Abstract
MYCN-amplified (MNA) neuroblastoma is an aggressive neural crest-derived pediatric cancer. However, MYCN is indispensable for development and transcriptionally regulates extensive network of genes. Integrating anti-MYCN ChIP-seq and gene expression profiles of neuroblastoma patients revealed the metabolic enzymes, MTHFD2 and PAICS, required for one-carbon metabolism and purine biosynthesis were concomitantly upregulated, which were more susceptible to metastatic neuroblastoma. Moreover, we found that MYCN mediated the folate cycle via MTHFD2, which contributed one-carbon unit to enhance purine synthesis, and further regulated nucleotide production by PAICS in response to cancer progression. Dual knockdown of the MYCN-targeted gene pair, MTHFD2 and PAICS, in MNA neuroblastoma cells synergically reduced cell proliferation, colony formation, migration ability, and DNA synthesis. By systematically screening the compound perturbagens, the gene expression levels of MTHFD2 and PAICS were specifically suppressed by anisomycin and apicidin across cell lines, and our co-treatment results also displayed synergistic inhibition of MNA neuroblastoma cell proliferation. Collectively, targeting a combination of MYCN-targeted genes that interrupts the interconnection of metabolic pathways may overcome drug toxicity and improve the efficacy of current therapeutic agents in MNA neuroblastoma.
Subject terms: Metabolomics, Paediatric cancer, Cell growth, Target identification, Transcriptomics
Introduction
Neuroblastoma is an embryonal tumor derived from the sympathetic nervous system and accounts for 15% of pediatric cancer mortality1,2. Although the outcome of high-risk patients has improved on the use of therapy, the majority eventually relapses and develops resistance with a 5-year event-free survival <50%3,4. High-risk (HR) neuroblastoma is characterized by V-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) amplification and segmental DNA copy number aberrations which are strongly correlated with poor prognosis, metastasis, and even treatment failure5–9. MYCN is a transcription factor that governs downstream signaling pathways for a broad range of fundamental processes during embryogenesis and development10,11. Amplification of MYCN is associated with tumor initiation and progression, and occurs in ~20% of all neuroblastoma cases, with a higher incidence in INSS stages 3 and 4 compared to stages 1, 2, and 4S, suggesting a contribution in regulating oncogenic pathways3,12,13.
Alteration of cellular metabolism to maintain energy status for rapid cell progression has been considered as a feature of cancer cells14,15. MYCN has been closely tied to the regulation of neuroblastoma cell growth, and confers the serine-glycine-one-carbon pathway to promote metabolic reprogramming in HR neuroblastoma16,17. MYCN-amplified (MNA) neuroblastoma increases cell growth by glutamine transport and metabolism enhancement, thus depletion of glutamine causes the limitation of TCA cycle intermediates and eventually cell death18. Moreover, MYCN has been shown to stimulate mitochondrial biogenesis of β-oxidation and glucose metabolism in cancer progression19,20.
Cellular metabolism relies on highly regulated sequential enzyme-catalyzed reactions to acquire the essential components such as purines to generate DNA and RNA molecules21. Other than nucleotide formation, purines are the constituents of important biomolecules as energy and signal transduction cofactors to drive cellular processes22. Interestingly, a co-localization of purinosomes with mitochondria was found under purine-depleted environment suggesting a functional interaction between mitochondria and purine biosynthesis23. The mitochondrial folate enzyme methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) is the most consistently overexpressed metabolic gene in cancer which plays a pivotal role in regulating the release of formate into cytoplasm24. In addition, serine is a precursor for the synthesis of glycine in one carbon pool by folate pathway at the mitochondria compartment which is further used up for purine biosynthesis25.
Collectively, we aimed to explore the relationship of MYCN-mediated pathways by integrating ChIP-seq and gene expression profiles to comprehensively dissect potential MYCN-targeted genes. Here, we elucidated that MNA neuroblastoma favors one carbon metabolism and purine biosynthesis by the up-regulation of MYCN-targeted metabolic genes MTHFD2 and PAICS. Our results also indicated that dual knockdown of MTHFD2/PAICS significantly reduced cell proliferation, colony formation, and migration which might diminish the aggressiveness and tumor progression ability in MNA neuroblastoma. Furthermore, combining the compound perturbagens to MYCN target gene pair displayed synergistic effects in MNA neuroblastoma cells which provides therapeutic opportunities for children with neuroblastoma.
Materials and methods
Differential expression analysis
Microarray datasets of neuroblastoma samples were acquired from NCBI Gene Expression Omnibus (GEO) with accession number GSE45547, GSE3446, GSE19274, GSE16254, GSE12460, and GSE16237, including 1065 neuroblastoma. For multiple probes corresponding to a single gene, median expression value of those probes was taken. Total 11,939 common genes of six datasets were considered for downstream analysis. The expression values were log transformed and normalized by quantile normalization across all samples. HR neuroblastoma (age at diagnosis >18 months and stage 4) were classified into HR-MNA (n = 67) and HR-non-MNA (n = 156) groups based on the MYCN status. Significance Analysis of Microarrays (SAM) was used to identify differentially expressed genes between HR-MNA and HR-non-MNA with false discovery rate (FDR) <0.00126.
Public data sources and bioinformatics analysis
MYCN-bound genes were obtained from our previous work27 which ChIP-seq was used for genome-wide identification of MYCN regulatory networks. Two independent neuroblastma cohorts (SEQC and TARGET) were used for survival and correlation analyses. SEQC cohort was download from GEO with accession number GSE47792 and TARGET cohort was queried via GDC data portal (https://portal.gdc.cancer.gov/). The H3K4me3 and H3K27ac epigenetic profiles were obtained from ENCODE project. KEGG enrichment analysis was performed using the R/Bioconductor package clusterProfiler28.
Cell culture
Human neuroblastoma cell lines SK-N-DZ (CRL-2149), SK-N-SH (HTB-11), SK-N-BE(2)-C (CRL-2268) were obtained from ATCC. SH-SY5Y, SK-N-AS, and SK-N-FI neuroblastoma cell lines were obtained from Dr. Yung-Feng Liao (Academia Sinica, Taipei, Taiwan). The conditional MYCN-expressing SHEP Tet21N cell line (RRID, CVCL_9812) was cultured in the presence of 1 μg/ml of tetracycline (Sigma) for 48 h and the Tet21N control cells were cultured in the presence of 1 μg/ml of 70% ethanol. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco) with 10% fetal bovine serum (Biological Industries) at 37 °C in a humidified incubator with 5% CO2.
RNA isolation of neuroblastoma cells and patient tissues, and cDNA synthesis
Neuroblastoma cells and tissue samples were homogenized by TRIzol reagent (Invitrogen). Total RNA of cells and tissue samples were prepared by Direct-zol™ RNA MiniPrep kit (ZYMO RESEARCH). RNA concentration and quality were determined by NanoDrop ND-1000 (NanoDrop Technologies). In all, 100 ng of total RNA of each tissue sample or 500 ng of total RNA template of each neuroblastoma cell line sample was reverse transcript to cDNA by using RevertAidTM H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) with oligo (dT) 18 primer. The cDNA samples were stored at −30°C until use. The 21 neuroblastoma tissues were kindly provided by Dr. Wen-Ming Hsu (National Taiwan University Hospital, Taipei, Taiwan; IRB number 201407043RINC).
DNA manipulation and plasmid construction
The MYCN gene was amplified from synthesized cDNA as described previously (Thermo Fisher Scientific). PCR was performed to generate pCMV6-XL4 plasmids (Invitrogen) with a full-length sequence of MYCN. The resulting plasmid (pCMV6-XL4-MYCN) was transformed into Escherichia coli (E. coli) strain DH5α and selected by ampicillin. The plasmid DNA (pCMV6-XL4-MYCN) was prepared and sequenced at the DNA Sequencing Facility (Genomics BioSci. & Tech., Taipei, Taiwan). In all, 2.5 × 105 SK-N-AS cells were seeded 24 h before transfection in a 6-well plate using Lipofectamine 3000 (Invitrogen). Transiently transfected cells were harvested at 48 h post-transfection.
RNA interference
The non-target siRNA or MYCN-siRNA (SMARTpool, Dharmacon, USA) were transfected into SK-N-DZ or SK-N-BE(2)-C cells to generate transient silencing of MYCN using Lipofectamine RNAiMAX (Invitrogen). In all, 4 × 105 SK-N-DZ or SK-N-BE(2)-C cells were seeded on six-well plates 24 h before transfection, and harvested at 48 h post-transfection.
qRT-PCR analysis
The cDNA sample was amplified and applied by using CFX Connect™ Real-Time PCR Detection System (Bio-Rad Laboratories). The mRNA expression values were measured by ΔΔCt and normalized to GAPDH. The qRT-PCR primer sequences were listed in Supplementary Table S1.
