Abstract
As an essential post-transcriptional modification, N7-methylguanosine (m7G) regulates nearly every step of the life cycle of mRNA. Accurate identification of the m7G site in the transcriptome will provide insights into its biological functions and mechanisms. Although the m7G-methylated RNA immunoprecipitation sequencing (MeRIP-seq) method has been proposed in this regard, it is still cost-ineffective for detecting the m7G site. Therefore, it is urgent to develop new methods to identify the m7G site. In this work, we developed the first computational predictor called iRNA-m7G to identify m7G sites in the human transcriptome. The feature fusion strategy was used to integrate both sequence- and structure-based features. In the jackknife test, iRNA-m7G obtained an accuracy of 89.88%. The superiority of iRNA-m7G for identifying m7G sites was also demonstrated by comparing with other methods. We hope that iRNA-m7G can become a useful tool to identify m7G sites. A user-friendly web server for iRNA-m7G is freely accessible at http://lin-group.cn/server/iRNA-m7G/.
Keywords: N7-methylguanosine, nucleotide chemical property, RNA secondary structure, pseudo nucleotide composition, feature fusion
Introduction
Besides N1-methyladenosine (m1A), N7-methylguanosine (m7G) is another kind of positively charged RNA modification.1 m7G is added to the 5′ end co-transcriptionally during transcription, and it is essential for efficient gene expression and cell viability.2 It has been found that m7G is required for nearly all phages of the mRNA cycles, such as RNA splicing,3 polyadenylation,4 nuclear export of mRNA,5 translation,6 and so on. Although studies on m7G have been carried out for a long time, the knowledge about its function is still limited. The key step of revealing the functions of m7G is to determine its accurate position in the transcriptome.
By using the mass spectrometry quantification and m7G-methylated RNA immunoprecipitation sequencing (MeRIP-seq) method,7 Zhang et al. not only detected the m7G sites in Homo sapiens and Mus. Musculus but also provided the base resolution m7G sites in human HeLa and HepG2 cells. However, the MeRIP-seq method still has its own limitations,7 and it is cost-ineffective for performing transcriptome-wide detections. Therefore, it is necessary to develop computational methods for identifying m7G sites.
To the best of our knowledge, there are no computational methods available for this aim. Inspired by the wide application of machine-learning methods for identifying RNA modification sites,8, 9 in this study, we developed a support vector machine (SVM)-based method, called iRNA-m7G, to identify m7G sites. To extract informative features to encode the RNA sequence, the feature fusion strategy was used to integrate three kinds of features, including nucleotide property and frequency, pseudo nucleotide composition, and secondary structure component. Experiments exhibited that the feature fusion strategy is superior to the single kind of features for identifying m7G sites. Moreover, a user-friendly web server for iRNA-m7G has been provided at http://lin-group.cn/server/iRNA-m7G/. We expect that the proposed predictor will speed up the detection of the m7G site.
Results and Discussion
Performance of Each Kind of Feature
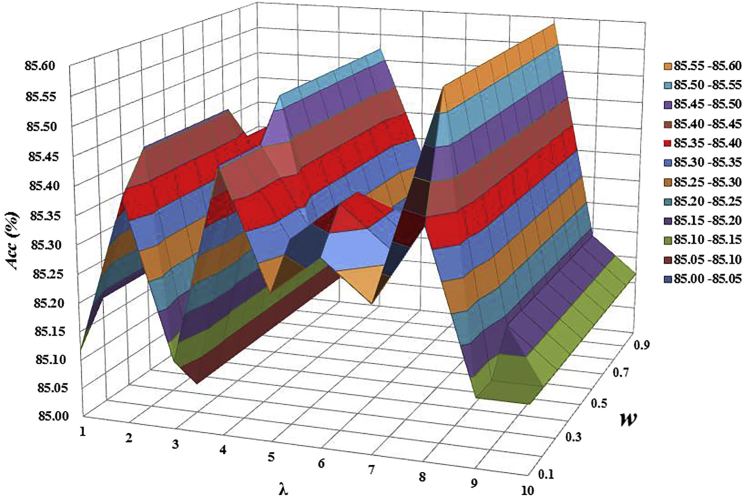
We built three models based on the three kinds of features (nucleotide property and frequency [NPF], pseudo nucleotide composition [PseDNC], and secondary structure component [SSC]), and we compared their performances for identifying m7G sites. As indicated in Equations 4 and 5, the PseDNC model is dependent on two parameters, w and λ. Hence, we first optimized the parameters of PseDNC. In general, the greater the λ value is, the more global sequence-order information the model contains. However, a larger λ would reduce the cluster-tolerant capacity so as to lower the cross-validation accuracy due to an overfitting problem. Therefore, the search ranges for w and λ were set in [0, 1] and [1, 10] with a step of 0.1 and 1, respectively. As shown in Figure 1, the PseDNC-based model yielded the best results when w = 0.8 and λ = 8.
