Abstract
Even though the RNAs contained in the small (40S) and large (60S) ribosomal subunits are cotranscribed, their assembly proceeds largely separately, involving entirely distinct machineries. Nevertheless, separation of the two subunits, an event that is critical for assembly of the small subunit, is delayed until domain I of the large subunit is transcribed, indicating crosstalk between the two assembly pathways. Here we show that this crosstalk is mediated by the assembly factor Rrp5, one of only three proteins required for assembly of both ribosomal subunits. Quantitative RNA binding and cleavage data demonstrate that early on, Rrp5 blocks separation of the two subunits, and thus 40S maturation by inhibiting the access of Rcl1 to promote cleavage of the nascent rRNA. Upon transcription of domain I of 25S rRNA, the 60S assembly factors Noc1/Noc2 bind both this RNA and Rrp5 to change the Rrp5 RNA binding mode to enable pre-40S rRNA processing. Mutants in the HEAT-repeat domain of Noc1 are deficient in the separation of the subunits, which is rescued by overexpression of wild-type but not inactive Rcl1 in vivo. Thus, Rrp5 establishes a checkpoint for 60S assembly during 40S maturation to ensure balanced levels of the two subunits.
Keywords: checkpoint, regulation, ribosome assembly
INTRODUCTION
Ribosomes are the most ancient and most conserved RNA–protein complexes. These macromolecular machines, which mediate protein synthesis in all cells, are composed of two subunits: the small subunit (40S in eukaryotes) decodes the messenger RNA (mRNA), and the large subunit (60S in eukaryotes) catalyzes formation of the peptide bond. These functions rely on an elaborate interaction network among four ribosomal RNAs (rRNAs) and 79 ribosomal proteins (r-proteins). Assembly of the r-proteins onto rRNA is a spatially and temporally coordinated pathway, errors in which are often deleterious (de la Cruz et al. 2015). To promote, regulate and quality control ribosome assembly, eukaryotes have evolved a ribosome biogenesis machinery involving over 200 ribosome assembly factors (AFs), of which ∼70 mediate 40S assembly and ∼130 are important for 60S biogenesis (Woolford and Baserga 2013). However, three AFs are essential for both 40S and 60S biogenesis: Rrp5, Has1, and Prp43 (Venema and Tollervey 1996; Emery et al. 2004; Lebaron et al. 2005; Combs et al. 2006; Leeds et al. 2006; Dembowski et al. 2013).
Ribosome assembly is initiated with transcription of the rRNA precursor, which undergoes cotranscriptional folding, modification, and processing in the nucleolus. Three of the four rRNAs are encoded in a single transcript, thus ensuring that the small and large ribosomal subunits are produced in similar amounts. Nonetheless, pre-rRNAs destined for the 40S and 60S subunits are separated early in assembly, at so-called site A2, located in the spacer sequence between 18S and 5.8S rRNAs (ITS1, Supplemental Fig. S1), resulting in separate assembly of 40S and 60S subunits. Interestingly, analysis of chromatin spreads and kinetic studies have revealed that this cleavage step, which is required for 40S production, is delayed for nearly 1 min after the cleavage site is transcribed, until domain I (and at least some of domain II) of 25S rRNA are also completed (Osheim et al. 2004; Koš and Tollervey 2010). How transcription of pre-60S rRNAs is sensed, and relayed to pre-40S subunits remains unknown.
While several early 60S AFs affect 40S maturation, Rrp5 is one of only three AFs absolutely required for assembly of both subunits (Venema and Tollervey 1996). We and others have previously shown that Rrp5 binds ITS1 at sequences around the A2 site, thus rationalizing its involvement in both 40S and 60S assembly (Young and Karbstein 2011; Lebaron et al. 2013; Young et al. 2013). Rrp5 binds the assembling 40S subunit early on and is required for recruitment of the kinase-regulated subcomplex UtpC and the DEAD-box protein Rok1 (Vos et al. 2004; Perez-Fernandez et al. 2007). Upon A2 cleavage and separation of pre-40S and pre-60S rRNAs, Rok1 releases Rrp5 from pre-40S subunits (Khoshnevis et al. 2016), and Rrp5 departs with pre-60S subunits (Lebreton et al. 2008; Jakovljevic et al. 2012; Gamalinda et al. 2014), where it binds the 60S AFs Noc1/Noc2 (Hierlmeier et al. 2013), and is required for the first pre-60S processing event (Venema and Tollervey 1996).
