Abstract
circRNAs arise from back splicing events during mRNA processing, and when deregulated can play an active role in cancer. Here we characterize a new circRNA (circPOK) encoded by the Zbtb7a gene (also kown as POKEMON, LRF) in the context of mesenchymal tumor progression. circPOK functions as a non-coding proto-oncogenic RNA independently and antithetically to its linear transcript counterpart, which acts as a tumor suppressor by encoding the Pokemon transcription factor. We find that circPOK regulates pro-proliferative and pro-angiogenic factors by co-activation of the ILF2/3 complex. Importantly, the expression of Pokemon protein and circRNA is aberrantly uncoupled in cancer through differential post-transcriptional regulation. Thus, we identify a novel type of genetic unit, the iRegulon, that yields biochemically distinct RNA products, circular and linear, with diverse and antithetical functions. Our findings further expand the cellular repertoire towards the control of normal biological outputs, while aberrant expression of such components may underlie disease pathogenesis including cancer.
Subject terms: Cancer genetics, Cancer microenvironment
Introduction
Research interest has recently been re-ignited in circRNAs, which result from an intriguing and specialized form of alternative splicing in which the 3′-tail of an exon back-splices and joins the 5′-head of the exon localized up-stream.1 Repetitive complementary sequences within introns up- and down-stream of the circularizing exons can favor back-splicing events,2,3 and in these cases multiple circRNAs consisting of one or more exons can arise from a single gene. Although some transcripts exist mainly in circular form, as in the case of CDR1as,4,5 many known circRNAs co-originate with linear transcripts from protein-coding mRNA precursors.1,6,4 Therefore the same genetic unit can produce functionally distinct RNA isoforms, linear and circular, protein coding and non-coding, which may have unique (in principle) and independent functions in both normal and pathological conditions, including cancer. In this respect, although circRNAs have been suggested as biomarkers or therapeutic targets, their contribution to cancer progression is still largely undefined. Indeed, while researchers have comprehensively studied the tumorigenic impact of several tumor suppressors and oncogenes encoded by linear mRNA transcripts, the roles of their circRNA counterparts, which derive from the same genetic locus but are the result of alternative back splicing, are mostly unknown.
Extensive study has been devoted to the capacity of the Zbtb7a gene to encode the protein Pokemon (also known as Lrf, FBI-1 or OCZF). Pokemon belongs to the POK/ZBTB family of transcription factors; it is made of a C-terminal Kruppel-type zinc finger domain, through which it can bind to DNA, and an N-terminal POZ/BTB domain by which it interacts with other proteins. Functionally, Pokemon is a transcriptional repressor that plays critical roles in stem cell differentiation as well as tumorigenesis.7,8 We have demonstrated that the Pokemon protein can function as a tumor suppressor in a tissue-specific manner, and that it specifically suppresses tumorigenesis in the prostatic epithelium,9 and the mesenchymal stem cell (MSC) compartment.8
We here report that non-coding circular elements derived from the Zbtb7a gene also contribute to the regulation of cancer progression, but through proto-oncogenic mechanisms that are distinct from its linear counterpart. Because of this dual functionality, the Zbtb7a locus epitomizes a new type of genetic unit that produces linear and circular RNAs with antagonistic and independent functions.
Results
CircPOK originates from the back splicing of the Zbtb7a exon 2
In the past few years RNA-sequencing assays have identified a large number of circRNAs, which have subsequently been cataloged in public databases.10 Among the thousands of these newly identified circRNAs, we were particularly intrigued by one generated by Zbtb7a, a key gene in the pathogenesis of tumors of different histology. According to previous studies, the contribution of Zbtb7a to cancer is tissue-specific7; and in the context of mesenchymal tumorigenesis, we recently demonstrated that Zbtb7a transcriptionally and functionally represses the proto-oncogenes Dlk1 and Sox9 by encoding the protein Pokemon, which in turn suppress sarcomagenesis.8 These previous data, together with the observation that circRNAs can originate from the Zbtb7a gene, prompted us to investigate whether the Zbtb7a-circRNA could be functional; we therefore initiated a comprehensive characterization of this circRNA in mesenchymal tissue.
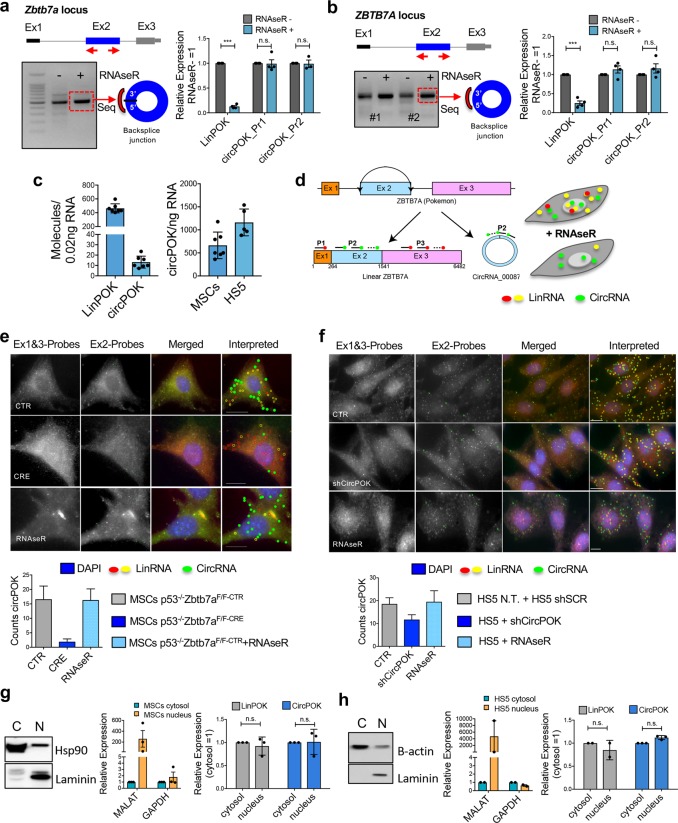
First, we designed divergent primers to detect the circRNA through PCRs and RT-qPCRs (Supplementary information, Fig. S1a); we then subjected the obtained PCR amplicons to Sanger-sequencing (Supplementary information, Fig. S1b). Although only one circRNA isoform was observed by RNA-sequencing, we designed a range of primers to detect whether multiple circRNA isoforms could be generated from the Zbtb7a gene (Supplementary information, Fig. S1b). As shown in Supplementary information, Fig. S1b, of all the samples analyzed, only the primers designed for Exon 2 were able to amplify a PCR product. Sanger sequencing of these amplicons revealed back-splicing at Zbtb7a Exon 2 (Supplementary information, Fig. S1b). These results, in line with previous observations, allowed us to conclude that the Zbtb7a gene gives rise to only one circRNA isoform, which is created by back-splicing events at its Exon 2. We labeled this new circRNA “circPOK”, and once identified, we proceeded to determine whether it possessed the features of a bona fide circRNA. In this respect, we subjected total RNA to RNase R treatment, which degrades linear transcripts but not circular ones (all transcripts were quantified by RT-qPCR before and after digestion). As shown in Fig. 1a, while the linear transcript (LinPOK) was dramatically reduced by RNase R treatment, the circPOK, as measured by two independent primer sets (circPOK_Pr1 and circPOK_Pr2), remained untouched. In addition to resisting RNase R treatment, circPOK showed higher stability within the cell compared to its linear counterpart (Supplementary information, Fig. S1c), another peculiar feature of circRNAs. Moreover, circPOK is conserved in mouse and human cells (Fig. 1b; Supplementary information, Fig. S1b) with a sequence similarity of 83%, and a comparative length of 1256 bp for the mouse, and 1277 bp for the human variants. Importantly, it is expressed in a range of cell types, including human mesenchymal cells and sarcoma cell lines (Fig. 1a, b; Supplementary information, Fig. S1d, and data not shown).
Fig. 1.
