Abstract
Patients with CYLD cutaneous syndrome (CCS; syn. Brooke-Spiegler syndrome) carry germline mutations in the tumor suppressor CYLD and develop multiple skin tumors with diverse histophenotypes. Here, we comprehensively profile the genomic landscape of 42 benign and malignant tumors across 13 individuals from four multigenerational families and discover recurrent mutations in epigenetic modifiers DNMT3A and BCOR in 29% of benign tumors. Multi-level and microdissected sampling strikingly reveal that many clones with different DNMT3A mutations exist in these benign tumors, suggesting that intra-tumor heterogeneity is common. Integrated genomic, methylation and transcriptomic profiling in selected tumors suggest that isoform-specific DNMT3A2 mutations are associated with dysregulated methylation. Phylogenetic and mutational signature analyses confirm cylindroma pulmonary metastases from primary skin tumors. These findings contribute to existing paradigms of cutaneous tumorigenesis and metastasis.
Subject terms: Cancer genetics, Skin cancer
CYLD cutaneous syndrome (also known as Brooke-Spiegler syndrome) is characterised by germline mutations in the tumor suppressor CYLD. Here, the authors highlight recurrent mutations in DNMT3A and BCOR, indicating a role for epigenetic dysregulation in this rare genetic skin disease.
Introduction
In human skin, benign tumors outnumber malignant tumors, yet genetic studies of these are limited1. Rare inherited skin tumor syndromes such as CYLD cutaneous syndrome (CCS) offer an opportunity to address this knowledge gap and novel molecular insights into cancer can be gained. They may reveal unexpected driver mutations2, highlight mechanisms that may be targetable with repurposed drugs developed for other cancers3, or refine models of tumor growth and patterning. CCS patients develop multiple skin tumors named cylindroma, spiradenoma, and trichoepithelioma4,5, a histophenotypic spectrum of hair follicle-related tumors consistent with the hypothesis that they arise in hair follicle stem cells6,7. These tumors occur both at sun-exposed and sun-protected sites. Infrequently, salivary gland tumors, pulmonary tumors8, malignant transformation9, and metastasis with lethal outcomes can occur.
CYLD encodes a ubiquitin hydrolase enzyme involved in deubiquitination of lysine 6310,11 and Met 1-linked ubiquitin chains12,13. In CCS families, germline mutations occur within the catalytic domains of CYLD and are frequently truncating14, predicting loss of function. Loss of the wild-type parental allele (loss of heterozygosity (LOH)) of CYLD is demonstrated in the majority of inherited cylindromas, consistent with its role as a recessive cancer gene15. Genetic analysis of sporadic spiradenomas, rare in the general population, has highlighted mutations in ALPK1 and MYB overexpression16,17. Taken together with the recent findings of upregulated MYB in CCS tumors, this supports MYB as a key downstream mediator of cylindroma pathogenesis following loss of CYLD18. However, beyond these drivers, CCS tumors studied using array-based comparative genomic hybridization demonstrate a paucity of DNA aberrations, restricted to copy-neutral LOH of CYLD15, incongruent with the diverse histophenotypes seen within and across tumor samples.
Arguably, CYLD loss alone may be sufficient for tumorigenesis, via its role in negatively regulating oncogenic pathways; CYLD depletion using RNA interference first revealed its role in negatively regulating nuclear factor-κB (NF-κB) signalling10,11,19. Corroborating this, murine CYLD-knockout models develop skin papillomas following chemical carcinogenesis that demonstrate increased expression of NF-κB target genes such as cyclin D1 (CCND1) mediated by dysregulation of BCL320. Furthermore, CYLD has been shown to negatively regulate various oncogenic signalling pathways that are also relevant in hair development in embryogenesis, including Wnt21, Notch, and TGF-β6.
In humans, recurrent loss of functional CYLD is reported in diverse cancers, including myeloma22, leukemia23,24, hepatocellular carcinoma25, neuroblastoma26, and pancreatic cancer27, consistent with its role as a tumor suppressor expressed ubiquitously in normal tissues. In CCS patients, increased Wnt signalling has been shown to be an oncogenic dependency in cylindroma and spiradenoma tumors7. Histologically organized cylindroma and histologically disorganized spiradenoma represent extremes of a spectrum of histophenotype of the same tumor. Transition from cylindroma to spiradenoma is associated with loss of expression of the negative Wnt signalling regulator Dickkopf 2 (DKK2)7. DNA methylation has been suggested as a mechanism to account for loss of DKK2 in a subset of samples studied7; however, comprehensive genomic and methylomic profiling of CCS tumors has not been performed. The inability of CYLD-knockout mouse models to recapitulate the human phenotype of cylindroma tumors has further limited characterization of the genetic drivers in CCS6.
