Abstract
Obesity is one of modifiable risk factors for clear cell renal cell cancer (ccRCC). We aim to identify the association between obesity-driven biomarkers and ccRCC risk. This is a retrospective, two-phase, case-control study involving 682 cases and 733 controls. Obesity-driven biomarkers [gastric inhibitory polypeptide (GIP), C-peptide, insulin, resistin, adipsin, peptide YY, pancreatic polypeptide, interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), plasminogen activator inhibitor-1, monocyte chemoattractant protein 1, lipocalin2, leptin, adiponectin] were measured using the Milliplex method. Multivariate logistic regression was used to assess the associations between biomarkers and ccRCC risk. Results revealed that GIP, C-peptide, IL-6 and TNF-α levels were consistently distinct between cases and controls. These markers were significantly associated with ccRCC risk in both phases (except C-peptide). In the combined population, compared with individuals with low levels of the biomarkers, individuals with high level of GIP [odds ratio (OR) = 0.52, 95% confidence interval (CI): 0.40–0.67] had lower risk, whereas individuals with high levels of C-peptide (OR = 1.46, 95% CI: 1.15–1.87), IL-6 (OR = 2.20, 95% CI: 1.50–3.22), TNF-α (OR = 1.90, 95% CI: 1.49–2.43) had significantly higher risk. Stratified analysis showed consistent associations with ccRCC risk in most subgroups (P < 0.05). The risk score based on the IL-6, TNF-α and GIP was positively associated with ccRCC risk in a dose-response manner (P for trend = 2.18E-13). Data from The Cancer Genome Atlas indicate that insulin signaling, IL-6 signaling and TNF-α signaling were enhanced in tumors. Collectively, our study demonstrates the integrative effect of insulin resistance and inflammation in ccRCC development, which may elucidate the basis of association between obesity and carcinogenesis. Further confirmation in prospective cohort studies are warranted for clinical applications in prevention and precision medicine of ccRCC.
In this two-phase, case-control study, we systematically evaluated the associations between 14 circulating obesity-driven biomarkers and ccRCC risk. GIP, TNF-α and IL-6 were significantly associated with ccRCC risk. We identified the predictive role of obesity-driven biomarkers in ccRCC susceptibility.
Introduction
Kidney cancer is one of the most lethal urologic cancer (1). Clear cell renal cell cancer (ccRCC) is the predominant form of kidney cancer among all subtypes. Obesity has been identified as one of the independent modifiable risk factors of ccRCC and studies have demonstrated that the risk of ccRCC is 1.5 times higher in obese population than that in normal weight population (2). However, the underlying mechanism remains largely unclear.
Dysfunction of adipose tissue is often linked to obesity. Increased volume of adipose tissue may lead to peripheral insulin resistance, abnormal adipokine secretion and inflammation, which may promote carcinogenesis (3,4). Insulin and insulin-like growth factor (IGF) related pathways have been reported to play important roles in renal cell cancer (RCC) risk and outcome (5). Genetic variations of IGF-binding protein-3 (IGFBP3) were associated with ccRCC risk in men (6). Several studies have examined the association of adiponectin and/or leptin levels with the risk of RCC with inconsistent findings, partly due to the difference in study population. For example, Liao et al. reported that higher circulating adiponectin levels were associated with reduced RCC risk in Finnish male smokers (7), whereas opposite results were observed in African American males (8). Inflammatory cytokines, including tumor necrosis factor-α (TNF-α) (9), interleukin-6 (IL-6) (10) and plasminogen activator inhibitor-1 (PAI-1) (11) were found significantly higher in obese individuals. These cytokines are mostly secreted by monocytes and other immune cells that infiltrate into the adipose tissues (12). Altered expressions of IL-6 and TNF-α were observed in RCC, which was mainly composed of ccRCC (13). Thus, insulin resistance, adipokines, inflammation and their cross talks have been proposed as major biological mechanisms linking obesity and ccRCC.
In this case-control study, we systematically evaluated the independent and cumulative associations between circulating levels of 14 obesity-driven biomarkers and ccRCC risk in a discovery phase and validated the findings in a validation phase. Further risk score and bioinformatic analysis were conducted to explore the predictive value and underlying biological mechanisms of our findings.
