Abstract
Reverse transcription of RNA is fallible, introducing biases and confounding the quantification of transcript abundance. We demonstrate that circular RNAs (circRNAs) are more subjective to overestimation of transcript abundance than cognate linear RNAs due to their covalently closed, circular form, producing multiple concatameric products from a single priming of reverse transcriptase. We developed SplintQuant, where custom DNA oligonucleotides are ligated by PBCV-1 DNA ligase only when bound to their target RNA. These circRNA-specific DNA oligonucleotides are terminally tagged with universal primers, allowing SplintQuant to accurately quantify even lowly abundant circRNAs through highly specific quantitative PCR (qPCR) in the absence of reverse transcription. SplintQuant is sensitive, specific, highly reproducible, and applicable to the quantification of canonical and noncanonical RNA transcripts including alternative splice variants, gene fusions, and offers a gold-standard approach for accurately quantifying circRNAs.
Keywords: splint ligation, circular RNA, qPCR, reverse transcription
INTRODUCTION
CircRNAs are a large family of covalently closed, single-stranded RNA molecules present in all eukaryotes and which display tissue- and disease-specific expression patterns (Wang et al. 2014). CircRNA molecules are formed by a cotranscriptional alternative splicing event, called backsplicing, most commonly linking the 3′-end of an exon to the 5′-end of the same or an upstream exon (Fig. 1A). While de novo identification of circRNAs is achieved by next-generation sequencing (NGS), a number of approaches exist for quantifying the abundance of candidate circRNAs, including northern blotting (Wang and Shan 2018) and quantitative real-time, reverse transcriptase PCR (qRT-PCR). However, northern blotting suffers from poor sensitivity and the need for large amounts of RNA, while qRT-PCR, the most widely used and cost-effective quantitative approach, for circRNAs has been proposed to be susceptible to template switching, rolling circle amplification, and PCR amplification bias (Szabo and Salzman 2016; Chen et al. 2018; Kristensen et al. 2018). While these factors have been proposed, they have yet to be demonstrated.
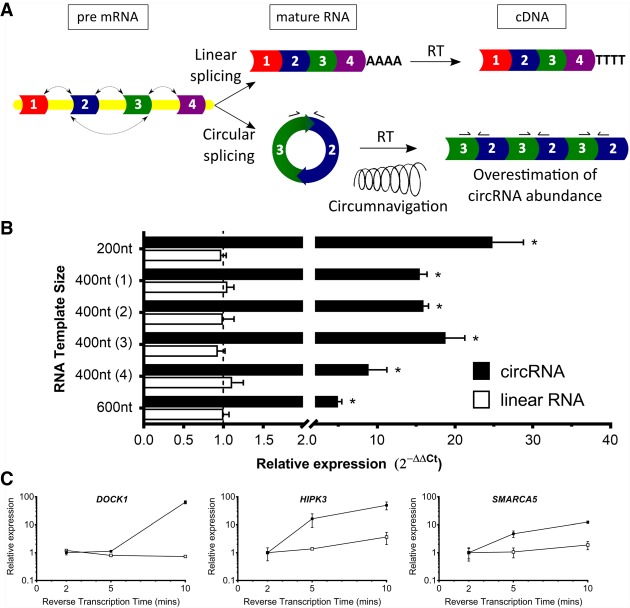
FIGURE 1.
Confounding of circRNA expression profiling by qRT-PCR. (A) circRNA and linear RNA splicing. Reverse transcription (RT) of circRNA yields concatameric backsplice junctions and multiple priming sites for qRT-PCR primers (arrows bound to exons 2 and 3) from circumnavigation of a single circRNA molecule, thereby overestimating circRNA transcript abundance. (B) Calculation of transcript abundance by qRT-PCR using 2−ΔΔCt method for six synthetic in vitro transcribed RNAs in either their linear form (unfilled boxes) or circular RNA form (filled boxes). n = 3 biological replicates, in technical triplicate. Mean ± SD. Linear RNA (600 nt) set as reference expression level of 1. (*) P < 0.05, Students t-test. (C) Calculation of transcript abundance by qRT-PCR of endogenous linear RNAs (unfilled symbols) and circRNAs (filled symbols) from HMLE RNA with varying reverse transcription time (2, 5, 10 min). Mean ± SD. n = 3 biological replicates, in technical triplicate.
CircRNA-specific qRT-PCR is achieved by designing divergent DNA oligonucleotides on the linear RNA, which flank or span the backsplice junction and become convergent upon circularization (Jeck and Sharpless 2014). As a result, when reverse transcribed by either random primers, or a gene-specific primer, this would cause continuous circumnavigation of the single circRNA molecule, producing concatameric cDNAs (Fig. 1A). Therefore, each of these cDNA molecules provide multiple priming targets for the qRT-PCR primers (or backsplice reads in the case of NGS), leading to overestimation of the actual abundance of the circRNA. Recently, the use of Nanostring nCounter technology—using color-coded DNA probes hybridized to cellular RNA in combination with high-content digital counting—was used to simultaneously enumerate 52 unique circRNAs on fresh, frozen tissue and formalin-fixed, paraffin-embedded tissue containing highly degraded RNA (Dahl et al. 2018). As the hybridization of the DNA probes is directly performed on RNA, not cDNA, this approach eloquently addresses the aforementioned biases of qRT-PCR. However, in situations where researchers seek to quantify only a small number of circRNA targets, the setup costs for this highly sensitive Nanostring nCounter method (probes, chips and equipment) may be prohibitive. Therefore, the majority of circRNA researchers would benefit from a more cost-effective option.