In this study, we use whole-genome sequencing (WGS) and whole-exome sequencing (WES) to delineate the mutational landscape of CCS. We demonstrate a relative paucity of mutations in benign CCS skin tumors, among which epigenetic modifiers DNA methyltransferase 3a (DNMT3A) and BCL6 co-repressor (BCOR) are recurrently mutated. Malignant tumors in CCS have distinct driver mutations to benign tumors, and we track the origin of pulmonary cylindromas to the skin using mutation signature analysis. These findings in CCS advance our understanding of cutaneous tumorigenesis, pulmonary metastases, and malignant transformation.
Results
Biallelic loss of CYLD drives CCS tumors
To delineate the genomic landscape of CCS (Fig. 1a and Supplementary Fig. 1a–d) in humans, we studied DNA from 11 fresh frozen tumors using WGS in two directly related patients who had been under clinical follow-up for 35 years (patients 1 and 2) (Fig. 1b and Supplementary Data 1). The average number of unique reads per tumor and normal sample for WGS was 374,496,607, generating 35.5 mean fold coverage for all samples. We detected on average 1381 substitutions per tumor sample (average 0.44 mutations per Mb), 72 small insertions and deletions (indels), and 1 rearrangement, using WGS. Biallelic mutations in CYLD were a recurrent driver mutation, and no MYB-NFIB fusions were found, consistent with previous studies (Fig. 1b)15. Tumors demonstrated neither recurrent structural rearrangements nor recurrent copy number aberrations (Supplementary Fig. 2).
Fig. 1.
The mutational landscape of CYLD cutaneous syndrome. a Distinct histophenotypes of benign organized cylindroma and disorganized spiradenoma seen within the same sample, a frequent finding in CCS (white scale bar = 50 μm). b Epigenetic modifiers are mutated in CCS tumors. Mutational burden is indicated in the bar graph with corresponding mutated genes shown below in the matrix. Matrix rows indicate mutated genes in each tumor and each matrix column represents a different sample (n = 42)
To validate these findings, we studied a further 31 tumors from 12 patients of 3 additional genotyped pedigrees using WES, given the lack of large structural rearrangements. We confirmed that CYLD biallelic loss was independent for each sample, reinforcing that each tumor arose independently: loss of the wild-type allele was observed either by LOH affecting 16q (31/42 tumors) or by a second mutation in CYLD (9/42), consistent with the loss of CYLD occurring across all benign and some malignant tumors in CCS.
DNMT3A and BCOR are mutated in CCS tumors
In addition to biallelic mutations in CYLD, we discovered multiple mutations in epigenetic modifiers DNMT3A (n = 6) and BCOR (n = 8) in 12 tumors (Figs. 1b, 2a, Supplementary Fig. 3a, Supplementary Table 1, and Supplementary Data 2). In two tumors, both genes were mutated. BCOR mutations have been reported to co-occur with DNMT3A in over 40% of BCOR-mutated cases of AML, and a future larger study of these tumors may offer insights as to whether there is mutational synergy in CCS tumorigenesis28. Mutations in DNMT3A were predominantly missense mutations in the methyltransferase domain, but mutations in the zinc-finger domains were also noted and have been reported previously in COSMIC (Fig. 2a)29. Mutations in BCOR were predominantly frameshift mutations. Notably, different DNMT3A and BCOR mutations were seen in disparate tumors in patients 1 and 4, suggesting that convergent evolution drives tumorigenesis through epigenetic mechanisms in this cutaneous syndrome.
Fig. 2.
Intratumoral heterogeneity of DNMT3A mutation in CCS tumors. a DNMT3A somatic mutation lollipop diagram for CCS tumors. b Spectrum of mutant variant allele fractions (VAF) of tumors in this study. c Sampling of additional, deeper slices from a single tumor (PD40537a) reveals intratumoral heterogeneity of DNMT3A mutations (tumor indicated with gray sphere, intratumoral clones with colored spheres). d Geographic sampling of distinct histophenotypes (of cylindroma and spiradenoma) within a single tumor section (PD40542e) highlights marked clonal heterogeneity particularly of DNMT3A mutations. e Protein expression of DNMT3A and Ki-67 is variable within a “cylinder” of CCS tumor and across cylinders. An adjacent outlined cylinder of cells shows loss of DNMT3A expression (white scale bar = 50 μm)
Interestingly, variant allele frequencies of DNMT3A and BCOR mutations ranged from 0.05 to 0.42—Fig. 2b), suggesting that intratumoral clonal heterogeneity may occur in these tumors. To explore this possibility, targeted deep sequencing (TDS; average coverage of >500×) of DNMT3A and BCOR was performed on additional material taken from further tissue sections of nine tumors studied above. This confirmed the presence of intratumoral heterogeneity of these putative driver mutations, with two distinct mutant clones or more found to co-occur within the same tumor in six samples (PD37330a, c, g, i, PD40536d, and PD40537a) (Fig. 2c and Supplementary Data 1).
