Coronaviruses encode an exoribonuclease (ExoN) that is important for viral replication, fitness, and virulence, yet coronaviruses with a defective ExoN (ExoN-AA) have not reverted under diverse experimental conditions. In this study, we identify multiple impediments to MHV-ExoN-AA reversion. We show that ExoN-AA reversion is possible but evolutionarily unfavorable. Instead, compensatory mutations outside ExoN-AA motif I are more accessible and beneficial than partial reversion. We also show that coevolution between replicase proteins over long-term passage partially compensates for ExoN-AA motif I but renders the virus inhospitable to a reverted ExoN. Our results reveal the evolutionary basis for the genetic stability of ExoN-inactivating mutations, illuminate complex functional and evolutionary relationships between coronavirus replicase proteins, and identify potential mechanisms for stabilization of ExoN-AA coronavirus mutants.
KEYWORDS: RNA virus, adaptive evolution, competitive fitness, coronavirus, exoribonuclease, plus-strand RNA virus, proofreading, replication fidelity
ABSTRACT
The 3′-to-5′ exoribonuclease in coronavirus (CoV) nonstructural protein 14 (nsp14-ExoN) mediates RNA proofreading during genome replication. ExoN catalytic residues are arranged in three motifs: I (DE), II (E), and III (D). Alanine replacement of the motif I residues (AA-E-D; four nucleotide substitutions) in murine hepatitis virus (MHV) and severe acute respiratory syndrome (SARS)-CoV yields viable mutants with impaired replication and fitness, increased mutation rates, and attenuated virulence in vivo. Despite these impairments, MHV- and SARS-CoV ExoN motif I AA mutants (ExoN-AA) have not reverted at motif I in diverse in vitro and in vivo environments, suggesting that profound fitness barriers prevent motif I reversion. To test this hypothesis, we engineered MHV-ExoN-AA with 1, 2, or 3 nucleotide mutations along genetic pathways to AA-to-DE reversion. We show that engineered intermediate revertants were viable but had no increased replication or competitive fitness compared to that of MHV-ExoN-AA. In contrast, a low-passage-number (passage 10 [P10]) MHV-ExoN-AA showed increased replication and competitive fitness without reversion of ExoN-AA. Finally, engineered reversion of ExoN-AA to ExoN-DE in the presence of ExoN-AA passage-adaptive mutations resulted in significant fitness loss. These results demonstrate that while reversion is possible, at least one alternative adaptive pathway is more rapidly advantageous than intermediate revertants and may alter the genetic background to render reversion detrimental to fitness. Our results provide an evolutionary rationale for lack of ExoN-AA reversion, illuminate potential multiprotein replicase interactions and coevolution, and support future studies aimed at stabilizing attenuated CoV ExoN-AA mutants.
IMPORTANCE Coronaviruses encode an exoribonuclease (ExoN) that is important for viral replication, fitness, and virulence, yet coronaviruses with a defective ExoN (ExoN-AA) have not reverted under diverse experimental conditions. In this study, we identify multiple impediments to MHV-ExoN-AA reversion. We show that ExoN-AA reversion is possible but evolutionarily unfavorable. Instead, compensatory mutations outside ExoN-AA motif I are more accessible and beneficial than partial reversion. We also show that coevolution between replicase proteins over long-term passage partially compensates for ExoN-AA motif I but renders the virus inhospitable to a reverted ExoN. Our results reveal the evolutionary basis for the genetic stability of ExoN-inactivating mutations, illuminate complex functional and evolutionary relationships between coronavirus replicase proteins, and identify potential mechanisms for stabilization of ExoN-AA coronavirus mutants.
INTRODUCTION
The rapid evolution of RNA viruses represents a significant challenge for preventing, treating, and eradicating RNA viral diseases. High mutation rates in RNA viruses generate extensive opportunities to overcome evolutionary hurdles, such as antiviral drugs, host immunity, or engineered attenuating changes (1). The evolutionary pathways traversed by RNA viruses are shaped by natural selection, which will favor some evolutionary trajectories more than others based on whether mutations are beneficial, deleterious, or neutral (2). Predicting the likely results of RNA virus evolution is an important step for anticipating viral emergence and for developing escape-resistant antiviral drugs and vaccines (3, 4).
Coronaviruses (CoVs) are a family of positive-sense RNA viruses that cause human illnesses ranging from the common cold to severe and lethal respiratory disease (5). All CoVs encode a proofreading exoribonuclease within nonstructural protein 14 (nsp14-ExoN) that is critical for replication, fidelity, fitness, and virulence, and ExoN inactivation has been proposed as a strategy for live attenuated vaccine development (6–15). As members of the DEDDh superfamily of exonucleases, CoV ExoNs hydrolyze nucleotides using four metal-coordinating amino acids arranged in three motifs: I (DE), II (E), and III (D) (16, 17). Alanine substitution in ExoN motif I (DE-to-AA) disrupts ExoN biochemical activity in both severe acute respiratory syndrome (SARS)-CoV and human CoV 229E (hCoV-229E) (16, 18, 19). The betacoronaviruses murine hepatitis virus (MHV) and SARS-CoV tolerate disruption of ExoN activity [ExoN(−)] but display mutator phenotypes accompanied by defects in replication, competitive fitness, and evasion of innate immune responses (10, 13, 14). ExoN active-site mutants in alphacoronaviruses, including transmissible gastroenteritis virus and hCoV-229E, have yet to be recovered and are proposed to be lethal for replication (19, 20).