Here we use a combination of genetic and biochemical experiments, in yeast and in fully reconstituted systems, to show that Rrp5 has two distinct modes for binding to pre-rRNA: an A2-cleavage compatible mode and an A2-cleavage incompatible mode, which misplaces the carboxyl terminus of the protein. In the A2-cleavage incompatible mode, S1 domain 7 (S1_7) of Rrp5 blocks premature pre-rRNA processing at site A2 in vivo and in vitro. Binding of the 60S AFs Noc1/Noc2 to Rrp5 S1_7 then unlatches this inhibitory structure to promote the A2-compatible conformation and eliminate its inhibition of A2 cleavage. RNA binding data, as well as rescue of Noc1 mutations by overexpression of Rcl1, both suggest that the different Rrp5•RNA conformations directly impinge on access of Rcl1 to the A2 site, thereby regulating A2 cleavage. Thus, our data support a model in which Rrp5 integrates the assembly of small and large subunits, by blocking premature A2 cleavage, until it senses 60S assembly through interaction with the 60S AFs Noc1/Noc2, ensuring equal amounts of both subunits are produced.
RESULTS
Rrp5 can be physically separated between S1 domains
Yeast Rrp5 is a conserved, essential 193-kDa protein with 12 tandem S1 domains followed by a TPR domain (Fig. 1A). Previous work has shown that the protein can be separated into two parts (Torchet et al. 1998; Eppens et al. 1999; Young and Karbstein 2011): the nine amino-terminal S1 domains (Rrp5_S1-9) and the three carboxy-terminal S1 domains together with the TPR domain (Rrp5_S10-12T; see Supplemental Fig. S2A for a description of the fragment nomenclature). These two domains, if coexpressed on two plasmids, largely complement growth in cells depleted of endogenous Rrp5 (Supplemental Fig. S2B; Torchet et al. 1998; Eppens et al. 1999; Young and Karbstein 2011). To use this system to systematically delete individual S1 domains and assay their roles in 40S and/or 60S assembly, we first confirmed the growth complementation of other pieces of Rrp5 by separating Rrp5 between different S1 domains. Separating Rrp5 between S1 domains 4 and 5 (Rrp5_S1-4 + Rrp5_S5-12T), 5 and 6 (Rrp5_S1-5 + Rrp5_S6-12T), 6 and 7 (Rrp5_S1-6 + Rrp5_S7-12T), and 7 and 8 (Rrp5_S1-7 + Rrp5_S8-12T) also resulted in nearly fully complementing fragments (Supplemental Fig. S2B). These findings provide us with a tool to study the function of individual S1 domains of Rrp5 in ribosome maturation by coexpressing amino- and carboxy-terminal fragments. For example, combining Rrp5_S1-4 and Rrp5_S6-12T is akin to deletion of S1 domain 5, and will be referred to as Δ5 (Fig. 1A).
FIGURE 1.
Deletion of S1 domain 5 blocks A2 cleavage, which can be rescued by deleting S1 domains 6–7. (A) Serial dilution growth assay of yeast containing Rrp5 under galactose-inducible control (YKK248), and supplemented with the indicated plasmids. (B) Northern blot analysis of detectable rRNA processing intermediates. U2 serves as loading control. To account for any effects from dividing Rrp5 into two separate molecules, we use the fully complementing fragments as our control, denoted as “FL.” (C) Quantification of two to three northern blots such as shown in B. 27SA2, 20S, 23, and 35S rRNA levels are normalized to U2 levels and to the “FL” construct. (D) Analysis of 5.8SL, 5.8SS, A4, and A2 cleavage levels by reverse transcription.
Rrp5's S1 domain 7 blocks early 18S rRNA processing in vivo
Δ5 or Δ5–6 produce a severe slow growth phenotype, which is partially rescued by additional removal of S1 domain 7 (Δ5–7, Fig. 1A). These data are consistent with previously published data that removal of S1 domains 3–5 is more deleterious to growth than deletion of S1 domains 6–8 (Eppens et al. 1999; Hierlmeier et al. 2013).
To garner insight into the function of these S1 domains in vivo, we studied the effect on rRNA processing by northern blotting. Both Δ5 and Δ5–6 cells show reduced levels of 20S and 27SA2 rRNAs (Fig. 1B,C), the product of cleavage at so-called site A2, which separates rRNAs destined for the small and large ribosomal subunits. However, 20S and 27SA2 levels are restored to near wild-type levels in Δ5–7 cells, demonstrating rescue of A2 cleavage in these cells. Δ7 alone has no effect on growth or rRNA processing (Fig. 1). These results suggest that S1 domain 7 negatively affects the early 40S maturation at site A2. Importantly, addition of LiCl to block exonucleolytic degradation of pre-rRNAs (Dichtl et al. 1997) does not affect the distribution of rRNA processing intermediates (Supplemental Fig. S2D), strongly suggesting that the loss of the A2 cleavage products (20S and 27SA2 rRNA) is due to blocked cleavage and not enhanced degradation in the Δ5 cells. Finally, we used primer extension to confirm that rescue of growth did not arise from the use of the so-called A4 site in the Δ5–7 cells. The data in Figure 1D show that growth was not correlated with use of the A4 site. Thus, the Northern and primer extension data show that S1 domain 5 promotes and S1 domain 7 opposes A2 cleavage.