Figure 1.
Determining the Optimal Values for the Two Parameters w and λ of PseDNC
The k-fold cross-validation test method was often used to examine the quality of various predictors.10 For saving computational time, in the current study, the 10-fold cross-validation test was used to evaluate the performance of these models. Their predictive results were reported in Table 1. Among the three models, the NPF-based model obtained the highest accuracy of 89.14%, which is approximately 5% and 14% higher than that of the PseDNC- and SSC-based models, respectively, for identifying m7G sites in the dataset.
Table 1.
Predictive Results for Identifying m7G Sites by Using Different Features
| Features | Sn (%) | Sp (%) | Acc (%) | MCC | auROC |
|---|---|---|---|---|---|
| NPF | 88.12 | 90.15 | 89.14 | 0.78 | 0.899 |
| PseDNC | 81.92 | 87.99 | 84.95 | 0.70 | 0.841 |
| SSC | 73.11 | 78.71 | 75.91 | 0.52 | 0.776 |
| Fusion | 88.66 | 90.96 | 89.81 | 0.80 | 0.946 |
Sn, sensitivity; Sp, specificity; Acc, accuracy; MCC, Mathew’s correlation coefficient; auROC, area under the receiver operating characteristic curve; NPF, nucleotide property and frequency; PseDNC, pseudo nucleotide composition; SSC, secondary structure component.
To objectively compare their performances, the area under the receiver operating characteristic curve (auROC) of these methods was also calculated. The NPF-based model obtained an auROC of 0.899, higher than the 0.841 and 0.776 obtained by the PseDNC- and SSC-based models, respectively.
Performance of Fusing Multiple Features
To investigate whether the feature fusion strategy could improve the performance, we built another model by fusing the NPF, PseDNC, and SSC features. The framework of how to build the model is shown in Figure 2. The model thus obtained was then evaluated by using the 10-fold cross-validation test. The detailed results are provided in the last row of Table 1. As indicated in Table 1, the sensitivity (Sn), specificity (Sp), accuracy (Acc), and Mathew’s correlation coefficient (MCC) were all improved compared with those obtained by the NPF-, PseDNC-, and SSC-based models.
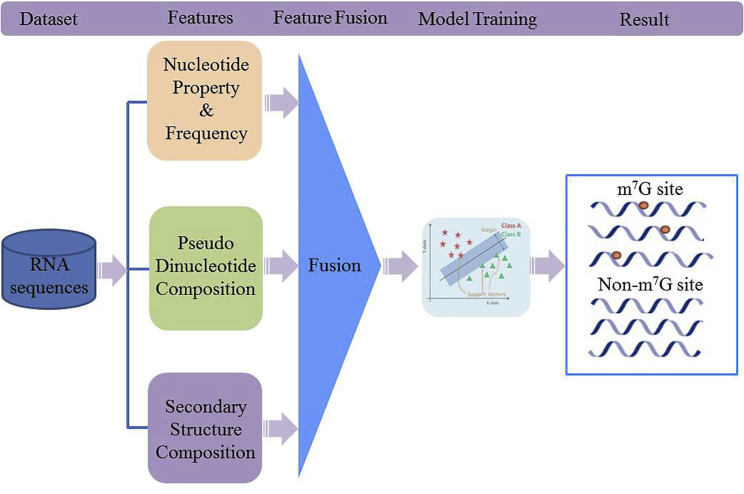
Figure 2.
Framework of Developing iRNA-m7G
For an RNA sequence, it is converted into a feature vector by fusing nucleotide property and frequency, pseudo nucleotide composition, and secondary structure component. The support vector machine was used to build the classification model.