Given the critical role of ExoN in CoV biology and the elevated mutation rate, we expected that natural selection would repeatedly drive reversion of the ExoN-inactivating substitutions. In line with this expectation, ExoN motif III mutants of SARS-CoV and MHV rapidly and repeatedly revert (14; also our unpublished observations). In contrast, we have never detected partial or complete reversion of ExoN motif I mutants (ExoN-AA) in SARS-CoV or MHV during 10 years of study and hundreds of experiments. More specifically, we have not detected consensus or minority variants of any kind at the motif I AA codons in either virus strain during acute infections and prolonged passage in tissue culture and following treatment with multiple nucleoside analogues (6–11, 13, 14). SARS-CoV-ExoN-AA also is stable during acute and persistent animal infections in immunocompetent and immunocompromised mice (12). The lack of primary reversion is not due simply to reduced adaptive capacity, as both SARS-CoV-ExoN-AA and MHV-ExoN-AA can adapt for increased replication (7, 14). Most strikingly, long-term passage of MHV-ExoN-AA (250 passages [P250]) yielded a highly fit population that had directly compensated for defective proofreading through evolution of a likely high-fidelity RNA-dependent RNA polymerase (RdRp) (7). Yet where primary reversion would have required just four total consensus mutations, MHV-ExoN-AA-P250 contained more than 170.
In this study, we sought to determine whether specific genetic or fitness barriers prevent primary reversion of ExoN motif I AA. To this end, we identified and engineered viable genetic pathways toward ExoN-AA motif I reversion in MHV (here, ExoN-AA). Our results show that partial reversion did not confer a selective advantage over ExoN-AA. Further, ExoN-AA adapted within 10 passages for greater fitness than any of the intermediate revertants. Finally, restoration of wild-type (WT)-ExoN-DE in the setting of passage-selected mutations in the nsp12 RdRp and nsp14-ExoN exacted profound fitness costs. Together, these data are the first observation of an ExoN(–) CoV genotype fitness landscape and identify multiple genetic barriers underlying ExoN(–) motif I stability in MHV. Further, they suggest extensive coevolution between MHV replicase proteins during adaptation and reveal potential strategies for stabilizing ExoN mutant CoVs.
(This article was submitted to an online preprint archive [21]).
RESULTS
Primary reversion of ExoN(–) motif I.
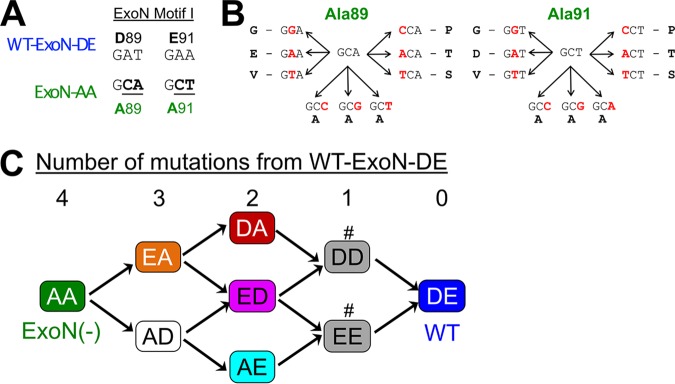
MHV-ExoN(–), here ExoN-AA, contains two engineered substitutions in each codon of motif I, such that complete reversion to WT-ExoN-DE requires mutations to all four sites (Fig. 1A). Viral mutation rates in the absence of proofreading range from 10−4 to 10−6 mutations per nucleotide per round of replication (μ) (1). Assuming an ExoN-AA mutation rate of 10−4 μ and accounting for codon degeneracy, the probability of restoring the native amino acid sequence in a single round of replication is 10−18. Only rarely do ExoN-AA titers exceed 106 PFU/ml, so it is unlikely that ExoN-AA could navigate this genetic barrier in a single infectious cycle. Thus, we hypothesized that ExoN-AA reversion, if possible, would proceed incrementally. To identify potential pathways toward ExoN-AA reversion, we examined the possible single-nucleotide substitutions surrounding A89 and A91 (Fig. 1B). Three mutations are synonymous, and five mutations yield amino acids unlikely to coordinate with the positively charged metals required for ExoN catalysis (glycine, valine, proline, threonine, and serine) (16, 19, 22, 23). One mutation per site can restore the acidic charge (i.e., AA-to-ED) but not the native amino acid. These variants have not been tested in a CoV ExoN, but biochemical studies of Escherichia coli DNA polymerase I ExoN mutants suggest that these conservative substitutions would not restore WT-like ExoN activity (24). We predicted stepwise pathways to ExoN-AA→DE reversion based on restoration of acidic charge, followed by reversion to native amino acids (Fig. 1C). We engineered and recovered variants in ExoN-AA requiring three mutations (3 nucleotides [nt]; ExoN-AD and ExoN-EA), two mutations (2 nt; ExoN-DA, ExoN-ED, and ExoN-AE), or one mutation (1 nt; ExoN-DD and ExoN-EE) for reversion to WT-ExoN-DE (Table 1). We will here refer to these mutants as intermediate revertants. All intermediate revertants generated viable progeny during recovery, demonstrating that reversion to WT-ExoN-DE along these pathways is theoretically possible. The 3-nt and 2-nt mutants were genetically stable during recovery, as confirmed by dideoxy sequencing. However, both 1-nt mutants (ExoN-DD and ExoN-EE) reverted to WT-ExoN-DE during three independent recovery attempts, suggesting that these two variants are less fit than WT-ExoN-DE and demonstrating that reversion by a 1-nt mutation is readily accessible. To test whether the 3-nt or 2-nt mutants would revert more rapidly than ExoN-AA (4 nt), we passaged three lineages of each mutant 10 times at multiplicities of infection (MOI) of 0.5 and 0.01 PFU/cell. We harvested supernatants and screened for reversion by visual inspection of plaque phenotypes at each passage. WT-ExoN-DE and WT-like viruses produce uniform, large plaques, while ExoN-AA-like viruses yield small, variably sized plaques (13). When we observed mixed plaque phenotypes, we sequenced three large plaques from each lineage to confirm reversion. The 3-nt (ExoN-AD and ExoN-EA) and 2-nt (ExoN-DA and ExoN-ED) intermediate revertants showed no evidence of reversion over 10 passages at either MOI (Table 1). In contrast, the 2-nt ExoN-AE contained WT revertants by P2 in all lineages at an MOI of 0.5 PFU/cell and in one lineage by P8 at an MOI of 0.01 PFU/cell. Once observed, WT revertants dominated the ExoN-AE population for the remaining passages. These data indicate that at least one 2-nt mutation pathway can lead to full reversion in tissue culture. The probability of ExoN-AE arising during a single infectious cycle of ExoN-AA is low but theoretically achievable (∼10−9), so ExoN-AA could conceivably revert within just two infectious cycles. However, complete reversion has never been observed even during prolonged passage or persistent infections, suggesting that additional barriers to the replication, fitness, or maintenance of intermediate revertants exist.
