Significance
The difference between brain age estimated from MRI and chronological age is thought to serve as an important biomarker reflecting pathological processes in the brain. Several recent studies showed the relation between accelerated brain aging and various disorders. However, until now, the utility of such an age difference for preclinical screening using longitudinal studies was absent. To fill this gap, we first built a deep learning model using brain MRI from a population-based study including 5,496 participants. And then, using follow-up information, we observed that this age difference was significantly associated with the risk of dementia. Therefore, our study shows that the difference between MRI-brain predicted and chronological age is potentially a biomarker for early dementia risk screening.
Keywords: deep learning, dementia, age prediction, magnetic resonance imaging, voxel-based morphometry
Abstract
The gap between predicted brain age using magnetic resonance imaging (MRI) and chronological age may serve as a biomarker for early-stage neurodegeneration. However, owing to the lack of large longitudinal studies, it has been challenging to validate this link. We aimed to investigate the utility of such a gap as a risk biomarker for incident dementia using a deep learning approach for predicting brain age based on MRI-derived gray matter (GM). We built a convolutional neural network (CNN) model to predict brain age trained on 3,688 dementia-free participants of the Rotterdam Study (mean age 66 ± 11 y, 55% women). Logistic regressions and Cox proportional hazards were used to assess the association of the age gap with incident dementia, adjusted for age, sex, intracranial volume, GM volume, hippocampal volume, white matter hyperintensities, years of education, and APOE ε4 allele carriership. Additionally, we computed the attention maps, which shows which regions are important for age prediction. Logistic regression and Cox proportional hazard models showed that the age gap was significantly related to incident dementia (odds ratio [OR] = 1.11 and 95% confidence intervals [CI] = 1.05–1.16; hazard ratio [HR] = 1.11, and 95% CI = 1.06–1.15, respectively). Attention maps indicated that GM density around the amygdala and hippocampi primarily drove the age estimation. We showed that the gap between predicted and chronological brain age is a biomarker, complimentary to those that are known, associated with risk of dementia, and could possibly be used for early-stage dementia risk screening.
The human brain continuously changes throughout the entire lifespan. These changes partially reflect a normal aging process and are not necessarily pathological (1). However, neurodegenerative diseases, including dementia, also affect brain structure and function (2, 3). Therefore, a better understanding and modeling of normal brain aging can help to disentangle these two processes and improve the detection of early-stage neurodegeneration.
Age prediction models based on brain MRI are a popular trend in neuroscience (4–7). The difference between predicted and chronological age is thought to serve as an important biomarker reflecting pathological processes in the brain. Several recent studies showed the relation between accelerated brain aging and various disorders, such as Alzheimer’s disease (8), schizophrenia, epilepsy, or diabetes (7, 9, 10).
In recent years, CNNs have become the methodology of choice for analyzing medical images. These models are able to learn complex relations between input data and desired outcomes. Recent studies (11, 12) were able to demonstrate that CNN models can be successfully applied in brain MRI-based age prediction (5, 6).
Although cross-sectional studies have suggested that the gap between predicted and chronological age may serve as a biomarker for dementia diagnosis, it remains unclear whether this is also the case for the years preceding dementia diagnosis (5, 7). It was shown in recent research that the brain age gap is associated with mortality risk (13). Longitudinal studies examining the link between such a gap and incident dementia are lacking and are crucial for validation of this biomarker for early-stage neurodegeneration detection. Using a deep learning (DL) model, we investigated the association of the GM age gap with incident dementia in a large population-based sample of middle-aged and elderly subjects.
Methods
Study Population.
Data were acquired from the Rotterdam Study, an ongoing population-based cohort study among the inhabitants of Ommoord, a suburb of Rotterdam, the Netherlands (14, 15). More details of the study design and population are described in SI Appendix, Methods 1.