Rrp5's S1 domain 7 modulates the RNA affinity of S10–12
To better understand the role of Rrp5's S1 domain 7 in early 40S maturation at site A2, we took advantage of an RNA binding assay we previously developed (Young and Karbstein 2011). In this assay different fragments of (or full length) Rrp5 are incubated with distinct rRNA mimics, protein-bound RNA separated from free RNA using native gels (Supplemental Fig. S3), and the fraction of bound RNA at different Rrp5 concentrations quantified and fit to a single binding model to obtain binding constants.
Rrp5_S10-12T binds about 10-fold more strongly to an rRNA mimic ending at the 3′-end of ITS1 (H44-ITS1) than to an rRNA mimic ending at the A2 cleavage site (H44-A2), indicating that the C-terminal S1 domains 10–12 interact with rRNA sequence between the A2 site and the end of ITS1 (Fig. 2A; Young and Karbstein 2011). This agrees with in vivo crosslinking data, which mapped the Rrp5_S10-12T binding site to the sequences between A2 and A3 cleavage sites (Lebaron et al. 2013). However, the full-length protein did not show stronger affinity for sequences 3′ to the A2 site (Young and Karbstein 2011), suggesting that an element within S1 domains 1–9 interferes with the binding of S1 domains 10–12 for the region between sites A2 and ITS1.
FIGURE 2.
S1 domain 7 modulates the RNA affinity of S1 domains 10–12. (A) The binding affinities of different Rrp5 fragments for pre-rRNA mimics were measured by native gel shift in three replicate experiments. The RNAs are depicted in the scheme on top. The plot reflects the difference in affinities of the indicated Rrp5 construct for the two RNAs. This difference represents the interaction strength between Rrp5 fragments with the RNA between the A2 and ITS1 sites. (B) Gel-shift assay to measure S1 domain 7-dependent release of Rrp5_S8-12T from H45-5.8S rRNA. Addition of Rrp5_S1-7, but not Rrp5_S1-5, displaces Rrp5_S8-12T from RNA. Noc1/Noc2 block this activity.
To delineate this element, we performed RNA binding experiments using different carboxy-terminal fragments of Rrp5, where we systematically added S1 domains. While both Rrp5_S10-12T and Rrp5_S8-12T have ∼10-fold higher affinity for H44-ITS1 than for H44-A2, Rrp5_S7-12T shows only an approximately twofold difference in binding to RNAs ending in ITS1 relative to A2 (Fig. 2A, last column). This ratio remains the same as more S1 domains are added (Rrp5_S6-12T, Rrp5_S5-12T, or Rrp5-FL). Thus, S1 domain 7 eliminates the preference for sequences between A2 and the end of ITS1, suggesting that it blocks the interaction of S1 domains 10–12 with pre-rRNA between sites A2 and ITS1.
To test directly if S1 domain 7 affects the interaction of S1 domains 10–12 with pre-rRNA, we developed a native gel-shift assay, coupled to western blotting. Importantly, while Rrp5_S8-12T is detected after native PAGE/western blotting, Rrp5_S1-7 is not (Fig. 2B), likely due to the fact that the Rrp5 antibody was raised against Rrp5_S9-12T. This simplifies the analysis of this complicated mixture, akin to RNA-binding experiments which also interrogate only a single component. Addition of H45-5.8S rRNA (RNA) to Rrp5_8-12T leads to an upshift in the band, as expected from Rrp5_8-12T binding to the RNA (see also Supplemental Fig. S3), and reproducibly makes it more smeary, indicative to RNA-dissociation during the extended gel run (Fig. 2B, lanes 3,4). Addition of the amino-terminal fragments (Rrp5_S1-5 or Rrp5_S1-7) to the carboxy-terminal Rrp5_S8-12T leads to a (smaller) upshift (lanes 3 vs. 7,8), suggesting the amino- and carboxy-terminal Rrp5 fragments can interact with each other, as expected from the yeast complementation. This upshift is smaller, because the RNA is about twice as large as Rrp5_S1-7 (Rrp5_S8-12T:94 kDa; Rrp5_S1-7:97 kDa; RNA: 174 kDa), thus allowing us to distinguish between Rrp5_S8-12T bound to RNA and Rrp5_S1-7.
When we assembled a Rrp5_S8-12T/RNA complex, and then added Rrp5_S1-7 (which contains S1 domain 7), the Rrp5_S8-12T/RNA complex was down-shifted to the position of the two proteins bound to each other (lanes 5 vs. 4,7), indicating that Rrp5_S1-7 binds Rrp5_S8-12T and leads to its release from the RNA. This depends on the presence of S1 domain 7, as addition of Rrp5_S1-5 retained the slower-migrating band of the Rrp5_S8-12T/RNA complex (Fig. 2B, lanes 4–6). Therefore, these data support a model of two rRNA-binding modes for Rrp5, one in which S1 domains 10–12 bind H45-5.8S rRNA, and one in which S1 domain 7 releases this interaction, by either remodeling the RNA structure, Rrp5, or both. Note that the electrophoretic shift differences are small due to the small relative size changes; nevertheless, the same changes were observed in three replicate experiments, leading us to have confidence in the data despite these small changes.