FIG 1.
Sequence landscape around ExoN-AA motif I. (A) ExoN motif I nucleotide sequences. (B) Landscape of single-nucleotide substitutions within ExoN-AA motif I. (C) Predicted pathways to reversion of ExoN-AA. Variants marked with a number sign (#) reverted to WT during three independent recovery attempts.
TABLE 1.
Recovery and passage of intermediate revertants
| Virus | No. of mutations to WT-ExoN-DE | Motif I sequencea | No. of reverted lineages by passage 10 at:b
|
|
|---|---|---|---|---|
| MOI = 0.01 (n = 3) | MOI = 0.5 (n = 3) | |||
| ExoN-AA | 4 | GCA…GCT | 0 | 0 |
| ExoN-AD | 3 | GCA…GAT | 0 | 0 |
| ExoN-EA | 3 | GAA…GCT | 0 | 0 |
| ExoN-DA | 2 | GAT…GCT | 0 | 0 |
| ExoN-AE | 2 | GCA…GAA | 1c | 3c |
| ExoN-ED | 2 | GAA…GAT | 0 | 0 |
| ExoN-EE | 1 | GAA…GAA | ND | ND |
| ExoN-DD | 1 | GAT…GAT | ND | ND |
| WT-ExoN-DE | 0 | GAT…GAA | ND | ND |
Bolded nucleotides must mutate to reach the WT-ExoN-DE genotype.
Recovered viruses were subjected to 10 passages at the indicated MOI. Samples were screened for wild-type revertants by plaque assay, and revertant lineages were sequence confirmed. ND, not done.
By P8 at an MOI of 0.01 and by P2 at an MOI of 0.5.
Partial reversion of MHV-ExoN(–) motif I does not confer a selective advantage.
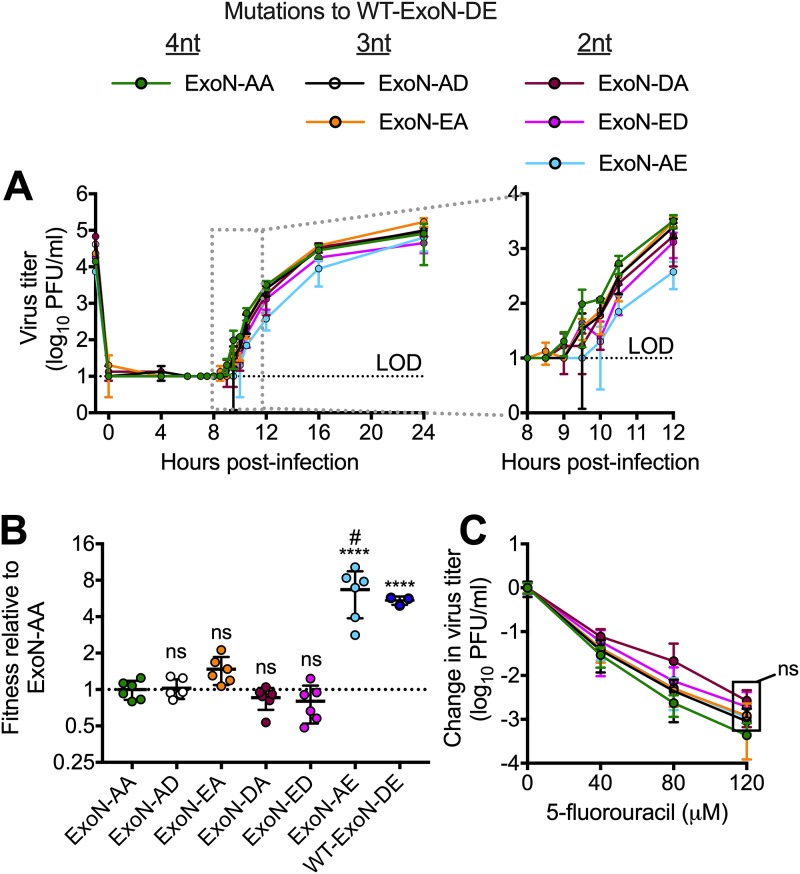
Because the intermediate revertants are viable as recombinants but are not found in ExoN-AA populations, we hypothesized that they confer no selective advantage over ExoN-AA (8, 9, 13). To test this hypothesis, we first analyzed replication of the 3-nt and 2-nt intermediate revertants (Fig. 2A). All variants achieved peak titers similar to the titer of ExoN-AA, but detailed examination of their kinetics suggested a potential delay of up to 1.5 h for all intermediate revertants compared to the kinetics of ExoN-AA. Of note, ExoN-AE was the most delayed, and we detected WT-ExoN-DE revertants in two of three replicates, suggesting increased selective pressure against this variant. We next measured the competitive fitness of each intermediate revertant relative to a recombinant ExoN-AA containing seven silent mutations in the nsp2 coding region (ExoN-AA-reference). Intermediate revertants were mixed with an equal titer of ExoN-AA-reference at a combined MOI of 0.05 PFU/cell and passaged four times. The ratio of each intermediate revertant to ExoN-AA-reference was quantified at each passage by reverse transcription-quantitative PCR (RT-qPCR), and the change in ratio over time was used to calculate their relative fitness levels. WT-ExoN-DE was significantly more fit than ExoN-AA, whereas the intermediate revertants (ExoN-AD, -EA, -DA, and -ED) had no increased fitness relative to that of ExoN-AA (Fig. 2B). The apparent increased fitness of ExoN-AE resulted from all lineages reverting to WT-ExoN-DE during the experiment. Finally, our previous studies have shown that adaptation of ExoN-AA includes partial compensation for the replication fidelity defect, as measured by reduced susceptibility to the mutagen 5-fluorouracil (5-FU) (7–11, 25). None of the intermediate variants demonstrated statistically significant differences in 5-FU sensitivity compared to that of ExoN-AA (Fig. 2C). Thus, with the exception of the ExoN-AE→DE revertants, no 3-nt and 2-nt intermediate genotypes along our predicted pathway demonstrated an advantage in replication, fitness, or fidelity that would favor their maintenance or expansion in the viral population. Thus, natural selection is unlikely to drive ExoN-AA down these pathways toward reversion.