Data from the Rotterdam Study are not publicly available due to informed consent and legal restrictions (e.g., General Data Protection Regulation law in the European Union). However, specific requests for access to the data can be addressed to the Rotterdam Study Management Team that assesses the proposals and adjudicates access—in line with national and international regulations—on a case-by-case basis.
Image Processing.
A 1.5 T GE Signa Excite MRI scanner was used to acquire multiparametric MRI brain data as previously reported (14). Voxel-based morphometry (VBM) was performed according to an optimized VBM protocol as was previously described (16, 17). First, all T1-weighted images were segmented into supratentorial GM, white matter (WM), and cerebrospinal fluid using a previously described k-nearest neighbor algorithm, which was trained on 6 manually labeled atlases (18). Functional MRI of the Brain’s Software Library software was used for VBM data processing (19). All GM density maps were nonlinearly registered to the standard Montreal Neurological Institute GM probability template with a 1 × 1 × 1 mm3 voxel resolution.
A spatial modulation procedure was used to avoid differences in absolute GM volume due to the registration. This involved multiplying voxel density values by the Jacobian determinants estimated during spatial normalization. We did not apply smoothing. While VBM smoothing procedures increase the signal to noise ratio, they can affect the features which the network learns from GM. The Gaussian smoothing is hardly invertible. Additionally, smoothing is only a subgroup of possible mathematical operations which the network filters in the convolutional layer can represent. Therefore, if the Gaussian smoothing is important for prediction, the neural network will incorporate this in one or more convolutional filters.
FreeSurfer 6.0 was used to segment the brain and estimate intracranial volume (ICV), GM volume, hippocampal volume, and WM hyperintensity (WMH) volume (20).
Other measurements.
APOE ε4 carriership was determined using a PCR on coded DNA samples. If these values were missing, Haplotype Reference Consortium imputed genotype values for rs7412 and rs429358 were used to define the APOE ε4 carrier status. Measurements on more characteristics are described in SI Appendix, Methods 2.
DL Model.
A full description of the applied DL model is presented in the SI Appendix, Methods 3. Briefly, a DL model takes a set of inputs and respective outputs from a training set and finds an optimal nonlinear relation between them. A CNN is a class of DL techniques which takes in multidimensional images as model input. These networks are generally used with a variety of different techniques and algorithms, which together define how the model optimizes the input–output relationship (21, 22). We describe this in detail in the model architecture.
Our 3-dimensional (3D) regression CNN model is designed to predict brain age using 3D GM density maps from VBM as input. It is inspired by ConvNet (23) and deep CNN (22) as shown in SI Appendix, Fig. S2. Besides GM brain images, we provide information about the sex of the subject. This allows the network to adjust for GM differences between male and female subjects.
The dataset, excluding subjects with incident dementia, was randomly split into 3 sets: training (3,688 subjects), validation (1,099 subjects), and test (550 subjects). Subjects with incident dementia (159 subjects) were put in a fourth independent dataset. The CNN was trained using the training set as described in SI Appendix, Methods 4. For training, we used all available scans for each subject. Prediction accuracy was assessed on the test set. Model accuracy was measured based on the absolute gap, or mean absolute error (MAE) of prediction, i.e., the difference between model output and real chronological age (gap = agebrain,predicted – agechronological). Given the design of the Rotterdam Study, several follow-up scans were available for some subjects. For training, we used all available scans for each subject. These training methods allowed us to increase the number of training images thereby introducing a natural type of data augmentation.
Attention mapping.
We retrieved attention maps from the trained networks using gradient-weighted class activation mapping (24). Attention maps show which areas on the subject GM image are more important for age prediction. More details about implementation of attention maps can be found in SI Appendix, Methods 5.
Statistical Analysis.
Reproducibility of the CNN age prediction was quantified using the intraclass correlation coefficient (ICC[3,1]), computed on a subset of 80 persons out of the test set who were scanned twice with a time interval of 1–9 wk (25).