To maximize accuracy of quantification and minimize experimental outlay costs, we developed SplintQuant using the enzyme SplintR ligase (New England Biolabs), a modified PBCV-1 DNA ligase—which ligates adjacent single-stranded DNA oligonucleotides when splinted by a complementary RNA strand (Lohman et al. 2014; Jin et al. 2016). By targeting the DNA oligonucleotides to be immediately flanking the circRNA backsplice junction we can detect specific circRNAs, unaffected by either the cognate linear RNA, circRNAs which comprise additional internal exons, or the innate size of the target circRNA. We adapted this methodology to incorporate flaking M13 universal primer sequences on the circRNA-specific oligonucleotides, permitting accurate quantification of even lowly abundant circRNA transcripts by quantitative PCR on the ligated DNA probes in the absence of reverse transcription (qPCR).
RESULTS
Reverse transcription and transcript size impact circRNA quantification
To demonstrate whether rolling circle amplification caused a significant artifact in quantifying circRNA abundance, we performed qRT-PCR on a panel of six synthetic RNAs that differ in their transcript length (200, 400 nt [four different RNAs], and 600 nt). These synthetic linear RNAs were generated through in vitro transcription and a subset of this RNA was circularized into circRNAs as per Kershaw and O'Keefe (2012a). Following individual reverse transcription of 0.2 fmols of the six linear RNAs and circRNAs, we performed Sanger sequencing of the qRT-PCR products and confirmed the expected backsplice junction for the circRNAs (Supplemental Fig. S1). While qRT-PCR targeting the six linear RNAs showed no difference in their estimated abundance, we found the circRNAs were overestimated by greater than fivefold in all instances. Higher rates of overestimation were found for smaller circRNAs, with the 200 nt circRNA showing ∼25-fold overexpression compared with its linear counterpart, the 400 nt circRNAs showing on average 14.8-fold overestimation and the 600 nt 5.1-fold (Fig. 1B). These observations support rolling circle amplification as a significant artifact in quantifying circRNAs, which is further confounded by smaller circRNAs being prone to higher rates of circumnavigation by reverse transcriptase compared with larger circRNAs. We performed qRT-PCR on RNA purified from human mammary epithelial cells (HMLE) and performed a timecourse of reverse transcription time at 2, 5, and 10 min. Endogenous human circRNAs from SMARCA5, HIPK3, and DOCK1 genes, but not their cognate linear RNAs, were seen to increase significantly along with increased reverse transcription times by qRT-PCR (Fig. 1C). This showed that circRNA abundance was more prone to overestimation than for linear RNAs, by an order of magnitude using qRT-PCR.
SplintQuant with direct fluorescence analysis is specific and quantitative
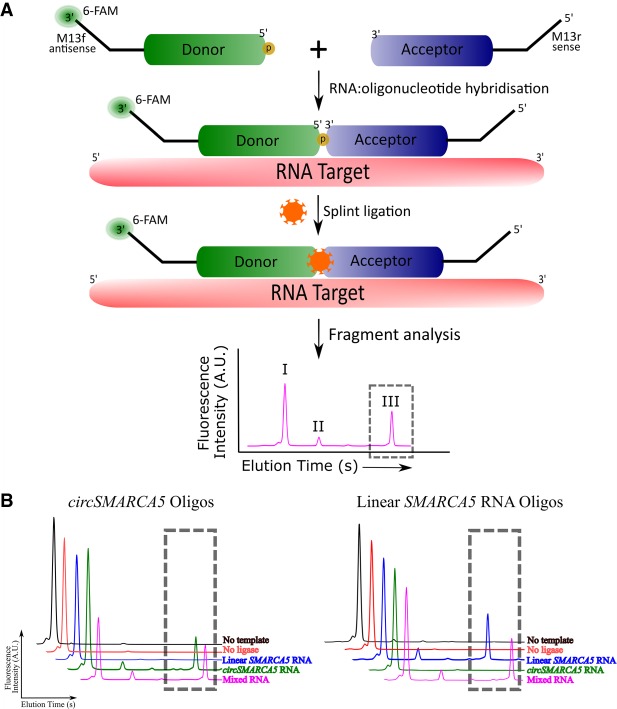
In order to resolve this confounding from reverse transcription of circRNAs, yet retain the ability to detect lowly abundant transcripts, we developed SplintQuant. This approach uses two, target-specific oligonucleotides (i) the “donor” DNA oligonucleotide, containing a 5′ phosphate, and (ii) the “acceptor” DNA oligonucleotide. Each oligonucleotide was distally tagged with M13 forward or M13 reverse sequences, permitting the use of M13 universal primers in qPCR for any target, without reverse transcription (Fig. 2A). Critically, the donor and acceptor oligonucleotides will only be ligated if they (a) bind perfectly complementary to the RNA splint and, are (b) immediately adjacent to each other on the RNA splint. As previously reported (Lohman et al. 2014), the splint reaction produces three products, the (I) unmodified donor, (II) adenylated donor and finally the largest product which is the (III) desired ligated product (Fig. 2A). To validate our SplintQuant approach, we added a 3′ FAM label to the donor DNA oligonucleotide to permit fluorescence quantification of these products by capillary electrophoresis (Fig. 2A). This reaction was performed on in vitro transcribed RNA corresponding to exons 16–17 of SMARCA5 (linear RNA) and circSMARCA5 (comprising exons 16–15) (Supplemental Fig. S1). We optimized the conditions and demonstrated that the ligated product was dependent on the presence of both the template and SplintR ligase and specific to the circSMARCA5 RNA, with no detectable product in the presence of the linear SMARCA5 RNA only (Fig. 2B). Conversely, using the linear SMARCA5 oligonucleotides, a roduct was seen in the presence of linear SMARCA5 RNA, but not circSMARCA5. A mixed template comprising equimolar circular and linear SMARCA5 RNA templates yielded products for both SplintQuant oligonucleotide pairs with similar quantitative output (peak area of ligated product) as using the RNAs in isolation. This shows that the quantitative output is strongly linked to the input RNA concentration.
