Abstract
In the last decade, researchers have been searching for innovative platforms, methods, and techniques able to address recurring problems with the current cancer detection methods. Early disease detection, fast results, point-of-care sensing, and cost are among the most prevalent issues that need further exploration in this field. Herein, studies are focused on overcoming these problems by developing an electrochemical device able to detect telomerase as a cancer biomarker. Electrochemical platforms and techniques are more appealing for cancer detection, offering lower costs than the established cancer detection methods, high sensitivity inherent to the technique, rapid signal processing, and their capacity of being miniaturized. Therefore, Au interdigital electrodes and electrochemical impedance spectroscopy were used to detect telomerase activity in acute T cell leukemia. Different cancer cell concentrations were evaluated, and a detection limit of 1.9 × 105 cells/mL was obtained. X-ray photoelectron spectroscopy was used to characterize the telomerase substrate (TS) DNA probe self-assembled monolayer on gold electrode surfaces. Atomic force microscopy displayed three-dimensional images of the surface to establish a height difference of 9.0 nm between the bare electrode and TS-modified Au electrodes. The TS probe is rich in guanines, thus forming secondary structures known as G-quadruplex that can be triggered with a fluorescence probe. Confocal microscopy fluorescence images showed the formation of DNA G-quadruplex because of TS elongation by telomerase on the Au electrode surface. Moreover, electrodes exposed to telomerase containing 2′,3′-dideoxyguanosine-5′-triphosphate (ddGTP) did not exhibit high fluorescence, as ddGTP is a telomerase inhibitor, thus making this device suitable for telomerase inhibitors capacity studies. The electrochemical method and Au microchip device may be developed as a biosensor for a point-of-care medical device.
Introduction
The development of microchips or electrochemical devices to detect and monitor diseases has been well studied and implemented in recent years. These electrochemical platforms are modified with biomolecules to guaranty fast and efficient detection of a specific analyte, target, or biomarker; offering additional advantages in terms of cost and lab-on-a-chip technology. For example, development of electrochemical devices that monitor glucose levels,1 detect cardiac attacks,2 viral3 and bacterial infections,4 and cancer,5 among other diseases, has been reported in the literature. Intriguingly, the development of biosensors that detect cancer has been one of the most challenging because of the complexity of the disease. As a critical matter, in the Review of Antimicrobial Resistance, for 2050, O’Neill projected 8.2 million diseases each year.6 This alarming number can be changed by ensuring the success of cancer treatment regularly achieved with early disease detection.7,8 Therefore, there is a need of low-cost devices that detect early stages of these lethal diseases in a fast and efficient way.
In prior efforts, biomarkers have played an important role in cancer research.9−12 Telomerase activity serves as a cancer biomarker because it is responsible for the uncontrolled growth of cancer cells in 80–90% of cancers.13,14 This enzyme is a ribonucleoprotein complex that synthesizes telomeres at the end of the chromosomal DNA, avoiding them to reach the Hayflick limit (maximum number of cell division) and thus cell apoptosis.15 Telomerase uses its components, the RNA matrix (TR) and telomerase reverse transcriptase (TERT), to perform telomerase substrate (TS) elongation. The structure of the catalytic subunit of telomerase bound to its RNA template and the telomeric DNA has a diameter of 10.6 nm.16 In addition, telomerase needs deoxyribonucleoside triphosphates (dNTPs) to synthesize the telomere. If the nature of the nucleotides is changed, the enzyme activity is affected. As it has been previously reported, the nucleotide 2′,3′-dideoxyguanosine-5′-triphosphate (ddGTP) is one of the molecules that have a better potential to inhibit telomerase activity.17 This type of inhibitor acts on TERT by stopping the telomere elongation because of the absence of the 3′-hydroxyl group (compared with the deoxyribonucleotide, dGTP). Telomerase inactivation is important because of its impact in cancer therapy.18 The implementation of this antitelomerase therapy has been challenging. Until 2016, just two antitelomerase drugs were used in clinical trials.19 Therefore, identification of new substances for cancer therapy and methods to determine their capacity are needed.
Currently, the assay used to detect telomerase activity in biological samples is the telomeric repeat amplification protocol assay, commercially called TRAPeze. This biological test cannot be miniaturized, is time-consuming, not sensitive, expensive, and requires careful handling.20,21 Opportunely, in the last years, few electrochemical studies have achieved telomerase detection using its recognition biomolecule, DNA (TS probe or telomere sequence).21−24
Telomere-modified electrodes have been used to detect telomerase activity.25−28 Since 1994, telomerase activity has been associated with cancer cells.29 Cancers such as breast,30 colorectal,31 bladder,32 cervical,33 leukemia,33 ovarian,34 among others, have shown high telomerase activity; thus, it has become a common biomarker for these types of cancers. Recently, a detection method for circulating tumor cells, using the telomerase-specific adenovirus OBP-401, has been presented.35 The development of electrochemical biosensors has been driven by considerable efforts in cancer sensing and monitoring.36,37 In spite of all achievements, it is still necessary to develop an electrochemical platform that can be implemented as a medical device.
Common DNA sensors are based on the correlation between surface modifications and the changes in the electrochemical impedance spectroscopy (EIS) measurements.22−24,38 EIS provides advantages in terms of selectivity, sensitivity, and fast response.39 EIS is usually combined with surface analysis techniques such as X-ray photoelectron spectroscopy (XPS) and atomic force microscopy (AFM) to characterize these sensors.40,41 Additionally, fluorescence spectroscopy and confocal microscopy are the techniques that can identify nucleic acid secondary structures.42−44 As the telomere repeat sequence is rich in guanine, a large fragment of this DNA forms a G-quadruplex. This secondary structure can intercalate positively charged electrochemical indicators, cationic porphyrins, and/or fluorescent dyes, allowing its detection by the spectroscopic techniques mentioned above.45−47
Previous research by our group has described the use of label-free/nonfaradaic methods to sense DNA hybridization, but this innovative report extends it to miniaturized sensors. Rivera-Gandía et al.48 managed to detect the double-layer capacitance of self-assembled monolayer hairpin probes before and after exposure to complementary strands at two distinct potentials: zero charge potential (PZC) and a higher potential to PZC. They noted that the ssDNA hairpin showed a lower Cdl, whereas dsDNA showed a higher Cdl at an applied potential versus PZC. On the other hand, our group has worked with DNA sensors based on single-wall carbon nanotube-modified Au surfaces using label-free EIS.41,49 Recently, Cunci et al.23 were able to develop this concept further by looking at DNA elongation in the presence of telomerase in a custom-made Au interdigital electrode device. Currently, our research group is focused on the development of an in situ, label-free, cost-effective, and less time-consuming DNA biosensor that can detect telomerase activity in Jurkar T-lymphoblastic leukemia cells. These cancer cells are responsible for acute lymphocytic leukemia, the most common cancer in children.50 In this work, a label-free electrochemical sensor for telomerase activity detection in real time, using an already scaled-up fabricated and disposable Au interdigital platform, was developed. Moreover, the investigation of TS modification on the electrode surface and the presence of enzyme in nuclear protein extraction is done by surface analysis and biochemical techniques, respectively. To the best of the authors’ knowledge, this is the first time that the telomerase inhibitor capacity is studied by label-free EIS. Others have used this electrochemical technique to measure telomerase activity, however, employing [Fe(CN)6]3–/4– as the redox probe in the supporting electrolyte solution.22 Additionally, this is the first time that TS elongation process on a surface is confirmed using confocal microscopy. The methods and equipment developed by our group in this and previous investigations are promising tools that hopefully merge in the development of a lab-on-a-chip technology that could be used at a point-of-care location.