In order to be able to compare our findings with previous studies, logistic regression models and Cox proportional hazard models were used to assess the association between the age gap and the incidence of dementia. We adjusted the regression models for biomarkers, which are known for their relation with dementia: age and sex (model I); additional GM volume, ICV, hippocampal volume, and WMHs (model II); and years of education and APOE ε4 carriership (model III) (26, 27). The logistic regression model used the occurrence of dementia development during follow-up as output. The proportional hazards and linearity assumption were met for the Cox proportional hazard models. Python and R were used to perform the statistical analyses (28–31).
Results
The study population characteristics are described in Table 1. The algorithm was trained and validated on random subsets of subjects with mean age 66.09 ± 10.76 y and 55% females; and mean age 64.84 ± 9.69 y and 54% females, respectively. The following results are reported for the test set (mean age 64.85 ± 10.82 y and 55% females).
Table 1.
Characteristics of data sets derived from the population-based Rotterdam Study
| Train | Validation | Test* | Incident dementia* | |
| Nsubj | 3688 | 1099 | 550 | 159 |
| Nimg | 5865 | 2353 | 550 | 159 |
| Mean age† (years ± SD) | 66.09 ± 10.76 | 64.84 ± 9.69 | 64.85 ± 10.82 | 77.33 ± 7.15 |
| Sex proportion† (female/male) | 0.55/0.45 | 0.54/0.46 | 0.55/0.45 | 0.58/0.42 |
| Education† (years ± SD) | 12.64 ± 3.89 | 12.63 ± 3.81 | 12.58 ± 4.00 | 11.43 ± 3.57 |
| GM volume† (liters ± SD) | 0.60 ± 0.06 | 0.60 ± 0.06 | 0.60 ± 0.06 | 0.55 ± 0.05 |
| ICV† (liters ± SD) | 1.48 ± 0.16 | 1.47 ± 0.16 | 1.48 ± 0.16 | 1.45 ± 0.17 |
| Hippocampal volume (mL ± SD) | 3.9 ± 05 | 3.8 ± 05 | 3.9 ± 05 | 3.3 ± 05 |
| WMH volume (mL ± SD) | 3.9 ± 4.6 | 3.6 ± 4.1 | 3.6 ± 3.8 | 7.8 ± 7.7 |
| APOE ε4 carriership† (0/1/2) | 0.72/0.26/0.02 | 0.72/0.25/0.02 | 0.74/0.23/0.03 | 0.57/0.36/0.06 |
| Follow-up time† (years ± SD) | 5.42 ± 2.81 | 4.93 ± 2.80 | 6.68 ± 2.29 | 4.29 ± 2.26 |
Milliliter (mL); number of subjects (Nsubj); number of images (Nimg); GM; ICV.
Selection only includes baseline image of subjects.
Values are based on Nimg.
Network Performance.
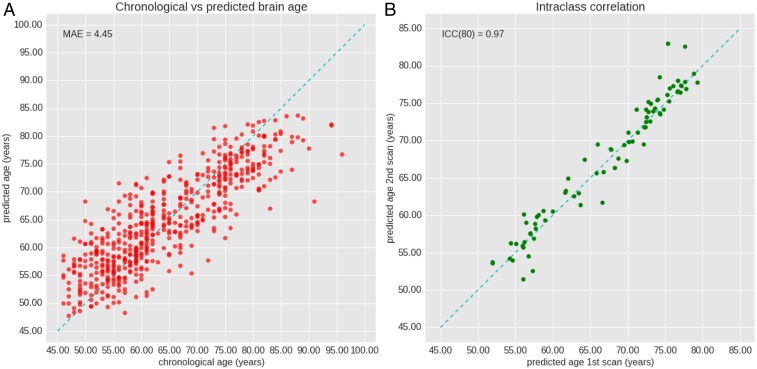
The overall performance measured on the test set was MAE = 4.45 ± 3.59 y (Fig. 1) with a correlation between chronological and predicted brain age of 0.85 (P value = 4.76 × 10−156). A reproducibility score of ICC = 0.97 (95% CI 0.96–0.98) was achieved. No significant difference in prediction was found between male and female subjects (P value = 0.34), and detailed numbers are provided in SI Appendix, Text 1.