FIGURE 2.
Validation of SplintQuant using fluorescence fragment analysis. (A) Schematic of SplintQuant-FAM. Donor is 5′ phosphorylated and 3′ 6-FAM-labeled (orange sun). Fragment analysis in FAM channel, producing three distinct products differing in size and elution time. (I) Unmodified donor, (II) adenylated donor, and (III) the ligated product (in gray dotted box). (B) Validation of SplintQuant, using 6-FAM labeled donor DNA oligonucleotide. (Left) Oligonucleotide pair for circSMARCA5 (targeting exons 16–15); (Right) Oligonucleotide pair for linear SMARCA5 RNA (targeting exons 16–17). Ligated product within gray dotted box, with representative traces listing color-coded reaction conditions.
Unlike the reverse transcription timecourse for qRT-PCR, increasing the length of ligation beyond 15 min showed no overestimation of circSMARCA5 (Supplemental Fig. S2A). To assess the detection limit of this method, we performed a dilution series on the circRNA template; we found that down to 0.2 pmol RNA the yield of ligated product was linearly correlated with the starting concentration of the target. However, at 0.1 pmol RNA, no product was detectable by fluorescent fragment analysis (Supplemental Fig. S2B). We attempted to quantify circSMARCA5 in a model of epithelial-mesenchymal transition (EMT) using human mammary epithelial cells (HMLE) and their mesenchymal equivalent (mesHMLE), the latter being enriched for circSMARCA5 (Conn et al. 2015). Given the low expression levels of circRNAs in cells, unsurprisingly, this fluorescence detection iteration of SplintQuant could not detect circSMARCA5 in 100 ng total RNA purified from either HMLE or mesHMLE cells (Supplemental Fig. S2C).
SplintQuant with universal qPCR is highly sensitive for circRNAs
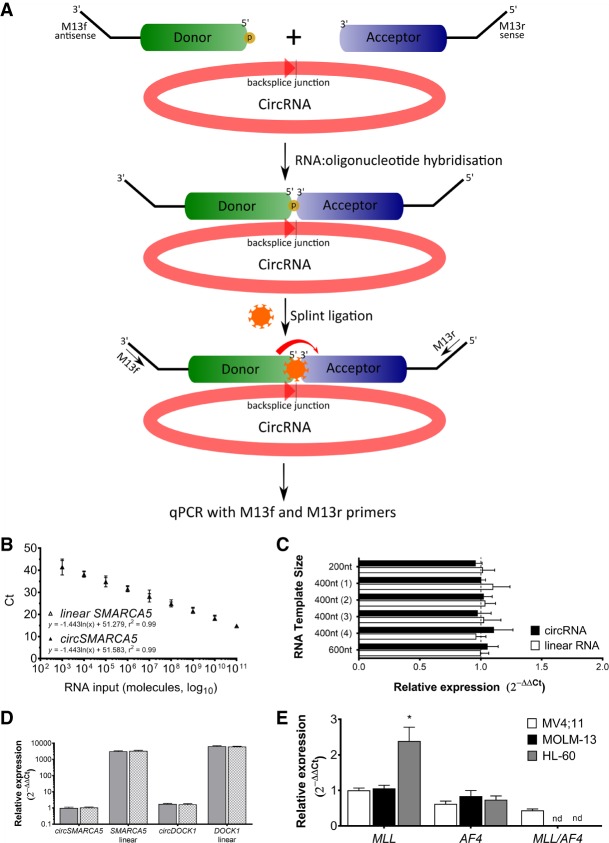
To enable highly sensitive detection of circRNAs with our SplintQuant method, we exploited the M13 primer-tags on the donor and acceptor oligonucleotides and performed qPCR with M13 forward and M13 reverse primers (Fig. 3A). Targeting the in vitro transcribed circSMARCA5 or linear SMARCA5 RNA (0.2 fmol RNA, one-thousand-times below the detection threshold of the fluorescence-based version), SplintQuant with this universal qPCR produced a single, specific product—confirmed by melt curve analysis and verified by Sanger sequencing for each target (Supplemental Fig. S3). Critically, no qPCR product was amplified in the absence of primers, template, or SplintR ligase after 45 cycles (Supplemental Fig. S3), demonstrating the specificity of the reaction. Furthermore, no off-target qPCR product was found when combining all circRNA splint oligonucleotides for the six synthetic circRNA or linear RNA targets, despite there being 72 possible acceptor/donor combinations in this subset (Supplemental Fig. S3F). We performed a dilution series, combining both linear SMARCA5 and circSMARCA5 in equimolar amounts and found that SplintQuant quantitative output was indistinguishable for these targets and strongly positively linked to input RNA concentration across eight orders of magnitude (r2circSMARCA5 = 0.99; r2linear SMARCA5 = 0.99) (Fig. 2B). We repeated the quantification of the six synthetic in vitro transcribed circRNAs and cognate linear RNAs, this time using SplintQuant in place of qRT-PCR. While still seeing no difference between linear RNAs of distinct sizes, SplintQuant eliminated the overestimation of circRNA abundance found with qRT-PCR, simultaneously eliminating any apparent confounding based on circRNA size (Fig. 3C).