Results and Discussion
Surface Analysis
X-ray Photoelectron Spectroscopy
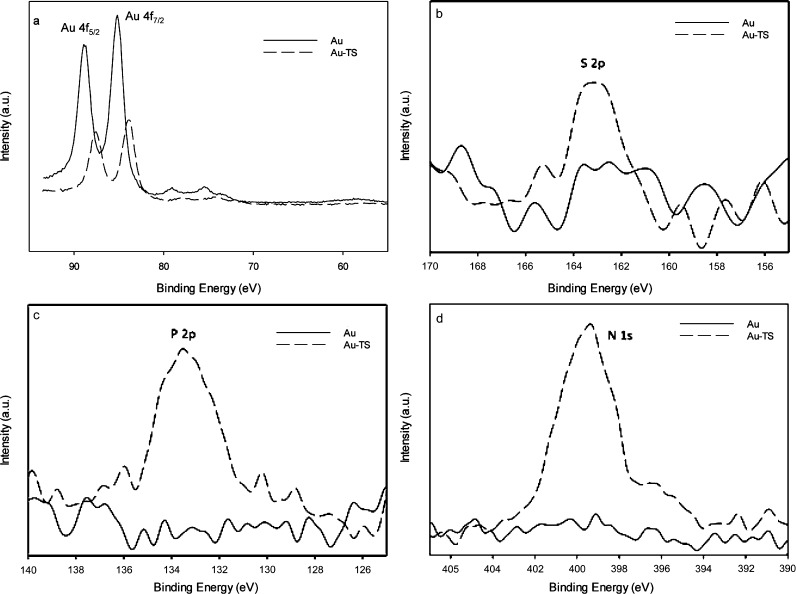
Comparing the XPS binding energy spectra obtained (Figure 1a–d) for the bare Au electrode and for the TS probe-modified electrode, a decrease in Au 4f binding energy peak intensity was observed because of the attachment of TS30 to the Au electrode surface (Figure 1a). This is due to the reduction of the Au sites exposed to the surface.51 Moreover, DNA characteristic XPS peaks were identified. Specifically, when thiol (S) on the 5′-terminal of the TS30 probe was immobilized forming a self-assembled monolayer on the Au electrode surface, a wide S 2p peak (from around 162 to 164 eV) appeared on the XPS spectra (Figure 1b).52 This is due to the formation of Au–S-DNA interaction and the remaining unbound sulfur possibly generated with the reduction of the disulfide bonds. Furthermore, N 1s and P 2p XPS peaks (Figure 1c,d) were observed, confirming the presence of the TS probe as phosphorus and nitrogen are the main components of the DNA structure. Therefore, the Au microchip surface modification, with the TS probe, was confirmed.
Figure 1.
XPS spectra for bare (solid line, Au) and DNA-modified (dashed line, Au-TS) Au electrodes for (a) Au 4f5/2 and 4f7/2 (b) S 2p, (c) P 2p, and (d) N 1s binding energy regions.
Atomic Force Microscopy
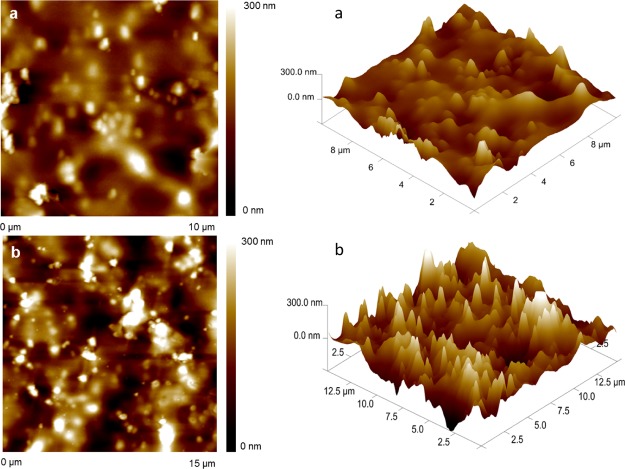
The AFM image of the bare Au electrode (Figure 2a) showed a minor surface height than the one of TS30-modified Au electrode (Figure 2b). This change in height is due to the formation of DNA self-assembled monolayers.53 Particularly, the roughness factor (Rq) or the standard deviation of the height attained for the bare electrode was 50 nm and for that for the modified Au electrode was 59 nm. This 9.0 nm height difference is attributed to the arrangement of the TS probe on the Au electrode surface that has a length of approximately 9.24 nm (CalcTool: DNA Strand Length Calculator). This tool assumes 3.3 Å as the distance between the nucleotides.
Figure 2.
AFM images for (a) bare and (b) TS probe-modified Au electrodes.