Fig. 1.
Performance of the CNN on the test dataset. (A) The plot depicts chronological age (x-axis) and brain-predicted age (y-axis) with MAE. The dashed line indicates the ideal case x=y. (B) The figure shows reproducibility of the CNN performance. Scans 1 and 2 are taken with an interval of 1-9 weeks. The dashed line indicates a perfect reproducibility and consistent predicting of the network.
Attention map.
SI Appendix, Fig. S5 shows the global attention map of the test set, indicating the areas contributing to age prediction in bright color, as well as the increase in attention map values over age. We found that the amygdala and hippocampus are not only important for predicting brain age, but also that these regions grow more important with increasing chronological age, which is shown in SI Appendix, Fig. S5B. A quantitative analysis per brain region is presented in Table 2, which shows that highest mean intensities were computed for the nucleus accumbens (0.89) and amygdala (0.71). Highest intensity quintiles were computed for the nucleus accumbens (0.99), amygdala (0.98), and subcallosal area (0.98).
Table 2.
Quantitative analysis of the attention map per brain region. Mean and fifth quintiles (lower boundary) of attention map intensity per brain region are listed. Brain regions are grouped by lobes
| Brain region | Size (voxels) | Attention map intensity | |
| Mean | Fifth quartile | ||
| Temporal lobe | |||
| Amygdala | 4,398 | 0.71 | 0.98 |
| Hippocampus | 6,687 | 0.61 | 0.80 |
| Anterior temporal lobe medial part | 22,842 | 0.54 | 0.78 |
| Superior temporal gyrus, anterior part | 14,369 | 0.54 | 0.74 |
| Lateral occipitotemporal gyrus (gyrus fusiformis) | 12,908 | 0.53 | 0.62 |
| Posterior temporal lobe | 143,237 | 0.52 | 0.68 |
| Superior temporal gyrus, central part | 42,794 | 0.52 | 0.68 |
| Gyri parahippocampalis and ambiens | 13,767 | 0.51 | 0.63 |
| Medial and inferior temporal gyri | 55,102 | 0.50 | 0.68 |
| Anterior temporal lobe lateral part | 11,999 | 0.49 | 0.65 |
| Insula and cingulate gyri | |||
| Cingulate gyrus anterior part (supragenual) | 24,751 | 0.53 | 0.63 |
| Cingulate gyrus posterior part | 24,235 | 0.52 | 0.64 |
| Insula | 44,328 | 0.51 | 0.64 |
| Frontal lobe | |||
| Subcallosal area | 788 | 0.70 | 0.98 |
| Posterior orbital gyrus | 15,061 | 0.54 | 0.72 |
| Straight gyrus (gyrus rectus) | 11,826 | 0.54 | 0.67 |
| Inferior frontal gyrus | 55,754 | 0.53 | 0.72 |
| Superior frontal gyrus | 166,766 | 0.52 | 0.77 |
| Precentral gyrus | 106,145 | 0.52 | 0.77 |
| Medial orbital gyrus | 18,554 | 0.52 | 0.77 |
| Presubgenual anterior cingulate gyrus | 2,451 | 0.52 | 0.61 |
| Middle frontal gyrus | 161,999 | 0.51 | 0.74 |
| Anterior orbital gyrus | 19,514 | 0.51 | 0.73 |
| Lateral orbital gyrus | 11,112 | 0.51 | 0.77 |
| Subgenual anterior cingulate gyrus | 4,287 | 0.50 | 0.71 |
| Occipital lobe | |||
| Cuneus | 28,209 | 0.57 | 0.67 |
| Lingual gyrus | 36,627 | 0.55 | 0.65 |
| Lateral remainder of occipital lobe | 131,852 | 0.54 | 0.73 |
| Parietal lobe | |||
| Superior parietal gyrus | 130,908 | 0.54 | 0.74 |
| Remainder of parietal lobe (including supramarginal and angular gyri) | 131,972 | 0.52 | 0.75 |
| Postcentral gyrus | 89,087 | 0.52 | 0.74 |
| Central structures | |||
| Nucleus accumbens | 888 | 0.89 | 0.99 |
| Thalamus | 20,953 | 0.61 | 0.79 |
| Putamen | 14,502 | 0.60 | 0.74 |
| Pallidum (globus pallidus) | 3,835 | 0.58 | 0.69 |
| Caudate nucleus | 12,229 | 0.56 | 0.67 |
Logistic Regression.