S1 domains 10–12 are required for 40S maturation at site A2
Deletion of S1 domain 7 unblocks 40S maturation at site A2 in vivo (Fig. 1) and modulates the interaction of S1 domain 10–12 for pre-rRNA sequences between the A2 site and the end of ITS1 (Fig. 2). We therefore asked if S1 domains 10–12 regulate A2 cleavage in vivo. To answer this question, we compared the rRNA processing intermediates in cells coexpressing Rrp5_S1-9 with either Rrp5_S10-12T (“FL”) or Rrp5-TPR (Δ10–12) using northern blotting. Δ10–12 cells showed reduced levels of 20S rRNA accompanied by an increase in 21S levels (Supplemental Fig. S5), indicative of a defect in cleavage at the A2 site. These results corroborate previous findings that S1 domains 10–12 play a role in A2 cleavage (Torchet and Hermann-Le Denmat 2000), although the borders of the Rrp5 fragments in the two experiments are slightly different.
S1 domain 7 inhibits the pre-40S cleavage activity of Rcl1
Rcl1 is required for the cotranscriptional 40S cleavage step at site A2 in vivo (Billy et al. 2000; Horn et al. 2011), and recombinant Rcl1 can bind and cleave rRNA containing the A2 site in vitro (Horn et al. 2011). Because S1 domains 10–12 are required for A2 cleavage in vivo (Fig. 2; Torchet and Hermann-Le Denmat 2000), and because S1 domain 7 disrupts the interaction of S1 domain 10–12 with rRNA, and blocks A2 cleavage in vivo, we wanted to test if S1 domain 7 blocks Rcl1 activity. We monitored Rcl1 activity by incubating Rcl1 with 5′-32P-labeled H45-ITS1 rRNA and followed the formation of the H45-A2 product over time. As shown in Figure 3, addition of Rrp5 blocks RNA cleavage. This effect from Rrp5 is specific for Rcl1, as Rrp5 does not affect Nob1-dependent cleavage at site D (Supplemental Fig. S6A) or Utp24-dependent cleavage near sites A0 and A1 (see below).
FIGURE 3.
S1 domain 7 inhibits A2 cleavage activity of Rcl1. (A) Rcl1 quantitatively cleaves H45-ITS1 to generate H45-A2 over time. Addition of Rrp5 fragments harboring S1 domain 7 inhibits this activity. 3 μM Rcl1 and 1 µM Rrp5 fragments were used in this experiment. (B) Quantification of three replicates of data such as in A. All data are normalized to the rRNA cleavage rate constant in the absence of Rrp5. Rrp5 fragments are depicted on the right.
In vivo data implicated Utp24 as the nuclease for A1 cleavage (Bleichert et al. 2006), but recent biochemical experiments have also suggested Utp24 as an alternative nuclease for A2 cleavage (Wells et al. 2016). To test if Utp24 is active as a nuclease at the A1 cleavage site, we cloned and expressed Utp24 and an active site mutant (Utp24_D38N), purified the proteins over three columns, while pooling away from nuclease contamination, and then incubated the purified proteins with rRNA mimics containing the A0, A1, and A2 cleavage sites to test for Utp24-dependent RNA cleavage at these sites. Importantly, primer extension analysis demonstrates cleavage adjacent to sites A0 and A1, on opposite sites of a putative duplex containing these cleavage sites (Supplemental Fig. S6B,C). These cleavage sites are not observed when the same RNAs are incubated with the Utp24_D38N mutant protein, thus suggesting that they arise directly from Utp24. We suspect that the cleavage adjacent to, but not at, the A1 site, and at site A0 arises because the many cofactors present in the early pre-40S precursor are not present in our assay, thus perhaps allowing the RNA to form a duplex in vitro. In contrast, recent EM structures demonstrate that Sof1, Utp7, and Utp14 directly interact with the A1 site RNA, thus keeping it single stranded (Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017). In addition, or alternatively, Utp24 might dimerize on this RNA as observed for other PIN-domain containing proteins (Arcus et al. 2004; Mattison et al. 2006).