Western blotting
Cells were lysed using lysis-C buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS and 0.0002% (v/v) bromophenol blue] containing protease inhibitor (Bioman). The cells were homogenized on ice using an ultrasonic homogenizer (LABSONIC M ultrasonic homogenizer) with 60% amplitude and 0.6 cycle duration for 2 min. Cell lysate was centrifuged at 16,000 × g for 30 min at 4 °C. The supernatants were collected and measured protein concentrations with protein assay dye reagent (Bio-Rad Laboratories). Protein extracts were separated by SDS-PAGE and transferred onto a PVDF membrane (Millipore) and immunoblotted with antibodies. The membrane was blocked in 5% non-fat milk/PBST and incubated overnight with primary antibody diluted in blocking buffer at 4 °C: mouse anti-MYCN (abcam; 1:1000), rabbit anti-MTHFD2 (Genetex; 1:1000), rabbit anti-PAICS (Genetex; 1:1000), mouse anti-β-actin (Millipore; 1:5000), and mouse anti-α-tubulin (Genetex; 1:1000). The membrane was then treated with secondary HRP-conjugated antibody anti-rabbit or anti-mouse IgG (Sigma-Aldrich; 1:100,000) for 2 h at room temperature. Images were acquired using ECL substrate (BioRad) and FluorChem M (ProteinSimple).
Luciferase reporter assay
Promoter regions of the MTHFD2 and PAICS genes were amplified using PCR and cloned into the pGL4.18 vector (Promega) flanked with NheI and HindIII sites. The sequences of the promoter region primers are listed in Supplementary Table S2. SK-N-AS cells were seeded at 2.5 × 105 per 6-well plate for 24 h. Then SK-N-AS cells were co-transfected with either 500 ng of MTHFD2, PAICS promoter luciferase reporters or pGL4.18 empty vector along with 10 ng of pGL4.74 Renilla luciferase plasmid DNA together with 500 ng of MYCN expression plasmid (pCMV6-XL4-MYCN) or control vector (pCMV6-XL4). At 5 h post-transfection, cells were recovered in completed DMEM for 1 h and then cells were maintained in completed DMEM containing 1 μl/ml 70% ethanol or 1 μg/ml tetracycline and incubated for 48 h. At 48 h post-transfection, cells were lysed with passive lysis buffer for 15 min at room temperature and the firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer’s instructions.
Generation of cell lines with stable knockdown of MTHFD2 and PAICS
SK-N-DZ cells were seeded at 4 × 105 cells per 6-well plate for 24 h, and then transfected with 2 μg shRNA plasmid (RNAi core, IBMS, Academia Sinica, Taipei, Taiwan) which inhibited MTHFD2 (shMTHFD2 #50 and #53), PAICS (shPAICS #74 and #75), MTHFD2/PAICS (shMTHFD2/PAICS) or LacZ (shRNA control) by lipofectamine 3000 (Invitrogen). Transfected cells were subsequently selected on 2 μg/ml puromycin to create the stable shRNA line. Stable cell subcultures were kept in DMEM medium containing 2 μg/ml puromycin (Supplementary Table S3).
Cell harvest and extraction for targeted metabolomics assay
Cells were grown in 15-cm culture dish, during which the medium was replaced every day (DMEM supplemented with 10% fetal bovine serum and 2 μg/ml puromycin for stable clones) at 37 °C with 5% CO2 before extraction. Cells were collected at 80% confluence and rapidly rinsed with warm 0.9% NaCl isotonic saline three times before quenching. Then, 1 ml of ice cold water was added and flash frozen in liquid nitrogen and detached using a cell scraper. Cell suspension were lysed by freeze-thaw cycles twice, followed by sonication. Protein concentration were measured by BCA Protein Assay Kit for normalization. Metabolism was quenched and metabolites were extracted by 75:25 methanol:water extraction buffer. In brief, 900 μl of −20 °C methanol was add to 300 μl cell lysate and vigorously vortexed and then centrifuged at 14,000 × g for 20 min at 4 °C. The supernatants were collected and dried using a centrifugal evaporator and stored at −80 °C prior to analysis29.
Instrumentation, method development, and data analysis for targeted metabolism analysis
The complete LC-MS platform consists of HPLC system (Agilent 1260 series, Germany) equipped with a time-of-flight mass spectrometer (micrOTOF-QII, Bruker Daltonik, Bremen, Germany), controlled by Bruker Daltonics Hystar 3.2 software. Liquid chromatography separation was achieved on a hypersphere C18 column (250 mm × 4.6 mm, 5 μm particle size, YMC, America), using reversed phase chromatography with the ion pairing agent tributylamine in the aqueous mobile phase to enhance retention and separation. The total run time was 30 min and the injection volume was 20 μl. Every dried sample was dissolved in 400 μl solution which contained H2O:ACN:MeOH at a ratio of 1:1:2 with 1% formic acid (FA). The flow rate was 500 μl/min. Solvent A was 3% methanol with 10 mM tributylamine and 15 mM acetic acid; solvent B was 100% acetonitrile/0.1% FA. The gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 20 min, 90% B; 25 min, 90% B; 25.1 min, 0% B; and 30 min, 0% B.
An electrospray ionization interface was used to direct column eluent to the mass spectrometer. Mass spectra in the range m/z 50–500 were obtained by electrospray ionization in negative-ion mode. Initial instrument optimization was performed by infusing a mixture of L-serine (m/z 104.0288; Cat. No.: SI-S4500, Sigma-Aldrich), AICAR (m/z 257.0790; Cat. No.: SI-A9978, Sigma-Aldrich), IMP (m/z 347.0398; Cat. No.: SI-I4625, Sigma-Aldrich), and GMP (m/z 362.0507; Cat. No.: SI-G8377, Sigma-Aldrich), using a syringe pump (KdScientific, Holliston, MA). Various instrumental settings were optimized to maximize the signal with the final parameters as follow: gas temperature 200 °C, drying gas flow rate 8.0 L/min, nebulizer gas pressure 4.0 bar, and capillary and endplate offset potentials were 2500 V and 500 V, respectively.
Cell cycle analysis
Stable shRNA expression cells were collected, fixed in 70% ethanol, and stored at −20 °C overnight. The cells were washed with PBS and resuspended in PBS containing 100 μg/ml RNase A and 0.1% Triton X-100, then incubated at 37 °C for 1 h. Cells were stained with 5 μg/ml propidium iodide (Santa Cruz). The DNA content of the cells was analyzed using a FACSCanto instrument (BD Biosciences Immunocytometry Systems). Ten thousand cells were collected for each measurement. The percentage of cells in different phases of the cell cycle was analyzed using ModFit LT (Verity Software House).
Cell proliferation, colony formation, and migration assays
For MTS analysis, stable shRNA expression cells (shLacZ, shMTHFD2#50, shMTHFD2#53, shPAICS#74, shPAICS#75, and shMTHFD2/PAICS) were seeded in 96-well plate at 4000 cells per well with DMEM containing 2 μg/ml puromycin. At each 24 h, 48 h, 72 h time point, 20 μl of MTS/PMS (Promega) were added and incubated for 2 h at 37 °C with 5% CO2. The cell viability was determined by OD 490 nm using an ELISA reader (BioRad). For colony formation assay, the stable shRNA expression cells were seeded in 6-well plates (600 cells/well) with DMEM containing 2 μg/ml puromycin and incubated for 2 weeks. After 2 weeks, the colonies were fixed with methanol overnight and then stained by 4% GIEMSA stain for 1 h and analyzed by ImageJ. For cell migration, 3 × 104 stable shRNA expression cells with 1% FBS medium were loaded into the inserts, and medium containing 10% FBS was loaded into the lower compartments of an 8-μM pore size Transwell plate (Corning). The cells were incubated at 37 °C with 5% CO2 for 6 h, then fixed for 30 min with 100% methanol and stained with 1% GIEMSA for 30 min. Cotton swabs were used to remove cells from the upper side of the inserts. Images of five different microscope fields of each insert were captured and counted.