FIG 2.
Intermediate revertants of ExoN-AA motif I do not have selective advantages. (A) Replication kinetics at an MOI of 0.01 PFU/cell plotted as means ± standard deviations (n = 3). (B) Competitive fitness of each variant relative to that of ExoN-AA. Viruses were competed with a tagged ExoN-AA-reference strain, and relative fitness was normalized to the mean of the ExoN-AA value. (C) 5-Fluorouracil sensitivity at an MOI of 0.01 PFU/cell. Statistical significance of each variant relative to the value of ExoN-AA was determined by one-way analysis of variance with multiple comparisons (D) and by two-way analysis of variance with Dunnett’s multiple-comparison test (C). ****, P < 0.0001; ns, not significant. LOD, limit of detection. Data in panels B and C represent means ± standard deviations (n = 6). Boxed values have the same significance. #, all lineages of ExoN-AE reverted to WT-ExoN-DE during the experiment.
Secondary adaptations outside ExoN-AA motif I increase fitness along alternative pathways.
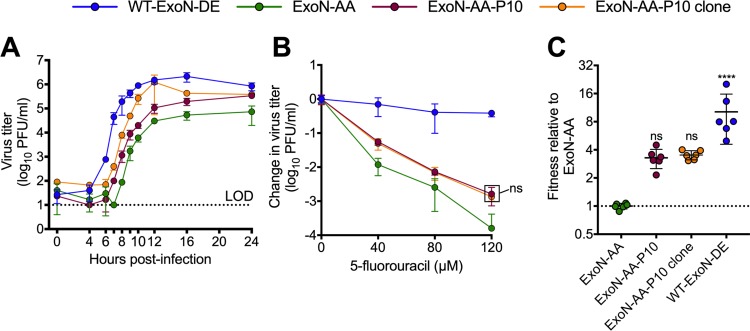
Although we did not find fitness advantages to intermediate revertants, we also did not identify profound fitness costs that would drive their immediate loss from populations. We have previously demonstrated that during 250 passages (P250), ExoN-AA can adapt for increased replication, fitness, and fidelity via secondary mutations outside motif I (7). We tested whether secondary adaptive mutations could exceed the fitness of ExoN-AA and its intermediate revertants. To examine the early adaptation of ExoN-AA, we studied passage 10 from the P250 passage series (Fig. 3). ExoN-AA-P10 retains the ExoN-AA motif I genotype but has increased replication and reduced susceptibility to 5-FU, altogether manifesting greater relative fitness (Fig. 3) (7). We identified only six total mutations within ExoN-AA-P10 by dideoxy sequencing (Table 2), indicating that rapid adaptation of and compensation for ExoN-AA requires relatively few genetic changes at the consensus level. To test whether interactions between multiple mutations or population level effects contribute to ExoN-AA-P10 fitness, we isolated a plaque-purified clone of ExoN-AA-P10. The clone replicated to higher titers than the ExoN-AA-P10 population but had identical 5-FU sensitivity and relative fitness levels (Fig. 3), indicating that genomes derived from a single virus plaque encode the adaptive changes required by the total population. Together, these data demonstrate that mutations outside ExoN(–) motif I can confer greater fitness advantages than intermediate revertants even at early passages. These early adaptive mutations likely reduce the selective pressure for motif I reversion and place the intermediate revertants at a selective disadvantage.
FIG 3.
ExoN-AA adapts for increased fitness within 10 passages. (A) Replication kinetics of the indicated viruses at an MOI of 0.01 PFU/cell plotted as means ± standard deviations (n = 3). (B) 5-Fluorouracil sensitivity at an MOI of 0.01 PFU/cell. (C) Competitive fitness of individual recombinants relative to that of ExoN-AA. Viruses were competed with a tagged ExoN-AA-reference strain, and relative fitness was normalized to the mean of the ExoN-AA value. Statistical significance of each virus relative to that of ExoN-AA was determined by two-way analysis of variance with Dunnett’s multiple-comparison test (B) or by one-way analysis of variance with multiple comparisons (C). ****, P < 0.0001; ns, not significant. LOD, limit of detection. Data in panels B and C represent means ± standard deviations (n = 6). Boxed values have the same significance.
TABLE 2.
Mutations in ExoN(−) at P10a
| Nucleotide change | Protein | Amino acid changed |
|---|---|---|
| G2520Ab | nsp2 | D524N |
| A3080Gb | nsp3 | Silent |
| T16017A | nsp12 | M814K |
| A17836Gb | nsp13 | I492M |
| G22673Ab | HEc | Noncoding |
| A29298Cb | M | Silent |
Data were derived from dideoxy sequencing.
Mutation present in approximately 50% of the population.
MHV hemagglutinin-esterase (HE) is not transcribed in tissue culture.
Amino acid numbers designate positions within cleaved nsps, not the polyprotein.
Adaptive mutations in nsp12 and nsp14 that increase ExoN-AA fitness confer significant fitness costs to WT-ExoN-DE.