Importantly, in a quantitative assay that relies on 5′-end labeled RNA, and which demonstrates that about half of the RNA is cleaved by Utp24, Rrp5 did not affect Utp24-mediated cleavage near the A0 site (Supplemental Fig. S6D), further demonstrating the specificity of the effect from Rrp5 on A2 cleavage, and not Nob1 or Utp24-mediated cleavage events. Note that in this quantitative assay A1 cleavage is obscured by prior or subsequent A0 cleavage, which removes the 5′-end 32P label, rendering the product “invisible.” Finally, we note that our highly purified Utp24 does not cleave the H45-5.8S rRNA mimic at the A2 site (Supplemental Fig. S6B), although the RNAs used differ from those used by Wells and coworkers, which could affect this outcome.
Next, we used our collection of Rrp5 fragments to test if, as expected, S1 domain 7 was required for Rcl1 inhibition. The data in Figure 3 show that Rrp5-FL, Rrp5_S5-12T, Rrp5_S6-12T, and Rrp5_S7-12T, which all contain S1 domain 7, inhibit between 95% (Rrp5-FL and Rrp5_S5-12T) and 85% (Rrp5_S7-12T) of Rcl1 activity. In contrast, addition of Rrp5_S8-12T, which lacks S1 domain 7, only provides for a moderate 25% reduction of Rcl1 activity. Thus, Rrp5 blocks cleavage at site A2 in a manner that is dependent on S1 domain 7.
Rrp5 blocks productive RNA binding by Rcl1
Interestingly, Rrp5 concentrations much below those of Rcl1 are sufficient for inhibition of Rcl1 (Supplemental Fig. S4C,D and data not shown), suggesting that the inhibition does not depend on a direct interaction between Rrp5 and Rcl1, and instead arises from blocking Rcl1's access to the RNA. To further investigate this hypothesis, we compared the inhibition by wild-type and the R921E mutant of Rrp5_6-12T. Quantitative RNA binding experiments demonstrate that wild-type and mutant Rrp5_6-12T bind rRNA with affinities of 130 and 297 nM, respectively (Supplemental Fig. S4A,B). Similarly, the Rcl1 inhibition constants for wild-type and mutant Rrp5_6-12T are 117 and 203 nM, respectively (Supplemental Fig. S4C,D). The observation that RNA binding and cleavage inhibition by Rrp5 follow quantitatively the same concentration dependences and are affected by mutations in the RNA binding site, strongly suggest that Rrp5's effect on Rcl1's activity is mediated by RNA. This conclusion is also supported by the absence of direct interactions between Rcl1 and Rrp5 (Supplemental Fig. S4E), as well as the recent cryo-EM structures of early pre-40S ribosomes, which show Rcl1 and the TPR domain of Rrp5 at some distance from each other (Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017). We thus suggest that Rrp5 blocks Rcl1 binding to the A2 cleavage site in a Noc1/Noc2-dependent manner.
S1 domain 7 interacts with the Noc1/Noc2 complex
Our data so far suggest opposing roles for S1 domains 7 and 10–12 in pre-rRNA processing, where the former inhibits cotranscriptional 40S processing at site A2 in vivo, while the latter promote it. Previous findings indicate that the A2 cleavage step is delayed for nearly 1 min until domains I and II of 25S rRNA are transcribed (Osheim et al. 2004; Koš and Tollervey 2010). We therefore hypothesized that Rrp5, via the activity of S1 domain 7, blocks A2 cleavage until it senses the correct initiation of 60S assembly via communication with a 60S AF. Previous work demonstrated that the Noc1/Noc2 complex directly interacts with Rrp5, and identified the middle domains of Rrp5 as the interaction surface (Hierlmeier et al. 2013). To further explore which of the middle S1 domains was involved in this interaction, we used protein–protein interaction assays. We coexpressed and copurified the (MBP-)Noc1/Noc2 complex, and added the complex, together with recombinant Rrp5 fragments, to amylose beads. As expected (Hierlmeier et al. 2013), full-length Rrp5 binds the Noc1/Noc2 complex (Supplemental Fig. S7A). Furthermore, Rrp5_S5-12T, Rrp5_S6-12T, and Rrp5_S7-12T bind Noc1/Noc2 in approximately stoichiometric complexes, but Rrp5_S8-12T does not (Fig. 4A, top; note that the intensity of Coomassie staining is proportional to the molecular weight. Thus, 160 kDa MBP-Noc1 will stain almost twice as strong as 100 kDa Rrp5_S7-12T). Similarly, Rrp5_S1-9 and Rrp5_S1-7 bind Noc1/Noc2, but Rrp5_S1-5 does not (Fig. 4A, bottom), demonstrating that S1 domain 7 is responsible for the interaction with Noc1/Noc2, consistent with previous yeast two hybrid data (Hierlmeier et al. 2013). Of note, Noc2 alone fails to pull down Rrp5, indicating that the interaction between Rrp5 and the Noc1/Noc2 complex occurs via Noc1 (Supplemental Fig. S7B). We were unable to test this directly as Noc1 alone was not expressed in Escherichia coli (data not shown).
FIGURE 4.
