Abstract
Background
Variants in the Structural Maintenance of Chromosomes flexible Hinge Domain-containing protein 1 (SMCHD1) can cause facioscapulohumeral muscular dystrophy type 2 (FSHD2) and the unrelated Bosma arhinia microphthalmia syndrome (BAMS). In FSHD2, pathogenic variants are found anywhere in SMCHD1 while in BAMS, pathogenic variants are restricted to the extended ATPase domain. Irrespective of the phenotypic outcome, both FSHD2-associated and BAMS-associated SMCHD1 variants result in quantifiable local DNA hypomethylation. We compared FSHD2, BAMS and non-pathogenic SMCHD1 variants to derive genotype–phenotype relationships.
Methods
Examination of SMCHD1 variants and methylation of the SMCHD1-sensitive FSHD locus DUX4 in 187 FSHD2 families, 41 patients with BAMS and in control individuals. analysis of variants in a three-dimensional model of the ATPase domain of SMCHD1.
Results
DUX4 methylation analysis is essential to establish pathogenicity of SMCHD1 variants. Although the FSHD2 mutation spectrum includes all types of variants covering the entire SMCHD1 locus, missense variants are significantly enriched in the extended ATPase domain. Identification of recurrent variants suggests disease-specific residues for FSHD2 and in BAMS, consistent with a largely disease-specific localisation of variants in SMCHD1.
Conclusions
The localisation of missense variants within the ATPase domain of SMCHD1 may contribute to the differences in phenotypic outcome.
INTRODUCTION
Next generation sequencing (NGS) has revolutionised the diagnostic workup of genetic diseases.1 One complicating factor of the use of NGS in diagnostics, however, is the interpretation of variants of uncertain significance (VUS).2 One way to deal with these VUS is to combine clinical, family and variant in silico prediction information with functional testing, if available. For epigenetic disorders, that is, disorders caused by defective chromatin modifiers, functional readouts may be readily available in the DNA itself through the appearance of aberrant epigenetic signatures. One such epigenetic disorder is facioscapulohumeral muscular dystrophy (FSHD), a myopathy with a prevalence of 1:8.500, clinically characterised by progressive and often asymmetric weakness and atrophy of the facial and upper extremity muscles.3–6 FSHD is caused by the misexpression of DUX4, a retrogene encoding for a germline and cleavage stage transcription factor.7,8 In FSHD, DUX4 becomes transcriptionally derepressed in skeletal muscle and ectopic DUX4 expression in myonuclei eventually leads to muscle cell death in vitro and in transgenic mice.9–11
A complete copy of the DUX4 retrogene is located in the distal end of the D4Z4 macrosatellite repeat on one of the two main variants of chromosome 4, 4qA. The D4Z4 repeat is polymorphic in size and normally varies between 8 and 100 units.12 For most cases, DUX4 derepression is caused by D4Z4 repeat contractions to a size of 1–10 repeat units (FSHD type 1; FSHD1; MIM 158900)13 Because of the repeat contraction in FSHD1, the D4Z4 chromatin structure relaxes and DUX4 becomes transcriptionally derepressed in somatic cells.14 Two major allelic variants of the subtelomere of chromosome 4q have been described, 4qA and 4qB, and a highly homologous D4Z4 repeat is also present in the subtelomere of chromosome 10 (10q). However, only D4Z4 repeat contractions on 4qA chromosomes are pathogenic.15 The explanation for this observation is that the DUX4 retrogene is incomplete on non-permissive chromosomes 4qB and 10q as they lack the most distal DUX4 exon or the somatic DUX4 polyadenylation signal, respectively.16,17
FSHD type 2 (FSHD2; MIM 158901), the rarer subtype of this myopathy,18 has a digenic inheritance pattern as it requires a heterozygous pathogenic variant in the Structural Maintenance of Chromosomes flexible Hinge Domain-containing 1 gene (SMCHD1) on chromosome 18p and an intermediate size D4Z4 repeat (8–20 units) on a 4qA chromosome.19,20 Pathogenic SMCHD1 variants are associated with DNA hypomethylation and other attributes of D4Z4 chromatin relaxation, resulting in somatic derepression of DUX4, like in FSHD1 (online supplementary figure 1). The SMCHD1 pathogenic variant spectrum in FSHD2 includes nonsense, missense, insertion-deletion (indel) variants and variants that affect pre-mRNA splicing, with 10% of variants affecting the 3’ splice site of exon 25 online supplementary table 1.19,21–31 The pathogenicity of SMCHD1 variants can be predicted by in silico programme, but the accuracy of these predictions is variable. Since SMCHD1 is a chromatin modifier that affects DNA methylation, methylation analysis seems a more reliable tool to establish the functional effect of SMCHD1 variants.