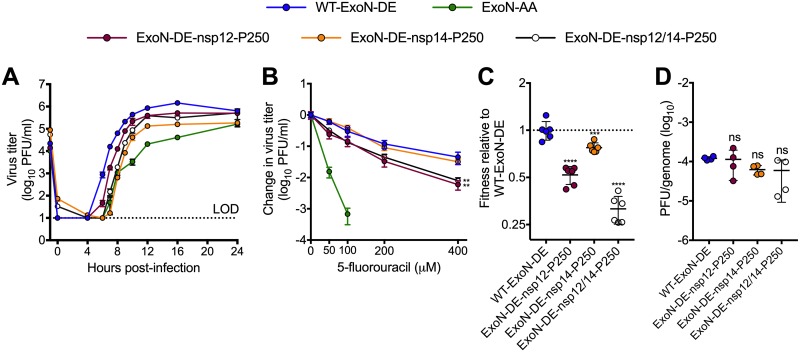
Mutational fitness effects are highly dependent upon the genetic background (26–28). In addition to reducing selective pressure for reversion, mutations conferring increased fitness to ExoN-AA might also reduce the benefits of motif I reversion. We previously reported that long-term passage (P250) of ExoN-AA selects for secondary adaptive mutations in the nsp12 RdRp and nsp14 (nsp12-P250 and nsp14-P250) (7). nsp12-P250 contains seven nonsynonymous mutations that partially compensate for defective proofreading and increase ExoN-AA fitness. nsp14-P250 contains six nonsynonymous mutations, including a conservative D-to-E substitution in ExoN motif III, and increases ExoN-AA fitness without compensating for defective proofreading. To test whether the fitness effects of passage-associated mutations in nsp12-P250 and nsp14-P250 depend upon the ExoN-AA genotype, we engineered a WT motif I (ExoN-DE) into viruses containing nsp12-P250 and nsp14-P250, alone and together, and analyzed replication, 5-FU sensitivity, and competitive fitness. Compared to WT-ExoN-DE, both ExoN-DE-nsp12-P250 and ExoN-DE-nsp14-P250 displayed delayed and decreased replication (Fig. 4A). In 5-FU sensitivity assays, ExoN-DE-nsp14-P250 was indistinguishable from WT-ExoN-DE, while both variants containing nsp12-P250 (ExoN-DE-nsp12-P250 and ExoN-DE-nsp12/14-P250) were significantly more sensitive to 5-FU (Fig. 4B). Finally, the nsp12-P250 and nsp14-P250 mutations significantly decreased fitness relative to that of WT-ExoN-DE (Fig. 4C). We detected no statistical differences between the specific infectivity of WT-ExoN-DE and that of any of the nsp12-P250 and nsp14-P250 variants in isolated infections (Fig. 4D). Thus, mutations in nsp12 and nsp14 that arose in the ExoN-AA background were detrimental to replication, mutagen sensitivity, and competitive fitness in the presence of a fully reverted ExoN-DE. These results support the conclusion that the adaptive pathways available to ExoN-AA may stabilize the ExoN-AA genotype, reducing both the selective pressure for, and the potential benefits of, primary reversion.
FIG 4.
Mutations that increase ExoN-AA fitness are detrimental in the presence of WT-ExoN-DE. (A) Replication kinetics of indicated viruses at an MOI of 0.01 PFU/cell plotted as means ± standard deviations (n = 3). (B) 5-Fluorouracil sensitivity at an MOI of 0.01 PFU/cell (means ± standard deviations of n = 6). (C) Competitive fitness of individual recombinants relative to WT-ExoN-DE. Viruses were competed with a tagged WT-ExoN-DE reference strain, and relative fitness was normalized to the mean value of the WT-ExoN-DE (means ± standard deviations of n = 6). (D) Specific infectivity (genomes per PFU) from isolated infections (means ± standard deviations of n = 4). Statistical significance of each virus relative to that of WT-ExoN-DE was determined with two-way analysis of variance with Dunnett’s multiple-comparison test (B) or by ordinary one-way analysis of variance with Dunnett’s multiple-comparison test (C and D). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. LOD, limit of detection.
DISCUSSION
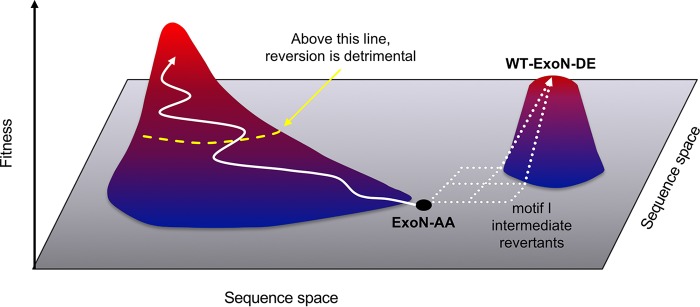
In this study, we demonstrate that the stability of the ExoN(–) motif I genotype in MHV (ExoN-AA) is a consequence of the limitations and opportunities of the genetic landscape it explores during replication (Fig. 5). Our results support a model in which the viable adaptive pathways leading to direct reversion of motif I from AA to DE are relatively flat on a fitness landscape, with intermediate revertants remaining phenotypically ExoN(–) and conferring no fitness advantage over ExoN-AA. In contrast, at least one alternative adaptive pathway is readily accessible and imparts immediate fitness gains over ExoN-AA. We propose that even minimal alternative pathway-adaptive fitness gains reduce the likelihood and benefits of motif I reversion until eventually the changing genetic background renders reversion detrimental. These data and this model suggest that selection during replication favors immediate, incremental fitness gains along the most accessible pathway rather than dramatic fitness increases across a larger genetic barrier. While this study focused on the issue of primary ExoN-AA reversion, it raises intriguing questions about the remaining topography of the ExoN-AA fitness landscape. Ongoing studies in our laboratory will determine whether ExoN-AA-P10 (and P250) represents a conserved pathway to compensate for defective proofreading or if CoV genomes accommodate multiple solutions to ExoN(–)-associated debilitations. Supporting the possibility of limited potential pathways, a recent study of vaccine-derived polioviruses determined that evolution for virulence in parallel epidemics proceeded through a limited set of genetic changes rather than a plethora of distinct trajectories (4).