FIGURE 3.
SplintQuant with Universal qPCR accurately quantifies circRNAs in total RNA. (A) Schematic of SplintQuant. Donor is 5′ phosphorylated and both acceptor and donor are tagged at their termini with M13R and M13F primers, respectively. Following Splint ligation, products are used as templates for qPCR with M13F/M13R primers. (B) Dilution series of circSMARCA5 and linear SMARCA5 showing high degree of correlation over 8-log dynamic range. Slope ± SEM calculated using linear regression and r2 value shown. (C) Calculation of transcript abundance by SplintQuant using 2−ΔΔCt method for six synthetic in vitro transcribed RNAs in either their linear form (unfilled boxes) or circular RNA form (filled boxes). n = 3 biological replicates, in technical triplicate. Linear RNA (600 nt) set as reference expression level of 1. Mean ± SD. (D) Abundance of circSMARCA5, SMARCA5 linear, circDOCK1, and DOCK1 linear RNA in HMLE cellular RNA using SplintQuant. Comparison of normalization with GAPDH SplintQuant (gray filled columns) or GAPDH qRT-PCR (gray checkered columns). Mean ± SD. n = 3 biological replicates, in technical duplicate. Expression shown relative to circSMARCA5, as a value of 1. (E) Abundance of MLL, AF4, and MLL/AF4 transcripts in three human cell leukaemic lines (MV4; 11, MOLM-13, and HL-60). Mean ± SD. n = 3 biological replicates, in technical duplicate; n.d., not detected. (*) P < 0.05 (Student's t-test).
SplintQuant with universal qPCR accurately quantifies circRNAs in total RNA purified from human cells
To streamline our methodology for quantifying transcript abundance in total RNA, we also designed SplintQuant oligonucleotides for a normalization (“housekeeping”) gene, GAPDH and compared the results to standard qRT-PCR on the same RNA for GAPDH, utilizing the 2−ΔΔCt method to calculate relative gene expression (Livak and Schmittgen 2001). We quantified circular and linear RNAs from SMARCA5 and DOCK1 in 100 ng total RNA from HMLE and mesHMLE cells. No significant differences were found in expression levels for these targets between normalization strategies, therefore we continued to use the SplintQuant GAPDH oligonucleotides (Fig. 3D).
We previously reported a 6.5-fold increase in circSMARCA5 between HMLE and mesHMLE cells using qRT-PCR (Conn et al. 2015), but with SplintQuant we found only a 3.7-fold increase (Supplemental Table S2). This diminished fold-change was also confirmed for circDOCK1, which is only decreased fivefold between HMLE and mesHMLE cells, rather than the 10-fold previously estimated by qRT-PCR (Conn et al. 2015). Calculating the absolute copy-number of these targets per cell by comparison to a standard curve, we found a 7.4-fold overestimation of circSMARCA5 in mesHMLE cells by qRT-PCR compared with SplintQuant. However, in HMLE cells where circSMARCA5 is less abundant, there was a reduced, but still fourfold overestimation, suggesting the abundance of the circRNA can also impact the extent of overestimation by qRT-PCR (Supplemental Table S2). A similar discrepancy between qRT-PCR and SplintQuant was observed for two other lowly abundant circRNAs from MLL and AF4. Therefore, even relative abundance comparisons for the same circRNA between samples using qRT-PCR is flawed, let alone absolute comparisons to other discrete circRNAs.
SplintQuant qPCR can amplify fusion transcripts
CircRNAs are an example of a noncanonical fusion transcript, thus we extended our testing of SplintQuant to detect a fusion mRNA transcript, in this case the leukemogenic fusion between mixed lineage leukemia (MLL) and MLLT2 (AF4), the most common translocation found in acute leukemia (Meyer et al. 2017). We utilized the MV4;11 cell line, a human infant acute monocytic leukemia with a characterized translocation between intron 8 of MLL and intron 4 of AF4, yielding an oncogenic fusion transcript (Felix et al. 1999). Targeting SplintQuant oligonucleotides to MLL, AF4, and the MLL/AF4 fusion at the junction between MLL exon 6 and AF4 exon 5 (Supplemental Fig. S4) identified that both wild-type transcripts and the fusion transcript were transcribed, in support of a lack of reciprocal translocations in this line (Fig. 3E). This fusion transcript was also undetectable in MOLM-13 (possessing a fusion transcript of MLL and AF9 Matsuo et al. 1997) and HL-60, lacking any MLL rearrangement (Kühn et al. 2015).
DISCUSSION
Recently, we have shown that the presence of circRNAs significantly skews transcript quantification by NGS, qRT-PCR, and microarray (Toubia et al. 2018). This is a direct result of reads emanating from the linear region of the circRNA itself contributing to the sum total of reads from the parental transcript and skewing the estimation of global transcript abundance by approximately twofold. Here we show that rolling circle amplification of circRNAs skews the estimation of circRNA abundance itself by increasing the number of backsplice reads, but this would concomitantly exacerbate the impact on global expression profiling as the linear component of the circRNAs would also be amplified.