Several D4Z4 methylation analysis strategies have been developed based on DNA digestion with methylation-sensitive endonucleases or on bisulfite sequencing.18,19,32–35 At the FseI restriction site in D4Z4, an average methylation level of 46.8% (with a SD of 14.1%) is found for all D4Z4 repeats in control individuals. Patients with FSHD2 have been defined by having D4Z4 methylation <1.5 SD of the average methylation in controls (<25%; online supplementary figure 2).24 As the methylation level at this site not only depends on variants in epigenetic modifiers of D4Z4 but also linearly correlates with the D4Z4 repeat size, the delta1 algorithm was developed to correct for the D4Z4 repeat size.24 The delta1 score has an average in controls of −0.2% with a SD of 10.0%. As the delta1 score is more precise than the FseI methylation, the threshold for FSHD2 is stricter and is defined to be <2 SD below the control delta1 score, that is, lower than −20% (online supplementary figure 2). To compare the methylation defect of different SMCHD1 variants, the delta2 score was created similarly for carriers of a pathogenic SMCHD1 variant.24
Pathogenic SMCHD1 variants have also been reported in the clinically unrelated Bosma arhinia microphthalmia syndrome (BAMS).36,37 Patients with BAMS typically do not show FSHD-like features but instead have severe hypoplasia or absence of the external nose.38 In contrast to FSHD2, all reported pathogenic BAMS variants are missense variants in the C-terminal extended ATPase domain of SMCHD1.36,37,39 Patients with BAMS typically have D4Z4 hypomethylation similar to FSHD2.37
The cause of the different clinical outcomes of pathogenic SMCHD1 variants is poorly understood. Some studies suggest that FSHD2 and BAMS variants have opposite functional effects on SMCHD1 ATPase activity.36,39 However, family studies have identified one BAMS family in which one family member was identified with a muscle phenotype without BAMS features.37 Moreover, identical pathogenic SMCHD1 variants have been described in BAMS and FSHD2, but within these families such variant is exclusively associated with only one of the two possible clinical outcomes.27
Our aim is to bring further clarity to the divergent clinical outcome of pathogenic SMCHD1 variants. We therefore explored pathogenic and non-pathogenic variants identified in patients with FSHD2 and BAMS and control individuals identified by a review of all cases previously reported and newly diagnosed patients. We thus expand the SMCHD1 mutation spectrum. Our hypothesis is that the location of specific variants determines the phenotype by affecting separate functions of the protein and will assess this with use of a computational model of SMCHD1.
MATERIALS AND METHODS
Patients and controls
All participants provided written consent. We included 101 families from previous publications19,21–31 and 86 newly identified FSHD2 families. All families have one or more patients with an age-corrected severity score (ACSS)40 ≥50 or a typical FSHD phenotype without documented ACSS. Altogether, we collected information on the SMCHD1 variant and D4Z4 methylation from 229 affected and 37 unaffected SMCHD1 mutation carriers from 187 different FSHD2 families. For BAMS, we collected information on SMCHD1 variants and D4Z4 methylation from 41 families from previous publications.36,37 Non-pathogenic SMCHD1 variants were identified in non-FSHD samples (unaffected controls, unaffected individuals and siblings/spouses of individuals with FSHD1 and individuals with a different established genetic condition). Non-pathogenicity was confirmed based on having normal D4Z4 methylation (<1 SD from the average FseI, or delta1 in controls, online supplementary figure 2) in blood-derived genomic DNA. For non-pathogenic variants, we established normal D4Z4 methylation in multiple carriers. In online supplementary figure 2, we used the methylation values of 89 unrelated patients with FSHD2 and 249 control individuals from a previous study.20
Genetic analysis of D4Z4 repeats, SMCHD1 sequencing and variant prediction and statistical analysis
These studies were done as described previously (online supplementary file 1).
Modelling of the SMCHD1 ATPase domain
The ATPase domain of SMCHD1 belongs to the protein family of GHKL ATPases that include proteins such as Gyrase B, Hsp90 and MutL. Sequence comparison of different family members showed closest sequence similarity to TRAP1, the mitochondrial paralog of Hsp90. The ATPase domain (residues 107–334) and downstream domain of SMCHD1 (residues 335–465) were modelled on the crystal structure of TRAP1.41 For this, 25 non-redundant SMCHD1 sequences (40%–95% sequence identity) were aligned with 25 non-redundant TRAP1 and Hsp90 sequences (38%–65% sequence identity) using MAFFT.42 Next, the aligned sequences of SMCHD1 and TRAP1 were extracted and manually adjusted, aided by secondary structure predictions of SMCHD1 calculated with the Quick2D tool in the MPI toolkit43 and by secondary structure elements from the crystal structure of TRAP1. The model for SMCHD1 ATPase domain was built using Modeller V.9.244 and manually inspected and adjusted in PyMOL (The PyMOL Molecular Graphics System, V.2.0 Schrödinger, LLC) and Coot45 and the sequence alignment adjusted where necessary.
RESULTS
Variants in FSHD2
We collected data from 187 families with at least one clinically affected relative. In the Leiden University Medical Center, FSHD2 patient selection was based on: having a phenotype consistent with FSHD and having a delta1 score less than or equal to −20%, or, if unavailable, having FseI site D4Z4 methylation ≤25% (online supplementary figure 2). All patients with FSHD2 identified have at least one permissive 4qA allele with a D4Z4 repeat of 8–20 units or a duplicated 4qA-type D4Z4 repeat.20 In 40 families, we included, next to the proband, other affected or unaffected carriers of the pathogenic SMCHD1 variant. Unaffected SMCHD1 variant carriers generally do not have a permissive 4qA allele or they have permissive alleles with a D4Z4 repeat >20 units. We also included SMCHD1 variants identified from independent studies, based on having an FSHD phenotype and D4Z4 methylation below the FSHD2 threshold.
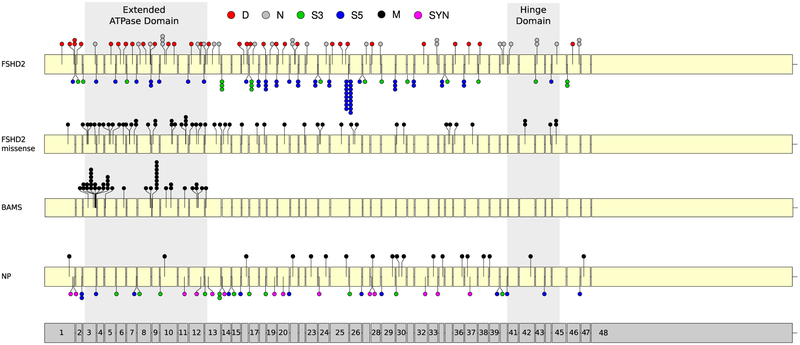
Currently, 101 pathogenic SMCHD1 variants have been reported in unrelated patients with FSHD2 and listed in the Leiden Open variant Database (LOVD) (https://www.lovd.nl/) as queried in January 2019 (online supplementary table 1).19,21–31 In this collaborative study, we identified 86 new FSHD2 causing variants, totaling 187 FSHD2 variants. This includes 28 nonsense, 35 indel and 54 missense variants (online supplementary table 2, 3). In three families, of which one has not been reported yet, one copy of the SMCHD1 locus was deleted entirely.25 The mutation spectrum further includes 70 variants that were shown or predicted to interfere with splicing, of which 52 are at the 5′ splice site and 18 at the 3′ splice site (online supplementary table 2, 3). Six of these splice-site variants are also predicted to cause an amino acid substitution. Consistent with previous studies,23,24 the variants are distributed over the entire SMCHD1 locus (figure 1). However, missense variants are significantly (p=3.61E–06) more often found in the extended ATPase domain than expected based on the size of this domain (figure 1, online supplementary table 4).