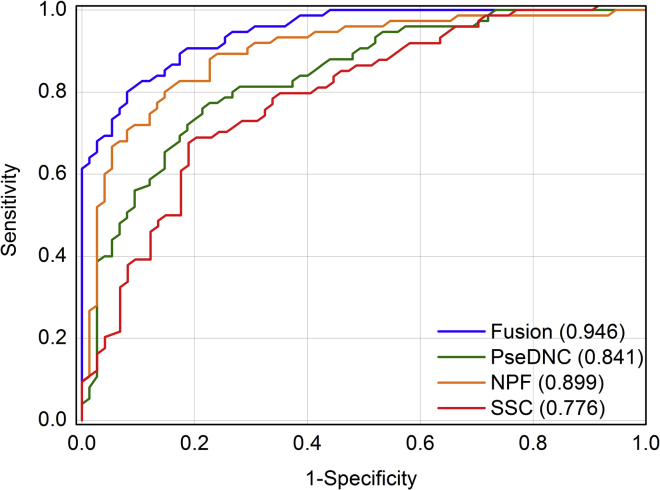
To intuitively compare the performance of the models based on different features, their ROC curves from the 10-fold cross-validation test were plotted in Figure 3. The fusion strategy-based model obtained an auROC of 0.946, which is higher than those of the NPF-, PseDNC-, and SSC-based models.
Figure 3.
The Receiver Operating Characteristic Curves of the Models Based on Different Features Identifying m7G sites
SSC is the abbreviation for secondary structure component, NPF is for nucleotide property and frequency, PseDNC is for pseudo nucleotide composition, and fusion is the combination of the abovementioned three kinds of features. The auROC values were provided in brackets.
Moreover, to further demonstrate its stability for identifying m7G sites, the fusion strategy-based model was also evaluated by the jackknife test, in which each sample in the training dataset is in turn singled out as an independent test sample, and all the properties are calculated without including the one being identified. In the jackknife test, the fusion strategy-based model obtained an accuracy of 89.88% with the sensitivity of 89.07%, specificity of 90.69%, and MCC of 0.80, which is comparable to those from the 10-fold cross-validation test. These results indicate that the feature fusion strategy is effective and the model is robust for identifying m7G sites.
Comparison of SVM and Other Classifiers
Since there is no computational method that has been proposed for identifying m7G sites, to demonstrate its effectiveness, we compared the performance of the current SVM-based model with those of the Naive Bayes-, Random Forest-, LogitBoost-, and BayesNet-based models. The Naive Bayes, Random Forest, LogitBoost, and BayesNet were implemented by using WEKA.11 For a fair comparison, all the models were built by using the the feature fusion strategy and tested on the same dataset. The 10-fold cross-validation test results of these models are reported in Table 2. As shown in Table 2, the SVM-based model obtained the best results in terms of the four metrics defined in Equation 9. The predictive accuracy of the SVM-based model is 9.7%, 3.3%, 6.1%, and 7.7% higher than those of the Naive Bayes-, Random Forest-, LogitBoost-, and BayesNet-based models, respectively. This result demonstrates that the SVM is more effective than other classification algorithms for identifying m7G sites.
Table 2.
Performance Comparison of Different Classifiers for Identifying m7G Sites by the 10-Fold Cross-Validation Test
| Classifiers | Sn (%) | Sp (%) | Acc (%) | MCC |
|---|---|---|---|---|
| Naive Bayes | 72.47 | 87.85 | 80.16 | 0.61 |
| Random Forest | 83.27 | 89.88 | 86.57 | 0.73 |
| LogitBoost | 81.38 | 86.23 | 83.81 | 0.68 |
| BayesNet | 77.19 | 87.04 | 82.12 | 0.65 |
| SVM | 88.66 | 90.96 | 89.81 | 0.80 |
Sn, sensitivity; Sp, specificity; Acc, accuracy; MCC, Mathew’s correlation coefficient; SVM, support vector machine.
Conclusions
In this study, we proposed iRNA-m7G, the first computational method to identify m7G sites. In this predictor, the feature fusion strategy was used to represent RNA sequences. Comparative results demonstrated that the feature fusion strategy is much more effective for identifying m7G sites than a single kind of feature.
Moreover, we also compared iRNA-m7G with the other four machine-learning algorithm-based methods, and we found that the SVM-based model achieves the best performance for identifying m7G sites.
For the convenience of the scientific community, a publicly accessible web server called iRNA-m7G that allows the prediction of m7G sites in RNA was established at http://lin-group.cn/server/iRNA-m7G/. We anticipate that iRNA-m7G will become a useful tool for identifying m7G sites. In future works, we will collect more m7G data and use powerful methods such as deep learning12, 13, 14, 15 to improve the performance of computationally identifying m7G sites.