We computed a logistic regression for the three models as shown in Table 3. The age gap was significantly associated with dementia incidence while age, sex, GM volume, ICV volume, hippocampal volume, WMH volume, years of education, and the APOE ε4 allele carriership were included in the model with model III: OR = 1.09 (95% CI 1.04–1.14) per year age gap.
Table 3.
Association of gap between brain age and chronological age with incident dementia assessed by logistic regression and Cox proportional hazards models, both in the total study sample and in a subsample with a minimum follow-up time of 5 years
| Logistic regression | Cox regression | |||||
| Model | n/N | OR (95% CI) | P value | n/N | HR (95% CI) | P value |
| Total sample | ||||||
| Model I | 159/1808 | 1.15 (1.10–1.20) | 2.66 × 10−10 | 159/1808 | 1.15 (1.11–1.20) | 1.02 × 10−12 |
| Model II | 154/1790 | 1.09 (1.04–1.14) | 4.77 × 10−4 | 154/1790 | 1.09 (1.05–1.15) | 2.27 × 10−5 |
| Model III | 150/1714 | 1.09 (1.04–1.14) | 7.97 × 10−4 | 150/1714 | 1.09 (1.04–1.14) | 9.62 × 10−5 |
| Sample follow-up time >5 y | ||||||
| Model I | 62/1366 | 1.11 (1.04–1.18) | 1.26 × 10−3 | 62/1366 | 1.13 (1.06–1.20) | 1.38 × 10−4 |
| Model II | 60/1352 | 1.08 (1.01–1.16) | 2.48 × 10−2 | 60/1352 | 1.10 (1.02–1.17) | 7.58 × 10−3 |
| Model III | 58/1305 | 1.08 (1.01–1.16) | 3.38 × 10−2 | 58/1305 | 1.09 (1.01–1.16) | 1.78 × 10−2 |
Model I: age + sex.
Model II: model I + GM volume + ICV + hippocampal volume + WMH volume.
Model III: model II + years of education + APOE ε4 carrier status.
CI; OR; HR; number of cases (n); total number of participants (N).
Survival Analysis.
As shown in Table 3 and Fig. 2, the age gap was significantly associated with the incidence of dementia with model III, HR = 1.09 (95% CI 1.04–1.14) per year age gap. These associations were similar in a subsample with a follow-up time for indecent dementia of more than 5 y, model III, HR = 1.09 (95% CI 1.01–1.16) per year age gap.
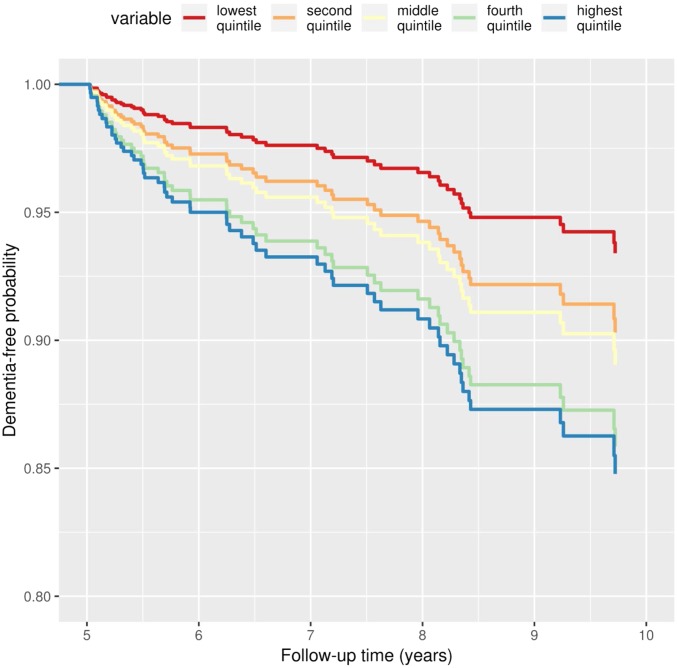
Fig. 2.