Figure 1.
Distribution of SMCHD1 variants in unrelated patients with FSHD2 and BAMS and in controls over the SMCHD1 locus. The top locus representation shows all indels (D, red), nonsense (N, white) and splice site (S3, green and S5, blue) variants identified in FSHD2. The marks S3 and S5 refer to the position of the splice variants with respect to the 3′ end or 5′ end splice site, respectively. The second representation shows all the missense (M, black) variants identified in FSHD2. The third representation shows all missense variants identified in 41 unrelated patients with BAMS. The fourth representation shows the non-pathogenic (NP) missense, SYN (pink) variants identified in control individuals. The grey box indicates the extended ATPase domain (left, exon 3–12) and the Hinge domain (right, exon 41–45). Identical indel and nonsense variants, missense variants for the same residue and splice variants for the same intronic region (S3 or S5) are indicated by a stack of dots at the same position. Exon numbering is shown at the bottom of the figure. An excess of missense variants in the extended ATPase domain is observed in BAMS and in FSHD2 coinciding with a paucity of NP missense variants in this domain. BAMS, Bosma arhinia microphthalmia syndrome; FSHD2, facioscapulohumeral muscular dystrophy type 2; NP, non-pathogenic; SMCHD1, Structural Maintenance of Chromosomes flexible Hinge Domain-containing protein 1; SYN, splice site and synonymous.
For 250 pathogenic variant carriers from 166 families, D4Z4 methylation data was available and for 156 carriers from 89 families full repeat size information was available facilitating the calculation of delta scores. In general, the delta scores are rather similar for carriers of the same variant within individual families and between families, with some exceptions (online supplementary figure 3). As two outliers represented the oldest members of their respective families (Rf975 and Rf1034), we excluded gonosomal mosaicism46 as possible explanation for the high D4Z4 methylation levels (data not shown). To compare the deleteriousness of variants, we selected one representative from each family, the proband or the oldest patient with FSHD2 with methylation information, for further analyses. The FseI methylation level in 89 independent SMCHD1 pathogenic variant carriers is 10.9% (±5.4%), the average delta1 score is −31.6% (±5.6%) and the average delta2 score is −0.7% (±4.6%), which is consistent with our previous study.24
Recurrent variants in SMCHD1
Some variants in unrelated families affect the same intron or the same residue: 9.1% (17/187) of variants were found in the mutation hotspot at the 5′ splice site of exon 25.24 Also, 14 other introns were affected in at least two unrelated families by variants in the same splice site consensus sequence and for four of these introns by an identical variant in unrelated families (figure 1). Similarly, one recurrent indel variant and five recurrent nonsense variants were found in 11 unrelated families. We identified three recurrent missense variants and three examples in which a different variant acted on the same amino acid. Interestingly, these missense variants are in the extended ATPase domain (n=4) and the Hinge domain (n=2) of SMCHD1.
Consequences of FSHD2-associated variants in SMCHD1
Pathogenic variants that preserve the open reading frame (ORF) of SMCHD1 are associated with more profound D4Z4 hypomethylation than those that disrupt the ORF.24 Analysis of nine splice site variants identified two outcomes: splice site variant resulted in a stable exon skip if this event did not disrupt the ORF while it resulted in RNA instability when the skipped exon causes a frame shift.24 Based on this observation, we predicted the effect for all newly identified splice variants. This yielded 37 unrelated ORF-disrupting (D-ORF) and 52 ORF-pre- serving (P-ORF) variants. For all studied variants, we find a significant lower (p=0.0037; unpaired t-test) delta2 value, that is, more reduced D4Z4 methylation, for P-ORF variants (mean −1.8%) compared with D-ORF variants (mean +0.9%) (online supplementary figure 4).
Based on the prediction tools Polyphen, SIFT and Align GVGD, 48/51 unique FSHD2 missense variants were predicted pathogenic in at least two prediction programmes and 3/51 were predicted non-pathogenic in at least two programmes (online supplementary table 2).
Non-pathogenic variants in SMCHD1
Non-pathogenic (benign or neutral) SMCHD1 variants were identified through whole exome sequencing analysis in unrelated non-FSHD individuals. If possible, D4Z4 methylation levels were determined for these variants. We considered a variant non-pathogenic if the FseI methylation was within 1 SD from the average methylation in controls (>32. 7%) (online supplementary figure 2). For the delta1 score, we considered values normal if they were within 1.5 SD from the average in controls (above −15%). Based on these thresholds, we identified 58 different non-pathogenic variants: 19 missense, 25 intronic and 14 synonymous variants in unaffected control individuals (online supplementary table 2). Intronic variants were almost equally prevalent in the 5′ and 3′ splice sites and, for these variants, highly conserved nucleotides in the splice consensus sequences were never involved. Based on three prediction tools, 14/19 missense variants were predicted non-pathogenic in at least two programmes, and 5/19 were predicted pathogenic in at least two programmes (online supplementary table 2). Interestingly, while non-pathogenic splice site and synonymous variants are equally distributed over the SMCHD1 locus, only a single missense variant (G396K) was found in the extended ATPase domain and this variant was predicted to be non-pathogenic by all prediction tools (figure 1).