PBCV-1 DNA ligase used in this study has been used to quantify miRNAs and mRNAs previously, unlike in this report, these target RNAs were not being overestimated with the standard methods for their quantification (Lohman et al. 2014; Jin et al. 2016). A conceptually similar approach targeting circRNAs utilized padlock probes ligated by T4 DNA ligase and rolling circle amplification permitting the in situ localization of circRNAs (Zaghlool et al. 2018). However, this approach would conceivably suffer from the same bias as qRT-PCR due to the dependence on reverse transcriptase. The breadth of diversity and functions of circRNAs are expanding and SplintQuant provides an easily adoptable approach to accurately assess the abundance of the circRNA. Accurate quantification of circRNAs is becoming more critical as circRNAs are commonly implicated as competing endogenous RNAs (ceRNAs) (Hansen et al. 2013; Memczak et al. 2013; Taulli et al. 2013). Like other noncoding RNAs, the stoichiometry between the circRNA and its target(s) are of paramount importance for inferring biological relevance (Fu 2014; Chiu et al. 2017). Therefore, a highly accurate estimate of circRNA abundance is required and NGS and qRT-PCR, both incorporating reverse transcription, are skewing these estimates. While Nanostring technology was exquisitely used to enumerate circular RNA molecules directly on RNA (Dahl et al. 2018), the disproportionate setup costs limit the use of this approach for the bulk of the circRNA researchers wanting to quantify a small number of circRNA targets.
Limitations
As demonstrated above, SplintQuant permits accurate quantification of candidate circRNA and fusion transcript abundance directly on purified RNA. However, this approach is similar to qRT-PCR in that it requires the design of custom target-specific oligonucleotides. Therefore, the desire to multiplex SplintQuant-qPCR in a single reaction may be challenging as it would require distinct fluorescent probes for each target in the master mix as well as, perhaps, distinct universal primers for the overhangs. Therefore, for those experiments requiring large scale profiling of circRNAs, the Nanostring method as per Dahl et al. (2018) would be recommended. A minor limitation of SplintQuant, also found in Nanostring and standard qRT-PCR approaches, is the inability to discriminate trans-spliced RNAs from bona fide circRNAs. Trans-spliced RNAs can resemble circRNAs as they possess the same backsplice junction sequence but are made from splicing two separate RNA molecules, rather than backsplicing within the single molecule, for circRNAs (Burgess 2013). However, trans-spliced RNAs account for <3% of noncanonical fusions, while circRNAs account for 60%–80% of these (Chuang et al. 2018). In each case, pretreatment of RNA with Ribonuclease R would degrade the trans-spliced RNAs, but not circRNAs, and eliminate this minor confounding from rare trans-spliced RNAs (Suzuki et al. 2006). Finally, the secondary structure of circRNAs and linear RNAs could conceivably impact the efficiency of the splint DNA oligonucleotides in annealing to their RNA targets. This SplintQuant method utilized single stranded binding protein in the reaction to enhance the accessibility of the oligonucleotides for their target, but increasing the reaction temperature (to 37°C) and/or altering the length of the oligonucleotides are strategies which can be utilized to mitigate these potential problems if they arise.
Conclusions
In conclusion, we have shown that the SplintQuant methodology is sensitive, specific, and highly accurate for quantifying circRNAs in RNA samples. SplintQuant offers a cost-effective solution while addressing the demonstrated artifacts inherent in qRT-PCR and has the potential to become the gold-standard method for circRNA quantification.
MATERIALS AND METHODS
Cell growth
The human mammary epithelial cell line (HMLE) was maintained and induced to form the mesenchymal version, called mesHMLE cells, as per Conn et al. (2015). HL-60 cells, MOLM-13, and MV4;11 were maintained in RPMI-1640 (Gibco), with 10% FBS and 1x penicillin-streptomycin-antimycotic (Sigma-Aldrich) at 37°C, with 5% carbon dioxide.
Harvesting RNA
Total RNA was extracted from cells with RNeasy Mini Kit (Qiagen) with on-column DNase treatment as per manufacturer's instructions. Alternatively, TRIzol (ThermoFisher Scientific) was used with DNase I digestions (Roche), ensuring post DNase-treatment cleanup with RNeasy MinElute Cleanup Kit (Qiagen).
For in vitro transcription of RNA substrates for use as linear RNA, we utilized overlapping long DNA oligonucleotides, with the forward primer tagged at its 5′ end with the T7 RNA polymerase promoter sequence (all oligonucleotides were ordered from Integrated DNA technologies and listed in Supplemental Table S1). The in vitro transcription reaction was performed using HiScribe T7 RNA polymerase kit (New England Biolabs), following manufacturer's instructions depending on size of RNA product: 10× Reaction Buffer (1.5 µL); 100 mM rATP (1.5 µL); 100 mM rCTP (1.5 µL); 100 mM rGTP (1.5 µL); 100 mM rUTP (1.5 µL); DNA template (100 ng); T7 RNA Polymerase Mix (2 µL); nuclease-free water to 20 µL. The reaction was incubated at 37°C in a thermal cycler for 16 h, followed by DNase I digestion (Roche) and cleanup with RNeasy MinElute Cleanup Kit (Qiagen).