Adjusted survival curves for dementia-free probability by age gap. Dementia-free probability is presented over time for participants with different age gap values, divided into quintiles. Lower gap values correspond to chronological ages surpassing brain age, whereas higher gap values correspond to chronological ages that are lower than the brain age. Plots are based on Cox proportional hazards models, adjusted for age, sex, total grey matter volume, intracranial volume, hippocampal volume, white matter hyperintensity volume, years of education and APOE ε4 carriership status, using a marginal approach.
Gap-Associated Features.
SI Appendix, Table S1 and Fig. S7 show a list of features that can affect the brain pathology and may be associated with the gap (10). Significantly lower values were found for GM volume, hippocampal volume, and WMH volume in the highest quintile.
Discussion
In a large sample of community-dwelling middle-aged and older adults, using a DL model for brain age prediction on MRI-derived GM tissue density, we found that the gap between predicted brain age and chronological age was related to an increased risk of dementia, independent of standard established risk factors for dementia.
Our trained CNN model showed a similar MAE value in age prediction compared to previous studies that use a multimodal data model (5) and DL-based approach (6), which achieved performances of MAE = 4.29 and MAE = 4.16, respectively. Previous studies looked cross sectionally (5, 6) at the association of the age gap and dementia occurrences, while in the current study, we evaluated associations in longitudinal data. As nonreversible pathological changes already occur years prior to diagnosis, identifying early-stage biomarkers for dementia is of importance. The age gap has the potential to be utilized alongside other clinical risk factors and biomarkers to separate the population into categories with sufficiently distinct degrees of risk to drive clinical or personal decision-making, e.g., dementia screening and informed life planning.
Moreover, we retrieved attention maps from the model, showing the relative importance of different brain regions for age prediction. While the network looks at the entire GM (SI Appendix, Fig. S6), the attention pattern is quite complex, which suggests that the gap holds more specific information than global measures of GM volume when predicting brain age. This was further established by the association found between the gap and the incident dementia, which remained significant after adjusting for total GM volume. Interestingly, based on the attention maps, the amygdala and hippocampus, in particular, are relatively more important for age prediction, also increasing in attention and map intensity with older subjects (SI Appendix, Fig. 5B). This is in accordance to literature where significant negative associations between GM volume and age have been reported for these regions (2, 26). Atrophy of these two structures has also shown to be more prevalent in dementia patients, including years before diagnosis (32, 33). Yet, even after adjusting for hippocampal volume, the association between the age gap and the risk of dementia remained significant. This shows that the features which the neural network extracts from images go beyond just global or local volumetric measurements. A more in-depth evaluation of the attention map can be found in SI Appendix, Text 2.
Limitations.
We were not able to perfectly predict the age for healthy subjects based only on MRI. We assume that, due to biological similarity of the brain within a range of several years, there will always be an according level of uncertainty in the age prediction.
While the MAE value of our model was comparable with previous research, the age range of our population-based study is more limited and shifted toward the elderly. Such a study design does not invalidate the subsequent dementia analysis, however, a model trained on an age range which covers the entire lifespan may increase the power for dementia associations.
We have added sex information as an additional input to our CNN to correct for prediction bias. It is known that there are volumetric differences in GM between females and males (34), i.e., for the same age range, there is a difference in male and female GM volume. Therefore, we adjusted for sex as a bias factor.