SMCHD1 variants of uncertain significance
In two affected individuals, we identified potentially pathogenic SMCHD1 variants associated with delta1 values above the established threshold for FSHD2. In an affected Asian woman (Rf653), we found missense variant E647K, which was predicted pathogenic in 2/3 prediction tools. She has a 10 D4Z4 units long 4qA allele with a delta1 score of −16%. The second patient is a European male (Rf668), who has a six D4Z4 units 4qA allele and a delta1 score of −11%. He also has an N-terminal SMCHD1 missense variant (c.4G>C, A2P), which was predicted pathogenic in 2/3 prediction tools. The patient has two mildly affected daughters who both carry a familial 16 D4Z4 units 4qA allele. They have delta1 methylation scores of −15% and −13%, but only one daughter inherited the SMCHD1 variant. Their mother is unaffected and has normal D4Z4 methylation (online supplementary figure 5).
Pathogenic sMCHD1 variants in BAMS versus FSHD2
Only missense variants in the extended ATPase domain of SMCHD1 have been identified in BAMS (online supplementary table 5). Remarkably, changes in only 20 different amino acids have been reported in 41 unrelated BAMS families, as nine amino acid positions are recurrently altered in unrelated patients (figure 1). As some of these amino acids were altered due to a different DNA variant and 15/25 carriers of an identical variant were de novo cases, accidental founder allele involvement is unlikely.36,37 A similar observation was made in FSHD2 for four amino acids in the extended ATPase domain that were recurrently altered. The nine recurrent BAMS variants were not overlapping with the 21 unique FSHD2 variants in the extended ATPase domain. Likewise, four recurrent FSHD2 missense variants in the extended ATPase domain did not overlap with the BAMS variants (figure 1 and online supplementary table 2). Although two variants (L107P and G137E) have been identified to cause BAMS and FSHD2, the high incidence of recurrent variants and the difference in mutation spectrum between both disorders suggests a variant-specific phenotype. This prompted us to study these variants in a three-dimensional model of the ATPase domain.
Modelling pathogenic variants in the ATPase domain of SMCHD1
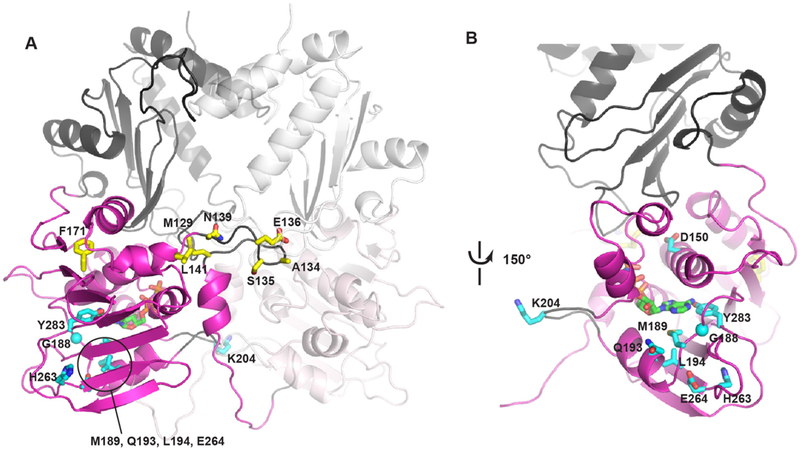
The ATPase and C-terminally extended, or downstream domain (residues 111–365),36,39 of SMCHD1 were modelled on the crystal structure of the GHKL ATPase TRAP1. The ATPase domain (residues 111–334) shows a strong degree of structural conservation between different members of the GHKL family, while the downstream domain (termed M-domain in TRAP1 or transducer domain in Gyrase B and MORC2) shows a much greater structural diversity (online supplementary figure 6). Therefore, variants were not mapped to this part of the model, and the extended domain is only shown to mark its position relative to the conserved ATPase domain. FSHD2 and BAMS variants that are located in the ATPase domain of SMCHD1 were mapped onto the model (figure 2A). Remarkably, FSHD2 variants almost exclusively locate around the ATP-binding site (figure 2B), with the exception of lysine 204 (K204) that is located on a loop adjacent to the ATP-binding pocket. This loop is longer than its equivalent loop in TRAP1 and therefore the position of this loop, and K204 on it, is not well defined. Yet, its close vicinity to the ATP-binding pocket, as well as the close location of all other FSHD2 variants to this pocket, suggest it may be positioned closer to the nucleotide in the real structure of SMCHD1. Interestingly, variant Q193P, recently described in a patient with FSHD2 and published after we finished this study,47 is also located around the ATP-binding site. In contrast to FSHD2 variants, the vast majority of BAMS variants localise to a loop that is positioned at the dimer interface (figure 2A). This loop is also involved in dimer contacts in other GHKL ATPases (online supplementary figure 6), suggesting that the BAMS variants may alter the dimerisation properties of SMCHD1.
Figure 2.
Computational model of the SMCHD1 extended ATPase domain. (A) Overview of the SMCHD1 dimer with conserved ATPase domain (residue 111–334) coloured in magenta and downstream domain in grey. Second monomer coloured in light magenta and light grey. FSHD2 variants in the ATPase domain (D150H, G188R, M189V, Q193P, L194F, K204E, H263D, E264K and Y283C) are shown in cyan sticks, BAMS variants (M129K/R, A134S, 135 C/N/I, E136G/D, N139H, L141F and F171V) in yellow sticks and AT P shown in green. Most BAMS variants are located at the dimer interface, while FSHD2 variants are primarily located around the AT P binding pocket. FSHD2 variants that are hidden behind the beta-sheet are highlighted in circle. (B) Close up of the nucleotide-binding site (rotated ~150° from the view in A). FSHD2 variants surround the AT P molecule. BAMS, Bosma arhinia microphthalmia syndrome; FSHD2, facioscapulohumeral muscular dystrophy type 2; SMCHD1, Structural Maintenance of Chromosomes flexible Hinge Domain-containing Protein 1.
