenza G.L. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin. Cancer Biol. 2009;19:12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Anderson M., Marayati R., Moffitt R., Yeh J.J. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2016;8:56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katabi M.M., Chan H.L., Karp S.E., Batist G. Hexokinase type II: a novel tumor-specific promoter for gene-targeted therapy differentially expressed and regulated in human cancer cells. Hum. Gene Ther. 1999;10:155–164. doi: 10.1089/10430349950018959. [DOI] [PubMed] [Google Scholar]
- 22.Mathupala S.P., Ko Y.H., Pedersen P.L. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patra K.C., Wang Q., Bhaskar P.T., Miller L., Wang Z., Wheaton W., Chandel N., Laakso M., Muller W.J., Allen E.L. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith T.A. Mammalian hexokinases and their abnormal expression in cancer. Br. J. Biomed. Sci. 2000;57:170–178. [PubMed] [Google Scholar]
- 25.Mathupala S.P., Ko Y.H., Pedersen P.L. Hexokinase-2 bound to mitochondria: cancer’s stygian link to the “Warburg effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adeva M., González-Lucán M., Seco M., Donapetry C. Enzymes involved in l-lactate metabolism in humans. Mitochondrion. 2013;13:615–629. doi: 10.1016/j.mito.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie H., Li J., Yang X.M., Cao Q.Z., Feng M.X., Xue F., Wei L., Qin W., Gu J., Xia Q., Zhang Z.G. Mineralocorticoid receptor suppresses cancer progression and the Warburg effect by modulating the miR-338-3p-PKLR axis in hepatocellular carcinoma. Hepatology. 2015;62:1145–1159. doi: 10.1002/hep.27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertino G., Demma S., Ardiri A., Proiti M., Gruttadauria S., Toro A., Malaguarnera G., Bertino N., Malaguarnera M., Malaguarnera M., Di Carlo I. Hepatocellular carcinoma: novel molecular targets in carcinogenesis for future therapies. BioMed Res. Int. 2014;2014:203693. doi: 10.1155/2014/203693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Yang N., Ekanem N.R., Sakyi C.A., Ray S.D. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv. Drug Deliv. Rev. 2015;81:62–74. doi: 10.1016/j.addr.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Reyes R.K., Motiwala T., Jacob S.T. Regulation of glucose metabolism in hepatocarcinogenesis by microRNAs. Gene Expr. 2014;16:85–92. doi: 10.3727/105221614X13919976902093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R., Hawkins C., Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robey R.B., Hay N. Is Akt the “Warburg kinase”?—Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W., Qiu Z., Wang Z., Wang Q., Tan N., Chen T., Chen Z., Huang S., Gu J., Li J. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 36.Maruyama M., Totsugawa T., Kunieda T., Okitsu T., Shibata N., Takesue M., Kurabayashi Y., Oshita M., Nakaji S., Kodama M. Hepatocyte isolation and transplantation in the pig. Cell Transplant. 2003;12:593–598. doi: 10.3727/000000003108747190. [DOI] [PubMed] [Google Scholar]
- 37.Meng F.Y., Chen Z.S., Han M., Hu X.P., He X.X., Liu Y., He W.T., Huang W., Guo H., Zhou P. Porcine hepatocyte isolation and reversible immortalization mediated by retroviral transfer and site-specific recombination. World J. Gastroenterol. 2010;16:1660–1664. doi: 10.3748/wjg.v16.i13.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Q., Xia Y., Ding S., Lu P., Sun L., Liu M. An alternatively spliced variant of CXCR3 mediates the metastasis of CD133+ liver cancer cells induced by CXCL9. Oncotarget. 2016;7:14405–14414. doi: 10.18632/oncotarget.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou C., Wang Y., Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J. Biochem. Biophys. Methods. 2005;64:207–215. doi: 10.1016/j.jbbm.2005.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.