Furthermore, we excluded subjects with dementia and stroke while training the model, but there are a number of other factors which can influence overall or local GM volume and affect the age prediction and gap (SI Appendix and Table 2). Although only total GM volume differed significantly between subjects with a high versus a low gap, effect estimates of some features differed substantially. Further research is needed to investigate gap-associated features, which may explain gap differences. These features can also introduce bias, which may be solved by adding the information as a covariate to the model. This, however, requires the respective information on the subjects, which can make the method less accessible for general use.
Additionally, brain age regression dilution (35) can affect the performance measurement. Therefore, following the suggestions from previous research (36), we have adjusted our dementia analysis models by chronological age to minimize such an influence on the incident dementia analysis.
The current CNN model is incapable of handling unfamiliar datasets, limiting its practical use. A drawback of the CNN is that the training data should be representative for the data for which the trained network is used. Thus, limiting the generalizability of our method. However, this can be addressed by training models on more diverse or new datasets. It would, therefore, be interesting to extend this model to another dataset and validate its use in a different context.
Lastly, the interpretation of the neural network attention maps should be performed with caution. Increased or decreased attention in specific brain regions might be due to various study specific factors, e.g., the image acquisition protocol, image preprocessing etc. Therefore, further research with an independent dataset is needed to confirm such findings. In general, better methods for neural network interpretation should be developed.
Conclusion
We showed that the gap between age predicted from brain MRI and chronological brain age is a biomarker associated with a risk of dementia development. DL visualization allows further investigation of the gap and neurodegeneration with respect to the human brain. This suggests that the age gap may be applicable for dementia risk screening, but there is still room for improvement in accuracy and for further research into the association between gap and dementia compared to other biomarkers.
Supplementary Material
Acknowledgments
Mr. Aleksei Tiulpin was supported by the KAUTE Foundation. The Rotterdam Study is funded by the Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (Directorate-General XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists. H.H.H.A. is supported by ZonMW Grant 916.19.151.
Footnotes
Competing interest statement: The authors declare no competing interest. W.J.N. is co-founder, scientific lead, and shareholder of Quantib BV. Other authors report no biomedical financial interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902376116/-/DCSupplemental.
References
- 1.Vinke E. J., et al. , Trajectories of imaging markers in brain aging: The Rotterdam study. Neurobiol. Aging 71, 32–40 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Manard M., Bahri M. A., Salmon E., Collette F., Relationship between grey matter integrity and executive abilities in aging. Brain Res. 1642, 562–580 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Abbott A., Dementia: A problem for our age. Nature 475, S2–S4 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Franke K., Luders E., May A., Wilke M., Gaser C., Brain maturation: Predicting individual BrainAGE in children and adolescents using structural MRI. Neuroimage 63, 1305–1312 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Liem F., et al. , Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage 148, 179–188 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Cole J. H., et al. , Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage 163, 115–124 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann T., et al. , Genetics of brain age suggest an overlap with common brain disorders. bioRxiv:10.1101/303164 (17 April 2018).
- 8.Gaser C., Franke K., Klöppel S., Koutsouleris N., Sauer H.; Alzheimer’s Disease Neuroimaging Initiative , BrainAGE in mild cognitive impaired patients: Predicting the conversion to Alzheimer’s disease. PLoS One 8, e67346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes G. L., Milh M. D. M., Dulac O., Maturation of the human brain and epilepsy. Handb. Clin. Neurol. 107, 135–143 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Franke K., Gaser C., Manor B., Novak V., Advanced BrainAGE in older adults with type 2 diabetes mellitus. Front. Aging Neurosci. 5, 90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herent P., Jegou S., Wainrib G., Clozel T., Brain age prediction of healthy subjects on anatomic MRI with deep learning: Going beyond with an “explainable AI” mindset. bioRxiv:10.1101/413302 (10 September 2018).