DISCUSSION
FSHD2-associated variants
Adding to 101 known SMCHD1 variants,19,21–31 we here introduce 86 new pathogenic variants totaling 187 FSHD2-causing SMCHD1 variants. These variants are distributed over the SMCHD1 locus and we note that the variant spectrum is compa- rable between the different studies (online supplementary table 3).
We confirm the mutation hotspot in the S5 splice site of exon 25 accounting for 17/187 variants. In this hotspot, we identified six unique variants, all shown or predicted to cause alternative splicing. Outside this hotspot, we also encountered one recurrent indel, five recurrent nonsense, three recurrent missense and 14 other recurrent splice site variants figure 1 online supplementary table 3 (figure 1). These variants have either arisen multiple times or the carriers of these variants are distantly related. We not only identified three identical missense variants but also three different variants modifying the same residue. Four of these recurrent amino acid changes were found in the extended ATPase domain and two in the Hinge domain.
For some recurrent variants, we documented de novo appearance in one of the families, but for most cases we cannot determine the origin of the variant in the absence of family information. Due to the digenic inheritance in FSHD2, it is not unlikely that identical variants can be found in apparently unrelated individuals: the combination of a pathogenic SMCHD1 variant and a permissive 4qA allele with a D4Z4 repeat of 8–20 units, or a permissive duplication allele, is rare as these permissive alleles have a prevalence of only 12% in the European population.20 Thus, individuals may carry a pathogenic SMCHD1 variant without developing FSHD, causing the segregation of pathogenic SMCHD1 variants in the population. Indeed, we have identified unaffected carriers with non-pathogenic chromosome 4 configurations in 21/187 families.
Currently, the diagnosis of FSHD2 is considered confirmed when SMCHD1 variants are associated with D4Z4 hypomethylation. Those that are not associated with D4Z4 hypomethylation are likely non-pathogenic. Pathogenicity prediction of SMCHD1 variants by computer-based algorithms is problematic: we identified 5/19 (20%) missense variants in controls that were predicted pathogenic in 2/3 algorithms, but which were not associated with D4Z4 hypomethylation. Likewise, we identified 3/54 missense variants in patients with FSHD2, predicted non-pathogenic in at least two algorithms, which were associated with disease presentation and D4Z4 methylation below the FSHD2 threshold.
Often, FSHD2 is first diagnosed by D4Z4 methylation analysis, followed by SMCHD1 sequence analysis. Some of these cases required extensive analysis and were shown to have far intronic SMCHD1 variants, gene deletions or pathogenic heterozygous variants in DNMT3B.25,48 Alternatively, SMCHD1 sequencing precedes methylation analysis. For these variants, we encountered quite some examples where the variant appeared not to be associated with D4Z4 hypomethylation. Most of these variants were also found in control individuals with normal D4Z4 methylation or in online databases of control populations and in silico analysis mostly predicted non-pathogenicity. All these cases were atypical FSHD and follow-up studies confirmed a different muscle disease. This observation indicates that SMCHD1 sequencing and software-based prediction alone is often not sufficient, but that confirmation of the epigenetic signature created by pathogenic SMCHD1 variants, D4Z4 hypomethylation, is required to establish variant pathogenicity.
SMCHD1 variants of uncertain significance
We identified 58 non-pathogenic SMCHD1 variants with normal D4Z4 methylation. Forty-eight out of 58 variants were found in the Exome Aggregation Consortium (EXAC) database, of which 27/48 have a minor allele frequency >0.001 in either the European, African or Asian population. For 13/48 less common EXAC variants and for nine variants absent from the EXAC database, five were predicted pathogenic in at least two prediction programmes. These established non-pathogenic SMCHD1 variants have now been added to the LOVD database to assist genetic counselling of FSHD2.
We also identified two variants in patients with FSHD which were predicted pathogenic in two prediction programmes, but for which the pathogenicity was not immediately clear. The Asian patient (Rf653) has a delta1 methylation score of −16% which is between the thresholds of controls and patients with FSHD2. She has a repeat of 10 D4Z4 units, which is at the border of the FSHD1 threshold in patients with an European ancestry. However, in Asia, the D4Z4 repeat size range for FSHD1 seems to be shifted to 1–8 units.49–51 This suggests that the SMCHD1 variant might have contributed to the clinical presentation in this individual. In a European family (Rf668), we identified an FSHD1 individual (six D4Z4 units) with two very mildly affected daughters who showed a rather low D4Z4 methylation, but still within the control range. In this family, the reduced D4Z4 methylation does not segregate with the SMCHD1 variant, as it was absent in the youngest daughter. This suggests that the variant is non-pathogenic. These examples emphasise careful judgement for SMCHD1 variants associated with methylation between thresholds of controls and FSHD2
BAMS, FSHD2 and non-pathogenic variants in SMCHD1
Pathogenic variants in the extended ATPase domain of SMCHD1 associated with D4Z4 hypomethylation have been identified in 41 unrelated patients with BAMS. It is unknown why only missense variants in the extended ATPase domain have been found in BAMS. In FSHD2, we already showed that ORF-preserving variants, like missense variants, are generally more deleterious than those variants causing ORF disruption. It is striking that pathogenic missense variants in FSHD2 also occur significantly more often in the extended ATPase domain than elsewhere, while non-pathogenic missense variants are almost absent from this region figure 1 online supplementary table 3 (figure 1). This suggests that in FSHD2, like in BAMS, variants in the extended ATPase domain are more pathogenic. As few extended ATPase domain pathogenic variants are common in both diseases, the position of the variant does not necessarily determine the phenotypic outcome.27 One explanation could be that, like FSHD2, BAMS also has a digenic inheritance with another yet unknown locus being required to cause disease. However, 9 amino acid positions are recurrently affected in 30 unrelated BAMS families. These positions are non-overlapping with 21 unique ATPase missense variants in FSHD2. Similarly, we found four recurrent FSHD2 missense variants in the extended ATPase domain that are not found in BAMS. This suggests that the position of the disease variant in the extended ATPase domain is important for the phenotypic outcome.