Electrochemical Analysis
Biosensor Characterization
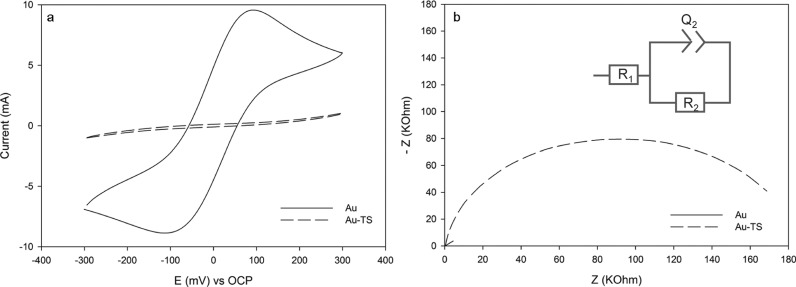
Cyclic voltammetry (CV) technique was employed to characterize the immobilization of the TS30 probe at the Au microchip surface. Figure 3a shows the voltammograms of the bare and TS30 probe-modified Au electrode surfaces in 2 mM K3Fe(CN)6/K4Fe(CN)6 in 0.1 M phosphate-buffered saline (PBS) at pH 7.0. A notable current decrease in the cathodic and anodic peaks occurs because the immobilization of ssDNA blocks the electron transfer, forming a barrier.22−24 Clarifying, the increased negative charge produced by the DNA probe hinders the charge transfer to the negatively charged redox molecules in solution. This causes a change in the charge-transfer resistance (Rct) measured using EIS because of its sensitivity (Figure 3b). As anticipated, the charge-transfer resistance increased with Au surface modification with the TS30 probe. The TS30 immobilization at the Au microchip was electrochemically confirmed. Au microchip exposition to enzyme extracts was followed.
Figure 3.
(a) Cyclic voltammograms and (b) Nyquist plots for bare (solid line, Au) and DNA-modified (dashed line, Au-TS) electrodes.
Electrochemical Telomerase Detection
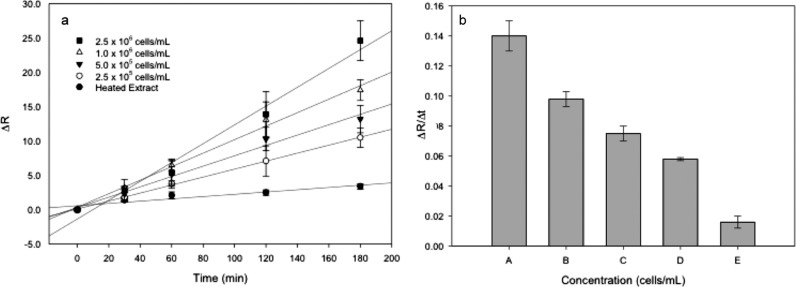
EIS was used as a sensing technique for telomerase in situ detection. To achieve a label-free detection, electroactive molecules contained in buffer C and in the nuclear extract (Supporting Information, Figure S1) were used, avoiding the use of the redox couple additive. Figure 4a showed changes in the charge-transfer resistance ((Rct(telo) – Rct(DNA))/Rct(DNA) or ΔR) for different extract concentration solutions and heated extract fraction (negative control) at various incubation times. The charge-transfer resistance was obtained performing a fitting for the equivalent electrical circuit model (R1 + Q2/R2).
Figure 4.
(a) Charge-transfer resistance changes (ΔR) of TS-modified Au electrodes as a function of telomerase enzymatic reaction time for (■) 2.5 × 106 cells/mL, (△) 1.0 × 106 cells/mL, (▼) 5 × 105 cells/mL, (○) 2.5 × 105 cells/mL, and (●) heated extract or negative control. (b) Relative change in charge-transfer resistance to enzymatic reaction time for (A) 2.5 × 106 cells/mL, (B) 1.0 × 106 cells/mL, (C) 5.0 × 105 cells/mL, (D) 2.5 × 105 cells/mL, and (E) heated extract.
Enzyme-assisted telomere elongation process is not instantaneous; it takes time for the enzyme to bind to the telomeres, synthesize it, relocate, and continue with replication.10 The elongation process was studied by changing the reaction time, and substantial changes in ΔR were observed, as shown in Figure 4a. At 0 min of exposure, there was no change for any concentration. On the other hand, more incubation time means synthesis of more telomere units, compacting the surface, resulting in an impedance increase. Generally, each concentration studied displayed an increase in ΔR as the enzymatic reaction time increased. Variations in the impedance are attributed to the possibility of DNA nonspecific adsorption and the interference of other nuclear extract residues.
Time study also showed a change, after 30 min, but for small concentrations, overlapping was observed. This result is in accordance with that of Cunci et al. in which they reported that for the Jurkat nuclear extract, 20 min was enough time to observe a quantitatively different change in label-free impedance measurements.23 Therefore, using the presented system, 30 min is enough time to make qualitative telomerase detection. Other modification times, 60, 120, and 180 min, showed more differences in ΔR, 3 h being the most accurate modification time for fundamental analysis.
Additionally, the change in ΔR for all the extract samples increased as the enzyme concentration increased. This occurs because more telomerase is able to synthesize DNA bases on the telomere-modified electrode surface contributing to the obstruction of the electron-transfer kinetics. As expected, there was no significant variation in ΔR for the electrodes incubated with negative control (heated extract). The detection limit attained from this calibration curve was 1.9 × 105 cells/mL. The detection limit was calculated according to the 3Snc/m criterion, where m is the slope of the linear range of the respective calibration curve (Supporting Information, Figure S2) and Snc is the standard deviation of the negative control. To the best the of the authors’ knowledge, there is no other EIS calibration curve for the nuclear extract of cancer samples to target telomerase, as reported in the literature.
Using Figure 4a data, ΔR/Δt for each cell concentration was obtained to acquire Figure 4b. As the cell concentration is raised, the related time sensitivity of detection increases. Therefore, telomerase activity was detected in a reliable way.
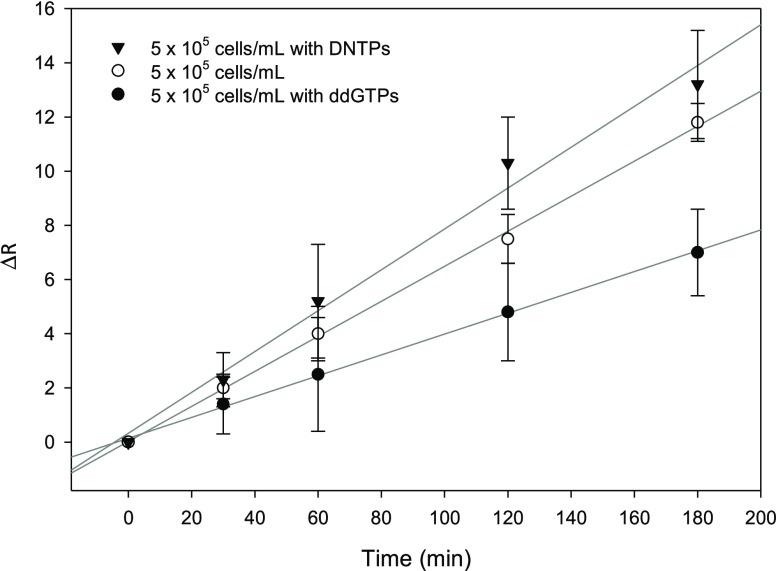
The nucleotide effect on telomerase activity was also evaluated in this study. Figure 5 shows the results for different extract solutions; when dNTPs are added to the extract or telomerase solution, an increase in ΔR and sensitivity is observed. Moreover, to confirm the reliability of the sensor, immobilization studies of the enzyme in the presence of the inhibitor nucleotide, ddGTP, were done. The observed results indicate a decrease in ΔR. This finding could be attributed to the possibility of an elongation process that is hampered when ddGTP is incorporated, thus, preventing the addition of further nucleotides.54
Figure 5.