Identification of potential compounds suppressing MTHFD2 or PAICS expression
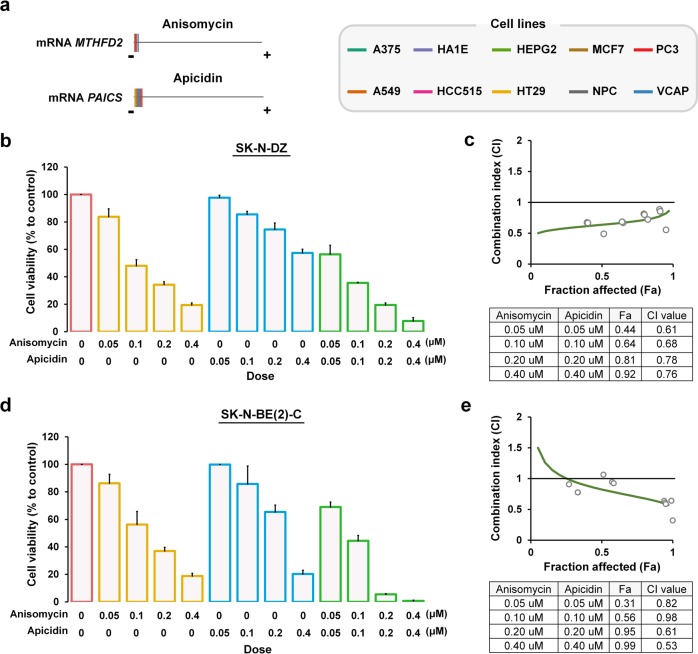
Perturbation datasets corresponding to the treatment of various drugs or compounds for ten cell lines, including A375, A549, HA1E, HCC515, HEPG2, HT29, MCF7, NPC, PC3, and VCAP, were obtained from the Library of Integrated Network-based Cellular Signatures (LINCS) project30. To identify the compounds that significantly suppress the gene expressions of MTHFD2 and PAICS based on the perturbation datasets, computational approach was performed as detailed in our previous methods31,32. Briefly, the expression changes of all measured transcripts (n = 12,494) were ordered from the most downregulated (negative) to the most upregulated (positive) cross ten cell lines and assessed if the gene of interest was significantly enriched on the down-regulation edge cross all cell types under the perturbation of the given compound. Finally, the expressions of MTHDF2 and PAICS were found to be suppressed by anisomycin and apicidin, respectively.
Drug combination assay
SK-N-DZ, SK-N-BE(2)-C, SK-N-AS, or SK-N-SH neuroblastoma cells were seeded at 5000 cells per well in 96-well plate for 24 h before drug treatments. To prepare the stock solution, anisomycin (CAS No.: 22862-76-6; Selleckchem) and apicidin (CAS No.: 183506-66-3; ApexBio) were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) to have a final concentration of 0.1, 0.2, and 0.4 µM respectively. To test the synergistic effect of anisomycin and apicidin combination, the corresponding combination solutions were prepared proportionally at 1:1 molar ratio. The control groups received an equivalent amount of DMSO as corresponding treatment groups. At the end of 24 and 48 h of treatments, 20 μl of MTS/PMS (Promega) were added and incubated for 2 h at 37 °C with 5% CO2. The cell viability was determined by OD 490 nm using an ELISA reader (BioRad) and normalized with the corresponding control groups. Drug synergy was evaluated using CompuSyn (http://www.combosyn.com) to calculate the combination index (CI) values: synergism (CI < 0.9), additive effect (CI = 0.9–1.1), and antagonism (CI > 1.1)33.
Cell apoptosis assessment
In all, 1 × 106 SK-N-DZ or SK-N-BE(2)-C cells were seeded in a 10-cm culture dish for 24 h before drug treatments. For apoptosis detection, 0.2 µM anisomycin, 0.2 µM apicidin, and the combination of solutions were prepared proportionally at 1:1 molar ratio. At the end of 24 h of treatment, cells underwent PBS washes before treated with the FITC Annexin V Apoptosis Detection Kit I (BD Pharmingen, USA) according to the manufacturer’s instructions. Ten thousand stained cells were acquired on a FACSCanto instrument (BD Biosciences Immunocytometry Systems) for each measurement.
Statistical analysis
Results were expressed as the mean ± standard deviation (SD). The Student’s t-test was used to analyze differences between two groups. The differences between groups were considered to be statistically significant when P < 0.05. All experiments were performed in triplicate.
Results
Integrated analysis of multi-layer omics data reveals higher cellular metabolism in MNA neuroblastoma
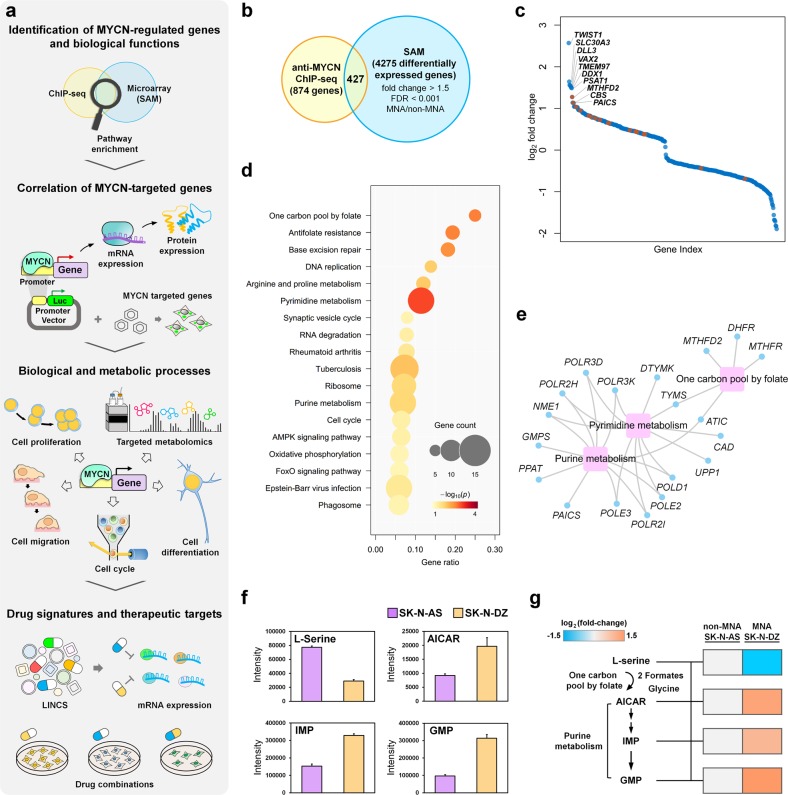
To identify highly confident MYCN-regulated genes, we combined genes that differentially expressed between MYCN status with MYCN-bound genes (Fig. 1a). Here, we compiled a neuroblastoma gene expression cohort from six independent studies and identified 4275 differentially expressed genes between HR-MNA and HR-non-MNA samples. By integrating with 874 MYCN-bound genes that identified in our previous anti-MYCN ChIP-seq experiment27, of which 427 common genes were found to be regulated by MYCN (Fig. 1b and Supplementary Table S4), and we termed these genes as potential MYCN-regulated genes. A few of these genes have been verified as direct transcriptional targets of MYCN, such as TWIST1, DLL3, and DDX1 (Fig. 1c)34–36. The functional enrichment analysis of the potential MYCN-regulated genes revealed cancer hallmark-associated biological processes such as cell cycle and metabolic pathways (Fig. 1d and Supplementary Table S5).
Fig. 1. Integrative analysis of anti-MYCN ChIP-seq and gene expression profile of neuroblastoma patients.
a Schematic representation of the experimental design. b Venn diagram showing the intersection of MYCN-bound genes of anti-MYCN ChIP-seq and differentially expressed genes (between HR-MNA and HR-non-MNA samples). c Scatter plot revealing fold-change expression rank of the 427 MYCN-regulated genes. Metabolic genes that illustrate in gene-pathway association networks (e) are highlighted in red. d The KEGG pathway enrichment of the 427 MYCN-regulated genes. The color and size of the node represent the p-value and the number of genes mapped to the indicated pathways, respectively. e The MYCN-regulated gene-pathway association networks, where the blue nodes and magenta rectangles correspond to genes and KEGG pathways, respectively. The gray lines represent the genes participating in the indicated pathways. f Targeted metabolomics assay was performed to measure the relative levels of L-serine, nucleoside intermediate (AICAR), and nucleoside monophosphates (IMP and GMP) between MNA SK-N-DZ and non-MNA SK-N-AS neuroblastoma cell lines; bar plot showing the intensity of each metabolite measured in technical replicates. g Relative abundance of metabolite levels in SK-N-DZ were normalized to SK-N-AS cells. The Log2 fold-change of L-serine, AICAR, IMP, and GMP are −1.47, 1.31, 1.00, and 1.49, respectively. At least two technical repeats for each of two biological replicates were performed
Previous studies have revealed that amplification of MYCN increases neuroblastoma cell growth through glutathione biosynthesis and glutamine uptake, thus suggesting MYCN reprograms neuroblastoma metabolism11,37,38. In our analysis, one carbon pool by folate, pyrimidine metabolism and purine metabolism were significantly altered by MYCN (Fig. 1e). One carbon pool by folate metabolism regulates the conversion of serine to glycine in mitochondria and releases the production of folate to cytosol for purine synthesis. Interestingly, a co-localization of purinosome and mitochondria was also reported under purine-depleted condition25. To investigate whether MNA neuroblastoma favors one carbon metabolism and purine metabolism, we first assessed the intrinsic differences of MNA and non-MNA neuroblastoma cells for serine consumption and nucleoside production. Targeted metabolic assay was performed to quantify the relative levels of serine, nucleoside intermediate (AICAR), and nucleoside monophosphate (IMP and GMP) between MNA SK-N-DZ and non-MNA SK-N-AS cell lines (Fig. 1f and Supplementary Fig. 1). The levels of AICAR, IMP, and GMP were significantly higher in MNA neuroblastoma compared to non-MNA neuroblastoma cells, suggesting MNA neuroblastoma may require higher demand in purine biosynthesis for rapid cell proliferation (Fig. 1g). In addition, we observed a lower level of serine in MNA neuroblastoma cells which could be converted into glycine in one carbon pool by folate pathway (Fig. 1g). Collectively, these data suggest that MYCN may play a role in regulating metabolism in neuroblastoma through potential MYCN-regulated genes.