Materials and Methods
Benchmark Datasets
By using the MeRIP-seq method, Zhang et al.7 detected 801 base-resolution m7G sites that appeared in human HeLa and HepG2 cells. By mapping these sites to the human genome (hg19), 801 m7G sites containing sequences were obtained. Preliminary tests indicated that the best predictive result was achieved when the sequence length is 41 bp with the m7G site in the center. To build a high-quality dataset, the CD-HIT software with the threshold of 80% was used to remove redundant sequences.16, 17 Accordingly, we obtained 741 m7G site-containing sequences.
The non-m7G site-containing sequences were obtained by choosing 41-bp-long sequences with the intermediate guanosine not detected as m7G by the MeRIP-seq method. By doing so, a huge number of negative samples is obtained. Since imbalanced datasets affect the performance evaluation of computational methods, to balance out the numbers between positive and negative samples in model training, we randomly picked out 741 non-m7G site sequences with the sequence similarity less than 80% to form the negative samples.
Sequence Representation
NPF
The NPF is an effective sequence-encoding scheme for computationally identifying nucleotide modification sites.18, 19, 20, 21 According to NPF, the i-th nucleotide ni in RNA sequence can be represented by a four-dimensional vector (xi, yi, zi, di), in which the elements are defined as follows:
| (Equation 1) |
where the x, y, and z coordinates stand for the ring structure, hydrogen bond, and chemical functionality, respectively; di is the accumulated frequency and is defined as
| (Equation 2) |
where l is the sequence length, and |Ni| is the length of the i-th prefix string {n1, n2, …, ni} in the sequence.
According to NPF, an RNA sequence with a length of l bp will be encoded by the following vector:
| (Equation 3) |
PseDNC
Besides the local sequence order information, the global sequence order effect is also important for computationally identifying RNA modification sites. Accordingly, in the current study, the PseDNC was also used to encode the RNA sequences,22 which can be calculated by using PseKNC23 and PseKNC-General.24 Based on PseDNC, the RNA sequence is converted into a discrete vector defined as follows:
| (Equation 4) |
where
| (Equation 5) |
is the occurrence frequency of the u-th non-overlapping dinucleotide in the RNA sequence, and
| (Equation 6) |
where is the j-tier correlation factor that reflects the sequence order correlation between all the j-th most contiguous dinucleotide, and is defined as
| (Equation 7) |
where μ is the number of RNA physicochemical properties considered, is the normalized numerical value of the g-th (g = 1, 2, 3, …, μ) RNA local structural property for the dinucleotide RiRi+1 at position i, and is the corresponding value for the dinucleotide Ri+jRi+j+1 at position i + j.
In the current work, the enthalpy, entropy, and free energy were used to define PseDNC, which have been used to identify other kinds of RNA modifications. The values for the three physicochemical properties of the 16 different RNA dinucleotides were obtained from previous works.25, 26 Thus, μ in Equation 7 is equal to 3.
SSC
The formation of RNA modification is affected by RNA structures. Hence, the RNAfold tool in the ViennaRNA package27 was used to predict the secondary structure of the RNA sequences in the dataset. For each position in the RNA, the paired nucleotide was represented by a parenthesis (“(” or “)”), while the unpaired one was represented by a dot (“.”). In the current study, we do not distinguish “(” and “)” and use “(” for both statuses. For a given tri-nucleotide, there are eight (23) possible structure statuses (i.e., “(((,” “((.,” “(..,” “(.(,” “.((,” “.(.,” “..(,” and “…”). Together with the first nucleotide of the tri-nucleotide, there will be 32 (4 × 8) possible sequence-structure modes denoted as “A-(((,” “A-((.,” “A-(..,” …, and “U-…”.28 Therefore, by using the sequence-structure mode, an RNA sequence can be represented as follows:
| (Equation 8) |
SVM
In the current study, the LibSVM package 3.18, which is available at https://www.csie.ntu.edu.tw/∼cjlin/libsvm/, was used to perform the classification task. The basic idea of SVM is to transform the input data into a high-dimensional feature space and then determine the optimal separating hyperplane. Because of its better performance, the radial basis kernel function (RBF) was used to obtain the separating hyperplane. The regularization parameter C and kernel parameter γ of the SVM operation engine were optimized in the ranges of [2−5, 215] and [2−15, 2−5] with the steps of 2 and 2−1, respectively. The final prediction was made according to the probability obtained by SVM.29, 30, 31, 32, 33 If its probability is >0.5, a guanine will be predicted as an m7G site.