Changes in the charge-transfer resistance (R) of the Au electrode as a function of enzymatic reaction time for (▼) 5 × 105 cell/mL with dNTPs, (○) 5 × 105 cell/mL, and (●) 5 × 105 cell/mL with ddGTP.
Nuclear Extraction: Western Blot
Immunoblotting analysis confirmed the presence of telomerase in the commercial positive control (Supporting Information, Figure S3a) and in the Jurkat nuclear cell protein extraction (Figure S3b) using Anti-hTERT antibody. The difference in signal intensity is attributed to the variations in telomerase concentration for these two samples. Detectable telomerase in the nuclear extract cytoplasmic fraction (Figure S3c) was not found, suggesting that telomerase is mostly present in the nuclei. GAPDH was used as a positive control, as it is a protein that can be found in the nuclear and cytoplasmic fractions. Collectively, the results demonstrated the presence of telomerase in the nuclear protein extracts that were used for electrochemical assays.
G-Quadruplex Formation: Emission Spectroscopy
Telomerase fluorescent light-up probes have been presented in the literature recently.32,44,55−57 This type of mechanism was used to confirm the formation of G-quadruplex structures in our systems by emission spectroscopy. Organic and/or pyridine (Py)-based molecules have been used to aim fluorescence in biological samples by aggregation-induced emission effect.56,57 Therefore, TMPyP4 was used as a ligand to intercalate in the G-quadruplex, thus acting as a fluorescence probe. Figure S4 shows two fluorescence peaks around 655 and 720 nm for TS30 and TS60; according to the literature, they are formed due to the splitting of the broad fluorescence band of the ligand.42 As expected, the spectra for the TS complementary probe (TSC; do not contain guanines) showed one broad peak (TMPyP4 fluorescence peak), suggesting that the intercalation did not occur. To confirm these results, fluorescence spectra for TS30, TSC, TS60 without TMPyP4 intercalation, and TMPyP4 itself were obtained (Supporting Information, Figure S4). Peaks for TS probes were not observed because there was no intercalation and thus no fluorescence emission. In TMPyP4 itself, the spectra show a broad fluorescence peak.
Confocal Microscopy
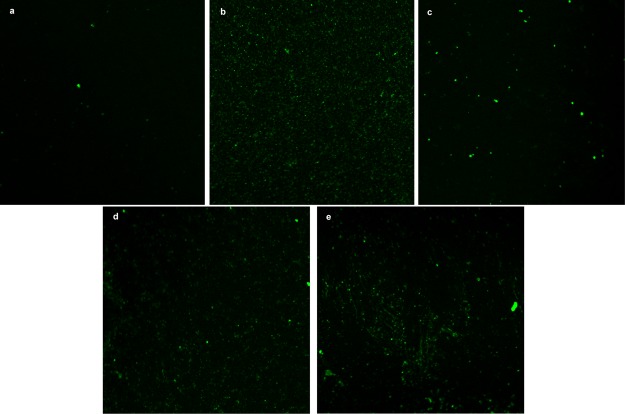
Confocal microscopy fluorescence was used to confirm the formation of TS G-quadruplex on the Au/TS30 electrode surface after being exposed to telomerase. Figure 6a corresponds to the Au electrode modified with the TS30 probe. Here, fluorescence of thiazole orange (TO) was not visible because the number of bases contained in the TS30 probe is not enough to achieve a noticeable TO intercalation [mean fluorescence intensity (MFI) = 94.35]. However, a TO MIF of 261 was observed when the TS30 probe was elongated and G-quadruplex was formed on the electrode surface (see Figure 6b). This phenomenon was expected because according to Zhuang et al. TS fluorescence increases as the telomerase concentration increases.32 In this direction, Figure 6b shows that compact modification of TS and the immobilization of the enzyme occur uniformly on the Au electrode surface. TO intercalation was confirmed using a larger TS probe (TS60) that, as can be observed in Figure 6c, exhibited a slightly more fluorescence than TS30 (MFI = 123). On the other hand, as expected, when the nature of the nucleotides (dNTPs) was changed, the enzyme activity was affected. Consequently, a less MFI (157) was observed when the TS30 electrode was exposed to the nuclear extract with ddGTP (Figure 6d). The negative control (Figure 6e) showed a slight fluorescence, probably because of the interaction of the 30 base probes with the extract components (MFI = 121).
Figure 6.
Confocal microscope images of the modified device with (a) TS30 (mean intensity fluorescence (MIF): 94), (b) after 3 h incubation with telomerase and dNTPs (MIF: 261), (c) TS60 (MIF: 123), (d) TS-30 after 3 h incubation with telomerase and ddGTPs (MIF: 157), (e) TS30 after 3 h incubation with negative control and dNTPs (MIF: 121).
A closer look into the images show a few noticeable dots (spots) of high fluorescence. This may be because of the nonspecifically absorbed residues of the nuclear extract (DNA and nuclear proteins) on the gold electrode surface that intercalate TO. In summary, the TS G-quadruplex structures were identified using confocal microscopy, confirming that the elongation process happens on the electrode surface. These results are in excellent agreement with the biosensor response, which show differences in ΔR; both are attributed to the telomerase-assisted DNA elongation.
Conclusions
In conclusion, the presence of telomerase in the nuclear protein extract of acute T leukemia cells using biochemical and electrochemical techniques was confirmed. To the best of authors’ knowledge, this is the first time that confocal microscopy is used to confirm the formation of G-quadruplex on an electrode surface after telomerase exposure. Moreover, specific telomerase label-free detection in real time was achieved using an electrochemical platform and EIS technique. Additionally, the developed telomerase activity-sensing microchip can be used to test the telomerase inhibitor capacity. This biosensor device offers several advantages over the existing technologies such as robustness, low cost, disposability, flexibility, compactness, and is less complicated than the existing technologies for telomerase detection. Enhancement of the sensitivity and clinical trial implementation should be the next steps for this system. All these features can have a tremendous impact on biomedical science as it will fill the existing gap of methods that can detect telomerase activity in a direct readout at the point-of-care location.