Materials and methods
Study population
All enrolled ccRCC patients were drawn from an ongoing case-control study at The University of Texas MD Anderson Cancer Center (Houston, TX) initiated in 2002. The study has been approved by MD Anderson Cancer Center Institutional Review Board. Details of the study have been described previously (14). In brief, all recruited cases were individuals with newly diagnosed (within 1 year of diagnosis), histologically confirmed and previously untreated ccRCC. Healthy control subjects without a history of cancer, except non-melanoma skin cancer, were identified and recruited via random digit dialing during the same time period as the cases, frequency matched according to age (±5 years) and gender. All participants provided written informed consent before participating in the study. Due to the small number of minority participants, we restricted our analysis to non-Hispanic whites.
Data collection
A 45-min structured in-person interview was conducted to collect epidemiological data by MD Anderson interviewers. Information on physical activity, weight at diagnosis (for cases) or recruitment (for control subjects), history of hypertension (yes/no), smoking status and pack-years of smoking was recorded. Body mass index (BMI, kg/m2) was calculated through self-reported usual height and weight. BMI was categorized according to the standard classifications of the World Health Organization (normal/underweight and overweight: <30 kg/m2; obese: ≥30 kg/m2). Smoking status and pack-years of smoking were defined as follows: an individual who had never smoked or had smoked <100 cigarettes in his or her lifetime was defined as a never smoker; an individual who had smoked at least 100 cigarettes in his or her lifetime but had quit at least 12 months before diagnosis (for cases) or before the interview (for controls) was classified as a former smoker. Current smokers were those who were currently smoking or quit <12 months before diagnosis (for cases) or before the interview (for controls). The number of pack-years was calculated as the average number of cigarettes smoked per day divided by 20 and then multiplied by smoking years. Immediately after interview, a 40 ml blood sample was collected from each participant and delivered to the laboratory for further analysis. The plasma was separated from blood and stored in MAPI straws (Cryo Bio System). All samples were then transferred into liquid nitrogen tanks for long-term storage.
Immunoassay for detecting plasma levels of various obesity-driven biomarkers
Plasma samples were thawed on ice prior to the assay. Samples were tested in duplicates using Milliplex® MAP Human Metabolic Hormone Magnetic Bead Panel kit (HMHEMAG-34K; EMD Millipore; using undiluted plasma) and Milliplex® MAP Human Adipokine Magnetic Bead Panel kit (HADK1MAG-61K; EMD Millipore; using 1:400 diluted plasma) in 96-well plate format to quantify the 14 selected obesity-driven biomarkers. The selection of obesity-related markers was based on systematic literature review (3,4) and the availability of Luminex assays. Among them, seven biomarkers were further assayed in the validation phase. Each plate contained positive and negative controls and samples for standard curve. Assay was conducted according to protocols provided by the manufacturer using Luminex 200™ instrument and xPONENT® software (Luminex Corp). All inter-assay and intra-assay coefficients of variation were below 15%.
Statistical analysis
To compare baseline characteristics between cases and controls, we used the Pearson’s chi-squared test for categorical variables, Student’s t-tests for continuous variables and Wilcoxon rank sum test for plasma biomarkers as they were not normally distributed. All biomarker levels were dichotomized using a logistic regression spline model (15). We estimated the associations between each biomarker and ccRCC risk using unconditional logistic regression models. Odds ratios (ORs) and 95% confidence intervals (CIs) were reported for gender- and age-adjusted as well as fully adjusted analysis by age (<60, ≥60), gender, smoking status (ever/never), history of hypertension (yes/no), diabetes mellitus (yes/no) and BMI (<30, ≥30). Combined analyses were performed based on the pooled populations.
Stratified analysis was applied in pooled population by known or suspected ccRCC risk factors, including age, gender, BMI, history of hypertension, diabetes mellitus and smoking status. We tested for multiplicative interaction by including a cross-product term and the significance of the interactions was determined using the Wald statistic for the cross-product term. The risk score was generated by a sum of the product of dichotomized expression level of each biomarker by its logistic regression coefficient obtained at the discovery stage as shown by the following equation:
where RS is the risk score for subject j; for each soluble obesity-driven marker (SOM) i I(SOMij ≥ cutoff value) is an indicator function which is 1 if SOMij level ≥ cutoff value and 0 otherwise. The bonferroni correction were applied in the multiple comparison of P-values. We assessed the association between the risk score (in quartiles) and ccRCC risk using unconditional logistic regression model.
In addition, messenger RNA expression data from 72 primary ccRCC samples and corresponding normal tissues were downloaded from The Cancer Genome Atlas (TCGA) Kidney Renal Clear Cell Carcinoma study to explore potential mechanism (16). The genes of signal pathways were selected from KEGG (http://www.genome.jp/kegg/) and Biocarta (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways). All the data were further analyzed and plotted using R software (v3.4.1).