To investigate whether DNMT3A mutational heterogeneity correlated with CCS tumor histophenotypes, we studied five tumors that contained intratumoral cylindroma and spiradenoma (Fig. 2d and Supplementary Fig. 4a, b). DNA was extracted from microdissected cylindroma and spiradenoma regions and TDS was performed. In three tumors, there was an identical DNMT3A mutation in both regions. In two tumors, there was heterogeneity between the histophenotypes, with private mutations in each regions, suggesting that multiple DNMT3A mutant clones of different sizes exist within tumors.
Mutated DNMT3A2 dysregulates methylation
To explore the functional relevance of mutations in DNMT3A and BCOR in CCS tumors, RNA-sequencing was performed in 16 tumors. This revealed increased expression of the short isoform of DNMT3A, called DNMT3A2, in 15 tumors compared to four perilesional skin controls (Fig. 3a). DNMT3A protein expression was also increased in CCS tumors compared to control skin and hair, and regions of heterogeneity were observed between islands of cylindroma (Fig. 2e, Supplementary Fig. 5a, b). It should be noted that while this confirms protein expression, a caveat of these data is that the expression of DNMT3A may not reflect mutational and functional status. BCOR was expressed at similar levels in both control and tumor tissue (Supplementary Fig. 3b).
Fig. 3.
DNMT3A2 is overexpressed in CCS tumors. a RNA-sequencing of 15 CCS tumors revealed that the short isoform of DNMT3A, DNMT3A2, is preferentially overexpressed in CCS tumors. b In a further eight CCS tumors, DNA and RNA were extracted from the same sections. Methylation profiling, followed by unsupervised clustering of the 500 most variably methylated probes revealed two clusters (DNMT3A mutant VAFs are indicated as pie charts; heatmap key demonstrates β-values; blue indicates a low β-value (hypomethylated) and red indicates a high β-value (hypermethylation). c Expression of Wnt-β-catenin target genes in the same samples demonstrate the same two clusters are distinguished by expression levels of these genes
To assess the impact of DNMT3A mutations on methylation patterns, eight samples genotyped by TDS were studied using genome-wide DNA methylation arrays. Unsupervised clustering of the 500 most variably methylated loci revealed two clusters, one comprising five tumors with DNMT3A2 isoform-specific mutations (DNMT3A2-mutated) (Fig. 3b). Comparison of these two clusters revealed 1512 differentially hypomethylated regions of contiguous probes in DNMT3A2-mutated tumors. Network analysis of these regions in DNMT3A2-mutated tumors identified the highest-ranked network to be functionally related to β-catenin (p < 1 × 10−45; Fisher’s exact test) (Supplementary Fig. 6 and Supplementary Data 3). Transcriptomic analysis of Wnt/β-catenin signalling pathway genes30 was performed on RNA extracted in parallel with DNA for the methylation analysis, as prior data in mouse skin showed DNMT3A loss is associated with dysregulation of multiple pathways including Wnt/β-catenin signalling pathway genes30 (Fig. 3c). This showed the same five tumors were distinguished as a cluster by Wnt/β-catenin target gene expression. This is an interesting preliminary finding in patient-derived tumors, and further functional studies will be needed to evaluate this association.
Malignant CCS tumors carry epigenetic modifier mutations
Malignant transformation although uncommon in CCS is well-recognized. We studied five malignant CCS tumors: basal cell adenocarcinoma-low grade (BCAC-LG), malignant spiradenocarcinoma, atypical spiradenocarcinoma, poorly differentiated adenocarcinoma, and basal cell carcinoma (BCC) (Supplementary Fig. 7)9. The case of malignant spiradenocarcinoma (PD36119a) presented at the age of 80 in patient 1. The tumor had a comparatively high number of coding substitutions (375 in the exome, corresponding to 8.4 per Mb), consisting largely of C > T transitions at CpG dinucleotides. This hypermutator phenotype has been reported previously in conjunction with germline methyl-binding domain 4 (MBD4) mutations31. Closer inspection confirmed a germline MBD4 mutation in the patient, with concomitant loss of the wild-type parental allele in the tumor. Cascade screening revealed other family members who also carried this variant (Supplementary Table 2), although their tumors did not have biallelic MBD4 loss and thus did not have the associated mutational signature. The observed burden and pattern of mutagenesis was consistent with MBD4’s role as a DNA glycosylase safeguarding the integrity of methylated CpGs from deamination. Notably, additional mutations detected included epigenetic modifiers, KDM6A and CREBBP. Tumor suppressors NOTCH2 and BAP1 were also noted to be mutated.