MTHFD2 and PAICS expressions are strongly correlated to MYCN status in neuroblastoma
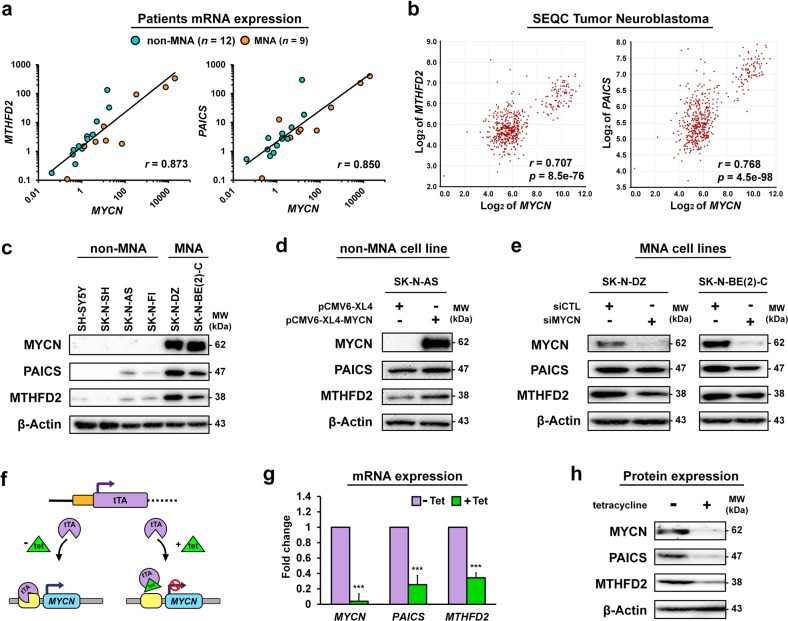
We speculate that high levels of cellular metabolism are linked to neuroblastoma progression, thus two highly expressed metabolic genes MTHFD2 and PAICS among one carbon pool by folate and purine metabolism pathways in MNA neuroblastoma prompted us to further examine their relationship with MYCN status (Fig. 1a, c, e). We first evaluated the mRNA expression levels of MTHFD2, PAICS, and MYCN in 21 tissue samples from neuroblastoma patients (MNA: n = 9; non-MNA: n = 12). The mRNA expressions of MTHFD2 and PAICS were positively correlated to MYCN by qRT-PCR with a Pearson correlation coefficient (r) of 0.873 and 0.850 respectively (Fig. 2a). In consistent with our finding, the SEQC cohort results showed that mRNA expressions of MTHFD2 and PAISC were positively correlated with MYCN, and highly expressed in MNA patients (Fig. 2b and Supplementary Fig. 2). Next, we assessed the protein expression levels in six neuroblastoma cell lines, where the MNA cell lines had markedly higher endogenous expressions of MTHFD2 and PAICS compared to non-MNA cell lines (Fig. 2c and Supplementary Fig. 3).
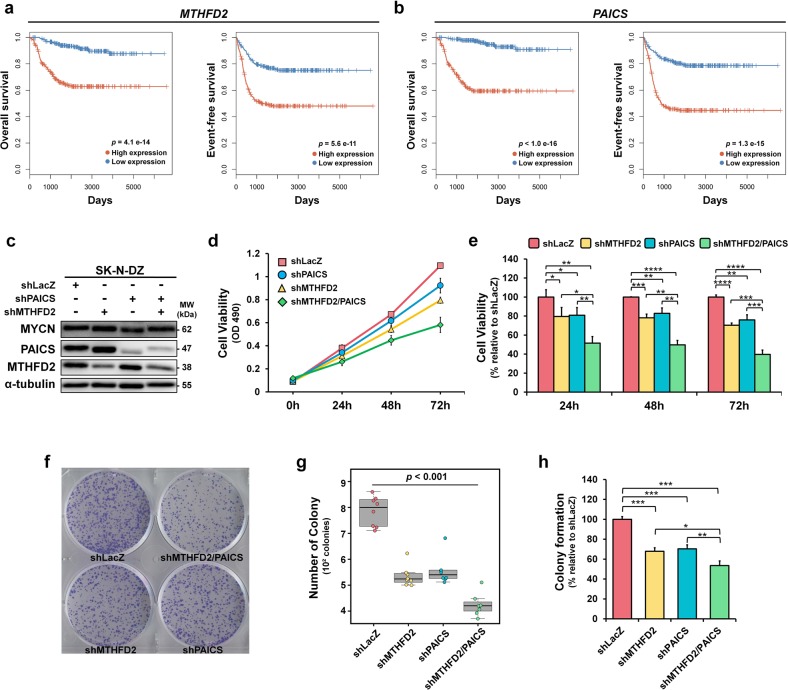
Fig. 2. Positive correlation of MTHFD2 and PAICS with MYCN in neuroblastoma.
a The MYCN, MTHFD2, and PAICS mRNA expression levels of MNA (n = 9) and non-MNA (n = 12) tissue samples from neuroblastoma patients were assessed by qRT-PCR analysis. MTHFD2 and PAICS were positively correlated to MYCN amplification with r = 0.873 and 0.850, respectively. b Correlation of MTHFD2 and PAICS to MYCN mRNA levels was assessed using the SEQC-NB498 dataset from R2 platform (http://r2.amc.nl/). c The results from western blot analyses demonstrating a positive correlation of MTHFD2 and PAICS with MYCN expression in six neuroblastoma cell lines. d Non-MNA SK-N-AS cells were transfected with pCMV6-XL4-MYCN, both PAICS and MTHFD2 were elevated along with MYCN. e MNA SK-N-DZ and SK-N-BE(2)-C cells were transfected with small interfering RNA (siRNA) to silence MYCN. Protein expression of MYCN was effectively suppressed where PAICS and MTHFD2 were concomitantly downregulated. f The Tet-21N cells express MYCN in the absence of tetracycline, where the addition of tetracycline inhibits tTA to bind to the promoter of MYCN thus gene expression is turned off. g, h After 48 h of tetracycline treatment, both MTHFD2 and PAICS were significantly downregulated following MYCN depletion at mRNA (g) and protein levels (h). The relative mRNA expressions were calculated by qRT-PCR analysis, and normalized to GAPDH gene. ***P < 0.001
To further confirm the regulation of MYCN with MTHFD2 and PAICS, we assessed the expression response upon MYCN perturbation in MNA and non-MNA cell lines. Overexpression of MYCN was introduced by pCMV6-XL4-MYCN in non-MNA SK-N-AS cells, both PAICS and MTHFD2 were elevated along with MYCN (Fig. 2d). Contrastingly, silencing MYCN in MNA SK-N-DZ and SK-N-BE(2)-C cells using small interfering RNA (siRNA) revealed that PAICS and MTHFD2 expressions were concomitantly downregulated with MYCN (Fig. 2e). Moreover, the tetracycline inducible system was also applied to evaluate the effects on neuroblastoma. The Tet-21N cells express MYCN in the absence of tetracycline, whereas the addition of tetracycline inhibits the tTA to bind to the promoter of MYCN thus gene expression is turned off (Fig. 2f). After 48 h of tetracycline treatment, both MTHFD2 and PAICS were significantly downregulated following MYCN depletion at mRNA and protein levels indicating that MYCN might influence the transcription efficiency of MTHFD2 and PAICS (Fig. 2g, h).