In vitro synthetic circular RNA constructs
For in vitro circularization experiments, we ordered synthetic DNA fragments (gBlocks gene fragments, Integrated DNA Technologies) tagged at the 5′ end with the T7 RNA polymerase promoter sequence. The in vitro transcription reaction was performed using HiScribe T7 RNA polymerase kit (New England Biolabs), following manufacturer's instructions depending on the size of RNA product: 10× Reaction Buffer (2.0 µL); 100 mM rATP (1.5 µL); 100 mM rCTP (1.5 µL); 100 mM rGTP (0.6 µL); 50 mM GMP (6 µL); 100 mM rUTP (1.5 µL); DNA template (100 ng); T7 RNA Polymerase Mix (2 µL); nuclease-free water to 20 µL. We utilized guanosine 5-monophosphate (GMP; Sigma-Aldrich) in a 5:1 ratio GMP:GTP to label 5′ ends of RNA with monophosphate and simplify subsequent circularization as per Kershaw and O'Keefe (2012b). RNA was incubated at 37°C for 16 h. Subsequently, 23 µL Nuclease-free water, 5 µL 10× DNase I buffer and 2 µL (4U) RNase-free DNase I (Roche) were added and incubate at 37°C for 15 min. Cleanup of RNA was achieved with the RNeasy MinElute Cleanup Kit (Qiagen), eluting in water. This was quantified by nanospectrophotometry and resuspended at 10 µM (10 µM for a 300 nt RNA is ∼1 µg µL−1). RNA sizing was visualized by LabChip (PerkinElmer).
RNA was circularized as per Kershaw and O'Keefe (2012a), with the following modifications. The DNA oligonucleotide bridge was hybridized to the RNA under the following conditions: 5× Oligonucleotide annealing buffer (50 mM Tris, pH 7.5; 100 mM NaCl in nuclease-free water; 70 µL); 10 µM RNA substrate (2 µL); 100 µM bridging oligonucleotide (0.2 µL) and nuclease-free water (285.8 µL). The reaction was heated to 90°C and cooled slowly to 25°C (0.1°C.sec−1 in thermal cycler, ∼11 min). To circularize the RNA, 40 µL of 10× T4 DNA ligase buffer, 1 µL (20 U) of RNaseOUT (ThermoFisher Scientific), and 1 µL (2000 U) of high-concentration T4 DNA ligase (New England Biolabs) were added into the hybridization mixture and incubated at 37°C for 1 h. The reaction was stopped with the addition of 20 µL of 0.5 M EDTA pH 8. These were subsequently phenol:chloroform purified and precipitated with 0.1 volumes of 3 M sodium acetate, pH 5.2 and 2.5 volumes of absolute ethanol. This was briefly vortexed and precipitated at −20°C for >30 min. The tubes were spun at 4°C at 17,900 rpm for 30 min, removing supernatant and resuspended in 75% ice-cold ethanol, vortex and spin at 10k rpm for 5 min. Pellets were resuspended in 10 µL nuclease-free water and DNase I-treated as above and cleanup with MinElute RNA cleanup column (Qiagen). Perform two rounds of Ribonuclease R (Epicentre) digestion and MinElute RNA Cleanup of RNA to purify circRNAs as per Conn et al. (2017).
Oligonucleotide design
All DNA oligonucleotides were synthesized by Integrated DNA Technologies and purified by standard desalting. Two custom DNA oligonucleotides (donor and acceptor) were designed per RNA target to be adjacent and reverse complementary to the region(s) of interest. The complementary regions are 13 nt in length and the donor oligonucleotide was 5′-phosphorylated and tagged at the 3′ end with the M13 forward primer in antisense orientation (add onto the 3′ end of the donor oligonucleotide 5′-CTG GCC GTC GTT TTA C-3′, making the donor oligonucleotide 29 nt in length). The acceptor oligonucleotide was tagged at its 5′-end with the M13 reverse primer in sense orientation (add onto the 5′ end of the acceptor oligonucleotide 5′-CAG GAA ACA GCT ATG AC-3′, making the acceptor oligonucleotide 30 nt in length). For validation experiments, the donor oligonucleotide was also 3′-end labeled with 6-FAM (Fluorescein). All oligonucleotides were resuspended in nuclease-free primer resuspension buffer (10 mM Tris, pH8.0, 0.1 mM EDTA, pH 8.0) at a concentration of 100 µM and stored at −20°C, with FAM-labeled donor oligonucleotides protected from light.
Hybridization of oligonucleotides to the RNA
Donor and acceptor oligonucleotides were diluted to 10 µM using nuclease-free water and mixed in RNase-free 0.2 mL PCR tubes on ice: 5× Oligonucleotide annealing buffer (2 µL), 10 µM donor oligonucleotide (1 µL), 10 µM acceptor oligonucleotide (1.1 µL), RNA (Total RNA [100 ng]) or 10 µM IVT RNA Splint (1.1 µL), nuclease-free Water to 10 µL. It was critical to include a no-template control reaction (no added RNA to act as a splint). The reaction mixture was heated in a thermal cycler with heated lid (>95°C) to 85°C for 2 min and then cooled to 25°C at a slow rate (−0.1°C.sec−1, ∼10 min in total). This annealed DNA/RNA hybrid (substrate) may be used immediately or, alternatively, can be stored at −20°C.
SplintQuant ligation
Reaction buffers and substrate were placed on ice and components added: 10× SplintR Ligase Reaction buffer (2 µL); 1 µM Substrate (RNA/DNA hybrid; 2 µL); 25 U.µL−1 SplintR Ligase (1 µL); nuclease-free water to 20 µL. An important control reaction is a ligase-negative reaction (no added SplintR ligase). This was incubated for 30 min at 25°C and the reaction stopped by heating at 65°C for 20 min.