CircPOK is generated from the Zbtb7a genomic locus through back splicing of the Exon 2. a Analysis of the expression of mouse circPOK in primary p53−/− MSCs. circPOK was detected by PCR with divergent primers, and the amplicon was subjected to Sanger sequencing. RT-qPCR analysis of LinPOK and circPOK before and after treatment with RNase R is shown in the chart on the right. In the chart, circPOK_Pr1 and circPOK_Pr2 refer to independent divergent primer sets used to identify mouse circPOK (n ≥ 3 independent experiments). b Analysis of the expression of human circPOK in the HS5 mesenchymal cell line. circPOK was detected by PCR with divergent primers, and the amplicon was subjected to Sanger sequencing. RT-qPCR analysis of LinPOK and circPOK before and after treatment with RNase R is shown in the chart on the right. circPOK_Pr1 and circPOK_Pr2 refer to independent divergent primer sets used to identify human circPOK (n ≥ 3 independent experiments). c Absolute quantification of mouse LinPOK and circPOK in p53−/− MSCs is shown in the chart on the left (n = 7). Absolute quantification of mouse and human circPOK in p53−/− MSCs (mouse, n = 7) and HS5 cells (human, n = 5) is shown in the chart on the right. d Schematic representation of the circFISH assay. e circFISH analysis of mouse circPOK in primary p53−/−Zbtb7a_Ex2F/F MSCs, transduced with a CTR vector (upper panels), with a vector expressing CRE (panels in the middle), or CTR cells treated with the RNase R (bottom panels). The chart at the bottom represents the number of molecules of circPOK per cell in the three different conditions analyzed. (n = 100 cells accounted for each condition, and the mRNA copy numbers were obtained using average counts per cell with 95% confidence interval). f circFISH analysis of human circPOK in HS5 mesenchymal cell line, transduced with a CTR vector (shSCR, upper panels), with an shCircPOK (panels in the middle), or CTR cells treated with RNase R (bottom panels). The chart at the bottom represents the number of molecules of circPOK per cell in the three different conditions analyzed. (n = 100 cells accounted for each condition, and the mRNA copy numbers were obtained using average counts per cell with 95% confidence interval). g Nucleus/Cytosol fractionation experiments to identify LinPOK and circPOK in mouse p53−/− MSCs. Western blot (Hsp90 and Laminin) and RT-qPCR (Malat and Gapdh) to confirm the fractionation are shown on the left, while RT-qPCR analyses of the transcripts are shown with the charts on the right. h Nucleus/Cytosol fractionation experiments to identify LinPOK and circPOK in human HS5 cell line. Western blot (b-actin and Laminin) and RT-qPCR (Malat and Gapdh) to confirm the fractionation are shown on the left, while RT-qPCR analyses of the transcripts are shown with the charts on the right
Although some RNAs exist exclusively in circular isoforms,4 back-splicing generally occurs at lower frequency than linear spicing. In order to assess the ratio of linear to back splicing events for Zbtb7a, we performed absolute quantifications of linear and circular transcripts within mouse MSCs. As shown in Fig. 1c, the circularization process was found to occur with a frequency of 2.8%, and generated an average of 15–20 molecules of circPOK in each primary mouse MSC or human HS5 cell (Fig. 1c).
We next aimed to investigate the subcellular localization of circPOK in mesenchymal cells. To this end, we developed a new methodology for circRNA visualization by in-situ hybridization (circFISH, Fig. 1d). Each of Zbtb7a’s three exons were tiled with probes labeled with a different fluorescent reporter. Probes annealing to Exons 1 and 3 (contained within the linear transcript but not the circular one) enabled visualization of the linear transcript (Red signals in Fig. 1), while probes targeting Exon 2 (Green signals in Fig. 1) allowed for discrimination between the linear and circular transcripts. By analyzing the co-localization of the two different reporters, we were able to differentiate between individual linear transcripts (Yellow signals in Fig. 1) and circular transcripts (Green signals only in Fig. 1) (Fig. 1d–f). circPOK-null cells (CRE cells in Fig. 1e) or circPOK knock-down cells (shCircPOK cells in Fig. 1f) were used as controls. Interestingly, while circRNAs usually accumulate in the cytosol,11 we observed that circPOK localized both in the cytosol and the nucleus by using this novel circFISH approach (Fig. 1e, f), as also confirmed by nucleus/cytosol fractionation followed by RT-qPCR analysis (Fig. 1g, h). Taken together, these experiments show that circPOK is a bona fide circRNA that is expressed in mesenchymal cells and tumors, and localizes in both the cytosol and nucleus, where it may play functional roles.
CircPOK and Pokemon protein are uncoupled in mesenchymal tumors, where they play independent roles
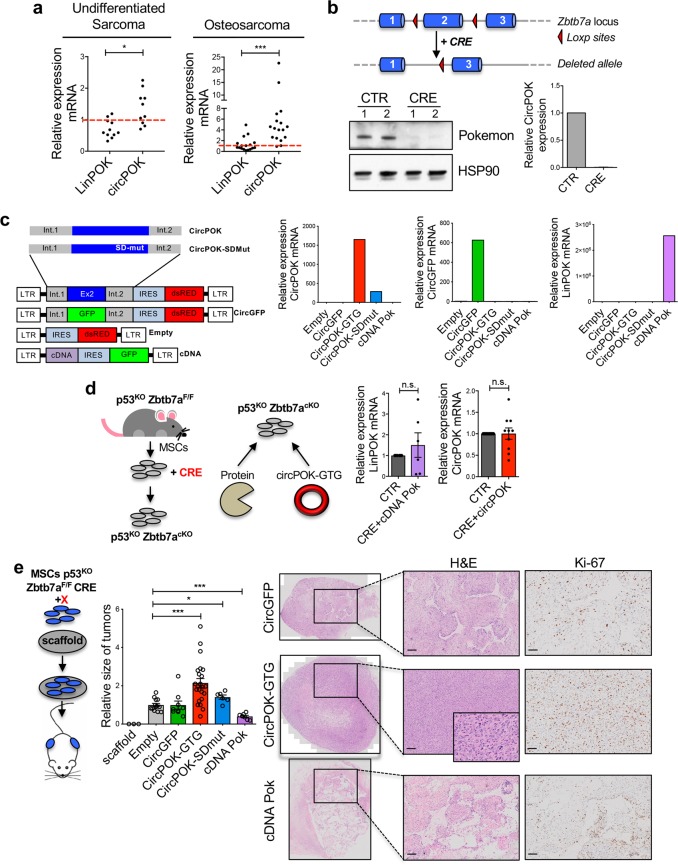
After analyzing the expression of circPOK in cultured cells, we next investigated whether circPOK was also expressed in primary human tumors. Although we previously reported that Pokemon protein significantly decreased in undifferentiated sarcoma samples compared to normal fibrous tissues,8 we had never analyzed the expression of circPOK in the context of mesenchymal tumors. We therefore analyzed undifferentiated sarcomas and osteosarcomas by RT-qPCR for the expression of both linear and circular Pokemon, and compared the results to normal tissue counterparts. Interestingly, as shown in Fig. 2a and Supplementary information, Fig. S2a, we observed a differential expression trend for LinPOK and circPOK in tumors: while LinPOK levels decreased compared to normal tissue, as expected due to the loss of Pokemon protein in these tumors,8 levels of circPOK increased. The expression of circRNAs, linRNAs, and proteins can be disentangled in normal and tumor cells due to post-transcriptional regulatory mechanisms in which miRNAs target sequences not shared by the linear and circular RNA species, such as the 3’-UTR of linear transcripts. Indeed, we have previously identified several miRNAs, such as those belonging to the miR-17 family, miR106, miR25 and miR93, which can target the 3’-UTR of the Pokemon linear transcript in turn affecting the translation of the Pokemon protein.12 To determine whether post-transcriptional mechanisms could be involved in the differential expression patterns of proteins and circRNAs in tumors, we analyzed the expression of MCM7, the host gene of mirR106b, miR93 and miR25, in tumor samples compared to their normal counterparts. Interestingly, MCM7 expression increased in tumor samples (Supplementary information, Fig. S2b), along with the expression of additional miRNAs targeting the 3’UTR portion of the Zbtb7a linear transcript, such as miRNAs belonging to the miR-17 family (Supplementary information, Fig. S2b). As a result, Pokemon protein is downregulated in tumors as well as the linear transcript, while the circPOK does not decrease (Fig. 2a). These data suggest that the uncoupling of the expression of different elements that originate from the same genetic locus might favor tumor development.
Fig. 2.