MYCN transcriptionally targets MTHFD2 and PAICS in neuroblastoma
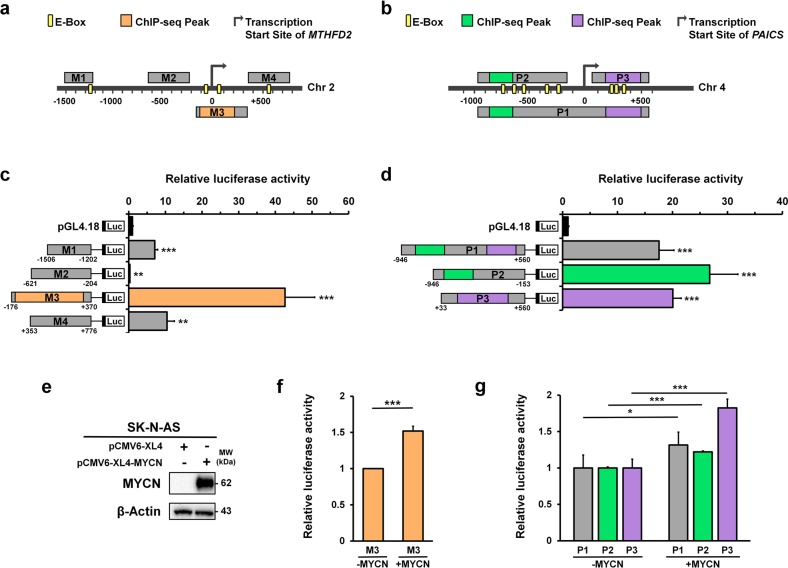
Our previous anti-MYCN ChIP-seq data revealed that MTHFD2 and PAICS had significant MYCN binding signals comparing with IgG control (Supplementary Fig. 4). To examine whether MYCN directly drives MTHFD2 or PAICS, we applied a dual luciferase reporter assay to determine the MYCN binding sites on MTHFD2 and PAICS promoters (Fig. 1a). According to the ChIP-seq results and examination of consensus E-box motifs, four MTHFD2 (M1, M2, M3, and M4) and three PAICS (P1, P2, and P3) promoter sequences were constructed using the pGL4.18 luciferase vector and co-transfected with pCMV6-XL4-MYCN in MNA SK-N-AS cell lines (Fig. 3a, b). The luciferase activity of M1, M3, and M4 were significantly enhanced, of which M3 that consisted the MYCN ChIP binding peaks resulted the highest activity with a 42‑fold change relative to the empty pGL4.18 vector (Fig. 3c). On the other hand, P1, P2, and P3 constructs containing the ChIP-seq binding peaks were increased by the presence of MYCN vector compared to empty pGL4.18 (Fig. 3d). Moreover, we compared the MYCN overexpressed SK-N-AS with SK-N-AS control (Fig. 3e), and also observed that the binding sequences of MTHFD2 and PAICS constructs were significantly upregulated by MYCN expression (Fig. 3f, g).
Fig. 3. MTHFD2 and PAICS are the MYCN target genes.
a Schematic representation of the four MTHFD2 DNA fragments including M1: the most upstream of transcription start site (TSS) containing E-box; M2: the upstream of TSS without E-box; M3: located across the up- and down- stream of TSS and overlapped with MACS_peak_36001 containing E-boxes; and M4: the downstream of TSS with a E-box. b Schematic representation of the three PAICS DNA fragments including P1: located across the up- and down- stream of TSS which contained both MACS_peak_50055 and MACS_peak_50056; P2: upstream of TSS with MACS_peak_50055; and P3: downstream of TSS with MACS_peak_50056. c, d Luciferase reporter assay was performed to measure the relative luciferase activity of MTHFD2 (c) and PAICS (d) constructs. e–g Non-MNA SK-N-AS cells were transfected with pCMV6-XL4 or pCMV6-XL4-MYCN. e The western blot analyses demonstrating the efficiency of transfection. f, g MYCN overexpressed SK-N-AS and SK-N-AS control cells that transfected with MTHFD2 and PAICS constructs indicating the binding sequences of MTHFD2 (f) and PAICS (g) are significantly upregulated by MYCN expression. *P < 0.05; **P < 0.01; ***P < 0.001
MTHFD2 and PAICS regulate purine biosynthesis in MNA neuroblastoma
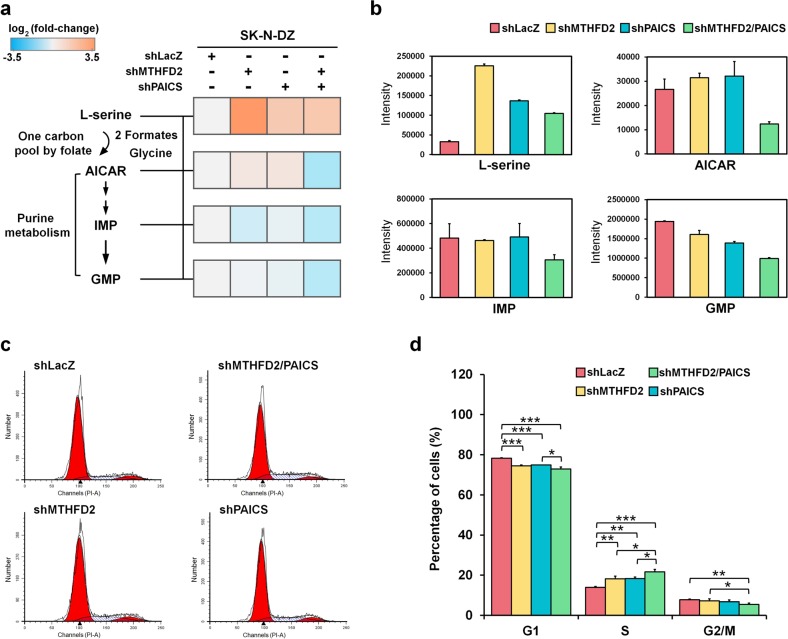
To explore the impact of the two metabolic enzymes on MNA neuroblastoma, we first constructed stable knockdown of LacZ (control), MTHFD2, PAICS, and MTHFD2/PAICS (dual-knockdown) by delivering shRNA hairpins into MNA SK-N-DZ cells followed by puromycin selection (Supplementary Fig. 5). In one carbon pool by folate pathway, serine is catabolized in mitochondria to produce formaldehyde, and then transforms into essential molecule, 10-formyl-THF, for purine biosynthesis by MTHFD239. To investigate the relationship of one carbon pool by folate and purine biosynthesis in neuroblastoma, we analyzed the relative abundance of metabolites using liquid chromatography mass spectrometry (Fig. 1a and Supplementary Fig. 6). We found shMTHFD2, shPAICS, and shMTHFD2/PAICS SK-N-DZ cells decreased the consumption of serine, and this observation implies that knockdown of either MTHFD2 or PAICS suppresses the metabolic activity in one carbon pool by folate pathway (Fig. 4a, b).
Fig. 4. Dual knockdown of MTHFD2 and PAICS depletes purine nucleoside production and interrupts cell cycle in MNA neuroblastoma.
a Metabolites levels of L-serine, AICAR, IMP and GMP were normalized to shLacZ SK-N-DZ cells. The log2 fold-change of L-serine in knockdown cells (shMTHFD2: 3.76; shPAICS: 1.65; and shMTHFD2/PAICS: 1.68), the log2 fold-change of AICAR in knockdown cells (shMTHFD2: 0.47; shPAICS: 0.52; and shMTHFD2/PAICS: −1.03), the log2 fold-change of IMP in knockdown cells (shMTHFD2: −0.48; shPAICS: −0.16; and shMTHFD2/PAICS: −0.87), and the log2 fold-change of GMP in knockdown cells (shMTHFD2: −0.02; shPAICS: −0.15; and shMTHFD2/PAICS: −0.83). At least two biological replicates for each metabolite were performed. b The intensity of L-serine, AICAR, IMP, and GMP in shLacZ, shMTHFD2, shPAICS, and shMTHFD2/PAICS SK-N-DZ cells. c The distribution of cells in the different phases of the cell cycle was analyzed by flow cytometry. d The percentage of cells in the S phase of shMTHFD2 and shPAICS cells increased, by 4.29% and 4.34%, respectively, and that of dual knockdown cells dramatically increased by 7.70% relative to shLacZ control of SK-N-DZ cells. The percentage of cells decreased in both G1 and G2/M phases, along with increased cell population of S phase, were observed in shMTHFD2, shPAICS, and dual knockdown. Of note, dual knockdown decreased by 5.33% and 2.36% of cell population in the G1 and G2/M phases, respectively. *P < 0.05; **P < 0.01; ***P < 0.001
On the other hand, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), the final intermediate for assembling inosine monophosphate (IMP), and guanosine monophosphate (GMP)40 were markedly reduced in shMTHFD2/PAICS relative to single knockdown. Thus, we suggest that the depletion of AICAR level limits the production of IMP and GMP when both MTHFD2 and PAICS are suppressed (Fig. 4a, b). Of note, the single knockdown of either MTHFD2 or PAICS elevated the levels of AICAR compared to the control. The results suggest that the synergistic effect of MTHFD2 and PAICS are related to the biosynthesis of nucleotides, as well as their interconnection in one carbon by folate and purine metabolism in MNA neuroblastoma cells.