All statistical tests were two sided with a significance cutoff at 0.05. All analyses were conducted using Stata 10.1 statistical software package (Stata Corp, College Station, TX).
Results
Host characteristics
This is a two-phase, case-control study. The discovery phase included 182 cases and 182 controls, whereas the validation phase consisted of 500 cases and 551 controls. The characteristics of study participants are described in Table 1, and the whole design of the study was depicted in Supplementary Figure S1. In discovery phase, the mean age was 59.0 ± 9.2 years in cases and 58.9 ± 9.0 years in controls. Over two-thirds of the subjects were males and around half were smokers in cases and controls with slightly more smokers among cases. BMI and hypertension were significantly different between cases and controls (P < 0.05). There were 36 (19.8%) cases and 25 (13.7%) controls with type 2 diabetes (T2D) mellitus. The distributions of patient characteristics in the validation phase were similar to those in the discovery phase.
Table 1.
Host characteristics
| Discovery | Validation | |||||
|---|---|---|---|---|---|---|
| Variables | Cases, n (%) | Controls, n (%) | P-value | Cases, n (%) | Controls, n (%) | P-value |
| Age, mean (SD) | 59.0 (9.2) | 58.9 (9.0) | 0.87 | 59.1 (10.9) | 59.7 (10.0) | 0.36 |
| BMI, mean (SD) | 30.4 (6.7) | 28.4 (5.6) | 2.28E-03 | 30.3 (6.2) | 28.5 (5.5) | 1.23E-06 |
| Sex | ||||||
| Male | 135 (74.2) | 135 (74.2) | 323 (64.6) | 381 (69.2) | ||
| Female | 47 (25.8) | 47 (25.8) | 1.00 | 177 (35.4) | 170 (30.8) | 0.12 |
| Age (years) | ||||||
| <60 | 87 (47.8) | 92 (50.6) | 245 (49.0) | 284 (51.5) | ||
| ≥60 | 95 (52.2) | 90 (49.4) | 0.60 | 255 (51.0) | 267 (48.5) | 0.41 |
| Ever smoking | ||||||
| Yes | 99 (54.4) | 88 (48.4) | 235 (47.0) | 266 (48.3) | ||
| No | 83 (45.6) | 94 (51.6) | 0.25 | 265 (53.0) | 285 (51.7) | 0.68 |
| BMI (kg/m2) | ||||||
| <30 | 102 (56.0) | 125 (68.7) | 246 (56.4) | 373 (67.9) | ||
| ≥30 | 80 (44.0) | 57 (31.3) | 0.01 | 190 (43.6) | 176 (32.1) | 2.02E-04 |
| Hypertension | ||||||
| Yes | 133 (73.1) | 72 (39.6) | 289 (57.8) | 236 (42.8) | ||
| No | 49 (26.9) | 110 (60.4) | 1.15E-10 | 211 (42.2) | 315 (57.2) | 1.25E-06 |
| Diabetes | ||||||
| Yes | 36 (19.8) | 25 (13.7) | 67 (13.5) | 77 (14.0) | ||
| No | 146 (80.2) | 157 (86.3) | 0.12 | 430 (86.5) | 474 (86.0) | 0.82 |
SD, standard deviation.
Obesity-driven biomarkers predict ccRCC risk
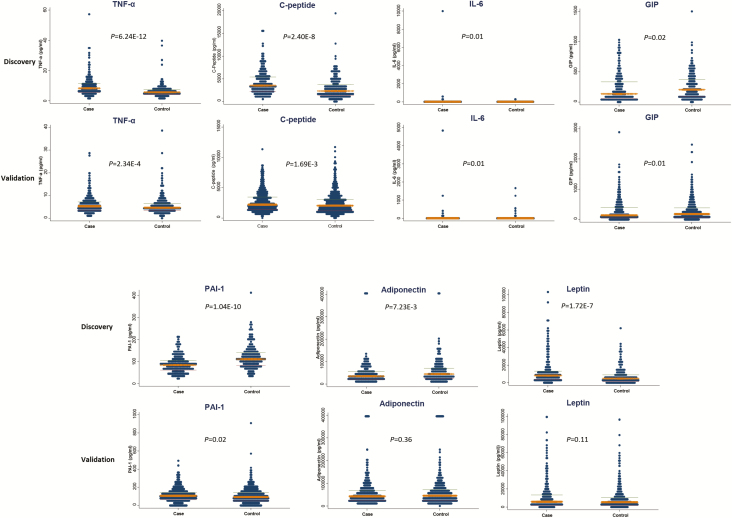
The distributions of all obesity-driven biomarkers (median and inter-quartile range) were listed in Supplementary Table S1. Among them, the levels of seven biomarkers were significantly different among cases and controls in the discovery phase (Figure 1, Supplementary Table S2). GIP levels were significantly lower in cases than in controls in both the discovery phase and the validation phase (P = 0.02 and 0.01, respectively), whereas C-peptide, IL-6 and TNF-α levels were higher in cases in both the discovery phase and the validation phase (P < 0.05). Leptin and adiponectin levels were significantly different between cases and controls in the discovery phase (P < 0.01) and demonstrated consistent trend in the validation phase (P = 0.10 and 0.36, respectively). PAI-1 levels showed opposite directions in the discovery and the validation phase.