Poorly differentiated adenocarcinoma (PD40536c) has not been reported in CCS and presented on the breast of a female CCS patient at age 47 years. The patient had extensive staging scans, mammograms, and biopsies of breast cylindromas, and has been followed up for 3 years with no evidence of a non-cutaneous primary tumor. This tumor had mutations in TP53 and the epigenetic modifier EP300. Strikingly, this did not demonstrate LOH for CYLD. The BCAC-LG (PD40545a) tumor demonstrated a frameshift mutation in BCOR. The atypical spiradenocarcinoma (PD40540a) did not show any changes apart from CYLD LOH. The BCC (PD45044c) demonstrated a PTCH driver mutation and CYLD LOH, consistent with genetic features of BCC32. It also demonstrated the highest number of coding substitutions (1287) in our cohort, comprising the ultraviolet (UV) signature, in contrast to benign trichoepithelioma also arising on the face of the same patient. In summary, malignant tumors in CCS appear to have specific mutational patterns, and it would be interesting to determine if these tumor-specific mutations are recurrent in additional tumors in future studies.
Pulmonary cylindromas originate from the skin
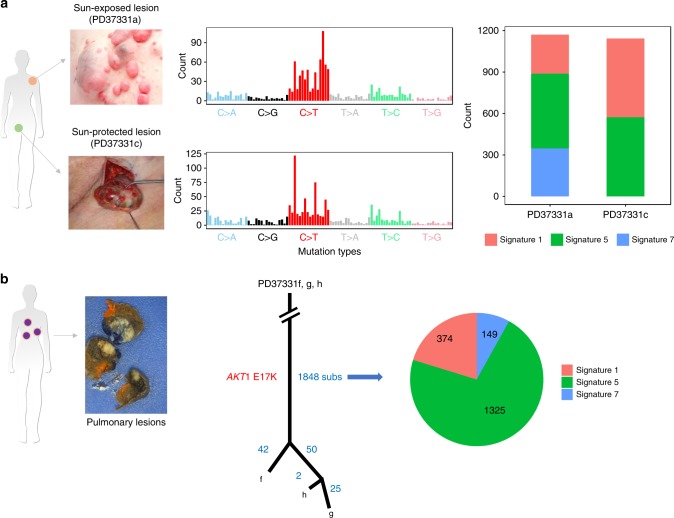
To investigate mutational mechanisms that may give rise to the mutations detected in CCS patients, we compared the mutational signatures in tumors with identical histological types at intermittently sun-exposed and typically sun-protected sites33 (Fig. 4a). Two tumors from the torso demonstrated substitution signature 7 (n = 2; PD37331a, i) consistent with UV exposure. By contrast, we did not find evidence of signature 7 and found the presence of mutational signatures 1 (associated with deamination of methylated cytosines) and 5 (unknown etiology) in sun-protected tumors from pubic and perianal sites (n = 4; PD37330c, e, g and PD37331c) and some intermittently sun-exposed tumors from the breast and torso (n = 2). We surmise that in CCS, additional mechanisms other than UV are relevant to development of skin cancer.
Fig. 4.
UV signature analysis reveals distinct mutational mechanisms in skin and tracks origin of lung tumors. a Examples of intermittently sun-exposed and sun-protected CCS tumors demonstrate differing mutational profiles. Mutational signature analysis reveals UV-related signature 7 in sun-exposed tumors only. b In one patient with three pulmonary lesions, a phylogenetic analysis reveals 1848 mutations were shared in common and showed a UV signature. Hence, these benign pulmonary lesions had a common origin, likely sun-exposed skin
We next used these data to investigate the concept of benign metastases seen in some patients with CCS, who develop multiple pulmonary cylindromas without typical features of malignancy8. We studied four pulmonary cylindromas that had benign histological features from patients 1 and 2, who were both ex-smokers. They did not have evidence of lymph node disease, hepatic, or bone metastases (Fig. 4b). Tumor phylogenetic analysis revealed that multiple pulmonary lesions from patient 2 shared 1848 substitutions, suggesting that these geographically separated lesions that seeded in the lung had a common origin. We found that the UV mutation signature 7 was present in the shared mutations, and thus tracked the origin of these pulmonary lesions to intermittently sun-exposed skin. Lastly, we found recurrent, E17K AKT1 oncogenic mutations in multiple lung cylindromas in each patient, and in both patients independently as well. This is interesting for two reasons: First, although the numbers are small, this suggests that AKT1 mutations likely arose prior to seeding in the lung. The AKT1 mutations may confer lung tissue tropism for cylindromas. Second, this recurrent E17K AKT1 mutation is clinically relevant and targetable. As drugs have been developed to target AKT1 mutations in a diverse range of solid tumors34, this finding further creates therapeutic opportunities for this limiting secondary complication of CCS. It is of interest to note that three sporadic cutaneous spiradenomas also have recently been reported to carry this identical AKT1 mutation (pulmonary status not reported) in the absence of a CYLD mutation17, suggesting that this finding may be relevant beyond CCS.