Since the targeted metabolomics analysis indicated an imbalance of purine nucleotides in dual knockdown cells, we hypothesized that the cell cycle perturbations might be induced. Flow cytometry for DNA content was conducted and the distribution of cells in different phases of the cell cycle was analyzed. The results showed that the percentage of cells in the S phase of shMTHFD2 and shPAICS cells increased, by 4.29% and 4.34%, respectively, and that of dual knockdown cells increased by 7.70% relative to shLacZ control of MNA SK-N-DZ cells, indicating an interruption of the transition from S to G2/M phases (Fig. 4c, d). The percentage of cells decreased in both G1 and G2/M phases, along with increased cell population of S phase, especially the dual knockdown cells which decreased by 5.33% and 2.36% of population in the G1 and G2/M phases, respectively (Fig. 4c, d). Thus, purine intermediates depletion and S phase arrest may be correlated with dysregulation of DNA synthesis.
Dual knockdown of MTHFD2 and PAICS suppresses neuroblastoma cell growth
To elucidate the prognostic implications of MTHFD2 and PAICS in neuroblastoma progression, we analyzed the association of MTHFD2 and PAICS expressions with patient survival. Poor overall and event-free survivals were found in highly expressed MTHFD2 or PAICS neuroblastoma patients (Fig. 5a, b). According to the impact of MTHFD2 and PACIS on neuroblastoma patient survival, we ascertain their stimulatory effects on cell proliferation by cell viability and colony formation assays in response to MTHFD2 and PAICS expression manipulation (Figs. 1a, 5c–h and Supplementary Fig. 7). Cell viability was measured at 24, 48, and 72 h by MTS assays, and the results indicated a noticeable decline in cell proliferation in response to MTHFD2 and PAICS. Cell proliferation was strongly affected by MTHFD2 and PAICS; the viability of shMTHFD2 cells declined by 20. 4%, 20.8%, and 29.7%, at 24, 48, and 72 h, and the viability of shPAICS cells also decreased by 19.2%, 17.2%, and 24.0% at 24, 48, and 72 h respectively (Fig. 5d, e). Either shRNA-mediated knockdown of MTHFD2 or PAICS reduced cell viability compared with shLacZ control (Supplementary Fig. 8). However, the most significant reduction in cell viability occurred in dual knockdown of MTHFD2 and PAICS (shMTHFD2/PAICS) where the cell viability declined 48.4%, 50.2%, and 60.3% at 24, 48, and 72 h respectively (Fig. 5d, e).
Fig. 5. MTHFD2 and PAICS are related to neuroblastoma cell growth.
a, b Kaplan–Meier curves showing significant poorer prognosis in overall and event-free survival for patients with high expressions of MTHFD2 (a) and PAICS (b) using SEQC NB-498 dataset. The samples were straitified into high and low expressions based on the median value of the given gene expression. The differences in survival time between two groups were assessed by the log-rank test. c Western blot analyses revealing the SK-N-DZ stable knockdown cell lines of shRNA control, MTHFD2, PAICS, and MTHFD2/PAICS. d Cell viability of the SK-N-DZ stable knockdown cell lines was measured by MTS assays. e The cell viability in dual knockdown of MTHFD2 and PAICS (shMTHFD2/PAICS) has declined 48.4%, 50.2%, and 60.3% at 24, 48, and 72 h respectively. f–h The effects of SK-N-DZ stable knockdown cell lines on colony formation ability in representative plates (f) and quantitative analysis (g). h Single knockdown of MTHFD2 or PAICS alone was associated with approximately 32.1% and 29.6% declension, respectively, whereas dual knockdown of both MTHFD2 and PAICS resulted in a decrease of colony number to 46.5% compared with shLacZ cells
Colony formation was monitored to investigate the long-term effects of MTHFD2 and PAICS on neuroblastoma cell proliferation (Fig. 5f–h). Single knockdown of MTHFD2 or PAICS alone was associated with ~32.1% and 29.6% declension of colony formation ability respectively, whereas dual knockdown of both MTHFD2 and PAICS resulted in a decrease of colony number to 46.5% compared with shLacZ cells (Fig. 5h). These observations suggest that simultaneous knockdown of MTHFD2 and PAICS further impair the MNA cell proliferation, and they may serve as a regulatory purpose in cell growth together.
Dual knockdown of MTHFD2 and PAICS suppresses neuroblastoma cell migration
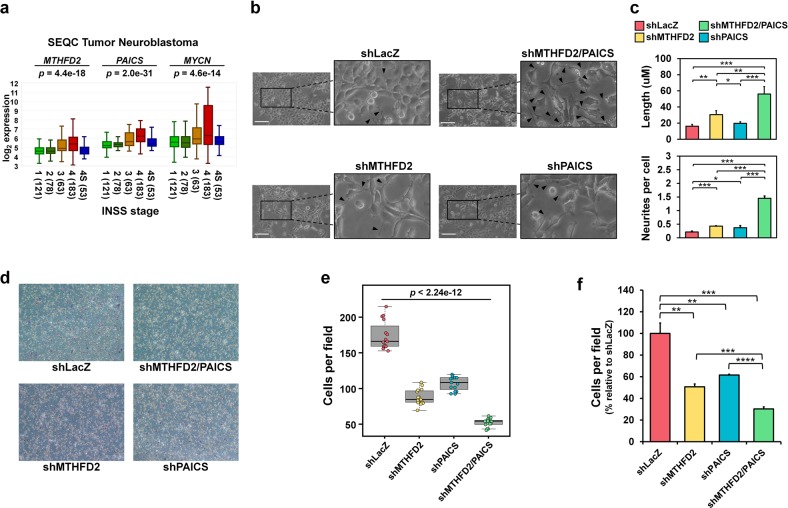
To further investigate the impacts of MTHFD2 and PAICS on tumor aggressiveness, we analyzed the relationship of MTHFD2 and PAICS expressions with tumor stages (INSS). A higher MTHFD2 or PAICS expression was found in stage 4 compared with stages 1–3 and 4S, which had a similar expression pattern as MYCN in INSS stages (Fig. 6a). The above results suggest that MTHFD2 and PAICS are correlated with metastatic stage and tumor progression. We also observed a differentiated cell morphology in dual knockdown shMTHFD2/PAICS cells in comparison with single knockdown or shLacZ control cells which is linked to a less aggressive neuronlike phenotype in neuroblastoma (Fig. 6b)4,11. The shLacZ control cells exhibiting typical rounder morphology compared with single or dual knockdown MNA neuroblastoma cells, whereas the dual knockdown shMTHFD2/PAICS cells exhibited a noticeable outgrowth of neurite lengths and numbers of branch points (Fig. 6b, c). These morphological changes, together with the correlation of MTHFD2 or PAICS expression to INSS, led us to probe whether MTHFD2 and PAICS play roles in malignant transformation and tumor aggressiveness.
Fig. 6. MTHFD2 and PAICS are correlated with metastatic tumor stage, neurite outgrowth, and cell migration in neuroblastoma.
a Expressions of MTHFD2, PAICS, and MYCN correlate with metastatic stage (4) as compared with localized stages (1–3) in neuroblastoma patients (http://r2.amc.nl/; Tumor Neuroblastoma –SEQZ – 498 – RPM – seqcnb1). The p-values were derived from ANOVA. b Phase contract microscopic images of stable single and dual knockdown of MTHFD2 and PAICS in MNA SK-N-DZ cells. A neuronlike cell morphology was observed in dual knockdown shMTHFD2/PAICS SK-N-DZ cells in comparison with single knockdown or control cells. Black arrow: outgrowth of neurites; Scale bar: 100 μm. c The average neurite length and number of neurites per cell were significantly increased in shMTHFD2/PACIS cells. d, e The images (d) and the quantitative analysis (e) showing the number of migrated cells in stable shLacZ, shMTHFD2, shPAICS, and shPAICS/MTHFD2 SK-N-DZ cells. f shMTHFD2/PAICS cells was dramatically decreased by 69.7% relative to shLacZ control. Single knockdown of MTHFD2 decreased by 49.3%, while that of PAICS decreased by 38.4% relative to shLacZ control
Cell migration is a crucial process during metastasis, thus we performed transwell assay to determine the migration ability on MTHFD2 and PAICS (Fig. 6d–f). Dual knockdown of MTHFD2 and PAICS had a significantly poorer migration ability compared with single knockdown of MTHFD2 and PAICS alone where shMTHFD2/PAICS was dramatically decreased by 69.7% compared to shLacZ control (Fig. 6f). As in the single knockdown results, shMTHFD2 cells decreased by 49.3%, while that of shPAICS cells decreased by 38.4% compared to shLacZ control, suggesting silencing of MTHFD2 and PAICS had synergistic effect on migration ability in MNA neuroblastoma (Fig. 6f).