Figure 1.
Distribution of seven obesity-driven biomarkers in discovery and validation stages. Among them, GIP, C-peptide, TNF-α and IL-6 levels were significantly different between controls and cases in both stages, whereas PAI-1, adiponectin and leptin were not.
Further multivariate logistic regression analysis was performed to assess the association between binary biomarker levels (i.e. low and high) and ccRCC risk adjusted for potential confounders (Table 2). All four biomarkers were significantly associated with ccRCC risk in the same direction in the discovery phase and the validation phase except C-peptide (P > 0.05 in the validation phase). In the combined population, compared with individuals with low levels of the biomarkers, individuals with high level of GIP (OR = 0.52, 95% CI: 0.40–0.67) had lower risk of ccRCC, whereas individuals with high levels of TNF-α (OR = 1.90, 95% CI: 1.49–2.43), IL-6 (OR = 2.20, 95% CI: 1.50–3.22) and C-peptide (OR = 1.46, 95% CI: 1.15–1.87) had significantly higher risk of ccRCC.
Table 2.
Multivariate logistic regression analysis of markers in cases and controls
| Discovery | Validation | Combined | ||||
|---|---|---|---|---|---|---|
| Markers | OR (95% CI)a | P | OR (95% CI)a | P | OR (95% CI)a | P |
| GIP | ||||||
| Low | 1 (reference) | 1 (reference) | 1 (reference) | |||
| High | 0.38 (0.22–0.66) | 5.83E-04 | 0.56 (0.42–0.76) | 1.19E-04 | 0.52 (0.40–0.67) | 6.63E-07 |
| TNF-α | ||||||
| Low | 1 (reference) | 1 (reference) | 1 (reference) | |||
| High | 4.32 (2.65–7.03) | 3.96E-09 | 1.39 (1.05–1.85) | 0.02 | 1.90 (1.49–2.43) | 1.85E-07 |
| IL-6 | ||||||
| Low | 1 (reference) | 1 (reference) | 1 (reference) | |||
| High | 3.13 (1.43–6.87) | 4.00E-03 | 1.93 (1.24–3.02) | 4.00E-03 | 2.20 (1.50–3.22) | 5.41E-05 |
| C-peptide | ||||||
| Low | 1 (reference) | 1 (reference) | 1 (reference) | |||
| High | 3.55 (1.96–6.43) | 2.82E-05 | 1.19 (0.91–1.57) | 0.21 | 1.46 (1.15–1.87) | 2.27E-03 |
Significant P-values in bold font.
aAdjusted by age, gender, smoking, BMI, diabetes and hypertension.