Noc1/Noc2 bind to S1 domain 7 of Rrp5 and change its structure. (A) Recombinant MBP-Noc1/Noc2 immobilized on amylose resin interacts with recombinant, purified fragments containing S1 domain 7 but not Rrp5_S8-12T (left top panel) as well as Rrp5_FL, S1-9, and S1-7, but not Rrp5_1-5 (left bottom panel). None of the Rrp5 fragments interact with MBP (right panel). The positions of Rrp5 fragments are marked with asterisks and Noc2 with a hashtag. (B) Glu-C limited proteolysis coupled to western blot analysis of Rrp5_S6-12T with and without Noc1/Noc2. (+) and (++) indicate a 1:100 ratio of glu-C either to Rrp5_S6-12T or Rrp5_S6-12T•Noc1/Noc2, respectively. Noc1/Noc2 induced fragments are marked.
Noc1/Noc2 binding changes Rrp5 structure
Above, we have shown that Noc1/Noc2 binds a critical structural element in Rrp5, S1 domain 7, which mediates a switch between an A2-cleavage inhibited and competent state of Rrp5. To test if binding of Noc1/Noc2 affects Rrp5 structure, we used limited proteolysis coupled to western blotting (Fig. 4B). These data show that while Noc1/Noc2 addition stabilizes the full-length protein (as expected for any binding partner), it leads to the appearance of a collection of novel Rrp5 fragments. These first arise at 5 min of protease treatment and are not observed in the absence of Noc1/Noc2 even after 10 min, thus indicating that they report on a novel structure of Rrp5 induced by the presence of Noc1/Noc2. Importantly, because the Rrp5 antibody requires the presence of S1 domains 10–12 (see Fig. 2B), these Rrp5 fragments must contain S1 domains 10–12, allowing us to estimate their boundaries to roughly correspond to Rrp5_S8-12T.
Binding of Noc1/Noc2 modulates Rrp5 RNA binding
The interaction between Noc1/Noc2 and S1 domain 7 of Rrp5 led us to hypothesize that binding of Noc1/Noc2 could neutralize the effect from this S1 domain on the RNA-binding interactions of S1 domains 10–12 (which are required for A2 cleavage), as well as on Rcl1 activity.
To test the former, we used quantitative RNA binding assays to measure the affinities of Rrp5_S6-12T, Noc1/Noc2 and Rrp5_S6-12T/Noc1/Noc2 for H44-A2 and H44-ITS1. Accurate RNA binding measurements require a range of protein concentrations from below to above the Kd value. However, Rrp5 binds RNA very tightly (Fig. 2A; Young and Karbstein 2011), such that at concentrations of Rrp5 that are subsaturating with respect to RNA, Rrp5 was no longer fully bound to Noc1/Noc2 and we would instead be measuring affinity of Rrp5_S6-12T alone (even though Noc1/Noc2 was present in the mixture, but not bound to Rrp5). Thus, we intentionally weakened the interaction of Rrp5_S6-12T with RNA, using the R921E mutation in S1 domain 8. Importantly, this mutation does not affect Rrp5 binding to MBP-Noc1/Noc2 (Supplemental Fig. S7C). While both Rrp5_S6-12TR921E and Noc1/Noc2 have approximately twofold higher affinity for H44-ITS1 over H44-A2, the complex of Rrp5_S6-12TR921E•Noc1/Noc2 binds ∼10-fold more tightly to H44-ITS1 than to H44-A2 (Fig. 5A). This is akin to Rrp5_S8-12T, which lacks S1 domain 7 (Fig. 2A), and strongly suggests that indeed Noc1/Noc2 binding to S1 domain 7 allows S1 domain 10–12 to interact with rRNA sequences near the A2 site.
FIGURE 5.
Noc1•Noc2 modulate Rrp5's RNA binding and Rcl1's cleavage activity. (A) Addition of Noc1•Noc2 changes the RNA binding affinity of Rrp5_S6-12T. (B) Addition of Noc1•Noc2 releases Rrp5-dependent inhibition of A2-cleavage. 0.2 µM Rrp5_S6-12T and Noc1/Noc2 were used with 5 µM Rcl1 and 0.1 µM D1 RNA. (C) Overexpression of Rcl1 rescues Noc1 mutations. Serial dilutions of a Gal::Noc1;Gal::Rrp5 strain (YKK821), supplemented with plasmids encoding wild-type Rrp5, wild-type Noc1 (WT), Noc1_E365K, or Noc1_S615A show severe growth defects for the Noc1 mutants. These are rescued by constitutive GPD-promoter-driven overexpression of Rcl1 (+Rcl1) from a plasmid. (D) Total cell northern blots from the strains in C.