Experimental Section
Reagents
Potassium hexacyanoferrate(III) (99.98%), potassium hexacyanoferrate(II) trihydrate (≥99.95%), ethylenediaminetetraacetic acid (EDTA) ACS reagent (99.4–100.6%), 2-propanol anhydrous (99.5%), sodium chloride (≥99.95%), acetic acid (≥99%), Tween 20, Trizma base (99.9%) Trizma hydrochloride (≥99%), methanol (≥99%), phenylmethanesulfonyl fluoride (PMSF), diethyl pyrocarbonate (DCEP) (≥97%), dl-dithiothreitol (DTT) (≥98%), glycine (≥99%), sodium dodecyl sulfate (SDS; ≥98.5%), and ddGTPs were purchased from Sigma-Aldrich (St. Louis, MO). Acrylamide (40%), ammonium persulfate, tetramethylethylenediamine, 4× Laemmli sample buffer, and precision plus protein standard were obtained from BioRad (Hercules, CA). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), sulfuric acid optima (93–98%), Coomassie Brilliant Blue G-250, HEPES buffer, detergent-compatible Bradford assay kit, and RPMI-164 medium + 2.05 mM glutamine (Hyclone) were purchased from Fisher Scientific (Fair-lawn, NJ). All synthetic oligonucleotides were purchased from Integrated DNA Technology (IDT) (San Diego, CA) with the following sequences:
TS30 (5′-S-S-(CH2)6-TTTTTTTTTTAATCCGTCGAGCAGAGTT-3′),
TS60 (5′-S-S-(CH2)6-TTTTTTTTTTAATCCGTCGAGCAGAGTTAGGGTTAGGGTTAGGGTTAGGGTTAGG-3′), and
TSC (5′-S-S-(CH2)6-AAAAAAAAAATTAGGCAGCTCGTCTCAA-3′).
dNTPs were purchased from Promega (Madison, WI, USA).
Biosensor Construction
The disposable interdigital Au electrode microstrips were modified following the steps shown in Figure 7. First, 1 μM TS30 was dissolved in an immobilization buffer (I-buffer: 10 mM Tris-HCl, 0.1 M NaCl, and 10 μM TCEP at pH 7.5) and incubated for 30 min at room temperature to reduce the disulfide bonds. Then, 10 μL of the reduced TS30 solution was placed on a cleaned microchip strip overnight (Supporting Information, Figure S5) at 4 °C. Finally, the sensing strip was carefully washed with Nanopure water and 0.05% SDS for 5 min to desorb nonspecific adsorptions and dried with N2. Nanopure water is treated with DCEP and autoclaved to inactivate RNase and DNase enzymes.
Figure 7.
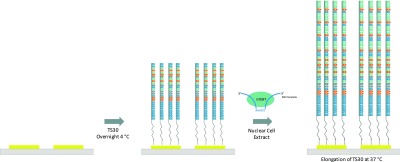
Schematic representation of surface modification and telomerase elongation process at Au interdigital electrodes.
Surface Analysis
XPS was done using a PHI 5600ci spectrometer with an Al K X-ray source at 15 kV and 350 W. The pass energy used was 187.85 eV for the survey analysis and 58.7 eV for the high-resolution studies. Binding energies were corrected with respect to the aliphatic hydrocarbon C 1s binding energy signal at 285.0 eV. A Nanoscope IIIa-Multimode atomic force microscope from Digital Instruments, with a scanning probe microscope controller equipped with a He–Ne laser (638.2 nm) and a type E scanner, was used for the AFM analysis. All samples were analyzed in tapping mode using a phosphorous-doped Si cantilever from Veeco Instrument, Inc. (Santa Clara, CA).
Electrochemical Measurements
Electrochemical measurements were done using a BioLogic Science Instruments VMP3 model. The Au microchip surface was electrochemically cleaned by cycling between a 0 and 1.5 V versus Ag/AgCl in 0.5 M sulfuric acid solution at a scan rate of 100 mV s–1 until a reproducible cyclic voltammogram was obtained. CV was done in a solution of 2 mM K3Fe(CN)6/K4Fe(CN)6 in 0.1 M PBS at pH 7.0 from −0.3 to 0.3 V versus an open-circuit potential (OCP) at a scan rate of 50 mV s–1. EIS experiments for the characterization of the Au microchip surface were performed from 1 × 106 to 0.1 Hz, taking 40 measurements in logarithmic scale, with an amplitude of 10 mV at 0 V versus OCP. EIS experiments to detect telomerase in cancer cells were performed from 2 × 105 to 0.1 Hz taking 40 measurements in logarithmic scale, with an amplitude of 10 mV at 0 V versus OCP every 5 min for 3 h. CV was done every hour from −0.3 to 0.3 mV versus OPC at a scan rate of 50 mV s–1 to remove nonspecific absorptions of molecules from the remaining extract. During the experiment, the microchip was incubated for 3 h in an Eppendorf Thermomixer C to keep the temperature at 37 °C with the desired extract sample supplemented with 0.2 mM dNTPs or ddGTPs.
Cell Culture and Nuclear Extraction
Jurkat T Cells (from the American Type Culture Collection of Manassas, VA, USA) were incubated at 37 °C with 5% CO2 using RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic that consisted of 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 0.25 μg/mL amphotericin B. Nuclear cell extraction was performed following an established protocol.23 Briefly, cells were collected in the exponential phase of growth and washed twice with ice-cold PBS (140 mM NaCl, 2.7 mM KCl, 10 mM NaH2PO4, and 1.8 mM KH2PO4). Then, the pellet was resuspended in buffer A [10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and 1 mM protein-sparing modified fast (PMSF)], incubated on ice for 10 min, and centrifuged at 1500 rpm for 10 min at 4 °C. Then, buffer A was added to the cell pellet and lysed in a cold 7 mL Dounce homogenizer (Kontes). The lysate was centrifuged at 6500 rpm for 2 min at 4 °C to separate the nuclei from the cytoplasm. The pellet (nuclei) was then resuspended in buffer C (20 mM HEPES pH 7.05, 0.42 M NaCl, 0.2 mM EDTA, 25% glycerol, 1.5 mM MgCl2, 1 mM DTT, and 1 mM PSMF) and placed under ultrasound six times for 10 s at maximum power. The nuclear fraction was centrifuged at 15 000 rpm for 20 min at 4 °C. Finally, the nuclear fraction (supernatant) was stored at −80 °C. The reproducibility of the extraction protocol was confirmed by obtaining the average protein concentration of 3118 ± 147 μg/mL for different extractions. Negative controls were obtained by heating the samples for 30 min at 100 °C.