Stratified analyses and risk score based on three obesity-driven biomarkers
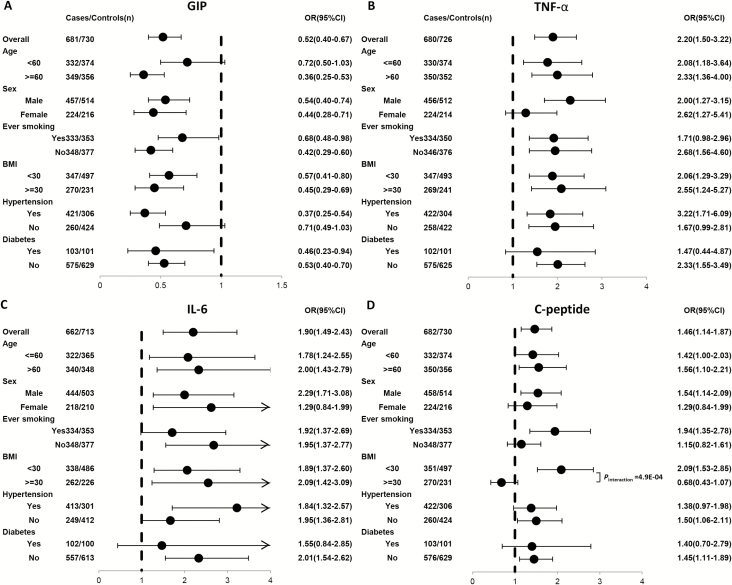
For the four biomarkers, we conducted stratified analysis in the combined population (Figure 2). The interaction between biomarkers and stratified factors were shown in Supplementary Table S3. High GIP (versus low) demonstrated significant and consistent associations with reduced ccRCC risk in most subgroups (Figure 2A). High TNF-α (Figure 2B) and IL-6 (Figure 2C) were consistently associated with increased ccRCC risk in all subgroups. Also, we showed the pairwise correlations among biomarkers in pooled population, GIP level was correlated with C-peptide level (rho = 0.52, P = 1.49E-99) (Supplementary Table S4). Additionally, we also investigated the association between C-peptide and ccRCC risk in subgroup. High C-peptide (versus low) demonstrated significant and consistent associations with increased ccRCC risk in most subgroups of pooled population (Figure 2D);C-peptide was significantly interacted with obese status (BMI ≥ 30; Pinteraction = 4.87E-04).
Figure 2.
Forest plot of ORs for obesity-driven biomarkers in stratified analyses based on selected covariates for (A) GIP, (B) TNF-α, (C) IL-6 and (D) C-peptide. All P-values of interaction between biomarker and selected covariates were Bonferroni corrected. The solid circles with horizontal lines corresponding to the multivariable-adjusted ORs and 95% CIs. The vertical dash line indicates an OR of 1.0.
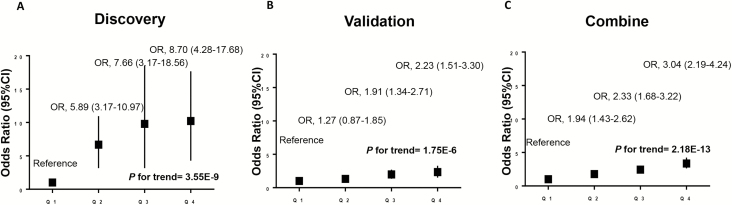
The combined effects of the three validated biomarkers were investigated by calculating a risk score using the linear combination of GIP, TNF-α and IL-6 in discovery, validation and combined population (Figure 3, Supplemental Table S5). The risk score was positively associated with ccRCC risk in a dose-response manner in all populations (P for trend <0.001). Compared with individuals in the first quartile, individuals in the second, third and fourth quartile had a 1.94-fold (95% CI: 1.43–2.62), 2.33-fold (95% CI: 1.68–3.22) and 3.04-fold (95% CI: 2.19–4.24) elevated risk of ccRCC in combined population, respectively.
Figure 3.
Risk score of obesity-driven biomarkers predicts ccRCC risk in (A) discovery, (B) validation and (C) combined populations. In combined population, the fourth (Q4), third (Q3) and second (Q2) quartile subgroups demonstrated significant increased cancer risk (3.04-fold, 2.33-fold and 1.94-fold) compared with first quartile (Q1) subgroup of risk score (P for trend = 2.18E-13) using multivariable logistic regression model adjusted by age, gender, smoking, BMI, diabetes and hypertension.
Enhanced IL-6, insulin and TNF signaling in ccRCC tissues
To further characterize the potential mechanisms underlying obesity-driven ccRCC carcinogenesis, we evaluated the obesity-related gene expressions in the TCGA database, which includes 72 ccRCC tumor tissues and paired adjacent 72 normal tissues. The genes in IL-6 signaling, TNF signaling and insulin signaling pathways were elevated in tumor tissues compared with normal tissues (except STAT3, IGF1, IGFR1, IGFR2), whereas GIPR was downregulated in tumor tissues (Supplementary Figure S2).