To confirm this conclusion, we took advantage of our native gel-shift assay and tested the effect from Noc1/Noc2 on S1 domain 7-dependent release of Rrp5_S8-12T from pre-rRNA. While addition of Rrp5_S1-7 to the Rrp5_S8-12•RNA complex leads to downshift (release) of Rrp5_S8-12T from the RNA as described above (Fig. 2B, lanes 4,5), in the presence of Noc1/Noc2 addition of Rrp5_S1-7 does not affect the migration of the Rrp5_S8-12T•RNA (lanes 9,10), as expected if Noc1/Noc2 blocks the effect from S1 domain 7.
Next, we tested if other Rrp5 interaction partners, such as Has1 and Rok1 (Young and Karbstein 2012; Khoshnevis et al. 2016), could similarly affect the binding of Rrp5_S6-12T to RNA. Importantly, neither addition of Rok1 nor Has1 affects the relative affinity of H44-ITS1 over H44-A2 (Supplemental Fig. S8), demonstrating the specificity of Noc1/Noc2 for affecting Rrp5•RNA structure in a manner that affects the interaction of S1 domain 10–12 with rRNA.
Binding of Noc1/Noc2 rescues the Rrp5-dependent inhibition of Rcl1 cleavage
To test if addition of Noc1/Noc2 to Rrp5 also rescues Rcl1 access to the A2 site and A2 cleavage, we compared the rate constants for A2 cleavage for Rrp5-bound RNA in the presence and absence of Noc1/Noc2. While Rcl1 is effectively blocked by Rrp5_S6-12T, addition of Noc1/Noc2 rescues the cleavage activity, indicating that the presence of Noc1/Noc2 suppresses the inhibitory effect of S1 domain 7 on Rcl1 (Fig. 5B). Intriguingly, in order for Noc1/Noc2 to relieve the Rrp5-mediated inhibition of Rcl1 cleavage, 25S D1 rRNA must be added (Supplemental Fig. S7E vs. Fig. 5B), consistent with the observation that transcription of D1 is required for A2 cleavage in vivo (Osheim et al. 2004; Koš and Tollervey 2010). This effect from the 25S D1 rRNA is specific, as addition of U3 RNA does not provide this effect (Supplemental Fig. S7G).
Together, these data strongly suggest that addition of the early 60S AFs Noc1/Noc2 allows for a change in the Rrp/RNA complex, remodeling the RNA, the protein, or both, such that S1 domains 10–12 are positioned near the A2 site to allow for Rcl1 access to that site.
Overexpression of Rcl1 can rescue Noc1 mutations
The data above indicate that Noc1/Noc2 binding to Rrp5 changes the interaction between Rrp5 and rRNA to allow Rcl1 to access pre-rRNA and cleave it at site A2. A prediction from this model is that overexpression of Rcl1 could (partially) compensate for inactivation of Noc1, as it might force access of Rcl1 to the rRNA by mass action even in the absence of (functional) Noc1. To test this prediction and thereby validate this model in vivo, we created a series of Noc1 mutants that were based on the previously published (Milkereit et al. 2001) Noc1-1 allele (P Milkereit, pers. comm.). Intriguingly, Noc1_E365K and Noc1_S615A, which are both located on the convex site of the HEAT repeat domain, show a very strong growth defect (Fig. 5C). Overexpression of wt Rcl1 rescues the deleterious effect from this mutation (Fig. 5C; Supplemental Fig. S7H), providing genetic evidence that the 60S AFs Noc1/Noc2 support Rcl1 activity in vivo. Rescue requires wild-type Rcl1 and is not observed with the inactive Rcl1_RDK mutant (Fig. 5C). This mutation does not affect pre-40S recruitment, but inactivates Rcl1 in vivo and in vitro, perhaps by weakening its RNA binding (Horn et al. 2011). Furthermore, it also affects Rcl1's interaction with the GTPase Bms1 (Delprato et al. 2014), which might further dysregulate Rcl1 and/or Bms1 activity in this mutant.
Importantly, in vitro pulldown data do not provide evidence for a direct interaction between Noc1/Noc2 and Rcl1 (Supplemental Fig. S7D), consistent with Noc1/Noc2 being 60S AFs, and, in contrast to Rcl1 and Rrp5, not part of pre-40S subunits (Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017).
Depletion of Noc1 blocks 60S maturation, but also has effects on A2 cleavage (Milkereit et al. 2001). We therefore used northern blotting to test if the Noc1_E365K and Noc1_S615A mutants similarly affect rRNA processing (Fig. 5D). As expected, both the Noc1_E365K and Noc1_S615A mutant showed decreased levels of the 20S and 27SA2 intermediates, indicative of defects in A2 cleavage, similar to what was previously observed for Noc1-1 (Milkereit et al. 2001). In addition, these mutants showed decreased levels of 18S rRNA and 25S rRNA. These defects in A2 cleavage are fully rescued by overexpression of wt Rcl1, which restores the levels of the 20S and 27SA2 intermediates, as well as the mature 18S and 25S rRNAs. In contrast, Rcl1_RDK does not rescue the deleterious effects from these Noc1 mutations. These data demonstrate a link between Noc1 and Rcl1 in vivo and are fully consistent with Rrp5 blocking Rcl1 access until relieved by Noc1.