Protein modelling indeed suggests a mostly disease-specific localisation in which FSHD2-specific variants are typically localised in the ATP binding pocket. In contrast, BAMS variants are most often positioned at the dimer interface, a region that might be important for the dimerisation of the ATPase domain. The different localisations of the FSHD2 and BAMS variants in the ATPase domain are remarkable and may provide an explanation for the different phenotypes observed in the two diseases. For example, variants in the nucleotide binding pocket could prevent ATP hydrolysis and keep the protein in a prolonged ATP-state, while the BAMS variants located at the dimer interface could interfer with the dynamics of the oligomer formation and its chromatin remodelling function.52 In support, for several variants the ATPase activity has been analysed showing a small but significant change compared with the rather low wildtype ATPase activity. Strikingly, all three analysed variants in the binding pocket (Q193P, L194F and H263D) showed a significant loss of ATPase activity, while 3/4 tested BAMS variants in the dimer interface (A134S, 135C and E136G) showed a significant gain of ATPase activity.36,39,47
Nevertheless, three positions in the extended ATPase domain have been reported to be affected in both conditions. Variants L107P and G137E have been identified in BAMS and FSHD2, while A242T has been reported in FSHD2 and A242G in BAMS. While L107P and A242G were identified in extended FSHD2 families in which carriers did not show signs or symptoms characteristic for BAMS, for the BAMS variants very little inheritance information is available but A242T was reported to have occurred de novo. Thus, the true outcome of the FSHD2 and BAMS variants on the function of SMCHD1 will require further structural and biochemical characterisation.
In summary, in this study, we highlight the utility of D4Z4 methylation analysis for the demonstration of variant pathogenicity in SMCHD1. As this can be directly tested in genomic DNA, it does not require elaborate functional tests and maximally takes advantage of an epigenetic signature that is left behind by a dysfunctional chromatin modifier. In addition, detailed analysis of the mutation spectrum provides a partial explanation for the different phenotypic outcomes of extended ATPase domain missense variants.
Supplementary Material
Acknowledgements
The authors thank all FSHD families members for their participation. this work was financially supported by grants from the lowa Wellstone Muscular Dystrophy cooperative research center, U54, NS053672 (SAM), US National institutes of Health (NIH) (National Institute of Neurological Disorders and Stroke (NINDS) P01NS069539 and National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) R01AR045203) and Prinses Beatrix Spierfonds (W.OR12–20, W.OP14–01 and W.OB17–01).
Funding This study was funded by lowa Wellstone Muscular Dystrophy Cooperative Research Center, U54 (NS053672) and National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR045203).
Footnotes
Competing interests None declared.
Patient consent for publication Not required.
Ethics approval This study was approved by the Medical Ethical Committee from the Leiden University Medical center
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information.
REFERENCES
- 1.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet 2014;30:418–26. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman-Andrews L. The known unknown: the challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci 2017;4:648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deenen JCW, Arnts H, van der Maarel SM, Padberg GW, Verschuuren JJGM, Bakker E, Weinreich SS, Verbeek ALM, van Engelen BGM. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 2014;83:1056–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mul K, Lassche S, Voermans NC, Padberg GW, Horlings CG, van Engelen BG. What’s in a name? the clinical features of facioscapulohumeral muscular dystrophy. Pract Neurol 2016;16:201–7. [DOI] [PubMed] [Google Scholar]
- 5.Padberg GW. Facioscapulohumeral disease: Leiden university, 1982. [Google Scholar]
- 6.Statland JM, Tawil R. Facioscapulohumeral muscular dystrophy. continuum (Minneap Minn).. (6, Muscle and Neuromuscular Junction Disorders) 1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De laco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 2017;49:941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, Tapscott SJ. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 2012;22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones T, Jones PL. A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS One 2018;13:e0192657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, Harper SQ. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol 2011;69:540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LH, Friedman SD, Shaw D, Snider L, Wong C-J, Budech CB, Poliachik SL,Gove NE, Lewis LM, Campbell AE, Lemmers RJFL, Maarel SM, Tapscott SJ, Tawil RN. MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum Mol Genet 2019;28:476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scionti I, Fabbri G, Fiorillo C, Ricci G, Greco F, D’Amico R, Termanini A, Vercelli L, Tomelleri G, Cao M, Santoro L, Percesepe A, Tupler R. Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J Med Genet 2012;49:171–8. [DOI] [PubMed] [Google Scholar]
- 13.Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ, Padberg GW, Frants RR. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 1992;2:26–30. [DOI] [PubMed] [Google Scholar]
- 14.Snider L, Geng LN, Lemmers RJLF, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet 2010;6:e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemmers RJLF, de Kievit P, Sandkuijl L, Padberg GW, van Ommen G-JB, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 2002;32:235–6. [DOI] [PubMed] [Google Scholar]