FIG 5.
Model for the in vitro evolution of MHV-ExoN-AA. MHV-ExoN-AA (black dot) is a low-fitness variant. Reversion to WT-ExoN-DE would dramatically increase fitness but can be achieved only by traversing a flat landscape and climbing a steep fitness cliff (dotted white arrows). However, secondary mutations that incrementally increase fitness are more accessible (solid white arrow). Eventually, the genetic background changes enough that reversion becomes detrimental (dotted yellow line).
A key finding of this study is that mutations compensating for ExoN-AA are detrimental to WT-ExoN-DE, yet the mechanisms underlying this disparity are not clear. We expected that compensatory mutations in nsp14-P250 would hobble WT-ExoN-DE by inducing structural changes in the active site. However, the replication and fitness defects observed in ExoN-DE viruses containing nsp14-P250 were not associated with altered proofreading, as measured by 5-FU sensitivity (Fig. 4). In fact, the nsp14-P250 mutations do not substantially affect 5-FU sensitivity in either the ExoN-AA or WT-ExoN-DE background (7), suggesting that these mutations enhance some other stage of the MHV replication cycle, perhaps through interactions with other replicase proteins or with viral RNA. If so, these results imply that the proteins of the MHV replication holoenzyme must evolve in a highly cooperative manner to compensate for defective proofreading while also maintaining their intricate and intimate functional relationships. In line with this hypothesis, the likely high-fidelity nsp12-P250 disrupted replication, 5-FU resistance, and fitness in the presence of an intact ExoN-DE catalytic motif. Given the inverse relationship between polymerase fidelity and replication speed (the slower the polymerase proceeds, the more time it has to discriminate incoming nucleotides) (29–32), nsp12-P250 could fracture the delicate kinetic balance between polymerization and nsp14-ExoN-mediated excision during replication. Support for this hypothesis comes from a recent study establishing that the SARS-CoV-nsp12 replicates with lower fidelity than the RdRp of dengue virus and from the observation that the likely high-fidelity RdRp variant MHV-nsp12-V553I delays replication of proofreading-capable MHV (9, 33). As with nsp14-P250, nsp12-P250 mutations could have additional effects on RdRp functions or protein-protein interactions that are incompatible with an active ExoN domain. Finally, it is possible that the adaptive mutations in nsp12-P250 and nsp14-P250, including synonymous changes, could modify RNA sequence features or secondary structures in ways that interfere with WT-ExoN-DE. Defining the molecular interactions underlying the strain specificity of ExoN-AA-compensatory mutations using recombinant viruses or reconstituted biochemical systems will provide a unique opportunity to explore the complex evolutionary and functional relationships that underpin the coronavirus replication machinery.
Our results also extend existing studies of CoV ExoN motif I. Motif I DE→AA mutations in the SARS-CoV nsp14-ExoN dramatically reduce nuclease activity in biochemical assays, but no study has examined the contributions of each residue independently (16, 18). While we cannot exclude the possibility that intermediate revertants of ExoN-AA retain some level of nucleolytic activity, the lack of consistent or statistical differences in replication, 5-FU sensitivity, or competitive fitness relative to levels for ExoN-AA supports previous studies demonstrating that motif I DE is essential for ExoN function (16, 18, 19). Given these results, we were surprised to observe repeated reversion of the ExoN-AE but not the other two 2-nt variants, ExoN-DA and ExoN-ED. One potential explanation is that the specific mutational bias of ExoN-AE makes the revertant mutations more accessible than in ExoN-DA or ExoN-ED. Alternatively, if ExoN-AE has profound replication or fitness defects, selection could drive primary reversion more quickly away from this genotype. Consistent with this hypothesis, ExoN-AE reverted more quickly at a higher MOI, where natural selection acts more efficiently on a larger population size (Table 1) (34). Biochemical studies of the ExoN-AA intermediate revertants will be valuable to determine whether nsp14-ExoN-AE differs from the other intermediate revertants and to define the structural and kinetic features of nsp14-ExoN catalysis.
Finally, our studies suggest that compensatory mutations identified through long-term passage could be used to stabilize the ExoN-AA genotype. In particular, the high-fidelity nsp12-P250 could reduce the probability of reversion by reducing mutational sampling within motif I (35), and both nsp12-P250 and nsp14-P250 render the MHV genome inhospitable to WT-ExoN-DE. Together, these studies argue that experimental evolution can generate reagents to define critical interactions involved in CoV replication and can identify new strategies for stabilizing attenuated CoVs.
MATERIALS AND METHODS
Cell culture.
Delayed brain tumor (DBT-9) cells (36) and baby hamster kidney 21 cells expressing the MHV receptor (BHK-R) (37) were maintained at 37°C in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% serum (HyClone Fetal Clone II [GE Healthcare] or fetal bovine serum [Invitrogen]), 100 U/ml penicillin and streptomycin (Gibco), and 0.25 μM amphotericin B (Corning). BHK-R cells were further supplemented with 0.8 mg/ml G418 selection antibiotic (Gibco). The infectious clone of the murine hepatitis virus strain A59 (MHV-A59; GenBank accession number AY910861) was used as the template for all recombinant viruses.
Determination of viral titer by plaque assay.
Virus samples were serially diluted and inoculated on subconfluent DBT-9 cell monolayers in either a 6- or 12-well format. Cells were overlaid with 1% agar in DMEM and incubated overnight at 37°C. Plates were fixed with 4% formaldehyde, and agar plugs were removed. The number of plaques per well was counted by hand and used to calculate titers (37).
Plaque purification of viral populations.