Discussion
This work delineates the mutational landscape of CCS. A strength of our study is that we have employed WGS to comprehensively profile tumors from carefully phenotyped CCS patients, where long-term clinical follow-up date is available. Our work highlights the presence of distinct DNMT3A and BCOR mutations in different tumor sites of the same patient (inter-tumor heterogeneity) and different geographic sites within the same tumor (intra-tumor heterogeneity), which suggests strong convergent evolution (Supplementary Fig. 8) towards epigenetic dysregulation in this orphan disease where no medical treatments are available. In addition, we have performed matched analysis of methylome and transcriptome data in a subset of tumors, which offers insights in the absence of transgenic mice that recapitulate the human CCS phenotype. Finally, we uncategorically demonstrate that the multiple benign pulmonary lesions in this syndrome have a clonal, cutaneous ancestral origin—reinforcing the concept of benign metastases as a clinical phenotype.
Our data support a model where DNMT3A2 isoform-specific mutations may selectively alter methylation in CCS tumors. We explored this in the context of Wnt/β-catenin pathway genes, as CCS tumor cells have a known Wnt dependency7; however, we could not conclusively prove a link between DNMT3A mutation and Wnt signalling using our models. It would be of interest to explore this potential association in mouse models in future studies, bearing in mind the caveat that existing CYLD mouse models fail to recapitulate the human phenotype of developing cylindromas. A separate limitation relating to the mutations detected in rare malignant CCS tumors is that future studies will be needed to demonstrate if the mutations found are recurrent.
Our findings may have clinical implications in the future. The AKT1 mutation we report is targetable34, and is relevant to patients with pulmonary cylindromas carrying this change. Also, due to the clinical interest in mutated epigenetic modifiers in leukemia, strategies used to target DNMT3A mutant hematological malignancies may be relevant to CCS35. The accessibility of CCS skin tumors lend themselves to direct drug delivery, which may be an attractive route avoiding systemic side effects, as suggested by the methodology of a recent early phase clinical trial in CCS3.
Materials and methods
Patients and samples
Retrospective review of the case notes and radiological data of 15 genotyped CYLD mutation carriers that were under follow-up between 1 July 2013 and 1 July 2017 was performed. Skin and lung samples were obtained from patients with signed, informed, consent, and details of samples are shown in Supplementary Data 1. The authors affirm that human research participants provided informed consent for publication of the images in Fig. 4, Supplementary Fig. 1a, and Supplementary Fig. 7. Research ethics committee approval was obtained from the Hartlepool Research Ethics Committee and North East—Newcastle & North Tyneside 1 Research Ethics Committee for this work (REC Ref: 06/Q1001/59; 08/H0906/95 + 5).
Histology and immunohistochemistry
Histological assessment was performed following standard hematoxylin and eosin (H + E) staining and in conjunction with a dermatopathologist (A.H.). Immunofluorescent labeling with antibodies against DNMT3A, β-catenin, and Ki-67 was performed7. Tissue sections from snap frozen skin tumor biopsies were fixed, blocked, and then probed overnight at 4 °C with primary antibodies. Antibodies against DNMT3A (#3598) and Ki-67 (#9449) were obtained from Cell Signalling, USA. β-Catenin antibody (#610153) was obtained from BD Transduction USA. Secondary fluorescent antibodies (Alexa Fluor #111-5451144 488-conjugated goat-anti-rabbit and #115-585-146 594-conjugated goat-anti-mouse) were applied the following day and visualized with a fluorescent microscope (Zeiss Axioimager Z2, with Apotome 2—Carl Zeiss, UK).
Whole-genome sequencing and whole-exome sequencing
DNA was extracted from 12 cases along with corresponding normal tissue and subjected to paired-end WGS on an Illumina HiSeq X Ten33,36. DNA for WES was extracted from blood and cyrosections of snap frozen tissue, and in five cases from formalin-fixed paraffin-embedded tissue (PD37330h, PD40536c, PD40540a, PD40545a, and PD40545c). Forty-two WES library samples were prepared using the Illumina Nextera DNA Exome Kit, prior to being sequenced on a S2 flowcell on an Illumina Novaseq machine. Three WES samples were enriched using the SureSelect Human All ExonV6 + UTR and 100 base paired-end sequencing performed on an Illumina Hiseq 2500 genome analyzers. For WES sequence depth was on average 255-fold. Resulting BAM files were aligned to the reference human genome (GRCh37) using Burrows-Wheeler Aligner, BWA-0.7.16a (r1181). Mutation calling was performed using CaVEMan (Cancer Variants through Expectation Maximization: http://cancerit.github.io/CaVEMan/) for calling somatic substitutions33. Indels in the tumor and normal genomes were called using a modified Pindel version 2.0 (http://cancerit.github.io/cgpPindel/) on the NCBI37 genome build. Structural variants were discovered using a bespoke algorithm, BRASS (BReakpoint AnalySiS; https://github.com/cancerit/BRASS) through discordantly mapping paired-end reads followed by de novo local assembly using Velvet to determine exact coordinates and features of breakpoint junction sequence. All mutations were annotated according to ENSEMBL version 75.