CircPOK and Pokemon protein play antithetical functions in mesenchymal tumors. a Expression of LinPOK and circPOK in human primary undifferentiated sarcoma samples (n = 10) compared to normal fibrous tissue (bar = 1, n = 6), and in human primary osteosarcoma (n = 17) compared to normal human osteoblasts (bar = 1, n = 1). b Schematic representation of the Zbtb7aF/F locus, and deletion of Pokemon protein and circPOK in the MSCs isolated from these mice, upon transduction with CRE-expressing vector. c Schematic representation of vectors used to express circPOK or cDNA-POK is shown on the left, along with the control vectors. Expression of circPOK-GFP, circGFP or LinPOK in all the conditions is shown in the charts on the right. d On the left, schematic representation of the strategy used to generate p53−/−Zbtb7a_Ex2F/F-CRE MSCs, which are null for the expression of both Pokemon-protein and circPOK, and to “add-back” specifically the protein or circPOK. The chart on the right show the expression of LinPOK or circPOK upon experiments of “adding-back” in null cells (CRE), compared to the endogenous expression of LinPOK and circPOK in wild-type MSCs (CTR). e Schematic representation of the in vivo tumorigenesis assays is depicted on the left; the chart in the middle represents the relative size of the tumors generated by p53−/−Zbtb7a_Ex2F/F-CRE MSCs expressing the following vectors: the empty vector, circGFP, cDNA POK-GFP, circPOK and circPOK-SDmut. Representative pictures of the tumor sections stained with H&E and Ki-67 are shown on the right
Having observed differential expression patterns between Pokemon protein and circPOK in mesenchymal tumors, we then sought to determine whether circPOK could play functional roles within the process of sarcomagenesis. To this end, we took advantage of a conditional Zbtb7a knock-out model previously generated in our lab, and employed it within a protocol aimed at modeling sarcomagenesis in vivo, as previously described.8 In this mouse model, Exon 2 of the Zbtb7a gene can be conditionally removed upon Cre-LoxP-mediated recombination13 (Fig. 2b). Because circPOK is generated from Exon 2, treatment with Cre-recombinase resulted in the inactivation of both circPOK and the linear mRNA that encodes the protein Pokemon (Fig. 2b). Thus, this situation allowed us to determine the functional role of circPOK through “add-back” experiments.
To determine the possible function of circPOK, we first isolated p53−/−Zbtb7a_Ex2F/F MSCs,8 and transduced them in vitro with a lentiviral vector that expressed Cre-recombinase, which deleted Zbtb7a_Ex2 (Fig. 2b). Subsequently, circPOK (circPOK-GTG vector) or Pokemon protein (cDNA POK) were added back to the null MSCs (Fig. 2c, d) in order to study the specific roles of circRNA and protein. The retroviral expression vector for circPOK was designed such that the Zbtb7a_Ex2 was flanked up- and down-stream by Zbtb7a intron sequences that allowed the circularization process to occur. Several additional control vectors were also generated, including: i) an empty vector, ii) a circPOK-expressing vector mutagenized at the splicing-donor site (circPOK-SDmut) in which the formation of the circRNA was impaired, and iii) a vector expressing a circular isoform of the GFP mRNA whose formation was allowed by the same intron regions up- and down-stream of the Zbtb7a-Exon2 (Fig. 2c). Importantly, by employing this add-back methodology, we were able to re-express the linear and circular transcripts of Zbtb7a in Zbtb7a-null cells, and achieve levels of expression for both linear and circular transcripts comparable to the normal endogenous levels measured in wild-type p53−/− MSCs (Fig. 2d). Additionally, even though endogenous circPOK is unlikely to encode protein, as we have extensively investigated (see below), because the Zbtb7a_Ex2 contains the Pokemon-protein translation initiation codon ATG, and because expression vectors generally contain polyadenylation sites, we excluded the possibility that this expression vector could generate an artifactual but functional polypeptide through site-directed mutagenesis of the ATG (Supplementary information, Fig. S2c). We subsequently used this ATG-mutagenized vector (circPOK-GTG) to express circPOK in all our experiments. Primary p53−/−Zbtb7a_Ex2F/F-CRE MSCs transduced with all these vectors were then tested in vitro and in vivo. We first tested the anchorage-independent growth potential of the cells in vitro using soft-agar assays. Surprisingly, MSCs that expressed circPOK-GTG formed more and larger colonies than controls (empty vector, circGFP-expressing vector, and circPOK-SDmut vector), while cells expressing Pokemon protein (cDNA-POK) demonstrated a decreased anchorage-independent growth (Supplementary information, Fig. S2d). Based on these results, we decided to further investigate whether and how circPOK could contribute in vivo to tumor formation. To this end we employed a protocol previously optimized to study sarcomagenesis in vivo, which is based on the implantation of MSCs within 3D scaffolds in syngenic mice8 (Fig. 2e). Remarkably, while the expression of Pokemon protein (cDNA POK) suppressed tumor formation of p53−/−Zbtb7a_Ex2F/F-CRE MSCs, in line with our previous data,8 the expression of circPOK-GTG markedly enhanced tumor progression and tumor cell proliferation when compared to control vectors, as shown in Fig. 2e. Thus, these experiments suggest that circPOK and Pokemon protein may play independent and opposed roles in the context of mesenchymal tumors. While the Pokemon protein acts as a tumor suppressor in mesenchymal tumors, circPOK may exert proto-oncogenic functions that promote tumor formation.
CircPOK plays a proto-oncogenic role in mesenchymal tumors
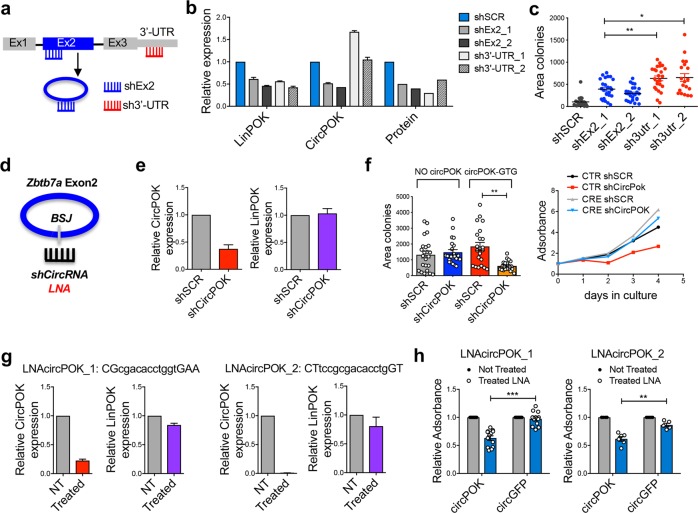
To test whether circPOK could indeed act as a proto-oncogenic element in mesenchymal tumors, we targeted circPOK in p53−/− MSCs in various experimental designs. In one experimental setting, we compared the tumorigenic potential of p53−/− MSCs in which both linear and circular Pokemon transcripts were concomitantly silenced through shRNAs designed to target Exon2 (shEx2), or in which only the linear transcript was knocked down through shRNAs designed to target its 3’UTR (sh3’UTR) (Fig. 3a). Three shRNAs for each target were designed (Supplementary information, Fig. S3a). Of these, two shRNAs with stronger on-target activity were then chosen for functional experiments (Fig. 3b). As shown in Fig. 3c, p53−/− MSCs that lost the expression of only the linear transcript (sh3’-UTR) were able to form larger colonies in anchorage-independent growth experiments than cells that had lost expression of both RNA isoforms (shEx2), suggesting that circPOK expression could add to the loss of the protein in contributing to tumor growth.
Fig. 3.
CircPOK plays proto-oncogenic functions in mesenchymal tumors. a Schematic representation of the experimental design used to selectively target circPOK or LinPOK. shRNAs targeting LinPOK transcript only are indicated as sh3′-UTR. ShRNAs targeting both linear and circular transcripts are indicated as shEx2. b Expression levels of LinPOK, circPOK or Pokemon protein upon the expression in p53−/− MSCs of the shRNAs targeting the 3’-UTR of the linear transcript (sh3′-UTR), or targeting Exon 2 (shEx2); two independent shRNAs for each conditions have been used. c Anchorage-independent proliferation, analyzed in soft-agar assays of p53−/−Zbtb7a_Ex2F/F-CTR MSCs transduced with shSCR, shEx2 or sh3′-UTR. One representative experiments out of 3 independent experiments. d Schematic overview of the strategy to block the expression of circPOK through shRNAs or LNAs directed at the back-splice junction of the circRNA. e Expression levels of circPOK and LinPOK upon knock-down of circPOK by shRNAs in mouse MSCs (shCircPOK). f Analysis of the proliferation of p53−/−Zbtb7a_Ex2F/F-CRE MSCs or p53−/−Zbtb7a_Ex2F/F-CTR MSCs, both silenced for the expression of circPOK is shown on the left. Measure of the anchorage-independent growth of p53−/−Zbtb7a_Ex2F/F-CRE MSCs expressing circGFP/circPOK or silenced for the expression of circPOK (shCircPOK) is shown on the right. g Expression levels of circPOK and LinPOK upon blocking of circPOK by 2 independent LNAs in mouse MSCs. LNAs’ sequence is shown. h Analysis of the proliferation of p53−/−Zbtb7a_Ex2F/F-CRE MSCs expressing either circGFP or circPOK and treated with LNAs targeting circPOK. Not treated cells were used as control. Each dot in the chart represents an independent experiment
In another experimental setting, we directly targeted circPOK by using shRNAs to block its back-splice junction (shCircPOK) (Fig. 3d). Cells transduced with ShCircPOK showed decreased levels of circPOK, but not of Pokemon protein (Fig. 3e; Supplementary information, Fig. S3b). When the circRNA was silenced in circPOK-expressing p53−/− MSCs, the cells showed a markedly impaired proliferation potential both in normal and anchorage-independent growth conditions (Fig. 3f). Importantly, similar results were also obtained by employing an independent shCircRNA (shCircPOK_2), and shCircRNAs were able to target circPOK both in the cytosol and in the nucleus (Supplementary information, Fig. S3c–f). Moreover, in addition to shRNAs targeting the back-splice junction, we used LNA-Gapmers to specifically target circPOK (Fig. 3g) in independent and complementary experiments. Because LNAs have a short half-life within cells, and allow only a transient reduction of the target, we measured the short-term proliferation of LNA-treated cells. In line with our shCircPOK observations, cells showed a decline in proliferation when the LNAs impaired the expression of circPOK (Fig. 3h), corroborating our hypothesis that specifically targeting circPOK can be beneficial in sarcoma treatment.