Discussion
To our knowledge, this is the first large-scale, two-phase case-control study to systematically evaluate obesity-driven biomarkers in ccRCC susceptibility. Obesity is a known risk factor for RCC, and possible biological mechanisms involving adipokines, chronic inflammation, insulin/IGF pathways and sex steroids have been implicated (17). In this study, we noted significant inverse association between circulating GIP and ccRCC risk, whereas circulating C-peptide, IL-6 and TNFα exhibited positive associations with ccRCC risk in both phases and the combined population (except C-peptide in validation phase). Stratified analysis in the combined population showed consistent associations in all subgroups except for C-peptide by BMI. High C-peptide was associated with significantly increased ccRCC risk in non-obese individuals but reduced ccRCC risk in obese individuals (Pinteraction < 0.001). Significant interactions were also observed for GIP with age and hypertension and TNF-α with sex. A risk score based on these three validated biomarkers could stratify the risk of ccRCC. Further bioinformatics analysis using TCGA data set indicated that insulin signaling, IL-6 signaling and TNF signaling were enhanced in tumor tissues. Therefore, our findings revealed the predictive role of obesity-driven biomarkers for ccRCC risk and highlighted potential obesity-related mechanisms during ccRCC carcinogenesis.
In this study, we observed that GIP, secreted by enteroendocrine K cells, was inversely associated with ccRCC risk for the first time. Previous publications indicated that GIP could induce insulin secretion in beta cells in obese or diabetic patients (18,19). As a notable regulator of insulin resistance, GIP was strongly associated with C-peptide in our study; however, GIP and C-peptide were associated with ccRCC risk in opposite direction (OR < 1 for GIP and OR > 1 for C-peptide). This could be resulted from the stimulatory effect of GIP on insulin secretion, which could also increase the C-peptide level (20). Intriguingly, GIP could also promote lipogenesis via stimulating lipoprotein lipase in adipocyte (19), which may promote obesity. GIP receptor (GIPR) expression in ccRCC tissues was significantly lower than normal tissues in TCGA data set. This suggested that GIP-GIPR signaling may be defective during ccRCC development. These findings indicate that circulating GIP may be protective in ccRCC development through abrogating insulin resistance, though it may also increase adiposity. However, more research is warranted in deciphering the mechanisms underlying GIP and ccRCC development.
C-peptide, a byproduct of insulin, is a surrogate marker of insulin resistance, which eventually results in the T2D (21). T2D has been reported as a risk factor of ccRCC in women (22). In this study, C-peptide is positively associated with ccRCC risk, which is consistent with previous studies (23,24). Interestingly, C-peptide demonstrated significant interaction with BMI for ccRCC risk for the first time, indicating higher level of C-peptide is associated with increased risk of ccRCC in non-obese patients, whereas higher C-peptide level showed opposite direction of OR in obese patients. Only one previous study demonstrated that C-peptide is independent of BMI in predicting colon cancer risk in men (25). However, our findings were focused on both genders in ccRCC. Moreover, it is reported that C-peptide could bind to a specific guanosine triphosphate-binding protein-coupled receptor, which could promote carcinogenesis through regulating multiple signaling pathways (26). However, these studies only partially explain the potential mechanisms of C-peptide in tumorigenesis. More studies are required to fully address the role of C-peptide in ccRCC development.
As critical cytokines actively participating in the obesity-induced inflammation (27), IL-6 and TNF-α were significantly associated with increased ccRCC risk in this study. This is consistent with a previous study reporting that enhanced IL-6 and TNF expression could promote liver inflammation and tumorigenesis (28). Our previous study also demonstrated that genetic variations of IL6 and TNFA were associated with risk of bladder cancer (29). Chronic inflammation resulting from obesity could induce secretion of TNF-α and related cytokines thereby promoting carcinogenesis in ccRCC as well as other cancer sites or chronic diseases (28–30). For instance, the IL-6 and TNF-α derived from chronic inflammation could impair host immunity and insulin signaling, resulting in immune dysfunction, insulin resistance and subsequent carcinogenesis (27,31). This is in line with our findings from TCGA database suggesting that IL-6 signaling, TNF signaling and insulin signaling were enhanced in ccRCC tissues. Therefore, we proposed that the impact of IL-6 and TNF-α on ccRCC risk may stem from integrative effect in chronic inflammation, insulin resistance and immune dysfunction.
Leptin and adiponectin are adipokines associated with susceptibility of multiple malignancies (3,9,32–35). Previous studies in RCC indicate that leptin is not associated with RCC risk, whereas adiponectin is only associated with the risk in male smokers (7) or African American males (8). However, we found that the leptin level and adiponectin level were not associated with ccRCC risk in our study. The divergence may be derived from various pathological subtypes of RCC, different racial groups or genders. To further elucidate the roles of the two biomarkers in ccRCC, more studies within independent cohort are warranted.