ASCAT copy number analysis
Allele-specific copy number analysis of tumors analyzed by WGS was performed using ASCAT (v2.1.1)33. ASCAT takes non-neoplastic cellular infiltration and overall tumor ploidy into consideration, to generate integer-based allele-specific copy number profiles for the tumor cells. Copy number values and estimates of aberrant tumor cell fraction provided by ASCAT were input into the CaVEMan substitution algorithm for WGS. In addition, ASCAT segmentation profiles were used to establish the presence of LOH across CYLD and relevant mutated cancer driver genes.
Identification of driver mutations
Somatic mutations present in known cancer genes (Cancer gene census https://cancer.sanger.ac.uk/census) were reviewed to identify those which were likely to be driver mutations. Mutations were deemed to be potential driver mutations if they were consistent with the type of mutations found in a particular cancer gene, that is, inactivating mutations in tumor suppressor genes (including nonsense, frameshift, essential splice site mutations, and recurrent missense) and recurrent mutations in dominant oncogenes. Recurrent mutations were determined by reference to reported mutation frequency in the COSMIC database (https://cancer.sanger.ac.uk/cosmic).
Mutational signature analysis
The contributions of substitution signatures for WGS samples were determined as follows: the substitution profile is described as a 96-channel vector. For each mutation, of which there are six substitution classes of C > A, C > G, C > T, T > A, T > C, and T > G, the flanking 5′ and 3′ sequence context is taken into account giving a total of 96 channels. A given set of mutational signatures was fitted into the mutational profile of each sample to estimate the exposure of each of the given signatures in that sample. The fitting algorithm detects the presence of mutational signatures with confidence, using a bootstrap approach to calculate the empirical probability of an exposure to be larger or equal to a given threshold (i.e., 5% of mutations of a sample). Here, we first used 30 COSMIC signatures (https://cancer.sanger.ac.uk/cosmic/signatures) to fit into each sample, and then chose the first three signatures with highest confidence, which are signature 1, 5, and 7, to do the final fitting.
For highly mutated malignant samples (the spiradenocarcinoma (PD36119a) and the BCC (PD40544c)), the mutation burden was orders of magnitude higher than other non-malignant tumors that were exome sequenced. We were able to use cosine similarity between the overall 96-channel profile and COSMIC signature to confirm the presence of particular mutational signatures in the relevant sample. The cosine similarity between each malignant sample and the suspected COSMIC signature was high: for PD36119a, cosine similarity to COSMIC signature 1 was 0.92 and for PD40544c cosine similarity to the UV light signature, COSMIC signature 7, was 0.98.
Targeted sequencing
The Truseq Myeloid panel (Illumina) was used to sequence DNMT3A and BCOR in 18 samples in accordance with the manufacturer’s protocol. A 20 pM library of the PhiX genome was added to achieve a 5% PhiX spike-in. This library was loaded onto a Miseq flowcell (600 cycles V3) for sequencing (Illumina, San Diego, CA, USA). Data were analyzed using BWA (v.0.7.15) to align reads to the reference sequence and Samtools used as a variant caller. Variant calls that passed strict filtering thresholds (“Filter” = PASS and “Qual” = 100) were included for the deep sequencing on sections in additional levels and in new samples. For five samples (PD37330k, PD37331k, PD37331m, PD40542e, and PD 40536e—Supplementary Fig. 4) where intratumoral clonal variation was studied across distinct histophenotypic regions, variant call thresholds were relaxed, and all non-synonymous variants called were confirmed by visualizing aligned read data using Integrated Genomics Viewer (IGV; v2.3). These variants were included if aligned reads supported the variant calls.
Transcriptomic analyses
RNA was extracted from 16 tumor samples and 4 control samples and stranded preparation was performed using the Illumina Stranded mRNA Kit3. Libraries were prepared and sequenced using an Illumina Hiseq 2500, giving 15 million paired-end reads per sample, which were 100 bp in length. For eight additional samples (PD37330a, c, e, k, PD40539d, e, and PD40542d, where DNA and RNA were extracted from the same cells), libraries were generated using the NEB Nextera Low Input RNA Library Prep Kit, and were sequenced using an Illumina Novaseq 6000. FASTQ files were aligned using the splice aware aligner program STAR to generate alignment files37. The read counts for each sample file were counted using the R package Subread38. Differential gene expression analysis was carried out using the package DeSeq239,40. Log-transformed count matrix values were used for heatmap generation using the gplots41 package.