Western Blot
Nuclear and cytoplasmic cell fractions were concentrated using centrifugal filter units. Approximately 40 μg of proteins was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using a 10% polyacrylamide gel and transferred onto a 0.45 μm polyvinylidene fluoride membrane. The membranes were cut into segments to allow immunoblotting with different antibodies. After blocking in Tris buffered saline containing 10% nonfat milk and 0.1% Tween 20 (TBS-T) for 1 h at room temperature, the blots were incubated overnight at 4 °C in blocking solution with an appropriate dilution of the primary antibodies. The following primary antibodies were used: antitelomerase reverse transcriptase antibody (rabbit polyclonal, ab191523; dilution 1:500; Abcam, Cambridge, MA, USA) and anti-GAPDH antibody GA1R (mouse monoclonal, ab125247; dilution 1:1000; Abcam, Cambridge, MA, USA). The blots were then washed three times for 5 min each in TBS-T and then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The following secondary antibodies were used: goat antirabbit IgG H&L (HRP) (ab205718; dilution 1:10000; Abcam, Cambridge, MA, USA) and goat antimouse IgG H&L (HRP) (12349; dilution 1:10000, EMD Millipore, Temecula, CA, USA). As positive control, Jurkat whole cell lysate (ab30128; Abcam, Cambridge, MA, USA) was used. After incubation with the secondary antibodies, the blots were again washed three times for 5 min with TBS-T, and the protein–antibody complexes were detected by WesternBright Sirus (K-12043-C20; Advansta, Menio Park, CA, USA) and visualized on a UVP Biospectrum Motorized Darkroom (Upland, CA, USA).
Emission Spectroscopy
To assess the formation of TS30, TS60, and TSC G-quadruplex and their binding affinity for the TMPyP4 solution (Tosylate ab120793; Abcam, Cambridge, MA, USA), fluorescence emission spectra were collected using a Tecan Infinite M200 Pro Microplate Reader, from 550 to 800 nm in energy wavelength. The 1 μM TMPyP4 solution was excited at 433 nm in the presence of 1 μM TS30, TS60, and TSC in buffer C. As negative control, the emission spectra of the ssDNA probes were collected without the TMPyP4 ligand.
Confocal Microscopy
Images were collected on a Nikon Eclipse Ti-E inverted microscope equipped with a Nikon A1 confocal laser microscope system. First, modified biosensor Au microchips with TS30 and TS60 were incubated with Jurkat nuclear cell fractions (5 × 105 cells/mL) with 0.2 mM of dNTPs or ddGTPs for 3 h at 37 °C to initiate the elongation process. Then, these sensor microchips were incubated in 10 μM TO solution for 1 h at room temperature. Excitation and emission wavelengths of 485 and 520 nm, respectively, were used to visualize the fluorescence. As negative control, we used the treated nuclear cell fraction.
Acknowledgments
This research work was supported by NSF-Chemistry grant no. CHE-1152940. D.C.D.-C. received financial support from Puerto Rico NASA Space Grant Consortium Grant Number NNX15AI11H. G.H.-C. was supported by the Puerto Rico Science, Technology and Research Trust. D.P.B.-R. likes to acknowledge the financial support of NIH-RISE grant number 5R25GM061151-16. The authors would like to thank the use of the Confocal Facility at the Molecular Sciences Research Center which is supported by the National Institute of General Medical Science (NIGMS) of the National Institute of Health (NIH) under the award number P20GM103642. D.C.D.-C. would like to thank Bismark Madera for confocal images acquisition and Dr. Ramonita Díaz-Ayala, BIDEA LLC, for helpful discussion.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b00783.
CV of nuclear cell extract using Au microchip; calibration curve for telomerase activity at 180 min of enzymatic reaction; nuclear extraction western blot; fluorescence spectra of TS probes; and change in charge-transfer resistance versus enzymatic reaction for telomerase activity (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Witkowska Nery E.; Kundys M.; Jeleń P. S.; Jönsson-Niedziółka M. Electrochemical Glucose Sensing: Is There Still Room for Improvement?. Anal. Chem. 2016, 88, 11271–11282. 10.1021/acs.analchem.6b03151. [DOI] [PubMed] [Google Scholar]
- Radha Shanmugam N.; Muthukumar S.; Chaudhry S.; Anguiano J.; Prasad S. Ultrasensitive nanostructure sensor arrays on flexible substrates for multiplexed and simultaneous electrochemical detection of a panel of cardiac biomarkers. Biosens. Bioelectron. 2017, 89, 764–772. 10.1016/j.bios.2016.10.046. [DOI] [PubMed] [Google Scholar]
- Radecka H.; Radecki J.. Label-free Electrochemical Immunosensors for Viruses and Antibodies Detection-Review. J. Mex. Chem. Soc. 2015, 59, 269––275., http://www.scielo.org.mx/pdf/jmcs/v59n4/v59n4a4.pdf. [Google Scholar]
- Roggo C.; van der Meer J. R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 2017, 45, 24–33. 10.1016/j.copbio.2016.11.023. [DOI] [PubMed] [Google Scholar]
- Chikkaveeraiah B. V.; Bhirde A. A.; Morgan N. Y.; Eden H. S.; Chen X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. 10.1021/nn3023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London, U.K.: Review on Antimicrobial Resistance, 2016, available at: https://amr-review.org/sites/default/files/160525_Finalpaper_withcover.pdf (accessed: Dec 1, 2018).