- 16.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Mattéotti C, van Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, Belayew A, Coppée F, Chen Y-W. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A 2007;104:18157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm Kr. Jan vO, Padberg GW, Miller dG, Tapscott SJ, Tawil R, Frants Rr, Van der Maarel Sm. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 2010;329:1650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Overveld PGM, Lemmers RJFL, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen G-JB, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 2003;35:315–7. [DOI] [PubMed] [Google Scholar]
- 19.Lemmers RJLF, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R, Sun Y, den Dunnen JT, Helmer Q, Donlin-Smith CM, Padberg GW, van Engelen BGM, de Greef JC, Aartsma Rus AM, Frants RR, de Visser M, Desnuelle C, Sacconi S, Filippova GN, Bakker B, Bamshad MJ, Tapscott SJ, Miller DG, van der Maarel SM. Digenic inheritance of an Smchd1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012;44:1370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmers RJLF, van der Vliet PJ, Vreijling JP, Henderson D, van der Stoep N, Voermans N, van Engelen B, Baas F, Sacconi S, Tawil R, van der Maarel SM. Cis D4Z4 repeat duplications associated with facioscapulohumeral muscular dystrophy type 2. Hum Mol Genet 2018;27:3488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaillard M-C, Puppo F, Roche S, Dion C, Campana ES, Mariot V, Chaix C, Vovan C, Mazaleyrat K, Tasmadjian A, Bernard R, Dumonceaux J, Attarian S, Lévy N, Nguyen K, Magdinier F, Bartoli M. Segregation between Smchd1 mutation, D4Z4 hypomethylation and facio-scapulo-humeral dystrophy: a case report. BMC Med Genet 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamanaka K, Goto K, Arai M, Nagao K, Obuse C, Noguchi S, Hayashi YK, Mitsuhashi S, Nishino I, Clinical NI. Clinical, muscle pathological, and genetic features of Japanese facioscapulohumeral muscular dystrophy 2 (FSHD2) patients with Smchd1 mutations. Neuromuscul Disord 2016;26:300–8. [DOI] [PubMed] [Google Scholar]
- 23.Larsen M, Rost S, EL Hajj N, Ferbert A, Deschauer M, Walter MC, Schoser B, Tacik P, Kress W, Müller CR. Diagnostic approach for FSHD revisited: Smchd1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur J Hum Genet 2015;23:808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmers RJLF, Goeman JJ, van der Vliet PJ, van Nieuwenhuizen MP, Balog J, Vos-Versteeg M, Camano P, Ramos Arroyo MA, Jerico I, Rogers MT, Miller DG, Upadhyaya M, Verschuuren JJGM, Lopez de Munain Arregui A, van Engelen BGM, Padbergg GW, Sacconi S, Tawil R, Tapscott SJ, Bakker B, van der Maarel SM. inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum Mol Genet 2015;24:659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemmers RJLF, van den Boogaard ML, van der Vliet PJ, Donlin-Smith CM, Nations SP, Ruivenkamp CAL, Heard P, Bakker B, Tapscott S, Cody JD, Tawil R, van der Maarel SM. Hemizygosity for Smchd1 in facioscapulohumeral muscular dystrophy type 2: consequences for 18p deletion syndrome. Hum Mutat 2015;36:679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsuhashi S, Boyden SE, Estrella EA, Jones TI, Rahimov F, Yu TW, Darras BT, Amato AA, Folkerth RD, Jones PL, Kunkel LM, Kang PB. Exome sequencing identifies a novel Smchd1 mutation in facioscapulohumeral muscular dystrophy 2. Neuromuscul Disord 2013;23:975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mul K, Lemmers RJLF, Kriek M, van der Vliet PJ, van den Boogaard ML, Badrising UA, Graham JM, Lin AE, Brand H, Moore SA, Johnson K, Evangelista T, Töpf A, Straub V, Kapetanovic García S, Sacconi S, Tawil R, Tapscott SJ, Voermans NC, van Engelen BGM, Horlings CGC, Shaw ND, van der Maarel SM. FSHD type 2 and Bosma arhinia microphthalmia syndrome: two faces of the same mutation. Neurology 2018;91:e562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen K, Puppo F, Roche S, Gaillard M-C, Chaix C, Lagarde A, Pierret M, Vovan C, Olschwang S, Salort-Campana E, Attarian S, Bartoli M, Bernard R, Magdinier F, Levy N. Molecular combing reveals complex 4q35 rearrangements in facioscapulohumeral dystrophy. Hum Mutat 2017;38:1432–41. [DOI] [PubMed] [Google Scholar]
- 29.Sacconi S, Lemmers RJLF, Balog J, van der Vliet PJ, Lahaut P, van Nieuwenhuizen MP, Straasheijm KR, Debipersad RD, Vos-Versteeg M, Salviati L, Casarin A, Pegoraro E, Tawil R, Bakker E, Tapscott SJ, Desnuelle C, van der Maarel SM. The FSHD2 gene Smchd1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet 2013;93:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Boogaard ML, Lemmers RJFL, Camaño P, van der Vliet PJ, Voermans N, van Engelen BGM, Lopez de Munain A, Tapscott SJ, van der Stoep N, Tawil R, van der Maarel SM. Double Smchd1 variants in FSHD2: the synergistic effect of two Smchd1 variants on D4Z4 hypomethylation and disease penetrance in FSHD2. Eur J Hum Genet 2016;24:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winston J, Duerden L, Mort M, Frayling IM, Rogers MT, Upadhyaya M. Identification of two novel Smchd1 sequence variants in families with FSHD-like muscular dystrophy. Eur J Hum Genet 2015;23:67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calandra P, Cascino I, Lemmers RJLF, Galluzzi G, Teveroni E, Monforte M, Tasca G, Ricci E, Moretti F, van der Maarel SM, Deidda G. Allele-specific DNA hypomethylation characterises FSHD1 and FSHD2. J Med Genet 2016;53:348–55. [DOI] [PubMed] [Google Scholar]