Evaluation Metrics
In this study, the four metrics,34, 35, 36, 37, 38, 39, 40 namely, Sn, Sp, Acc, and MCC, were used to measure the performance of the proposed methods, which are defined as follows:
| (Equation 9) |
where represents the m7G site-containing sequence, while is the number of m7G site-containing sequences incorrectly predicted to be of false m7G site-containing sequences; is the total number of false m7G site-containing sequences, while is the number of the false m7G site-containing sequences incorrectly predicted to be of m7G site-containing sequences.
Moreover, by plotting the sensitivity against (1-specificity) with the varying of the threshold, the ROC curve41, 42 was generated to evaluate the performance of the proposed method. The auROC is an indicator of the performance of the method. An auROC value of 0.5 is equivalent to random prediction while an auROC of 1 represents a perfect one.
Author Contributions
W.C. and H. Lin conceived and designed the study. W.C., P.F., X.S., and H. Lin conducted the experiments. P.F., W.C., and X.S. implemented the algorithms. H. Lv established the web server. W.C., P.F., X.S., H. Lv, and H. Lin performed the analysis and wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Nature Scientific Foundation of China (31771471 and 61772119) and the Natural Science Foundation for Distinguished Young Scholar of Hebei Province (C2017209244).
Contributor Information
Wei Chen, Email: chenweiimu@gmail.com.
Hao Lin, Email: hlin@uestc.edu.cn.
References
- 1.Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2015;91:394–409. doi: 10.2183/pjab.91.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowling V.H. Regulation of mRNA cap methylation. Biochem. J. 2009;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., 3rd, Hartzog G.A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drummond D.R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis J.D., Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 6.Murthy K.G., Park P., Manley J.L. A nuclear micrococcal-sensitive, ATP-dependent exoribonuclease degrades uncapped but not capped RNA substrates. Nucleic Acids Res. 1991;19:2685–2692. doi: 10.1093/nar/19.10.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., He C. Transcriptome-wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell. 2019;74:1304–1316.e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W., Feng P., Yang H., Ding H., Lin H., Chou K.C. iRNA-3typeA: Identifying Three Types of Modification at RNA’s Adenosine Sites. Mol. Ther. Nucleic Acids. 2018;11:468–474. doi: 10.1016/j.omtn.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y., Zeng P., Li Y.H., Zhang Z., Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao W., Zhou Y., Cui Q., Zhou Y. PACES: prediction of N4-acetylcytidine (ac4C) modification sites in mRNA. Sci. Rep. 2019;9:11112. doi: 10.1038/s41598-019-47594-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 12.Hou J., Wu T., Cao R., Cheng J. Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. Proteins. 2019 doi: 10.1002/prot.25697. Published online April 15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel S., Tripathi R., Kumari V., Varadwaj P. DeepInteract: Deep Neural Network Based Protein-Protein Interaction Prediction Tool. Curr. Bioinform. 2017;12:551–557. [Google Scholar]
- 14.Cao R., Bhattacharya D., Hou J., Cheng J. DeepQA: improving the estimation of single protein model quality with deep belief networks. BMC Bioinformatics. 2016;17:495. doi: 10.1186/s12859-016-1405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson N., Shane E., Chase J., Rowland J., Ries D., Justice N., Zhang J., Chan L., Cao R. Survey of Machine Learning Techniques in Drug Discovery. Curr. Drug Metab. 2019;20:185–193. doi: 10.2174/1389200219666180820112457. [DOI] [PubMed] [Google Scholar]
- 16.Zou Q., Lin G., Jiang X., Liu X., Zeng X. Sequence clustering in bioinformatics: an empirical study. Brief. Bioinform. 2018 doi: 10.1093/bib/bby090. Published online September 18, 2018. [DOI] [PubMed] [Google Scholar]
- 17.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W., Lv H., Nie F., Lin H. i6mA-Pred: identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics. 2019;35:2796–2800. doi: 10.1093/bioinformatics/btz015. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Yang H., Feng P., Ding H., Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z.C., Feng P.M., Yang H., Qiu W.R., Chen W., Lin H. iRNAD: a computational tool for identifying D modification sites in RNA sequence. Bioinformatics. 2019:btz358. doi: 10.1093/bioinformatics/btz358. [DOI] [PubMed] [Google Scholar]