Finally, to determine whether targeting circPOK might have similar effects in human sarcoma cells, we employed 3 different mesenchymal/sarcoma cell lines (HS5, MFH and HT1080, Supplementary information, Fig. S3g), silenced their expression of circPOK using human shCircPOK (Supplementary information, Fig. S3h), and measured their proliferation rate. Importantly, all the three cell lines showed impaired proliferation upon circPOK silencing (Supplementary information, Fig. S3i), in line with what had been observed, and previously described, in the mouse cells. Taken together, these results further support the notion that circPOK plays proto-oncogenic roles in mesenchymal tumors, and that targeting circPOK in combinatorial therapies, for example by using LNAs, could be beneficial.
CircPOK interacts with nuclear RNA-binding proteins
Having observed a functional role for circPOK as a proto-oncogene in mesenchymal tumors, we next sought to determine the molecular mechanism behind this role. It has recently been shown that circRNAs are enriched in the cytosol, where they can function as competing endogenous RNAs for miRNAs, and thereby positively regulate the expression of those genes that share the same miRNA recognition elements (MREs).14–17 Because linear and circular transcripts share the same genetic sequence (Exon 2 of Zbtb7a), we first tested whether circPOK could affect the expression of Pokemon protein through competition for shared miRNAs. While this would have been counterintuitive in view of their opposing functions, we did not observe any differences in the expression levels of Pokemon protein when circPOK-GTG was overexpressed in p53−/− MSCs (Supplementary information, Fig. S4a), nor did circPOK-GTG affect the expression of known Pokemon-protein targets when overexpressed in wild-type p53−/− MSCs (Supplementary information, Fig. S4a). This in turn suggested that circPOK may function in a Pokemon-independent manner.
It has also been reported that circRNAs can be translated into proteins.18,19 In this respect, circPOK contains a unique open reading frame (ORF), starting with the first ATG codon (in common with the linear transcript) and terminating with an in-frame STOP codon that is created only upon circularization after the back-splicing junction (Supplementary information, Fig. S4b). Therefore a protein of about 50 KDa, which has the same N-terminus as the protein generated from the linear transcript but a different C-terminus, might be encoded from this ORF. However, by performing western blot analysis with two independent antibodies that cross-react with the N-terminal portion of the Pokemon protein encoded by the linear mRNA transcript, we were unable to detect any additional polypeptides either of the predicted or other sizes (Supplementary information, Fig. S4c). Additionally, by performing polysomal fractionation, followed by RT-qPCR, we did not detect circPOK within the polysomal fractions (fractions #10–13 of the Supplementary information, Fig. S4d), suggesting that this circRNA is not translated (Supplementary information, Fig. S4d). Thus we excluded the possibility that circPOK could function in the cytosol through the Pokemon protein, or that it could encode for new oncogenic proteins, which is also supported by the fact that a circPOK devoid of ATG (circPOK-GTG) still retained its proto-oncogenic function (Fig. 2; Supplementary information, Fig. S2).
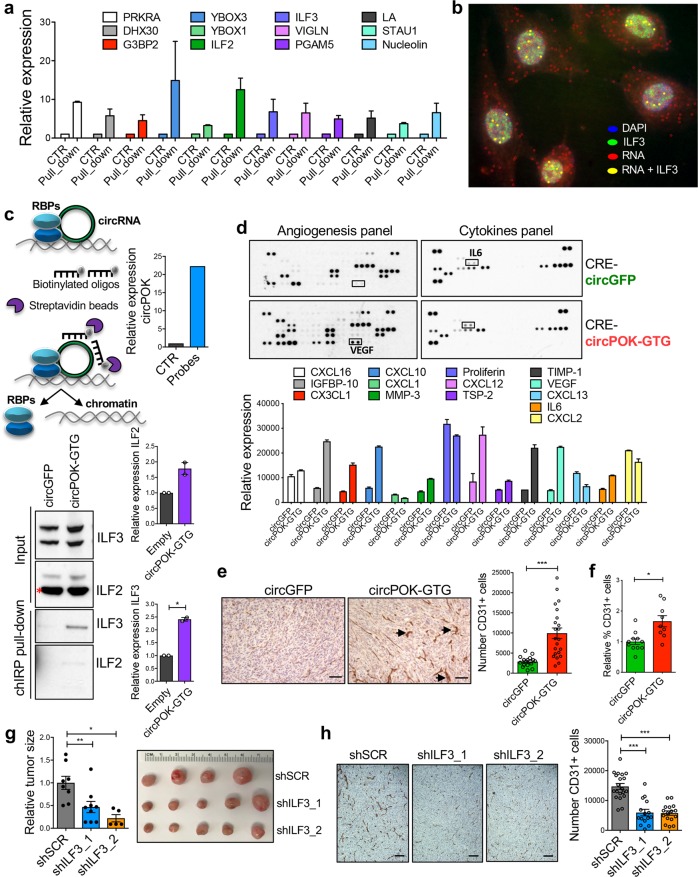
In addition to sponging for miRNAs and encoding new proteins, circRNAs could also regulate the functionality of RNA-binding proteins through direct binding.20,21 To investigate this possibility for circPOK, we performed experiments in which we biotinylated the Zbtb7a Exon 2 through assays of in vitro transcription (which originates circPOK), pulled it down using anti-biotin beads, and subjected the pull-downs to mass spectrometry analysis in order to identify putative circPOK-interacting proteins (see MATERIALS AND METHODS). Several proteins were identified from this analysis (Fig. 4a). Intriguingly, a number of them showed dual DNA and RNA-binding activity, a capability that can play a role in regulating mRNA stability and transcription (Fig. 4a). Among the top candidates identified, ILF2 and ILF3 most sparked our interest. ILF proteins interact with each other in a complex, and have recently been identified as key oncogenes of different types of tumor.22–24 Thus we hypothesized that circPOK could function through the modulation of the ILF2 and ILF3 oncogenic functions. Next, we further confirmed the interaction between circPOK and ILF2/3 by running additional independent experiments. First we performed RIP (RNA Immuno-Precipitation) assays, where we pulled-down ILF2 and ILF3 proteins and confirmed that endogenous circPOK expressed in MSCs interacts with both ILF2 and ILF3 (Supplementary information, Fig. S4e). Second, we coupled the circFISH protocol for circRNA visualization with an immunofluorescence staining for ILF3 protein, and in so doing demonstrated that circPOK can co-localize with the ILF2/3 complex in the nucleus (Fig. 4b; Supplementary information, Fig. S4f). Finally, we pulled down the endogenous circPOK through ChIRP (chromatin isolation by RNA purification) assays,25,26 and subjected the pull-down material to western blot analysis to detect the ILF2 and ILF3 proteins (Fig. 4c). Taken together, these complementary set of experiments consistently showed that endogenous circPOK can interact with ILF2 and ILF3 in MSCs. These interactions take place mainly in the nucleus, where ILF2 and ILF3 mostly reside.
Fig. 4.