DBT cells were infected with serial dilutions of virus and overlaid with 1% agar in DMEM. Single plaques were isolated with glass Pasteur pipettes, resuspended in phosphate-buffered saline (PBS) containing calcium and magnesium, and inoculated onto fresh DBT cells. This process was completed three times before experimental stocks were generated.
Cloning and recovery of recombinant viruses.
Site-directed mutagenesis in MHV genome fragments was performed using “round-the-horn” PCR (originally described in Ho et al. [38]). Briefly, adjacent primers containing the mutation of interest were 5′-phosphorylated using T4 polynucleotide kinase (M0201S; NEB) using the buffer from the T4 DNA ligase, which contains ATP (M0202S). PCR was performed on a plasmid template using Q5 High-Fidelity 2× master mix (M0492L; NEB), with primers at a final concentration of 500 nM. The linear amplification product was purified using a Promega Wizard SV Gel and PCR Clean-up System (A9282; Promega Corporation), and 4 μl was ligated at 16°C overnight with T4 DNA ligase (M0202S; NEB). After transformation into chemically competent Top10 E. coli (lab-derived) and expansion in liquid culture, the MHV segment of each plasmid was sequenced. Viruses were constructed, rescued, and sequenced as described previously (7, 13, 37). Experimental stocks were generated by infecting a subconfluent 150-cm2 flask of DBT-9 cells at an MOI of 0.01 PFU/cell. Flasks were frozen at –80°C when monolayers were fully involved, which was approximately 20 to 28 h postinfection depending on the variant. After thawing, the supernatant was clarified by centrifugation at 4,000 × g (Sorvall RC 3B Plus with an HA-6000A rotor) for 10 min at 4°C. For intermediate revertants, stocks were generated in serum-free DMEM and processed as described above before being concentrated roughly 10-fold by centrifugation at 4,000 × g using Amicon Ultra-15 centrifugal filter units (100-kDa cutoff) (UFC910008; EMD Millipore). The virus titer of each stock was determined by plaque assay using DBT-9 cells as described above.
Passage of ExoN intermediate revertants.
Intermediate revertants of ExoN-AA were passaged 10 times on subconfluent DBT-9 cell monolayers in 24-well plates at an estimated MOI of either 0.01 or 0.5 PFU/cell. Supernatants were harvested at 24 and 20 h postinfection for an MOI of 0.01 and 0.5 PFU/cell, respectively, and screened for WT reversion by plaque assay. At least three WT-like plaques were sequenced for each lineage to confirm motif I reversion.
Replication kinetics.
Viral replication kinetics in DBT-9 cells were determined at an MOI of 0.01 PFU/cell as described previously (11). Replicates were synchronized by 30-min incubation at 4°C before being transferred to the 37°C incubator. Supernatant (300 μl) was harvested, and titers were determined by plaque assay.
Determination of specific infectivity.
Subconfluent monolayers of DBT-9 cells in 24-well plates were infected with the indicated virus at an MOI of 0.05 PFU/cell, and supernatant was harvested at 16 h postinfection. Genomic RNA in supernatant was quantified using one-step reverse transcription-quantitative RT-PCR (RT-qPCR) on TRIzol-extracted RNA as described previously (9). Briefly, genomic RNA was detected with a 5′ 6-carboxyfluorescein (FAM)- and 3′ black hole quencher 1 (BHQ-1)-labeled probe targeting nsp2 (Biosearch Technologies, Petaluma, CA), and RNA copy number was calculated by reference to an RNA standard derived from the MHV A fragment. Samples were plated in technical duplicates to minimize well-to-well variation. Titers were determined by plaque assay in DBT-9 cells, and specific infectivity was calculated as the number of PFU per supernatant genomic RNA copy.
5-Fluorouracil sensitivity assays.
Stock solutions of 5-fluorouracil (F6627; Sigma) were prepared in dimethyl sulfoxide (DMSO). Sensitivity assays were performed in 24-well plates at an MOI of 0.01 PFU/cell, as previously described (7). Cells were incubated with drug for 30 min prior to infection. Supernatants were harvested at 24 h postinfection, and titers were determined by plaque assay.
Competitive fitness assays.
ExoN-AA-reference and WT-ExoN-DE-reference viruses were marked with seven consecutive silent mutations within nsp2 (wild type, 1301-TTCGTCC-1307; reference, 1301-CAGCAGC-1307) by round-the-horn PCR, as described above. Competitions were performed in triplicate on DBT-9 cells in 12-well plates, plated at a density of 1 × 105 cells per well 24 h prior to infection. Cells were infected at a total MOI of 0.1 PFU/cell (MOI of 0.05 PFU/cell each for competitor and reference virus). Supernatants were harvested at 15 and 16 h postinfection for experiments with ExoN-AA-reference and WT-ExoN-DE-reference, respectively, and passaged four times. Titers of samples were determined between all passages to maintain a total MOI of 0.1 PFU/cell. RNA was extracted from 70 μl of supernatant using a QIAamp 96 virus QIAcube HT kit on a QIAcube HT System (Qiagen). Each RNA sample was analyzed by one-step RT-qPCR with two SYBR green assays. Reference viruses were detected with forward primer SS-qPCR-Sil-F (5′-CTATGCTGTATACGGACAGCAGT-3′; 200 nM final concentration) and reverse primer SS-qPCR-R2 (5′-GGTGTCACCACAACAATCCAC-3′; 200 nM final concentration). Competitors were detected with forward primer SS-qPCR-WT-F (5′-CTATGCTGTATACGGATTCGTCC-3′, 450 nM final concentration) and reverse primer SS-qPCR-R2 (5′-GGTGTCACCACAACAATCCAC-3′, 450 nM final concentration). RNA samples were diluted 1:100 prior to RT-qPCR with a Power SYBR green RNA-to-Ct 1-step kit (Applied Biosystems) according to the manufacturer’s protocol. Duplicate wells were averaged, and values were excluded from subsequent analysis if the duplicate wells differed by >0.5 threshold cycle (CT). The relative abundances of competitor and reference were determined by subtracting CT values (ΔCT competitor = CT competitor – CT reference) and converted to reflect the fold change in the ratio (Δratio = 2−Δ CT competitor). The log10(Δratio) was plotted against passage number, and the change in log10(Δratio) (i.e., slope of linear regression) is the relative fitness. Note that regressions were fit only for P1 to P4 as slight deviations in the 1:1 ratio in the input (P0) can skew the slope.