Furthermore, risk score based on linear combination of three identified biomarkers demonstrated ideal risk stratification of ccRCC risk in this study. This finding highlighted synergic effect of insulin signaling, TNF signaling and IL-6 signaling pathways on ccRCC susceptibility, suggesting potential integrative mechanisms underlying obesity-driven carcinogenesis.
Although our study has distinct strengths including the two-phase study design, careful matching of controls to cases by age, gender and sample storage time, integrative obesity markers panel and relatively large sample size, we acknowledge several limitations. First, this is a retrospective, case-control study; therefore, reverse causation is always a possibility to account for the observed associations. Blood samples were collected at the time of diagnosis, so we cannot establish the temporality and causality between obesity-driven markers and the risk of developing ccRCC. Whether the obesity-driven genes play causative roles in RCC development require further experimental evidence in animal and human studies (36). To minimize the potential impact of disease and treatment on obesity-driven biomarkers, we only included newly diagnosed ccRCC patients (within 1 year of diagnosis) for which blood samples were drawn at recruitment and before treatment. Future prospective studies are warranted. Second, we only evaluated the most representative obesity-driven biomarkers in this study; other obesity-related factors, such as IGF-1, were not included. Third, an independent cohort is warranted to validate the risk score as it was constructed based on discovery stage. Fourth, we restricted our ethnicity on non-Hispanic white subjects, thus the results may not be applicable to other ethnic groups. Finally, our results were based solely on patients treated at a single institution; further validation in large, independent cohorts is required.
In summary, we investigated the associations between obesity-driven biomarkers and ccRCC susceptibility. GIP, TNF-α and IL-6 were significantly associated with the risk of ccRCC, which implicates the integrative effect of insulin resistance and chronic inflammation in the development of ccRCC. These findings may shed light on the preventive approaches and precision medicine of ccRCC.
Funding
This work was funded by NCI CA170298 and Nature Science Foundation of Zhejiang province, China Q16H160010 (Qinchuan Wang).
Author contributions
The conception and design were done by X.W. The development of methodology was performed by Q.W. and X.W. The acquisition of data (acquired and managed patients, provided facilities, etc.) was done by Q.W., M.Z., H.T., D.W.C., Y.L., D.L. and X.Wu. The analysis and interpretation of data (e.g. statistical analysis, biostatistics, computational analysis) were done by Q.W., Y.Y., D.W.C. and H.T. All authors contributed to the writing, review, and/or revision of the manuscript. Data management and analysis were done by Y. Y. The study was supervised by X.W.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- ccRCC
clear cell renal cell cancer
- CI
confidence interval
- IGF
insulin-like growth factor
- IL-6
interleukin-6
- OR
odds ratio
- PAI-1
plasminogen activator inhibitor-1
- RCC
renal cell cancer
- T2D
type 2 diabetes
- TNF-α
tumor necrosis factor-α
References
- 1. Siegel R.L., et al. (2018) Cancer statistics, 2018. CA Cancer J. Clin., 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. McGuire B.B., et al. (2011) BMI and the risk of renal cell carcinoma. Curr. Opin. Urol., 21, 356–361. [DOI] [PubMed] [Google Scholar]
- 3. Khandekar M.J., et al. (2011) Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer, 11, 886–895. [DOI] [PubMed] [Google Scholar]
- 4. Trinchieri G. (2012) Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol., 30, 677–706. [DOI] [PubMed] [Google Scholar]
- 5. Solarek W., et al. (2015) Insulin and IGFs in renal cancer risk and progression. Endocr. Relat. Cancer, 22, R253–R264. [DOI] [PubMed] [Google Scholar]
- 6. Safarinejad M.R. (2011) Insulin-like growth factor binding protein-3 (IGFBP-3) gene variants are associated with renal cell carcinoma. BJU Int., 108, 762–770. [DOI] [PubMed] [Google Scholar]
- 7. Liao L.M., et al. (2013) Prediagnostic circulating adipokine concentrations and risk of renal cell carcinoma in male smokers. Carcinogenesis, 34, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao L.M., et al. (2013) Serum leptin and adiponectin levels and risk of renal cell carcinoma. Obesity (Silver Spring)., 21, 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hotamisligil G.S., et al. (1995) Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Invest., 95, 2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fried S.K., et al. (1998) Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J. Clin. Endocrinol. Metab., 83, 847–850. [DOI] [PubMed] [Google Scholar]