- 33.Gaillard M-C, Roche S, Dion C, Tasmadjian A, Bouget G, Salort-Campana E, Vovan C, Chaix C, Broucqsault N, Morere J, Puppo F, Bartoli M, Levy N, Bernard R, Attarian S, Nguyen K, Magdinier F. Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology 2014;83:733–42. [DOI] [PubMed] [Google Scholar]
- 34.Hartweck LM, Anderson LJ, Lemmers RJ, Dandapat A, Toso EA, Dalton JC, Tawil R, Day JW, van der Maarel SM, Kyba M. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology 2013;80:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones TI, Yan C, Sapp PC, McKenna-Yasek D, Kang PB, Quinn C, Salameh JS, King OD, Jones PL. Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin Epigenetics 2014;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon CT, Xue S, Yigit G, Filali H, Chen K, Rosin N, Yoshiura K-I, Oufadem M, Beck TJ, Mcgowan R, Magee AC, Altmüller J, Dion C, Thiele H, Gurzau AD, Nürnberg P, Meschede D, Mühlbauer W, Okamoto N, Varghese V, Irving R, Sigaudy S, Williams D, Ahmed SF, Bonnard C, Kong MK, Ratbi I, Fejjal N, Fikri M, Elalaoui SC, Reigstad H, Bole-Feysot C, Nitschké P, Ragge N, Lévy N, Tunçbilek G, Teo ASM, Cunningham ML, Sefiani A, Kayserili H, Murphy JM, Chatdokmaiprai C, Hillmer AM, Wattanasirichaigoon D, Lyonnet S, Magdinier F, Javed A, Blewitt ME, Amiel J, Wollnik B, Reversade B. De novo mutations in Smchd1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat Genet 2017;49:249–55. [DOI] [PubMed] [Google Scholar]
- 37.Shaw ND, Brand H, Kupchinsky ZA, Bengani H, Plummer L, Jones TI, Erdin S, Williamson KA, Rainger J, Stortchevoi A, Samocha K, Currall BB, Dunican DS, Collins RL, Willer JR, Lek A, Lek M, Nassan M, Pereira S, Kammin T, Lucente D, Silva A, Seabra CM, Chiang C, An Y, Ansari M, Rainger JK, Joss S, Smith JC, Lippincott MF, Singh SS, Patel N, Jing JW, Law JR, Ferraro N, Verloes A, Rauch A, Steindl K, Zweier M, Scheer I, Sato D, Okamoto N, Jacobsen C, Tryggestad J, Chernausek S, Schimmenti LA, Brasseur B, Cesaretti C, García-Ortiz JE, Buitrago TP, Silva OP, Hoffman JD, Mühlbauer W, Ruprecht KW, Loeys BL, Shino M, Kaindl AM, Cho C-H, Morton CC, Meehan RR, van Heyningen V, Liao EC, Balasubramanian R, Hall JE, Seminara SB, Macarthur D, Moore SA, Yoshiura K-I, Gusella JF, Marsh JA, Graham JM, Lin AE, Katsanis N, Jones PL, Crowley WF, Davis EE, FitzPatrick DR, Talkowski ME. Smchd1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat Genet 2017;49:238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosma JF, Henkin RI, Christiansen RL, Herdt JR. Hypoplasia of the nose and eyes, hyposmia, hypogeusia, and hypogonadotrophic hypogonadism in two males. J Craniofac Genet Dev Biol 1981;1:153–84. [PubMed] [Google Scholar]
- 39.Gurzau AD, Chen K, Xue S, Dai W, Lucet IS, Ly TTN, Reversade B, Blewitt ME, Murphy JM. FSHD2- and BAMS-associated mutations confer opposing effects on SMcHD1 function. J Biol Chem 2018;293:9841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Overveld PGM, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, Winokur ST, Frants RR, Padberg GW, Van Der Maarel SM. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol 2005;58:569–76. [DOI] [PubMed] [Google Scholar]
- 41.Sung N, Lee J, Kim JH, Chang C, Tsai FTF, Lee S. 2.4 Å resolution crystal structure of human traP1nM, the Hsp90 paralog in the mitochondrial matrix. Acta Crystallogr D Struct Biol 2016;72:904–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018;34:2490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. A completely reimplemented Mpi bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 2018;430:2237–43. [DOI] [PubMed] [Google Scholar]
- 44.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol 2017;1654:39–54. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemmers RJLF, Van Overveld PGM, Sandkuijl LA, Vrieling H, Padberg GW, Frants RR, van der Maarel SM. Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am J Hum Genet 2004;75:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dion C, Roche S, Laberthonnière C, Broucqsault N, Mariot V, Xue S, Gurzau AD, Nowak A, Gordon CT, Gaillard M-C, El-Yazidi C, Thomas M, Schlupp-Robaglia A, Missirian C, Malan V, Ratbi L, Sefiani A, Wollnik B, Binetruy B, Salort Campana E, Attarian S, Bernard R, Nguyen K, Amiel J, Dumonceaux J, Murphy JM, Déjardin J, Blewitt ME, Reversade B, Robin JD, Magdinier F. Smchd1 is involved in de novo methylation of the DUX4-encoding D4Z4 macrosatellite. Nucleic Acids Res 2019;47:2822–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Boogaard ML, Lemmers RJLF, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, van der Vliet PJ, Straasheijm KR, van den Akker RFP, Kriek M, Laurense-Bik MEY, Raz V, van Ostaijen-Ten Dam MM, Hansson KBM, van der Kooi EL, Kiuru-Enari S, Udd B, van Tol MJD, Nishino I, Tawil R, Tapscott SJ, van Engelen BGM, van der Maarel SM. Mutations in Dnmt3b modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet 2016;98:1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemmers RJLF, van der Wielen MJR, Bakker E, Frants RR, van der Maarel SM. Rapid and accurate diagnosis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord 2006;16:615–7. [DOI] [PubMed] [Google Scholar]
- 50.Park HJ, Hong J-M, Lee JH, Lee HS, Shin HY, Kim SM, Ki C-S, Lee JH, Choi Y-C. Low D4Z4 copy number and gender difference in Korean patientswith facioscapulohumeral muscular dystrophy type 1. Neuromuscul Disord 2015;25:859–64. [DOI] [PubMed] [Google Scholar]
- 51.Lin F, Wang Z-Q, Lin M-T, Murong S-X, Wang N. New insights into genotypephenotype correlations in chinese facioscapulohumeral muscular dystrophy: a retrospective analysis of 178 patients. Chin Med J 2015;128:1707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C-Y, Jégu T, Chu H-P, Oh HJ, Lee JT. Smchd1 merges chromosome compartments and assists formation of Super-Structures on the inactive X. Cell 2018;174:406–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.