Methylation assay and analysis
We assessed genome-wide DNA methylation in eight tumor samples with the Illumina Methylation EPIC microarray (Illumina, San Diego, CA, USA). DNA methylation assays were performed as per the standard manufacturer’s protocol by MWG (Aros, Denmark). Briefly, these are eight CCS tumors in which detailed analysis was performed as follows. DNA and RNA were extracted from the same cells, and mutation status of DNMT3A and methylation profiling were performed. Methylation array processing, functional normalization42, and quality control checks were implemented using the R package minfi43. Differentially methylated probes were identified using minfi. Differentially methylated regions spanning multiple probes were identified using bumphunter;44 these regions were visualized using Gviz45. When these methylation profiles were assessed, the 500 most variably methylated probes were subject to unsupervised hierarchical clustering. The study of the 500 most variable probes is an accepted approach to help distinguish methylation profiles of tumors46. A Euclidean distance matrix was constructed and hierarchical clustering was subsequently performed using the “complete” agglomeration method. The 500 probes with the highest standard deviation were selected for visualization. This analysis demonstrated that the majority of DNMT3A2-mutant tumors clustered separately from DNMT3A2 wild-type tumors (Fig. 3b). We then studied these two groups and assessed all genes related to probes that were significantly differentially methylated between these two clusters with a p value of <0.05. Network analysis of these genes using Ingenuity Pathway Analysis47 revealed networks of genes related by function that were ranked by p value (Supplementary Data 3).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are indebted to the patients and families who took part in this study. We are grateful for helpful discussions with Debbie Hicks, Nick Reynolds, Joris Veltman, and Muzlifah Haniffa. The female silhouette in Fig. 4 was designed by Freepik. N.R.’s work was supported by a Wellcome Trust funded Intermediate Clinical Fellowship-WT097163MA. N.R.’s research is also supported by the Newcastle NIHR Biomedical Research Center (BRC) and the Newcastle MRC/EPSRC Molecular Pathology Node. K.H. is supported by a Ph.D. studentship from the British Skin Foundation. H.R.D. is funded by a CRUK Grand Challenge Award (C60100/A25274) and S.N.-Z. is funded by a CRUK Advanced Clinician Scientist Award (C60100/A23916). S.N.-Z.’s research is also funded by a Wellcome-Beit Award, Wellcome Strategic Award (101126/Z/13/Z), CRUK Grand Challenge Award (C60100/A25274), and Josef Steiner Award 2019.
Author contributions
N.R., E.S., H.R.D., K.H., N.S., J.C., X.Z., S.C., S.N.-Z. and A.H. contributed to the experiments, scientific hypotheses, data analysis, and compiling of the manuscript. J.C. and A.H. contributed to the experiments and data analysis. N.R. and S.N.-Z. designed the experiments. N.R., H.R.D., S.N.-Z. wrote the manuscript.
Data availability
The WGS data have been deposited in the European Genome-phenome Archive (EGA) database under the accession code EGAD00001004573. The WES data have been deposited in the EGA database under the accession code EGAD00001005305. RNA-sequencing data have been deposited in the EGA database under the accession code EGAD00001005305. TDS data were deposited in the EGA database under the accession code EGAD00001005305. Methylation data have been deposited in the EGA database under the accession code EGAD00010001755. All the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding author upon reasonable request.
Code availability
No new code was produced for the analysis of the data in this manuscript. Details of the computer code used are included in the Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Serena Nik-Zainal, Email: snz@mrc-cu.cam.ac.uk.
Neil Rajan, Email: neil.rajan@ncl.ac.uk.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-12746-w.