- Hazelton W. D.; Luebeck E. G. Biomarker-Based Early Cancer Detection: Is It Achievable?. Sci. Transl. Med. 2011, 3, 109fs9. 10.1126/scitranslmed.3003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman J. D.; Fisher P. G.; Gibbs P. Early Detection of Cancer: Past, Present, and Future. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, 57–65. 10.14694/edbook_am.2015.35.57. [DOI] [PubMed] [Google Scholar]
- Wu L.; Qu X. Cancer biomarker detection: recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. 10.1039/c4cs00370e. [DOI] [PubMed] [Google Scholar]
- Wu R. A.; Upton H. E.; Vogan J. M.; Collins K. Telomerase Mechanism of Telomere Synthesis. Annu. Rev. Biochem. 2017, 86, 439–460. 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson V. Biomarkers for early detection of breast cancer: What, when, and where?. Biochim. Biophys. Acta, Gen. Subj. 2007, 1770, 847–856. 10.1016/j.bbagen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Topkaya S. N.; Azimzadeh M.; Ozsoz M. Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges. Electroanalysis 2016, 28, 1402–1419. 10.1002/elan.201501174. [DOI] [Google Scholar]
- Narwaria M.; Shrivastav A.; Shrivastav B. R.. Mini review telomerase-a biomarker in the carcinogenesis and diagnosis. ARPN J. Sci. Technol. 2012, 2, 733––737., http://www.ejournalofscience.org/archive/vol2no8/vol2no8_7.pdf. [Google Scholar]
- Kim K. W.; Shin Y.; Perera A. P.; Qing L.; Kee J. S.; Han K.; Yoon Y.-J.; Park M. K. Label-free, PCR-free chip-based detection of telomerase activity in bladder cancer cells. Biosens. Bioelectron. 2013, 45, 152–157. 10.1016/j.bios.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Hayflick L.; Moorhead P. S. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res. 1961, 25, 585–621. 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Mitchell M.; Gillis A.; Futahashi M.; Fujiwara H.; Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010, 17, 513–518. 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Pai R. B.; Pai S. B.; Kukhanova M.; Dutschman G. E.; Guo X.; Cheng Y. C.. Telomerase from human leukemia cells: properties and its interaction with deoxynucleoside analogues. Cancer Res. 1998, 58, 1909––1913., http://cancerres.aacrjournals.org/content/canres/58/9/1909.full.pdf. [PubMed] [Google Scholar]
- Savelyev N.; Baykuzina P.; Dokudovskaya S.; Lavrik O.; Rubtsova M.; Dontsova O. Comprehensive analysis of telomerase inhibition by gallotannin. Oncotarget 2018, 9, 18712–18719. 10.18632/oncotarget.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016, 6, 584. 10.1158/2159-8290.cd-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsov D. A.; Zvereva M. E.; Shpanchenko O. V.; Dontsova O. A.. Assays for detection of telomerase activity. Acta Natur. 2011, 3, 48––68., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3347595/pdf/AN20758251-08-048.pdf. [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Lou X.; Xia F. Advances in the detection of telomerase activity using isothermal amplification. Theranostics 2017, 7, 1847–1862. 10.7150/thno.18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Zhu X.; Liu Q.; Lin Z.; Qiu B.; Chen G. Label-free detection of telomerase activity in HeLa cells using electrochemical impedance spectroscopy. Chem. Commun. 2011, 47, 3129–3131. 10.1039/c0cc05230b. [DOI] [PubMed] [Google Scholar]
- Cunci L.; Martinez Vargas M.; Cunci R.; Gomez-Moreno R.; Perez I.; Baerga-Ortiz A.; Gonzalez C. I.; Cabrera C. R. Real-time detection of telomerase activity in cancer cells using a label-free electrochemical impedimetric biosensing microchip. RSC Adv. 2014, 4, 52357–52365. 10.1039/c4ra09689d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Cartagena D. C.; Hernández G.; Bracho-Rincon D.; González-Feliciano J. A.; Cunci Perez L.; González C. I.; Cabrera C. R. Development of an Electrochemical Impedimetric Biosensor for the Detection of Telomerase Activity in Cancer Cells. ECS Trans. 2017, 77, 1833–1840. 10.1149/07711.1833ecst. [DOI] [Google Scholar]
- Teller C.; Shimron S.; Willner I. Aptamer–DNAzyme Hairpins for Amplified Biosensing. Anal. Chem. 2009, 81, 9114–9119. 10.1021/ac901773b. [DOI] [PubMed] [Google Scholar]
- Heaphy C. M.; Meeker A. K. The potential utility of telomere-related markers for cancer diagnosis. J. Cell. Mol. Med. 2011, 15, 1227–1238. 10.1111/j.1582-4934.2011.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Meng T.; Liang L.; Sun J.; Wu S.; Wang H.; Yang X.; Zhang Y. Fabrication of amine-functionalized metal-organic frameworks with embedded palladium nanoparticles for highly sensitive electrochemical detection of telomerase activity. Sensor. Actuat. B-Chem. 2019, 278, 133–139. 10.1016/j.snb.2018.09.034. [DOI] [Google Scholar]
- Li C.-c.; Hu J.; Lu M.; Zhang C.-y. Quantum dot-based electrochemical biosensor for stripping voltammetric detection of telomerase at the single-cell level. Biosens. Bioelectron. 2018, 122, 51–57. 10.1016/j.bios.2018.09.049. [DOI] [PubMed] [Google Scholar]
- Kim N.; Piatyszek M.; Prowse K.; Harley C.; West M.; Ho P.; Coviello G.; Wright W.; Weinrich S.; Shay J. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kulić A.; Plavetic N. D.; Gamulin S.; Jakic-Razumovic J.; Vrbanec D.; Sirotkovic-Skerlev M., Telomerase activity in breast cancer patients: association with poor prognosis and more aggressive phenotype. Med. Oncol. 2016, 33 (). 10.1007/s12032-016-0736-x [DOI] [PubMed] [Google Scholar]
- Fernández-Marcelo T.; Sánchez-Pernaute A.; Pascua I.; De Juan C.; Head J.; Torres-García A.-J.; Iniesta P. Clinical Relevance of Telomere Status and Telomerase Activity in Colorectal Cancer. PLoS One 2016, 11, e0149626 10.1371/journal.pone.0149626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.; Huang F.; Xu Q.; Zhang M.; Lou X.; Xia F. Facile, Fast-Responsive, and Photostable Imaging of Telomerase Activity in Living Cells with a Fluorescence Turn-On Manner. Anal. Chem. 2016, 88, 3289–3294. 10.1021/acs.analchem.5b04756. [DOI] [PubMed] [Google Scholar]