Statistical analysis.
GraphPad Prism, version 6 (La Jolla, CA), was used to perform statistical tests. Only the comparisons marked within the figures or legends were performed. In many cases the data were normalized to levels of the untreated controls. This was performed using GraphPad Prism, version 6. The number of replicate samples is indicated within each figure legend.
ACKNOWLEDGMENTS
We thank members of the Denison laboratory and Seth Bordenstein for valuable discussions, as well as Andrea Pruijssers for critical reviews of the manuscript.
This work was supported by U.S. Public Health Service awards R01-AI108197 (M.R.D), T32-GM007347 (K.W.G), F30-AI129229 (K.W.G), T32-AI089554 (N.R.S.), F31-AI133952 (M.L.A.), and T32-AI089554 (M.L.A.), all from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolan PT, Whitfield ZJ, Andino R. 2018. Mapping the evolutionary potential of RNA viruses. Cell Host Microbe 23:435–446. doi: 10.1016/j.chom.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern A, Yeh Te M, Zinger T, Smith M, Wright C, Ling G, Nielsen R, Macadam A, Andino R. 2017. The evolutionary pathway to virulence of an RNA virus. Cell 169:35–46.e19. doi: 10.1016/j.cell.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, Ray AS, Cihlar T, Siegel D, Mackman RL, Clarke MO, Baric RS, Denison MR. 2018. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9:e00221-18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graepel KW, Lu X, Case JB, Sexton NR, Smith EC, Denison MR. 2017. Proofreading-deficient coronaviruses adapt for increased fitness over long-term passage without reversion of exoribonuclease-inactivating mutations. mBio 8:e01503-17. doi: 10.1128/mBio.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. 2013. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog 9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sexton NR, Smith EC, Blanc H, Vignuzzi M, Peersen OB, Denison MR. 2016. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J Virol 90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Case JB, Li Y, Elliott R, Lu X, Graepel KW, Sexton NR, Smith EC, Weiss SR, Denison MR. 2017. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J Virol 92:e01531–17–38. doi: 10.1128/JVI.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith EC, Case JB, Blanc H, Isakov O, Shomron N, Vignuzzi M, Denison MR. 2015. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J Virol 89:6418–6426. doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, Baric RS. 2012. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med 18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol 81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH III, Leist SR, Yount BL Jr, McAnarney ET, Graham RL, Waters KM, Baric RS. 2018. Combination attenuation offers strategy for live-attenuated coronavirus vaccines. J Virol 92:e00710-18. doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, Wu L, Shaw N, Gao Y, Wang J, Sun Y, Lou Z, Yan L, Zhang R, Rao Z. 2015. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci U S A 112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. 2012. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A 109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becares M, Pascual-Iglesias A, Nogales A, Sola I, Enjuanes L, Zuñiga S. 2016. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol 90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graepel KW, Agostini ML, Lu X, Sexton NR, Denison MR. 2019. Fitness barriers limit reversion of a proofreading-deficient coronavirus. bioRxiv 10.1101/618249. [DOI] [PMC free article] [PubMed]
- 22.Steitz TA, Steitz JA. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A 90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P, Jiang M, Hu T, Liu Q, Chen XS, Guo D. 2007. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol 40:649–655. [DOI] [PubMed] [Google Scholar]
- 24.Derbyshire V, Grindley ND, Joyce CM. 1991. The 3′→5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J 10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case JB, Ashbrook AW, Dermody TS, Denison MR. 2016. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J Virol 90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, Russ G, Gibbs JS, Bennink JR, Yewdell JW. 2013. Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe 13:314–323. doi: 10.1016/j.chom.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima K, Nobusawa E, Nagy A, Nakajima S. 2005. Accumulation of amino acid substitutions promotes irreversible structural changes in the hemagglutinin of human influenza AH3 virus during evolution. J Virol 79:6472–6477. doi: 10.1128/JVI.79.10.6472-6477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koel BF, Burke DF, van der Vliet S, Bestebroer TM, Rimmelzqaan GF, Osterhaus ADME, Smith DJ, Fouchier RAM. 2018. Epistatic interactions can moderate the antigenic effect of substitutions in hemagglutinin of influenza H3NII virus. bioRxiv 10.1101/506030. [DOI] [PMC free article] [PubMed]
- 29.Fitzsimmons WJ, Woods RJ, McCrone JT, Woodman A, Arnold JJ, Yennawar M, Evans R, Cameron CE, Lauring AS. 2018. A speed-fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol 16:e2006459. doi: 10.1371/journal.pbio.2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regoes RR, Hamblin S, Tanaka MM. 2013. Viral mutation rates: modelling the roles of within-host viral dynamics and the trade-off between replication fidelity and speed. Proc Biol Sci 280:20122047–20122047. doi: 10.1098/rspb.2012.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. 1996. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry 35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 32.Elena SF, Sanjuán R. 2005. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol 79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferron F, Subissi L, Silveira De Morais AT, Le NTT, Sevajol M, Gluais L, Decroly E, Vonrhein C, Bricogne G, Canard B, Imbert I. 2018. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A 115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolan PT, Whitfield ZJ, Andino R. 2018. Mechanisms and concepts in RNA virus population dynamics and evolution. Annu Rev Virol 5:69–92. doi: 10.1146/annurev-virology-101416-041718. [DOI] [PubMed] [Google Scholar]
- 35.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem 280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W, Baric RS. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol 70:3947–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/jvi.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]