- 12.Li H., Satterthwaite T. D., Fan Y., “Brain age prediction based on resting-state functional connectivity patterns using convolutional neural networks” in Proceedings–International Symposium on Biomedical Imaging (IEEE, 2018), pp. 101–104. [DOI] [PMC free article] [PubMed]
- 13.Cole J. H., et al. , Brain age predicts mortality. Mol. Psychiatry 23, 1385–1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikram M. A., et al. , The Rotterdam scan study: Design update 2016 and main findings. Eur. J. Epidemiol. 30, 1299–1315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikram M. A., et al. , The Rotterdam study: 2018 update on objectives, design and main results. Eur. J. Epidemiol. 32, 807–850 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good C. D., et al. , A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 14, 21–36 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Roshchupkin G. V., et al. , Fine-mapping the effects of Alzheimer’s disease risk loci on brain morphology. Neurobiol. Aging 48, 204–211 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Vrooman H. A., et al. , Multi-spectral brain tissue segmentation using automatically trained k-Nearest-Neighbor classification. Neuroimage 37, 71–81 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Smith S. M., Nichols T. E., Threshold-free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44, 83–98 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Fischl B., et al. , Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 21.LeCun Y., Bottou L., Bengio Y., Haffner P., Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2323 (1998). [Google Scholar]
- 22.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks” in Advances in Neural Information Processing Systems 25, Pereira F., Burges C. J. C., Bottou L., Weinberger K. Q., Eds. (Curran Associates, Inc., 2012), pp. 1097–1105. [Google Scholar]
- 23.Simonyan K., Zisserman A., Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 (4 September 2014).
- 24.Selvaraju R. R., et al. , Grad-CAM: Visual explanations from deep networks via gradient-based localization. Proc. IEEE Int. Conf. Comput. Vis. 2017, 618–626 (2016). [Google Scholar]
- 25.Shrout P. E., Fleiss J. L., Intraclass correlations: Uses in assessing rater reliability. Psychol. Bull. 86, 420–428 (1979). [DOI] [PubMed] [Google Scholar]
- 26.Matsuda H., Voxel-based morphometry of brain MRI in normal aging and Alzheimer’s disease. Aging Dis. 4, 29–37 (2013). [PMC free article] [PubMed] [Google Scholar]
- 27.Roses A. D., Saunders A. M., APOE is a major susceptibility gene for Alzheimer’s disease. Curr. Opin. Biotechnol. 5, 663–667 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Van Rossum G., Drake F. L., Python Reference Manual (Python Software Foundation, 2001). [Google Scholar]
- 29.Ascher D., Dubois P., Hinsen K., Hugunin J., Oliphant T., Numerical Python (Lawrence Livermore National Laboratory, 2001). [Google Scholar]
- 30.Chollet F., Data from “Keras library.” GitHub. https://github.com/keras-team/keras. Accessed 20 September 2019.
- 31.R Core Team , R: A Language and Environment for Statistical Computing (Version 3.4.4, R Foundation for Statistical Computing, 2018).
- 32.Aylward E. H., et al. , MRI volumes of the hippocampus and amygdala in adults with Down’s syndrome with and without dementia. Am. J. Psychiatry 156, 564–568 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Wachinger C., Salat D. H., Weiner M., Reuter M.; Alzheimer’s Disease Neuroimaging Initiative , Whole-brain analysis reveals increased neuroanatomical asymmetries in dementia for hippocampus and amygdala. Brain 139, 3253–3266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie S. J., et al. , Sex differences in the adult human brain: Evidence from 5216 UK biobank participants. Cereb. Cortex 28, 2959–2975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith S. M., Vidaurre D., Alfaro-Almagro F., Nichols T. E., Miller K. L., Estimation of brain age delta from brain imaging. Neuroimage 200, 528–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le T. T., et al. ; Tulsa 1000 Investigators , A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front. Aging Neurosci. 10, 317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.