Quantitative PCR (qPCR)
We performed qPCR on the ligated product (template) using M13 universal primers (M13f: GTAAAACGACGGCCAG, M13r: CAGGAAACAGCTATGAC), according to the following reaction mix: 2× Quantitect SYBR Master Mix (5 µL), 500 nM of each primer, 2 nM template, and nuclease-free water to 10 µL. We performed qPCR on a suitable real-time PCR instrument (i.e., RotorGene Q [Qiagen]) using the following conditions: 95°C 10 min; 35–45 cycles of (95°C for 10 sec, 58°C for 10 sec, 72°C for 10 sec) followed by a melt curve from 70–90°C. Fluorescence snapshots were acquired at 72°C. A single product was confirmed by observing the melt curve, and Sanger sequencing of PCR products after gel purification with QiaQuick Gel extraction kit (Qiagen) and cloning into pGEM-T Easy vector (Promega). Normalization between total RNA samples was performed using GAPDH as a normalization gene(s) by SplintQuant, or standard qRT-PCR using Quantitect Reverse transcription kit (Qiagen) to make cDNA. Relative and absolute expression abundances were calculated using the standard 2−ΔΔCt method and interpolation from standard curves.
Gene fragment analysis
Following hybridization and ligation of FAM-labeled oligonucleotides, samples were submitted for separation on the ABI 3730 fragment analyser. The neat splint ligation mix (1–4 µL) was mixed with an appropriate size marker (GeneScan 120 LIZ dye Size Standard, ThermoFisher Scientific. LIZ fluorophore occupies the orange channel) with the resultant chromatograms (FAM fluorophore occupies the blue channel) analyzed with GeneMarker v.2.7.4 software (SoftGenetics, LLC). The area of the peak corresponding to the ligated product was quantified and compared between samples.
Key reagents
DNA oligonucleotides, 100 nmol, desalted (Integrated DNA Technologies)
SplintR Ligase (New England Biolabs, Cat. #M0375L)
Extreme Thermostable Single-Stranded DNA Binding Protein (ET SSB) (MCLAB, Cat. # ETSSB-200)
5X oligonucleotide annealing buffer (50 mM Tris, pH 7.5; 100 mM NaCl in RNase-free water)
Primer resuspension buffer (10 mM Tris, pH 8.0, 0.1 mM EDTA, pH 8.0 in RNase-free water)
Nuclease free water (ThermoFisher Scientific, Cat. # 10977023)
HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, Cat. #E2030)
TRIzol reagent (ThermoFisher Scientific, Cat. #15596018)
DNase I, recombinant, RNase-free (Roche, Cat. #04 716 728 001)
QuantiTect SYBR Green PCR Kit (Qiagen, Cat. #204145)
QuantiTect Reverse Transcription Kit (Qiagen, Cat. #205313)
pGEM-T Easy vector system (Promega, Cat. # A1360)
GeneScan 120 LIZ dye Size Standard, ThermoFisher Scientific, Cat. #4324287
Consumables
Strip Tubes and Caps for qPCR, 0.1 mL (Qiagen, Cat. # 981106)
1.5 mL micro-centrifuge tube (Ambion, Cat. #AM12450)
0.2 mL PCR tube, 8-strip (Ambion, Cat. #AM12230)
Qiaquick Gel extraction kit (Qiagen, Cat. #28706)
RNeasy Mini Kit (Qiagen, Cat. #74014)
RNeasy Minelute Cleanup Kit (Qiagen, Cat. #74204)
Optional (quality control only)
T4 DNA Ligase (New England Biolabs, Cat. #M0202T)
Guanosine 5′-monophosphate disodium salt hydrate (Sigma-Aldrich, Cat. #G8377-1G)
Ribonuclease R (Epicentre, Cat. #RNR07250)
Equipment
SimpliAmp Thermal cycler (ThermoFisher Scientific).
Rotor-Gene Q real-time PCR machine (Qiagen)
LabChipHT (PerkinElmer)
3730 DNA Analyzer (Applied Biosystems)
Timing
The RNA extraction steps take 30–120 min to complete, depending on the method. In vitro transcription (if control RNAs are required) takes up to 20 h to complete. The hybridization reaction takes ∼10–20 min per sample, but multiple reactions can be set up concurrently. The splint ligation setup and reaction takes ∼45 min and the subsequent qPCR setup and reaction will take ∼2 h. In total, the SplintQuant experiment can be completed in 3–4 h.
Troubleshooting
If multiple products are produced following qPCR, addition of ET SSB (MCLAB, Cat # ETSSB-200) has also been shown to increase activity and reduce off-target ligation when ligating DNA probes to RNA using SplintR Ligase.
If no products are obtained even with positive control RNA, consider running validation step, using 3′-end FAM-labeled donor oligonucleotide and running ligated products on DNA fragment analyzer.
If no products are detected, ensure good RNA handling procedures and purify oligonucleotides by RNase-free HPLC, instead of standard desalting.
If no products are detected, revisit oligonucleotide design, ensuring donor is 5′-phosphorylated, and donor/acceptor are adjacent and reverse complementary to the RNA target. The SplintR ligase enzyme tolerates all base pair combinations at the ligation junction, but is partially inhibited by dC/G and dG/C base pairs at the donor (phosphorylated) side ligation junction, particularly when the +2 base was also a C/G base pair, which are known to reduce efficiency of splint ligation. If backpslice junction of circRNA contains these bases, shift the junction point along, so the donor or acceptor will bridge the backsplice junction.
If a weak signal is obtained, consider increasing reaction temperature to 37°C and supplementing with Mn2+> 5 mM.