Compound perturbagens suppressing MTHFD2 and PAICS show synergistic effects on MNA neuroblastoma
Anti-metabolic agents have been used in cancer treatment for decades, however, a relatively low number of metabolic inhibitors developed for cancer therapy due to the adverse effects on patients by blocking the synthesis of vital cellular constituents from metabolic pathways41. Our strategy for identifying anti-cancer agents is to screen for compounds that particularly target MYCN-regulated metabolic genes across cell lines by using transcriptomic profiles31,32. By matching gene signatures with drug signatures, we systematically identified the gene expressions of MTHFD2 and PAICS were significantly suppressed by anisomycin and apicidin across ten cell lines (Fig. 7a and Supplementary Fig. 9). Anisomycin is a bacterial antibiotic which inhibits protein biosynthesis by blocking peptide bond formation in ribosomes, and activates kinase cascades including JNK, p38 MAPK signaling pathways42–44. Apicidin is a fungal metabolites shown to inhibit histone deacetylase, and induce p21(WAD1/Cip1), caspase-3 and caspase 9, and DNA fragmentation45,46.
Fig. 7. Combinatorial targeting of MTHFD2 and PAICS inhibits MNA neuroblastoma cell proliferation.
a The relative transcriptional changes of MTHFD2 and PAICS after anisomycin or apicidin treatment for 24 h across ten cell lines. For each cell type, the expression changes of measured transcripts (n = 12,494) were ordered from the most downregulated (the negative side) to the most upregulated gene (the positive side), from which MTHFD2 (upper left panel) or PAICS (lower left panel) was indicated by a colored line. b–e The combined effects of anisomycin and apicidin on MNA neuroblastoma cell lines. Percent cell growth of MNA SK-N-DZ (b) and SK-N-BE(2)-C (d) cells relative to corresponding controls treated with anisomycin, apicidin, or the combined treatment at indicated dosage for 48 h. c, e Combination index (CI) plot for anisomycin and apicidin in SK-N-DZ (c) and SK-N-BE(2)-C (e). CI values are plotted as a function of the fractional inhibition of cell viability by computer simulation using CompuSyn, where the gray circles represent the actual experimental points. CI values: synergism (CI < 0.9), additive effect (CI = 0.9–1.1), and antagonism (CI > 1.1), and the Fa value indicates cell fraction affection by the combination of anisomycin and apicidin. The mean CI and Fa values are listed in the table (c, e). Three biological replicates were performed with two technical replicates each. Data depicted are the mean ± SEM
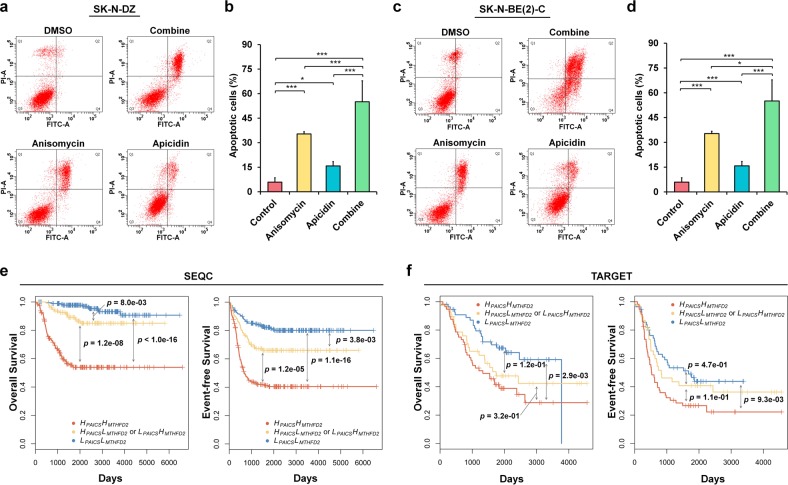
To investigate the effects of anisomycin combined with apicidin on MNA neuroblastoma, drug combination assays were performed using SK-N-DZ and SK-N-BE(2)-C cells with various concentrations for 24 and 48 h (Fig. 7b–e and Supplementary Fig.10). The drug combination assay showed synergistic effects on the inhibition of MNA neuroblastoma cell proliferation, as compared with either anisomycin or apicidin treatment alone (Fig. 7b, d). Furthermore, the mean CI values were 0.71 and 0.74 in SK-N-DZ and SK-N-BE(2)-C, respectively, which indicated synergistic drug combinations (Fig. 7c, e). In addition, the non-MNA SK-N-AS and SK-N-SH neuroblastoma cells displayed a less sensitive effects to the anisomycin and apicidin treatments (Supplementary Fig. 11). Anisomycin and apicidin have been reported to induce apoptosis in certain cell types46,47, however, the effects of drug combination on apoptosis in neuroblastoma remains unidentified. Therefore, we performed apoptosis assays and found that both anisomycin and apicidin treatments induced apoptosis in MNA neuroblastoma (Fig. 8a–d). Notably, the co-treatment results exhibited the most significant cell apoptosis (Fig. 8b, d).
Fig. 8. Combinatorial targeting of MTHFD2 and PAICS induces cell apoptosis, and a low-low expression of MTHFD2 and PAICS suppresses patient survival in neuroblastoma.
a–d Apoptotic cell death was measured by flow cytometry for drug combination assay. a, c Representative flow cytometry profiles using Annexin V-FITC/PI staining for apoptosis in SK-N-DZ (a) and SK-N-BE(2)-C (c) cells. b, d Quantification of apoptotic cells between anisomycin, apicidin, and the combined treatment in SK-N-DZ (b) and SK-N-BE(2)-C (d) cells. Experiments are performed in triplicate and data are expressed as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. e, f Kaplan-Meier survival curves of overall and event-free survival of patients with neuroblastoma. Neuroblastoma patients with high-high expression of MTHDF2 and PAICS (HMTHFD2HPAICS); low-low expression of MTHDF2 and PAICS (LMTHFD2LPAICS), and high-low or low-high expression of MTHFD2 and PAICS (HMTHFD2LPAICS or LMTHFD2HPAICS) from SEQC (e) and TARGET (f) neuroblastoma cohorts. The median of the gene expression was used as the cutoff to straitify patients into high and low groups. The differences in survival time between groups were assessed by the log-rank test
Next, we examined the MTHFD2/PAICS gene combinations with synergistic effects on neuroblastoma patient survival. First, the median expression value of the given gene was used to define the high and low expressions, and then we split neuroblastoma patients into three groups: high-high expression of MTHDF2 and PAICS (HMTHFD2HPAICS); low-low expression of MTHDF2 and PAICS (LMTHFD2LPAICS), and high-low or low-high expression of MTHFD2 and PAICS (HMTHFD2LPAICS or LMTHFD2HPAICS). Interestingly, the HMTHFD2HPAICS group had the worst overall and event-free survival against the other groups, and the HMTHFD2LPAICS or LMTHFD2HPAICS group had poorer survival than LMTHFD2LPAICS, but better than the HMTHFD2HPAICS (Fig. 8e, f). Altogether, the patient survival yielded consistent results as the drug combination assays that simultaneously suppressing MTHFD2 and PAICS expressions showed a synergism in MNA neuroblastoma patients.
Discussion
MYCN serves as an oncogenic regulator that mediates aberrant signaling pathways and leads to HR neuroblastoma. Recent studies achieved some clinical success to indirectly target MYCN, since developing direct MYCN-target agent encounters difficulties and has not yet been established38,48,49. Here, we addressed highly confident MYCN-targeted genes which may provide unbiased and genome-wide mapping of MYCN-gene interactions. Altogether, we revealed 427 potential MYCN-targeted genes, contributing to characterize downstream signaling pathways.
Cancer metabolism is a complex enzyme-catalyzed reaction that involves multiple factors to overcome any single disruption50. Thus, we aim to investigate the MYCN-regulated interconnection of metabolic pathways and biological processes. The potential MYCN-targeted genes are found to be involved in purine biosynthesis which consists ten steps and six enzymes from phosphoribosyl pyrophosphate (PRPP) to inosine monophosphate (IMP), guanosine monophosphate (GMP), and adenosine monophosphate (AMP)51. Purinosome was coined to describe the reversible six-enzyme complex for purine biosynthesis that assembled and co-localized on mitochondria while the purine levels of cytosol depleted23,52. Besides, one carbon pool by folate was also identified to be associated with MNA neuroblastoma, which generated formates, the precursor to 10-formyltetrahydrofolate in the mitochondria and exported into cytosol for the need of transformylase enzymes, GRAT and ATIC in purine biosynthesis21. On the other hand, loss of MTHFD2 would cause folate trapping within the mitochondria, therefore, folate-dependent pathways may be altered24,53,54.