CircPOK interacts with ILF2/3, and it regulates the expression of several cytokines and angiogenesis-related factors. a Mass-spectrometry analysis of the RNA-binding proteins that interact with the Zbtb7a_Ex2, which was generated through assays of in vitro transcription. b Co-localization of Zbtb7a_Ex2 RNA and ILF3 visualized by coupling circFISH assay with ILF3 Immunofluorescence staining. RNase R untreated HS5 cells have been used in the experiments. c ChIRP experiments aimed at pulling down the endogenous circPOK and analyze its interactions with ILF2/3. The schematic representation of the ChIRP assay is shown in the upper left panel. The upper right chart shows the functionality of the assay, and the capacity to effectively isolate circPOK by using biotinylated probes (oligonucleotides). The lower part of the panel shows the interaction between circPOK, ILF2 and ILF3. Representative blots are shown in the lower left panel, while the quantification of two independent experiments is shown in the lower right panel. d Analysis of the “secretome” of p53−/−Zbtb7a_Ex2F/F-CRE MSCs expressing either circGFP or circPOK, assayed with ELISA arrays. Differentially expressed cytokines and soluble factors are shown in the chart at the bottom. e IHC detection of CD31+ endothelial cells within tumors expressing circGFP or circPOK. Representative pictures are shown on the left, while the quantification of CD31+ cells is shown in the chart on the right. Dots in the chart represent the independent sections analyzed. f Flow-cytometry analysis of CD31+ endothelial cells within tumors expressing circGFP or circPOK. Dots in the chart represent the independent tumors analyzed. g Knock-down of ILF3 in p53−/−Zbtb7a_Ex2F/F-CRE MSCs expressing circPOK, and measure of their in vivo sarcomagenesis. Two independent shRNAs were used to knock down ILF3. h IHC detection of CD31+ endothelial cells in tumors expressing circPOK, with or without ILF3 knock-down. Representative pictures are shown on the left, while the quantification of CD31+ cells is shown in the chart on the right. Dots in the chart represent the independent sections analyzed
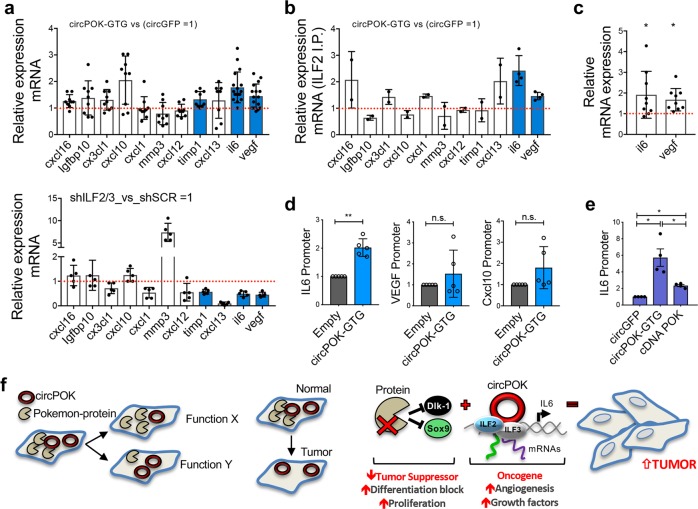
Based on these findings, we then aimed to investigate the possibility that circPOK could function through the regulation of ILF2/3 complex. Published data have already reported several functions for ILF2/3, including their capacity to increase the stability of messenger RNAs,27 and, in some cases, to function as transcription factors (TFs).28,29 Interestingly, several of the RNAs regulated by ILF2/3, transcriptionally or post-transcriptionally, encode pro-tumorigenic and pro-angiogenic cytokines.28–31 Based on these findings, we hypothesized that circPOK could regulate the “secretome” of mesenchymal tumor cells through ILF2/3. To investigate this possibility, we performed ELISA arrays, and assayed the expression of >90 known soluble factors that have been reported to play key roles in tumor growth, immunity and angiogenesis (Fig. 4d). Interestingly, the expression of many pro-angiogenic and growth factors, some of which are known to be regulated by ILF2/3 (e.g., IL6 and VEGF),32,33 was markedly increased in circPOK-expressing cells (circPOK-GTG) compared to controls (Fig. 4d). Accordingly, circPOK-GTG-expressing tumors displayed enhanced angiogenesis in vivo compared to circPOK-null tumors, as assessed by quantitating CD31+ endothelial cells within the tumor burdens (Fig. 4e, f; Supplementary information, Fig. S4g). Next, in order to investigate whether the increased angiogenesis of circPOK-expressing tumors was caused by the engagement of ILF2/3, we used shRNAs to knock down one component of the ILF2/3 complex in circPOK-GTG cells (Supplementary information, Fig. S4h), and then ran assays for in vivo. As shown in Fig. 4g, cells with silenced expression of ILF3 generated smaller tumors compared to controls; importantly, IHC analysis revealed a reduction in angiogenesis (Fig. 4h). Anchorage-independent proliferation assays in vitro were in line with what was observed in vivo, and showed that circPOK-expressing cells exhibited proliferation disadvantages when the expression of ILF3 or ILF2 was lost (Supplementary information, Fig. S4i). Taken together, these experiments suggest that circPOK can bind ILF2/3 complex within the nucleus, and promote the oncogenic functions of the complex, through altering the expression of cytokines that lend proliferative advantages, and promote angiogenesis.
CircPOK is a co-activator of the ILF2/3 complex
Once it was clear that circPOK functions at least in part through the ILF2/3 complex, we began to investigate the mechanisms behind this function. First we aimed to test the possibility that, in addition of binding to ILF2/3, circPOK could also modulate transcriptionally the expression of ILF2/3. RT-qPCR analysis of cells expressing or lacking circPOK excluded this possibility. Indeed, ILF2 and ILF3 showed similar levels of expression regardless of the presence of circPOK (Supplementary information, Fig. S4j). Another possibility was that the formation of the ILF2/3 complex could be regulated by circPOK. In order to assess this hypothesis, we performed co-immuno-precipitation of ILF2 and ILF3 in the presence or absence of circPOK. No differences in terms of interaction between ILF2 and ILF3 were observed (Supplementary information, Fig. S4k). Moreover, no differences were detected in the localization of ILF2/3 in cells expressing or lacking circPOK (Supplementary information, Fig. S4l), suggesting that circPOK affects neither the formation of the complex, nor the sub-cellular localization of ILF2/3 proteins, which localize predominantly in the nucleus (Supplementary information, Fig. S4l).
After these investigations, we hypothesized that circPOK could affect the functionality of the ILF2/3 complex, rather than its formation. It has been already reported that ILF2/3 can play several roles within cells, either at the level of transcription or post-transcriptionally (i.e., through mRNA stability).27 CircPOK could potentially strengthen ILF2/3 in mediating both these activities, and so produce a differential expression of those mRNAs under the control of ILF2/3. To investigate this possibility, we first analyzed whether the mRNAs of the proteins identified in ELISA arrays as de-regulated by circPOK (Fig. 4d), could be controlled by ILF2/3. To this end, ILF2/3 expression in MSCs was silenced by shRNAs, and the expression of these mRNAs was then analyzed. Interestingly, the mRNA and protein levels of the cytokines IL6 and VEGF decreased in the absence of ILF2/3 (Fig. 5a; Supplementary information, Fig. S5a), but increased in the presence of circPOK (Fig. 5a). Accordingly, in a complementary experiment, Il6 and Vegf mRNA expression decreased by targeting circPOK with LNA GapmeRs (Supplementary information, Fig. S5b). We then sought to determine whether the circPOK-ILF2/3 complex could affect these mRNAs transcriptionally or post-transcriptionally. To this end, we first pulled down ILF2 in cells expressing circPOK (circGFP cells were used as controls), and looked at the binding of ILF2/3 with the cytokines’ mRNAs. Interestingly, Il6 and Vegf RNAs showed increased binding affinity for ILF2 where circPOK was expressed (Fig. 5b). Then, we treated cells with Actinomyosin-D to block the transcription, and in this context we measured the stability of mRNAs Il6 and Vegf. As shown in Fig. 5c, the presence of circPOK increased the expression of both Il6 and Vegf mRNAs upon treatment. Taken together, these two experiments suggested that circPOK can promote the ability of ILF2/3 to bind mRNAs, particularly of Il6 and Vegf, and stabilize their mRNAs.
Fig. 5.