- Li X. Y.; Wang X.. The role of human cervical cancer oncogene in cancer progression. Int. J. Clin. Exp. Med. 2015, 8, 8363––8368., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4538137/pdf/ijcem0008-8363.pdf. [PMC free article] [PubMed] [Google Scholar]
- Sun P.-M.; Wei L.-H.; Luo M.-Y.; Liu G.; Wang J.-L.; Mustea A.; Könsgen D.; Lichtenegger W.; Sehouli J. The telomerase activity and expression of hTERT gene can serve as indicators in the anti-cancer treatment of human ovarian cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 2007, 130, 249–257. 10.1016/j.ejogrb.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Yabusaki M.; Sato J.; Kohyama A.; Kojima T.; Nobuoka D.; Yoshikawa T.; Sawada Y.; Gohda K.; Okegawa T.; Nakamura M.; Takamatsu K.; Ito M.; Kaneko K.; Nakatsura T. Detection and preliminary evaluation of circulating tumor cells in the peripheral blood of patients with eight types of cancer using a telomerase-specific adenovirus. Oncol. Rep. 2014, 32, 1772–1778. 10.3892/or.2014.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Wang W.; Huang S.; Li J.; Rui K.; Zhang J.-R.; Zhu J.-J. Coupling a DNA-Based Machine with Glucometer Readouts for Amplified Detection of Telomerase Activity in Cancer Cells. Sci. Rep. 2016, 6, 23504. 10.1038/srep23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S.; Su Q.; Gutiérrez-Sánchez C.; Nöll G. Critical View on Electrochemical Impedance Spectroscopy Using the Ferri/Ferrocyanide Redox Couple at Gold Electrodes. Anal. Chem. 2016, 88, 4383–4390. 10.1021/acs.analchem.5b04814. [DOI] [PubMed] [Google Scholar]
- Chen X.; Chen W.; Tang L.; Hu W.; Wang M.; Miao P. Electrochemical impedance spectroscopic analysis of nucleic acids through DNA tetrahedron self-walking machine. Electrochem. Commun. 2019, 101, 1–5. 10.1016/j.elecom.2019.02.006. [DOI] [Google Scholar]
- Moses S.; Brewer S. H.; Lowe L. B.; Lappi S. E.; Gilvey L. B. G.; Sauthier M.; Tenent R. C.; Feldheim D. L.; Franzen S. Characterization of Single- and Double-Stranded DNA on Gold Surfaces. Langmuir 2004, 20, 11134–11140. 10.1021/la0492815. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodríguez L.; Sánchez-Pomales G.; Cabrera C. R. Electrochemical DNA Sensing at Single-walled Carbon Nanotubes Chemically Assembled on Gold Surfaces. Electroanalysis 2010, 22, 2817–2824. 10.1002/elan.201000305. [DOI] [Google Scholar]
- Wei C.; Han G.; Jia G.; Zhou J.; Li C. Study on the interaction of porphyrin with G-quadruplex DNAs. Biophys. Chem. 2008, 137, 19–23. 10.1016/j.bpc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Di Antonio M.; Rodriguez R.; Balasubramanian S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods 2012, 57, 84–92. 10.1016/j.ymeth.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Dai J.; Min X.; Yu Z.; Cheng Y.; Huang K.; Yang J.; Yi X.; Lou X.; Xia F. DNA-Conjugated Amphiphilic Aggregation-Induced Emission Probe for Cancer Tissue Imaging and Prognosis Analysis. Anal. Chem. 2018, 90, 8162–8169. 10.1021/acs.analchem.8b01456. [DOI] [PubMed] [Google Scholar]
- Zhang F.-T.; Nie J.; Zhang D.-W.; Chen J.-T.; Zhou Y.-L.; Zhang X.-X. Methylene Blue as a G-Quadruplex Binding Probe for Label-Free Homogeneous Electrochemical Biosensing. Anal. Chem. 2014, 86, 9489–9495. 10.1021/ac502540m. [DOI] [PubMed] [Google Scholar]
- Yaku H.; Murashima T.; Miyoshi D.; Sugimoto N. Specific Binding of Anionic Porphyrin and Phthalocyanine to the G-Quadruplex with a Variety of in Vitro and in Vivo Applications. Molecules 2012, 17, 10586–10613. 10.3390/molecules170910586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasikuttan A. C.; Mohanty J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: promising fluorescence sensors. Chem. Commun. 2015, 51, 7581–7597. 10.1039/c4cc10030a. [DOI] [PubMed] [Google Scholar]
- Rivera-Gandía J.; del Mar Maldonado M.; De La Torre-Meléndez Y.; Ortiz-Quiles E. O.; Vargas-Barbosa N. M.; Cabrera C. R. Electrochemical Capacitance DNA Sensing at Hairpin-Modified Au Electrodes. J. Sensors 2011, 2011, 735279. 10.1155/2011/735279. [DOI] [Google Scholar]
- Santiago-Rodríguez L.; Sánchez-Pomales G.; Cabrera C. R. DNA-Functionalized Carbon Nanotubes: Synthesis, Self-Assembly, and Applications. Isr. J. Chem. 2010, 50, 277–290. 10.1002/ijch.201000034. [DOI] [Google Scholar]
- Ward E.; DeSantis C.; Robbins A.; Kohler B.; Jemal A. Childhood and adolescent cancer statistics, 2014. CA: A Cancer J. Clin. 2014, 64, 83–103. 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- Petrovykh D. Y.; Kimura-Suda H.; Whitman L. J.; Tarlov M. J. Quantitative Analysis and Characterization of DNA Immobilized on Gold. J. Am. Chem. Soc. 2003, 125, 5219–5226. 10.1021/ja029450c. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y.; Canavan H. E.; Gamble L. J.; Castner D. G. Evidence of Impurities in Thiolated Single-Stranded DNA Oligomers and Their Effect on DNA Self-Assembly on Gold. Langmuir 2005, 21, 5134–5141. 10.1021/la0472302. [DOI] [PubMed] [Google Scholar]
- Sánchez-Pomales G.; Cabrera C. R. Vertical attachment of DNA-CNT hybrids on gold. J. Electroanal. Chem. 2007, 606, 47–54. 10.1016/j.jelechem.2007.04.010. [DOI] [Google Scholar]
- Strahl C.; Blackburn E. H. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol. Cell. Biol. 1996, 16, 53–65. 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.; Shang C.; Lou X.; Xia F. Construction of AIEgens-Based Bioprobe with Two Fluorescent Signals for Enhanced Monitor of Extracellular and Intracellular Telomerase Activity. Anal. Chem. 2017, 89, 2073–2079. 10.1021/acs.analchem.6b04696. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Sun C.; Liu R.; Yang J.; Dai J.; Zhai T.; Lou X.; Xia F. A Multifunctional Peptide-Conjugated AIEgen for Efficient and Sequential Targeted Gene Delivery into the Nucleus. Angew. Chem., Int. Ed. 2019, 58, 5049–5053. 10.1002/anie.201901527. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Dai J.; Sun C.; Liu R.; Zhai T.; Lou X.; Xia F. An Intracellular H2O2-Responsive AIEgen for the Peroxidase-Mediated Selective Imaging and Inhibition of Inflammatory Cells. Angew. Chem., Int. Ed. 2018, 57, 3123–3127. 10.1002/anie.201712803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.