Previous studies indicate that amino acid metabolism, nucleic acid metabolism, and mitochondrial metabolism play important roles in cancer progression55,56. Biosynthetic pathways are essential for cancer metabolism since they generate energy and macromolecules that are required for replicative cell division and tumor growth56. Our targeted metabolomics analysis indicated that the levels of IMP and GMP were significantly higher in MNA neuroblastoma compared to non-MNA neuroblastoma cells, suggesting MNA neuroblastoma may require higher demand in purine biosynthesis for rapid cell proliferation. The MYCN-targeted gene pair results implied that MYCN could regulate the interconnection of metabolic pathways by MTHFD2 and PAICS in two directions. MYCN transcriptionally regulates the mitochondrial metabolic enzyme MTHFD2 for formate exportation and the bifunctional enzyme PAICS of the purinosome to generate SAICAR for purine biosynthesis.
Treatment that combines two or more specific therapeutic targets might reduce the dosage and side-effect for aggressive therapy in children57. Our dual knockdown and co-treatment assays that target MTHFD2 and PAICS had synergistic effects on MNA neuroblastoma which might diminish the aggressiveness and tumor progression ability. In conclusion, establishing MYCN downstream signaling pathways in neuroblastoma may improve our capacity in identifying molecular interactions, disease pathways, and even targeted drug discovery.
Supplementary information
Acknowledgements
The instrument supports from NTU Mass Spectrometry Platform and Technology Commons in College of Life Science were acknowledged. We thank Chiao-Hui Hsieh for consulting data analysis. The results published here are in whole or part based upon data generated by TCGA managed by the NCI and NHGRI. This work was supported by the Ministry of Science and Technology (MOST 105-2320-B-002-057-MY3, MOST 106-2320-B-002-053-MY3, MOST 107-2221-E-010-017-MY2, and MOST 107-2321-B-006-020), the Aim for the Top University Project and the National Health Research Institutes (NHRI-EX108-10709BI) in Taiwan.
Author contributions
C.H.Y.C., H.C.H. and H.F.J. conceptualized and designed research, developed the methodology, administrative, technical or material supported including reporting or organizing data, constructing databases. W.M.H., H.Y.W. and C.C.H. acquired the data including animals, managed patients, and provided facilities. C.H.Y.C., C.L.H., C.Y.T., T.T.K., C.T.H. and Y.H.C. analyzed and interpreted the data including statistical analysis, biostatistics, and computational analysis. C.T.H. helped with revision. C.H.Y.C., C.L.H., H.C.H. and H.F.J. wrote, reviewed, and/or revised the manuscript. H.C.H. and H.F.J. supervised this project.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by N. Bazan
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hsuan-Cheng Huang, Phone: +886-2-2826-7357, Email: hsuancheng@ym.edu.tw.
Hsueh-Fen Juan, Phone: +886-2-3366-4536, Email: yukijuan@ntu.edu.tw.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-2033-z).
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Olsen RR, et al. MYCN induces neuroblastoma in primary neural crest cells. Oncogene. 2017;36:5075–5082. doi: 10.1038/onc.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn SL, et al. TheInternational Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat. Rev. Clin. Oncol. 2014;11:704–713. doi: 10.1038/nrclinonc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seeger RC, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 6.Bown N, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 7.Attiyeh EF, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 8.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 9.Maris JM. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathsyaraja H, Eisenman RN. Parsing Myc paralogs in oncogenesis. Cancer Cell. 2016;29:1–2. doi: 10.1016/j.ccell.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Perez MV, Henley AB, Arsenian-Henriksson M. The MYCN protein in health and disease. Genes. 2017;8:E113. doi: 10.3390/genes8040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irwin MS, Park JR. Neuroblastoma: paradigm for precision medicine. Pediatr. Clin. North Am. 2015;62:225–256. doi: 10.1016/j.pcl.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Qing G, et al. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1alpha. Cancer Res. 2010;70:10351–10361. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 15.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, et al. Transcriptional profiling reveals a common metabolic program in high-risk human neuroblastoma and mouse neuroblastoma sphere-forming cells. Cell Rep. 2016;17:609–623. doi: 10.1016/j.celrep.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Y, et al. Metabolic reprogramming by MYCN confers dependence on the serine-glycine-one-carbon biosynthetic pathway. Cancer Res. 2019;79:3837–3850. doi: 10.1158/0008-5472.CAN-18-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren P, et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J. Pathol. 2015;235:90–100. doi: 10.1002/path.4429. [DOI] [PubMed] [Google Scholar]
- 19.Zirath H, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl Acad. Sci. USA. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman BJ, et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 2015;22:1009–1019. doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedley AM, Benkovic SJ. A New view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy B, Depaix A, Perigaud C, Peyrottes S. Recent trends in nucleotide synthesis. Chem. Rev. 2016;116:7854–7897. doi: 10.1021/acs.chemrev.6b00174. [DOI] [PubMed] [Google Scholar]
- 23.French JB, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson R, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu CL, et al. Unveiling MYCN regulatory networks in neuroblastoma via integrative analysis of heterogeneous genomics data. Oncotarget. 2016;7:36293–36310. doi: 10.18632/oncotarget.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, Wang LG, Han Y, He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi H, et al. Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Anal. Bioanal. Chem. 2013;405:5279–5289. doi: 10.1007/s00216-013-6927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452 e1417. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang CT, et al. Therapeutic targeting of non-oncogene dependencies in high-risk neuroblastoma. Clin. Cancer Res. 2019;25:4063–4078. doi: 10.1158/1078-0432.CCR-18-4117. [DOI] [PubMed] [Google Scholar]
- 32.Huang CT, et al. Perturbational gene-expression signatures for combinatorial drug discovery. iScience. 2019;15:291–306. doi: 10.1016/j.isci.2019.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 34.Selmi A, et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett. 2015;357:412–418. doi: 10.1016/j.canlet.2014.11.056. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev. Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Defferrari R, et al. Concomitant DDX1 and MYCN gain in neuroblastoma. Cancer Lett. 2007;256:56–63. doi: 10.1016/j.canlet.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Korshunov A, et al. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathol. 2012;123:515–527. doi: 10.1007/s00401-011-0918-8. [DOI] [PubMed] [Google Scholar]
- 38.Carter DR, et al. Glutathione biosynthesis is upregulated at the initiation of MYCN-driven neuroblastoma tumorigenesis. Mol. Oncol. 2016;10:866–878. doi: 10.1016/j.molonc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stover PJ. One-carbon metabolism-genome interactions in folate-associated pathologies. J. Nutr. 2009;139:2402–2405. doi: 10.3945/jn.109.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ducker GS, et al. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 42.Grollman AP. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 1967;242:3226–3233. [PubMed] [Google Scholar]
- 43.Torocsik B, Szeberenyi J. Anisomycin uses multiple mechanisms to stimulate mitogen-activated protein kinases and gene expression and to inhibit neuronal differentiation in PC12 phaeochromocytoma cells. Eur. J. Neurosci. 2000;12:527–532. doi: 10.1046/j.1460-9568.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 44.Stadheim TA, Kucera GL. c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) is required for mitoxantrone- and anisomycin-induced apoptosis in HL-60 cells. Leuk. Res. 2002;26:55–65. doi: 10.1016/S0145-2126(01)00099-6. [DOI] [PubMed] [Google Scholar]
- 45.Han JW, et al. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 2000;60:6068–6074. [PubMed] [Google Scholar]
- 46.Kwon SH, et al. Apicidin, a histone deacetylase inhibitor, induces apoptosis and Fas/Fas ligand expression in human acute promyelocytic leukemia cells. J. Biol. Chem. 2002;277:2073–2080. doi: 10.1074/jbc.M106699200. [DOI] [PubMed] [Google Scholar]
- 47.Li JY, et al. Anisomycin induces glioma cell death via down-regulation of PP2A catalytic subunit in vitro. Acta Pharmacol. Sin. 2012;33:935–940. doi: 10.1038/aps.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beltran H. The N-myc nncogene: maximizing its targets, regulation, and therapeutic potential. Mol. Cancer Res. 2014;12:815–822. doi: 10.1158/1541-7786.MCR-13-0536. [DOI] [PubMed] [Google Scholar]
- 49.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H, French JB, Fang Y, Benkovic SJ. The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem. Commun. 2013;49:4444–4452. doi: 10.1039/c3cc41437j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 53.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 54.Nishimura T, et al. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene. 2019;38:2464–2481. doi: 10.1038/s41388-018-0589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westhoff MA, et al. Cell death-based treatment of childhood cancer. Cell Death Dis. 2018;9:116. doi: 10.1038/s41419-017-0062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.