CircPOK is a co-activator of ILF2/3 in mesenchymal tumors. a Expression analysis of cytokines and angiogenic factors regulated by the expression of circPOK (compared to circGFP) (upper panel), and regulated by ILF2/3 (lower panel). Timp1, Il6 and Vegf were differentially expressed in the conditions analyzed, with statistical relevance. Dots show independent replicates of the experiment; blue bars identify cytokines that are significantly different than controls. b Relative expression of several mRNAs of cytokines and angiogenesis factors associated to ILF2 (pulled-down from MSCs expressing either circGFP or circPOK). Dots show independent replicates of the experiment; blue bars identify cytokines that are significantly different than controls. c Expression of Il6 and Vegf mRNA, measured upon treatment with Actinimyosin-D in cells expressing circGFP (control cells, represented as a bar = 1), or circPOK. Dots show independent replicates of the experiment. d ChIRP assays to measure the occupancy of the endogenous circPOK at the promoter regions of Il6, Vegf, and Cxcl10. Dots show independent replicates of the experiment. e ChIP assay to measure the binding of ILF2 at the promoter region of Il6 in cells expressing circGFP, circPOK-GFP or cDNA-POK. Dots show independent replicates of the experiment. f Schematic representation of the antithetical role of Pokemon protein and circPOK in normal conditions or within the process of tumorigenesis (on the left). Schematic representation of the functions played by Pokemon protein (tumor suppressor) and circPOK (proto-oncogene) in the process of mesenchymal tumorigenesis, with their relative mechanisms of action (on the right)
Because the ILF2/3 have been reported to be involved in transcriptional regulation of gene expression, we wanted to investigate whether Il6 and Vegf were also regulated by ILF2/3 at the level of their transcription, and whether circPOK could play a role in this regulation. In this respect, we first investigated whether circPOK itself is present at the promoter-regions of Il6 and Vegf (whose RNAs were found to be affected by the circPOK-ILF2/3 complex) by performing ChIRP assays. Accordingly, endogenous circPOK was pulled down, and its binding to the promoter-regions of Il6 and Vegf was tested. As an internal negative control for this experiment, we analyzed the binding of circPOK to the promoter region of Cxcl10 (not affected by the circPOK-ILF2/3 complex). As shown in Fig. 5d, ChIRP experiments revealed that circPOK is able to bind the proximal promoter regions of Il6, but not those of Vegf and Cxcl10. In addition, to confirm that circPOK could function as a transcriptional co-activator of ILF2/3 over the expression of Il6, we performed ChIP analyses to compare the occupancy of ILF2/3 on the promoter region of Il6 in the presence or absence of circPOK. Interestingly, the results clearly showed that ILF2/3 can bind the proximal promoter region of Il6, and that, importantly, their occupancy of the promoter can be enhanced by circPOK (Fig. 5e). In sum, these experiments suggest that circPOK may function as a co-activator for the ILF2/3 complex, and can potentiate the activity of ILF2/3 in mediating both mRNA transcription and stability.
Discussion
In this manuscript, we characterize the unexpected functional roles and molecular mechanisms of a circRNA generated from a genetic locus already directly implicated in tumorigenesis in its protein-coding dimension. This study increases our knowledge of circRNA biology, and offers a number of important conclusions.
First, we have demonstrated, in vivo, the functional relevance of a new circRNA to mesenchymal tumor growth by employing a novel “dual KO/add-back” genetic approach. While the aberrant expression of normal genes and proteins, even when not mutated, is known to drive cancer, whether deregulated circRNA could additionally contribute to cancer progression remained to be proven. This finding now provides direct evidence that normal circRNAs not only exert important functional roles within the cell, but also contribute to the pathology of diseases including cancer, when over- or under-expressed. This “dual KO/add-back” experimental approach will be of general relevance to determining the function of circRNAs and linear RNAs produced by the same genetic locus. Because linear and circRNAs are generated from the same RNA precursor, it is very difficult to engineer a genetic locus that favors one RNA species without affecting the other. In contrast, our dual approach allows functional in vitro and in vivo studies by restoring the physiologic level of circRNA or linear mRNA, and complements additional approaches designed to knock down a circRNA or linear mRNA selectively. While we have previously demonstrated the Pokemon protein to be a tumor suppressor in sarcomagenesis, by taking advantage of these complementary approaches we have defined a new proto-oncogenic pathway for this disease, and have identified the circPOK-ILF2/3 complex as a proto-oncogenic component that promotes the expression of pro-proliferative and pro-angiogenic factors. Furthermore, we show that circPOK affects the expression of multiple secreted cytokines and chemokines in tumor cells. It is also worth noting that, although these interactions are not directly investigated in the current study, circPOK could also affect the composition of the tumor-microenvironment by regulating the “secretome” of tumor cells, and thereby re-shape cancer-directed immune responses. This suggests that circRNAs may also play indirect non-cell-autonomous roles within tumors. Additionally, circPOK could synergize in a context-dependent manner with the Pokemon protein in tumors where, as a transcriptional repressor, the Pokemon protein suppresses the ARF tumor suppressor.34
Secondly, our studies identify a novel role for circRNAs through their ability to bind directly to, and regulate the function of, nuclear transcription factors. To date, circRNAs have mainly been attributed active roles in the cytosol, where they can be translated into peptides18,19 or act as ceRNAs.4,6 We have now found that, circPOK can bind RNA/DNA-binding proteins such as ILF2 and ILF3, and potentiate their regulatory activities both at the transcriptional and post-transcriptional levels. This role could be shared by other nuclear circRNAs. Furthermore, the observation that circRNAs can regulate the activity of nuclear factors also suggests the possibility of developing transcriptional therapies through circRNA targeting. This targeting could be achieved by using RNA-therapy approaches directed at the circRNA back-splice junction through the use of LNAs or GapMers. Although we focused our investigation on the non-coding role of the nuclear circPOK, further studies will be required to illuminate whether the cytosolic circPOK can also play functional roles in tumors. In this regard, although we could not detect any endogenous circPOK-associated proteins, circPOK does contain an ORF, which makes its translation theoretically possible. In this case, cytosolic rather than nuclear circPOK would be the translated species.
Finally, we have here identified a new type of intragenic regulon (iRegulon for brevity) that performs different biological functions through the production of biochemically and functionally distinct RNA products: linear and circular. The iRegulon may offer to the cells multiple ways to fine-tune biological outputs, by differentially regulating the expression of linear and circular components. For example, miRNAs, which target the 3′UTR region of the mRNA may affect the linear transcript, while leaving the circular RNA unaffected. In the specific case of Zbtb7a, the aberrant expression of a number of established proto-oncogenic miRNAs such as the miR-17 family, miR106, miR25 and miR93 represent a pervasive mechanism at the root of the pathological linear/circular uncoupling observed in cancer. The fact that the overexpression of these miRNAs is pleiotropic in cancers of multiple histologies suggests that the aberrant uncoupling of the Zbtb7a genetic unit may be a very common event in human tumorigenesis. However, miRNAs may not be the sole players at stake. Indeed, while the increased levels of miRNAs in tumor can explain the down-regulation of linear transcripts and protein, it does not explain differential expression of the circRNA isoform in tumor cells, compared to their normal counterpart. Accordingly, all the elements critical for the Pokemon circRNA biogenesis need to be investigated with additional experiments. Moreover, because sarcomas are rare tumors, we could collect only a small number of human samples during the timeframe of this work. Therefore, analysis of additional samples will help to corroborate the results.
Finally, the identification of iRegulon has important implications for cancer genetics, as it may represent a new class of genes whose natural selection in disease and cancer differs dramatically from that of the prototypical tumor suppressor or oncogene. This natural selection would now hinge on molecular uncoupling at the level of transcription or RNA processing rather than classical amplification or deletion at the genomic level. Indeed, these studies point to a novel class of disease gene, and highlight the need to consider the relevance of distinct elements derived from a single gene locus in an era of renewed enthusiasm for genomic engineering for therapy.
Materials and methods
Mice, Cell Lines and Human sarcoma primary samples
Transgenic mice Zbtb7aF/F were generated as described13; p53KO mice were purchased from The Jackson Lab. All the experimental animals were kept in C57Bl/6J background. Animal experiments were performed in accordance with the guidelines of Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
The following sarcoma-cell lines were used in PCR and functional assays: HS5, MFH, HT1080. HS5 cells were purchased from ATCC, while MFH and HT1080 have been gently provided by Carlos-Cordon-Cardo’s laboratory.35 In specific analysis, human primary MSCs were used. The cells were purchased from Lonza (PT-2501), and kept in culture with specific media, as indicated in the manufacturer’s instructions.
Human undifferentiated sarcoma primary samples and normal fibrous tissues as controls were collected, cryopreserved, and provided by the bio-repository of the Mount Sinai School of Medicine, NY. Samples were homogenized, and the RNA was extracted with TRIZOL (Invitrogen) according to the manufacturer’s instructions.
Osteosarcoma specimens used were from the Osteosarcoma Tissue Bank at the Massachusetts General Hospital under an IRB approved protocol for collection and use of archived tissue. Specimens were derived from patients with classic primary high grade osteosarcoma of the long bones. Slides from these specimens were reviewed by expert pathologists to verify histology and a tissue cut was taken from each sample for RNA isolation. After homogenization, RNA was isolated with the RNeasy Plus Universal Mini Kit (QIAGEN) per standard manufacturer instructions. Human Osteoblasts Total RNA was purchased from Applied StemCell.
The PCR reactions were then run with the primers indicated in the Primers Table (Supplememntary Information, Table S1).