- 21.He W., Jia C., Zou Q. 4mCPred: machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics. 2019;35:593–601. doi: 10.1093/bioinformatics/bty668. [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Qiu W.R., Liu G., Guo F.B., Chen W., Chou K.C., Lin H. iRSpot-Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC. Int. J. Biol. Sci. 2018;14:883–891. doi: 10.7150/ijbs.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W., Lei T.-Y., Jin D.-C., Lin H., Chou K.-C. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014;456:53–60. doi: 10.1016/j.ab.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Chen W., Zhang X., Brooker J., Lin H., Zhang L., Chou K.-C. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics. 2015;31:119–120. doi: 10.1093/bioinformatics/btu602. [DOI] [PubMed] [Google Scholar]
- 25.Freier S.M., Kierzek R., Jaeger J.A., Sugimoto N., Caruthers M.H., Neilson T., Turner D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia T., SantaLucia J., Jr., Burkard M.E., Kierzek R., Schroeder S.J., Jiao X., Cox C., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue C., Li F., He T., Liu G.P., Li Y., Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H., Chen W., Lin H. Identification of immunoglobulins using Chou’s pseudo amino acid composition with feature selection technique. Mol. Biosyst. 2016;12:1269–1275. doi: 10.1039/c5mb00883b. [DOI] [PubMed] [Google Scholar]
- 30.Zhu P.P., Li W.C., Zhong Z.J., Deng E.Z., Ding H., Chen W., Lin H. Predicting the subcellular localization of mycobacterial proteins by incorporating the optimal tripeptides into the general form of pseudo amino acid composition. Mol. Biosyst. 2015;11:558–563. doi: 10.1039/c4mb00645c. [DOI] [PubMed] [Google Scholar]
- 31.Ding H., Li D. Identification of mitochondrial proteins of malaria parasite using analysis of variance. Amino Acids. 2015;47:329–333. doi: 10.1007/s00726-014-1862-4. [DOI] [PubMed] [Google Scholar]
- 32.Manavalan B., Shin T.H., Lee G. PVP-SVM: Sequence-Based Prediction of Phage Virion Proteins Using a Support Vector Machine. Front. Microbiol. 2018;9:476. doi: 10.3389/fmicb.2018.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao R., Wang Z., Wang Y., Cheng J. SMOQ: a tool for predicting the absolute residue-specific quality of a single protein model with support vector machines. BMC Bioinformatics. 2014;15:120. doi: 10.1186/1471-2105-15-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X.J., Feng C.Q., Lai H.Y., Chen W., Lin H. Predicting protein structural classes for low-similarity sequences by evaluating different features. Knowl. Based Syst. 2019;163:787–793. [Google Scholar]
- 35.Tan J.X., Li S.H., Zhang Z.M., Chen C.X., Chen W., Tang H., Lin H. Identification of hormone binding proteins based on machine learning methods. Math. Biosci. Eng. 2019;16:2466–2480. doi: 10.3934/mbe.2019123. [DOI] [PubMed] [Google Scholar]
- 36.Lv H., Zhang Z.M., Li S.H., Tan J.X., Chen W., Lin H. Evaluation of different computational methods on 5-methylcytosine sites identification. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz048. Published online June 3, 2019. [DOI] [PubMed] [Google Scholar]
- 37.Manavalan B., Subramaniyam S., Shin T.H., Kim M.O., Lee G. Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy. J. Proteome Res. 2018;17:2715–2726. doi: 10.1021/acs.jproteome.8b00148. [DOI] [PubMed] [Google Scholar]
- 38.Tang H., Cao R.Z., Wang W., Liu T.S., Wang L.M., He C.M. A two-step discriminated method to identify thermophilic proteins. Int. J. Biomath. 2017;10:1750050. [Google Scholar]
- 39.Liu B., Han L., Liu X., Wu J., Ma Q. Computational prediction of sigma-54 promoters in bacterial genomes by integrating motif finding and machine learning strategies. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019;16:1211–1218. doi: 10.1109/TCBB.2018.2816032. [DOI] [PubMed] [Google Scholar]
- 40.Wei L., Xing P., Zeng J., Chen J., Su R., Guo F. Improved prediction of protein-protein interactions using novel negative samples, features, and an ensemble classifier. Artif. Intell. Med. 2017;83:67–74. doi: 10.1016/j.artmed.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Feng C.Q., Zhang Z.Y., Zhu X.J., Lin Y., Chen W., Tang H., Lin H. iTerm-PseKNC: a sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics. 2019;35:1469–1477. doi: 10.1093/bioinformatics/bty827. [DOI] [PubMed] [Google Scholar]
- 42.Dao F.Y., Lv H., Wang F., Feng C.Q., Ding H., Chen W., Lin H. Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics. 2019;35:2075–2083. doi: 10.1093/bioinformatics/bty943. [DOI] [PubMed] [Google Scholar]