If poor amplification occurs with M13F/M13R primers, consider other universal primers. We have validated CS1/CS2 primers.
Anticipated results
The abundance of the RNA target (circRNA or linear RNA) in the sample should be reflected in the quantitative output from qPCR, unaffected by the size of the product and other RNA splice variants. Negative control reactions (no template control, or no-ligase control) should not display any product after 45 cycles.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joel Geoghegan, Wendy Parker, and colleagues at the ACRF Genomics Facility (SA Pathology) for discussions on this technology and performing the fragment analysis. We thank Oliver van Wageningen for performing Sanger Sequencing for this project. This work was supported by the Australian Research Council (FT160100318 to S.J.C.); the National Health and Medical Research Council (GNT1144250 to S.J.C.); and the Flinders Medical Centre Foundation (EMCR seed grant to V.C.). Funding for open access charge: Australian Research Council.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070953.119.
REFERENCES
- Burgess DJ. 2013. Gene expression: controls and roles for trans-splicing. Nat Rev Genet 14: 822 10.1038/nrg3618 [DOI] [PubMed] [Google Scholar]
- Chen D-F, Zhang L-J, Tan K, Jing Q. 2018. Application of droplet digital PCR in quantitative detection of the cell-free circulating circRNAs. Biotechnol Biotechnol Equip 32: 116–123. 10.1080/13102818.2017.1398596 [DOI] [Google Scholar]
- Chiu H-S, Martínez MR, Bansal M, Subramanian A, Golub TR, Yang X, Sumazin P, Califano A. 2017. High-throughput validation of ceRNA regulatory networks. BMC Genomics 18: 418 10.1186/s12864-017-3790-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T-J, Chen Y-J, Chen C-Y, Mai T-L, Wang Y-D, Yeh C-S, Yang M-Y, Hsiao Y-T, Chang T-H, Kuo T-C, et al. 2018. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res 46: 3671–3691. 10.1093/nar/gky032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S, Pillman K, Toubia J, Conn V, Salmanidis M, Phillips C, Roslan S, Schreiber A, Gregory P, Goodall G. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, et al. 2017. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 3: 17053 10.1038/nplants.2017.53 [DOI] [PubMed] [Google Scholar]
- Dahl M, Daugaard I, Andersen MS, Hansen TB, Grønbæk K, Kjems J, Kristensen LS. 2018. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest 98: 1657–1669. 10.1038/s41374-018-0108-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix CA, Hosler MR, Slater DJ, Megonigal MD, Lovett BD, Williams TM, Nowell PC, Spinner NB, Owens NL, Hoxie J, et al. 1999. Duplicated regions of AF-4 intron 4 at t(4;11) translocation breakpoints. Mol Diagn 4: 269–283. 10.1016/S1084-8592(99)80002-2 [DOI] [PubMed] [Google Scholar]
- Fu X-D. 2014. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev 1: 190–204. 10.1093/nsr/nwu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32: 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Vaud S, Zhelkovsky AM, Posfai J, McReynolds LA. 2016. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res 44: e116 10.1093/nar/gkw399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw CJ, O'Keefe RT. 2012a. Splint ligation of RNA with T4 DNA ligase. Methods Mol Biol 941: 257–269. 10.1007/978-1-62703-113-4_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw CJ, O'Keefe RT. 2012b. Splint ligation of RNA with T4 DNA ligase. Methods Mol Biol 941: 257–269. 10.1007/978-1-62703-113-4_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Hansen TB, Venø MT, Kjems J. 2018. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37: 555–565. 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn MW, Hadler MJ, Daigle SR, Koche RP, Krivtsov AV, Olhava EJ, Caligiuri MA, Huang G, Bradner JE, Pollock RM, Armstrong SA. 2015. MLL partial tandem duplication leukemia cells are sensitive to small molecule DOT1L inhibition. Haematologica 100: e190–e193. 10.3324/haematol.2014.115337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lohman GJS, Zhang Y, Zhelkovsky AM, Cantor EJ, Evans TC. 2014. Efficient DNA ligation in DNA–RNA hybrid helices by Chlorella virus DNA ligase. Nucleic Acids Res 42: 1831–1844. 10.1093/nar/gkt1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, MacLeod RA, Uphoff CC, Drexler HG, Nishizaki C, Katayama Y, Kimura G, Fujii N, Omoto E, Harada M, et al. 1997. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia 11: 1469–1477. 10.1038/sj.leu.2400768 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS, et al. 2017. The MLL recombinome of acute leukemias in 2017. Leukemia 32: 273–284. 10.1038/leu.2017.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. 2006. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34: e63 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Salzman J. 2016. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 17: 679–692. 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R, Loretelli C, Pandolfi PP. 2013. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 20: 541–543. 10.1038/nsmb.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubia J, Conn VM, Conn SJ. 2018. Don't go in circles: confounding factors in gene expression profiling. EMBO J 37: e97945 10.15252/embj.201797945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shan G. 2018. Nonradioactive northern blot of circRNAs. Methods Mol Biol 1724.: 135–141. 10.1007/978-1-4939-7562-4_11 [DOI] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. 2014. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9: e90859 10.1371/journal.pone.0090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A, Ameur A, Wu C, Westholm JO, Niazi A, Manivannan M, Bramlett K, Nilsson M, Feuk L. 2018. Expression profiling and in situ screening of circular RNAs in human tissues. Sci Rep 8: 16953 10.1038/s41598-018-35001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.