Mouse Mesenchymal Stem Cell isolation, maintenance, and in vivo tumorigenesis
Long bones were collected, crushed and digested with collagenase II (1 mg/mL) for 1 h and shaken at 37 °C. Recovered cells were stained and FACS-sorted as: CD45−CD31−Ter119−Sca1+PDGFRα+, and cultured using complete MesenCult medium (STEMCELL Technologies). MSCs were maintained in humidified chamber with 5% CO2 and 1% O2, half medium was changed every 3 days. After 7 days in culture at 1% O2 cells formed visible CFU-F colonies, after this point cells were split once they reached 80% of confluency. Experiments, aimed at measuring in vivo tumorigenesis, were carried out following the protocol previously described.8 Briefly, 3D scaffolds made with reticulated polycarbonate polyurethane urea matrix (5 mm × 2 mm) were seeded with MSCs at a concentration of 1 × 105 cells/scaffold. Cells were allowed to adhere to the scaffolds for a minimum of 6 h. Scaffolds were then implanted sub-cutaneously into mice flanks.
Flow cytometry
Cells were analyzed using LRSII (BD, Pharmingen) and sorted using FACS-ARIA II (BD, Pharmingen). The following antibodies were used: anti-CD45 FITC, anti-CD31 FITC, anti-Ter119 FITC, anti-Sca1 Pacific Blue, anti-PDGFRα PE, anti-CD31 PE/APC (all purchased from Biolegend).
RNA extraction, PCR and RT-qPCR
Total RNA was extracted using TRIZOL (Invitrogen) according to the manufacturer’s instructions. To enrich circRNA isoforms, RNase R treatment was carried out for 15 min at 37 °C using 2U RNase R (Epicenter) per 1 μg of RNA. Treated RNA was directly reverse transcribed using the RETROscript System (Life Technologies) with random decamer primers according to the manufacturer’s instructions. In 10–20 ng of RNA were used for the PCR and RT-qPCR analysis. The PCR reactions were performed using HotStarTaq Master Mix (Qiagen). PCR products were visualized after electrophoresis in a 1.5% ethidium bromide-stained agarose gel. For sequencing, PCR products were cut from the gel and purified using QIAquick gel extraction kit (Qiagen) according to the manufacturer’s instructions. RT-qPCRs were carried out using SybrGreen reaction mix and StepOnePlus real-time PCR system (Applied Biosystems). Primer sequences are provided in Primers Table (Supplememntary Information, Table S1).
Absolute quantifications of linear and circular transcripts were performed by RT-qPCR in respect to their standard curves. In order to originate the standard curve, the PCR products of LinPOK and circPOK amplifications were cloned into the pGEM-T easy vector system II (Promega), following manufacturer’s instructions. The number of RNA molecules was then calculated by quantifying the PCR bands and by plotting them to serially diluted standard curves.
IHC staining
IHC was performed on 5-mm paraffin sections with the avidin-biotin-peroxidase method. The following primary antibodies were used: anti-CD31 (abcam #28364), anti-Ki-67. Antigen retrieval was performed using citrate buffer.
Retroviral and lentiviral plasmids
All the viral particles were produced in 293 T cells. Retroviral vectors (backbone: pCMMP-MCS-IRES-mRFP) were co-transfected with the pECO packaging plasmid, while lentiviral vectors (plko.1 for shRNAs and shCircRNAs) were co-transfected with packaging vectors for lentiviral vectors of second generation. After 48 h of transfection, the viral supernatant was collected; viral particles were concentrated through centrifugation and used to transduce target cells, together with polybrene. The retroviral vectors expressing circPOK or circGFP were generated with the Gibson Assembly kit (Nebs), following the manufacturer’s instructions. When indicated, circPOK-expressing retroviral vectors were subjected to single-base mutagenesis, which was performed by using the QuikChange XL Site-Directed mutagenesis kit (Agilent). Plko.1-TCR cloning vectors (Addgene 10878) were used to express shRNAs. Vectors were cloned following the instructions: http://www.addgene.org/tools/protocols/plko/. A scramble shRNA was used as control. CRISPR sgRNAs that guide to knock down Zbtb7a gene were designed according to the following: (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), and cloned into the lentiCRISPRv2, Addgene (plasmid #52961).
LNA GapMers
The LNA targeting the back-splice junction of circPOK was designed in the laboratory of Dr. Kauppinen. The sequences are provided in the figure (upper case: LNA, lower case: DNA). The LNA was transfected into cells by using DharmaFECT transfection reagent (3 μL/mL) at the concentrations indicated. The down-regulation of the circPOK was measured 48 h after the transfection.
CircFISH
The Circular RNA was imaged using an adaptation of the single molecule fluorescent in situ hybridization method as previously described in.36,37 Unique sets of probes (20 nt in length) were designed for different regions of the Pokemon RNA (6 for Exon 1, 35 for Exon 2 and 29 for Exon 3) using LGC bio search technologies and ordered with a 3′ amino modified group. The 35 probes specific for Exon 2 were pooled together and labeled with Cy5 and the probes specific for Exon 1 and 3 (total 35) were pooled and labeled with Texas red. The labeled probes were purified using reverse phase HPLC to get the labeled pool.36 The cells were cultured on gelatin coated glass coverslips, fixed with 4% p-formaldehyde, permeabilized with 70% ethanol and kept at 4 °C until ready to use. The coverslips were incubated overnight at 37 °C with hybridization mix containing 1 ng/μL of each probe mix. The unbound probes were washed off the next day and cells were mounted after staining with DAPI. Images were acquired using ×100 oil objective in a Nikon TiE inverted fluorescence microscope equipped with pixies 1024b camera and Metamorph imaging software. The image analysis was performed using custom written programs in MATLAB (Mathworks Inc).
Western blot, immunoprecipitation, and fractionation experiments
For western blot, cell lysates were prepared with RIPA buffer (1 × PBS, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche)). The following antibodies were used for western blotting: mouse polyclonal antibody to β-actin (Sigma-aldrich), mouse monoclonal anti-HSP90 (BD Biosciences), hamster anti-Pokemon/Lrf antibody (clone 13E9), rabbit polyclonal anti-NF90/NF110 antibody (Bethyl), rabbit polyclonal anti-NF45 antibody (Bethyl). For ILF2/ILF3 co-immunoprecipitation experiments, cell lysates were prepared with the following NP-40 buffer: 50 mM Tris-Cl pH 7.5, 10% glycerol, 5 mM MgCl2, 150 mM NaCl, 0.2% NP-40 and complete protease inhibitor cocktail (Roche). Protein-G magnetic beads (Invitrogen) were conjugated with the anti-ILF2 antibody indicated above, and rotated for 1 h at 4 °C. Lysate was firstly pre-cleared by using Protein-G magnetic beads (Invitrogen), and then incubated for 1 h with Protein-G magnetic beads conjugated with the anti-ILF2 antibody. Proteins were then eluted from beads by boiling them with SDS-sample buffer (Boston BioProducts) for 10 min. Western blot was then performed for detecting ILF3 with the antibody indicated above. The lysis buffer used for performing co-IP experiments, was also used for RNA-immunoprecipitation (RIP) experiments. In this case RNase inhibitors were supplemented to the buffer. RIP experiments were performed following the protocol published in.38 Nucleus/cytosol fractionation experiments were performed by using the PARIS kit (Thermo Fisher). Samples were then partially used for western blot to control the efficiency of the fractionation, and partially included in TRIZOL for RNA extraction. In this case the following antibodies were used: mouse monoclonal anti-β-actin (Sigma-aldrich), mouse monoclonal anti-HSP90 (BD Biosciences), rabbit polyclonal anti LamininB1 (Abcam).
miRNA expression profile
Total RNA was isolated from cells using TRIAZOL Reagent, 1 μg of total RNA was poly-adenylated using E. coli Poly(A) Polymerase (New England Biolabs M0276L). cDNA was synthesized starting from 250 ng of Poly-adenylated RNA and using RETROscript kit from Invitrogen (AM1710) and a specific RT primer. cDNA was diluted 1:10 and 1 μL was used to perform qPCR. Each miRNA was amplified using a common reverse and specific forward primers. GAPDH was used as internal control and was amplified with specific primers (see Primers Table, Supplememntary Information, Table S1).