- 11. Sawdey M.S., et al. (1991) Regulation of murine type 1 plasminogen activator inhibitor gene expression in vivo. Tissue specificity and induction by lipopolysaccharide, tumor necrosis factor-alpha, and transforming growth factor-beta. J. Clin. Invest., 88, 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weisberg S.P., et al. (2003) Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest., 112, 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. König B., et al. (1999) The differential expression of proinflammatory cytokines IL-6, IL-8 and TNF-alpha in renal cell carcinoma. Anticancer Res., 19, 1519–1524. [PubMed] [Google Scholar]
- 14. Clague J., et al. (2009) Family history and risk of renal cell carcinoma: results from a case-control study and systematic meta-analysis. Cancer Epidemiol. Biomarkers Prev., 18, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosenberg P.S., et al. (2003) Quantifying epidemiologic risk factors using non-parametric regression: model selection remains the greatest challenge. Stat. Med., 22, 3369–3381. [DOI] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research Network. (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature, 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gild P., et al. (2017) Effect of obesity on bladder cancer and renal cell carcinoma incidence and survival. Curr. Opin. Urol., 27, 409–414. [DOI] [PubMed] [Google Scholar]
- 18. Sadry S.A., et al. (2013) Emerging combinatorial hormone therapies for the treatment of obesity and T2DM. Nat. Rev. Endocrinol., 9, 425–433. [DOI] [PubMed] [Google Scholar]
- 19. Campbell J.E., et al. (2013) Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab., 17, 819–837. [DOI] [PubMed] [Google Scholar]
- 20. Kulkarni R.N. (2010) GIP: no longer the neglected incretin twin? Sci. Transl. Med., 2, 49ps47. [DOI] [PubMed] [Google Scholar]
- 21. Robbins D.C., et al. (1984) Biologic and clinical importance of proinsulin. N. Engl. J. Med., 310, 1165–1175. [DOI] [PubMed] [Google Scholar]
- 22. Joh H.K., et al. (2011) Type 2 diabetes and the risk of renal cell cancer in women. Diabetes Care, 34, 1552–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liao L.M., et al. (2017) Circulating levels of obesity-related markers and risk of renal cell carcinoma in the PLCO cancer screening trial. Cancer Causes Control, 28, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L., et al. (2013) Circulating C-peptide level is a predictive factor for colorectal neoplasia: evidence from the meta-analysis of prospective studies. Cancer Causes Control, 24, 1837–1847. [DOI] [PubMed] [Google Scholar]
- 25. Ma J., et al. (2004) A prospective study of plasma C-peptide and colorectal cancer risk in men. J. Natl. Cancer Inst., 96, 546–553. [DOI] [PubMed] [Google Scholar]
- 26. Hills C.E., et al. (2008) Intracellular signalling by C-peptide. Exp. Diabetes Res., 2008, 635158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osborn O., et al. (2012) The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med., 18, 363–374. [DOI] [PubMed] [Google Scholar]
- 28. Park E.J., et al. (2010) Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell, 140, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leibovici D., et al. (2005) Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. J. Clin. Oncol., 23, 5746–5756. [DOI] [PubMed] [Google Scholar]
- 30. Al-Lamki R.S., et al. (2015) TNF receptors: signaling pathways and contribution to renal dysfunction. Kidney Int., 87, 281–296. [DOI] [PubMed] [Google Scholar]
- 31. de Vivar Chevez A.R., et al. (2014) The role of inflammation in kidney cancer. Adv. Exp. Med. Biol., 816, 197–234. [DOI] [PubMed] [Google Scholar]
- 32. Crisostomo J., et al. (2016) Hyperresistinemia and metabolic dysregulation: a risky crosstalk in obese breast cancer. Endocrine, 53, 433–442. [DOI] [PubMed] [Google Scholar]
- 33. Gunter M.J., et al. (2015) Circulating adipokines and inflammatory markers and postmenopausal breast cancer risk. J. Natl. Cancer Inst., 2015 Sep; 107: djv169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaaks R., et al. (2000) Serum C-peptide, insulin-like growth factor (IGF)-I, IGF-binding proteins, and colorectal cancer risk in women. J. Natl. Cancer Inst., 92, 1592–1600. [DOI] [PubMed] [Google Scholar]
- 35. Howard J.M., et al. (2010) Leptin and gastro-intestinal malignancies. Obes. Rev., 11, 863–874. [DOI] [PubMed] [Google Scholar]
- 36. Carbone M., et al. (2004) Modern criteria to establish human cancer etiology. Cancer Res., 64, 5518–5524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.