References
- 1.Marino-Enriquez A, Fletcher CD. Shouldn’t we care about the biology of benign tumors? Nat. Rev. Cancer. 2014;14:701–702. doi: 10.1038/nrc3845. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson IP, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 3.Danilenko M, et al. Targeting tropomyosin receptor kinase in cutaneous CYLD defective tumors with Pegcantratinib: the TRAC randomized clinical trial. JAMA Dermatol. 2018;154:913–921. doi: 10.1001/jamadermatol.2018.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajan N, et al. Tumor mapping in 2 large multigenerational families with CYLD mutations: implications for disease management and tumor induction. Arch. Dermatol. 2009;145:1277–1284. doi: 10.1001/archdermatol.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bignell GR, et al. Identification of the familial cylindromatosis tumor-suppressor gene. Nat. Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 6.Rajan N, Ashworth A. Inherited cylindromas: lessons from a rare tumor. Lancet Oncol. 2015;16:e460–e469. doi: 10.1016/S1470-2045(15)00245-4. [DOI] [PubMed] [Google Scholar]
- 7.Rajan N, et al. Transition from cylindroma to spiradenoma in CYLD-defective tumors is associated with reduced DKK2 expression. J. Pathol. 2011;224:309–321. doi: 10.1002/path.2896. [DOI] [PubMed] [Google Scholar]
- 8.Brown SM, et al. Inherited pulmonary cylindromas: extending the phenotype of CYLD mutation carriers. Br. J. Dermatol. 2018;179:662–668. doi: 10.1111/bjd.16573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazakov DV, et al. Morphologic diversity of malignant neoplasms arising in preexisting spiradenoma, cylindroma, and spiradenocylindroma based on the study of 24 cases, sporadic or occurring in the setting of Brooke–Spiegler syndrome. Am. J. Surg. Pathol. 2009;33:705–719. doi: 10.1097/PAS.0b013e3181966762. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumor suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 11.Trompouki E, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 12.Draber P, et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 2015;13:2258–2272. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupka S, et al. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 2016;16:2271–2280. doi: 10.1016/j.celrep.2016.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy N, Farkas K, Kemeny L, Szell M. Phenotype–genotype correlations for clinical variants caused by CYLD mutations. Eur. J. Med. Genet. 2015;58:271–278. doi: 10.1016/j.ejmg.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Rajan N, et al. Dysregulated TRK signalling is a therapeutic target in CYLD defective tumors. Oncogene. 2011;30:4243–4260. doi: 10.1038/onc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehr A, et al. The MYB-NFIB gene fusion—a novel genetic link between adenoid cystic carcinoma and dermal cylindroma. J. Pathol. 2011;224:322–327. doi: 10.1002/path.2909. [DOI] [PubMed] [Google Scholar]
- 17.Rashid M, et al. ALPK1 hotspot mutation as a driver of human spiradenoma and spiradenocarcinoma. Nat. Commun. 2019;10:2213. doi: 10.1038/s41467-019-09979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajan N, et al. Overexpression of MYB drives proliferation of CYLD-defective cylindroma cells. J. Pathol. 2016;239:197–205. doi: 10.1002/path.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovalenko A, et al. The tumor suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 20.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 21.Tauriello DV, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol. Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Jenner MW, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 23.Braun T, et al. Targeting NF-kappaB in hematologic malignancies. Cell Death Differ. 2006;13:748–758. doi: 10.1038/sj.cdd.4401874. [DOI] [PubMed] [Google Scholar]
- 24.Hahn M, et al. Aberrant splicing of the tumor suppressor CYLD promotes the development of chronic lymphocytic leukemia via sustained NF-kappaB signaling. Leukemia. 2018;32:72–82. doi: 10.1038/leu.2017.168. [DOI] [PubMed] [Google Scholar]
- 25.He G, Karin M. NF-kappaB and STAT3—key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T, Masoumi KC, Massoumi R. Deubiquitinating activity of CYLD is impaired by SUMOylation in neuroblastoma cells. Oncogene. 2015;34:2251–2260. doi: 10.1038/onc.2014.159. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 28.Tiacci E, et al. The corepressors BCOR and BCORL1: two novel players in acute myeloid leukemia. Haematologica. 2012;97:3–5. doi: 10.3324/haematol.2011.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinaldi L, et al. Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-gamma. Elife. 2017;6:1350. doi: 10.7554/eLife.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues, M. et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat. Commun.9, 1–6 (2018). [DOI] [PMC free article] [PubMed]
- 32.Bonilla X, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 33.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyman DM, et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. J. Clin. Oncol. 2017;35:2251–2259. doi: 10.1200/JCO.2017.73.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rau RE, et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood. 2016;128:971–981. doi: 10.1182/blood-2015-11-684225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warnes, G. R., Bolker, B., Bonebakker, L. & Gentleman, R. gplots: various R programming tools for plotting data. R package version 3.0. 1. 2016. The Comprehensive R Archive Network (2016). https://cran.r-project.org/web/packages/gplots.
- 42.Fortin JP, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaffe AE, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahne, F. & Ivanek, R. in Statistical Genomics: Methods and Protocols (eds. Mathé, E. & Davis, S.) 335–351 (Springer, New York, 2016).
- 46.Schwalbe EC, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18:958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The WGS data have been deposited in the European Genome-phenome Archive (EGA) database under the accession code EGAD00001004573. The WES data have been deposited in the EGA database under the accession code EGAD00001005305. RNA-sequencing data have been deposited in the EGA database under the accession code EGAD00001005305. TDS data were deposited in the EGA database under the accession code EGAD00001005305. Methylation data have been deposited in the EGA database under the accession code EGAD00010001755. All the other data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding author upon reasonable request.
No new code was produced for the analysis of the data in this manuscript. Details of the computer code used are included in the Methods section.