Polysome fractionation
Subconfluent cultures of MSCs were used for polysome analysis. Cells were incubated with cycloheximide (Sigma) at a final concentration of 100 μg/mL for 15 min. Plates were then washed with ice-cold PBS containing 100 μg/mL cycloheximide (PBS/CHX), scraped, and collected in ice-cold PBS/CHX. Cells were pelleted by centrifugation and subsequently lysed in polysome lysis buffer (20 mM Tris-HCl (pH 7.4); 5 mM MgCl2; 150 mM NaCl; 1% Triton X-100; 1% deoxycholate; 2.5 mM DTT; 200 U/mL RNasin; 100 μg/Ml cycloheximide; EDTA-free protease inhibitor (Roche); α1-antitrypsin (EMD Biosciences)) and incubated on ice for 10 min. Lysates were centrifuged at 7500 rpm. for 5 min at 4 °C, and the supernatant was carefully removed. Lysates were loaded on a 15–50% sucrose gradient containing 100 μg/mL cycloheximide, 0.2 mg/mL heparin, and 1 mM DTT. Gradients were centrifuged at 36,000 rpm for 3 h at 4 °C in a Beckman SW40 rotor and subsequently fractionated using an ISCO-Foxy Jr fraction collector. Polysome profiles were measured using a UA-6 absorbance detector connected to the fraction collector and measuring absorbance at 254 nm.
Anchorage-independent cell growth (soft-agar assays)
Soft agar colony formation assay was carried out seeding 1 × 104 MSCs in DMEM containing 0.3% low-melting agarose and 10% FCS. The cells were then plated in 6-well plates previously coated with DMEM containing 0.6% of low-melting agarose and 10% FCS. The number of colonies was scored 3 weeks later, and quantification was completed using ImageJ.
RNA-pull down and mass spectrometry analysis
Biotin-labeled RNA was in vitro transcribed (IVT) with the Megascript kit (Ambion) following the manufacturer’s protocol, and by adding not labeled or biotin-labeled UTP at a ratio 1:5. Control RNA was in vitro transcribed and not labeled with biotin. 3 μg of RNA were then heated at 90 °C for 2 min, put in ice for another 2 min, supplied with the RNA structure buffer (10 mM Tris pH 7, 0.1 M KCl, 10 mM MgCl2), and then incubated at RT for 20 min. The RNA was then incubated with 1 mg of protein lysate, which was derived from MSCs treated with the following extraction buffer: 150 mM KCl, 25 mM Tris pH 7.4, 0.5 mM DTT, 0.5% NP40, 1 mM PMSF and protease inhibitor. Prior to incubation the lysate was pre-cleared with magnetic beads; the incubation of the RNA and protein lysate was performed at RT for 1 h in rotation. Anti-biotin-magnetic beads (GE Pharmingen) were then added to the samples and remained for 1 h at RT in rotation. Beads were then recovered through a magnet, washed 4 times with the protein lysate buffer and the proteins were detached through incubation with 6 × SDS-Sample buffer for 10 min at 90 °C. Proteins were then run in a SDS-PAGE gels. Gels were stained with comassie blue, cut at 60 kDa, and then subjected to mass spectrometry analysis (Results of the analysis are shown in Supplementary Information, Table S2).
ChIRP assays
Chromatin Isolation through RNA Precipitation assays were adjusted from the method described in.25 Briefly, cells were crosslinked with 1% formaldehyde for 10 min; the cross-linking was quenched with 0.125 M glycine. Lysis buffer was added directly on the plate; cells were then collected and pelleted for 6 min at 1200 rpm. The lysate was then sonicated to generate chromatin with average size of 500 bp. Pre-clearing was performed using anti-biotin beads for 1 h, then the biotinylated probes were added to the lysate, and incubated in rotation overnight at 37 °C. Anti-biotin magnetic beads were then added for 1 h at 37 °C. After incubation the magnetic beads were collected and washed for at least 5 times. For western blot analysis, beads were pelleted, and the proteins eluted by adding LB-SDS buffer, and boiling for 10 min. For chromatin assays, beads were incubated overnight in de-crosslinking buffer, and then DNA was extracted by using Qiagen kit, following manufacturer’s procedures. For RNA extraction, beads were put in TRIAZOL, and total RNA was extracted as per manufacturer’s protocol.
ChIP assays
Chromatin immunoprecipitation was performed using anti-ILF2 and anti-ILF3 Rabbit antibodies (Bethyl). Primer sequences used for PCR are included in the Primers Table (Supplememntary Information, Table S1). Briefly, MSCs were crosslinked with formaldehyde for 5 min and terminated with 0.125 M glycine. Cells were then sonicated to generate chromatin with average size of 500 bp. Samples were first pre-cleared with Protein-A magnetic beads for 1 h at 4 °C. Cleared chromatin was then incubated with the antibodies overnight at 4 °C, and then with Protein-A magnetic beads for 4 h at 4 °C. Beads were then recovered, washed multiple times, and de-cross-linked by incubating them overnight at 65 °C in 1% SDS, 0.1 M NaHCO3. DNA was then purified by using the QUIAGEN kit. The promoters of the specific genes were then assayed in RT-qPCR.
ELISA measurement and ELISA arrays
To detect soluble protein, p53-null MSCs expressing circGFP or circPOK were plated in the same numbers, and the supernatant was collected after a few days of culture in hypoxic conditions. Collected supernatants were analyzed within Mouse Cytokine array panel-A array kit and Angiogenesis array kit (R&D Systems), following manufacturer’s instructions.
Supplementary information
Acknowledgements
We thank Lauren Southwood and Elizabeth Stack for their insightful editing, and all members of the Pandolfi lab for critical discussion. Jlenia Guarnerio was supported by the K99/R00 grant (CA212200) for the execution of this work. Mona Batish was supported by the grant 5 DP5 OD012160. This work was supported by the grant NIH/NCI-R35CA197529 and the grant GP009/2014 from the Human Frontier Science Program Organization to Pier Paolo Pandolfi. The collection of osteosarcoma samples was possible thanks to the NIH grant R01CA178908 to Dr. Spentzos Dimitrios.
Authors contributions
P.P.P. and J.G. conceived the hypothesis and designed the experiments. J.G., Y.Z., G.C., R.P. and J.M.K. performed the experiments. P.P.P., J.G., and J.G.C. wrote the paper. M.S. and M.B. performed the circFISH presented in the paper. A.M., A.P. and C.L. helped to perform the experiments. A.P. and S.K. helped with LNA-GapmeRs. C.G., G.P.N., V.D. and S.D. provided osteosarcoma samples; S.D. helped to design experiments with osteosarcoma samples. M.C.M. and C.C.C. provided undifferentiated sarcoma samples.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41422-019-0192-1.
References
- 1.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Piwecka M, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP. Role of LRF/Pokemon in lineage fate decisions. Blood. 2013;121:2845–2853. doi: 10.1182/blood-2012-11-292037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guarnerio J, et al. A genetic platform to model sarcomagenesis from primary adult mesenchymal stem cells. Cancer Discov. 2015;5:396–409. doi: 10.1158/2159-8290.CD-14-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poliseno L, et al. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karreth FA, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legnini I, et al. Mol Cell. 2017. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis; pp. 22–37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pamudurti NR, et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21 e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du WW, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudekula DB, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchesini M, et al. ILF2 Is a Regulator of RNA Splicing and DNA Damage Response in 1q21-Amplified Multiple Myeloma. Cancer Cell. 2017;32:88–100 e106. doi: 10.1016/j.ccell.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, et al. Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene. 2015;34:4460–4470. doi: 10.1038/onc.2014.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Q, et al. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32:3933–3943. doi: 10.1038/onc.2012.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu, C., Quinn, J. & Chang, H. Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp.10.3791/3912 (2012). [DOI] [PMC free article] [PubMed]
- 26.Chen YG, et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238 e225. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell Biol. 2008;28:772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J. Exp. Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiesler P, et al. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J. Biol. Chem. 2010;285:8256–8267. doi: 10.1074/jbc.M109.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tominaga-Yamanaka K, et al. NF90 coordinately represses the senescence-associated secretory phenotype. Aging. 2012;4:695–708. doi: 10.18632/aging.100497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakadai T, Fukuda A, Shimada M, Nishimura K, Hisatake K. The RNA binding complexes NF45-NF90 and NF45-NF110 associate dynamically with the c-fos gene and function as transcriptional coactivators. J. Biol. Chem. 2015;290:26832–26845. doi: 10.1074/jbc.M115.688317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauer J, Denson JL, Bruning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36:92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat. Rev. Cancer. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 35.Mills J, et al. Characterization and comparison of the properties of sarcoma cell lines in vitro and in vivo. Hum. Cell. 2009;22:85–93. doi: 10.1111/j.1749-0774.2009.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batish M, Raj A, Tyagi S. Single molecule imaging of RNA in situ. Methods Mol. Biol. 2011;714:3–13. doi: 10.1007/978-1-61779-005-8_1. [DOI] [PubMed] [Google Scholar]
- 37.Markey FB, Ruezinsky W, Tyagi S, Batish M. Fusion FISH imaging: single-molecule detection of gene fusion transcripts in situ. PLoS ONE. 2014;9:e93488. doi: 10.1371/journal.pone.0093488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peritz T, et al. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.