Noc1/Noc2 bind pre-60S rRNA
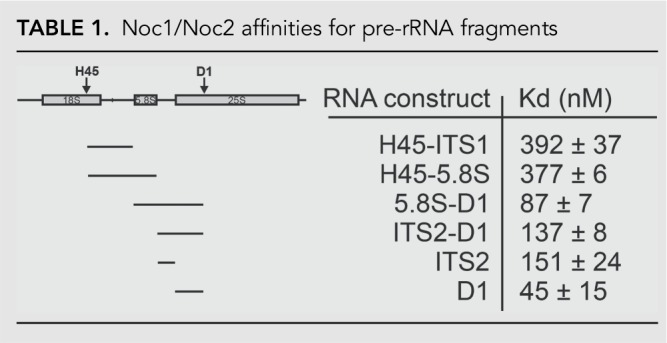
A2 cleavage is delayed until 5.8S rRNA, ITS2, and domain 1 (D1) of 25S rRNA are transcribed (Osheim et al. 2004; Koš and Tollervey 2010). Here we show that Noc1/Noc2 binding to Rrp5 regulates its structure and function, to allow access of Rcl1 to the A2 site. Importantly, this effect requires the addition of D1 of 25S rRNA. We thus hypothesized that the temporal regulation of A2 cleavage after transcription of D1 is due to Noc1/Noc2 binding to these elements. To test this hypothesis, we performed RNA binding studies using purified recombinant Noc1/Noc2 and different pre-rRNAs fragments, encompassing rRNAs from H45, ITS1, 5.8S, ITS2, and D1 (Table 1). Relative to the other tested RNAs, Noc1/Noc2 does not bind strongly to H45-ITS1 (encompassing the A2 cleavage site), and addition of 5.8S rRNA (H45-5.8S) does not substantially increase the binding. In contrast, binding to 5.8S-D1 and ITS2-D1 is strong, suggesting that Noc1/Noc2 binds within the early pre-60S complex, as hypothesized. The strongest binding interactions were found with D1 alone (Kd = 45 nM), suggesting strongly that D1 comprises the Noc1/Noc2 binding site, consistent with the observations that it is required for Noc1/Noc2-dependent rescue of RNA cleavage as described above. Importantly, this finding is consistent with recent affinity purifications demonstrating that Noc1/Noc2 and Rrp5 are bound to D1-containing assembly intermediates (Chen et al. 2017). Intriguingly, while no RNA is bound as strongly as D1, linking D1 to ITS2 to give ITS2-D1 weakens the interactions with this RNA (Kd = 45 nM and 137 nM for binding to D1 and ITS2-D1, respectively). Similarly, 5.8S-D1 binds weaker than D1 alone (Kd = 45 nM and 87 nM for binding to D1 and 5.8S-D1, respectively). Together, these observations indicate that structural differences arising from the ITS2-D1 linkage affect Noc1/Noc2 binding to D1, perhaps because Noc1/Noc2 bind near the junction of ITS2 and D1 of 25S.
TABLE 1.
Noc1/Noc2 affinities for pre-rRNA fragments
DISCUSSION
Rrp5 introduces a 60S-assembly dependent checkpoint to 40S subunit maturation
Rrp5 is one of only three AFs required for assembly of both ribosomal subunits (Venema and Tollervey 1996; Emery et al. 2004; Lebaron et al. 2005; Combs et al. 2006; Leeds et al. 2006; Dembowski et al. 2013). In addition, A2 cleavage and thus 40S maturation in vivo are delayed for nearly a minute until D1 of 25S rRNA is transcribed (Osheim et al. 2004; Koš and Tollervey 2010). The data herein describe the mechanism for the coupling of 40S maturation to 60S transcription, and demonstrate an essential role for Rrp5 in this process.
Specifically, the in vivo rRNA processing data show that an interaction between the three carboxy-terminal S1 domains of Rrp5 and rRNA near the A2 cleavage site is required for A2 cleavage in vivo but is antagonized by the presence of S1 domain 7. In vitro RNA binding and cleavage experiments show that in this conformation, Rcl1 cannot access (and cleave) the A2 site. Furthermore, our data demonstrate that the early 60S AFs Noc1/Noc2, which are recruited to nascent ribosomes by binding to domain I (D1) of 25S rRNA, bind Rrp5 at S1 domain 7 to neutralize the inhibitory effect from this S1 domain. Finally, overexpression of wild-type but not mutant Rcl1 can rescue A2 cleavage in yeast strains where Noc1 is inactivated.
Based on these observations, as well as others in the literature, we suggest the following model for the regulation of A2 cleavage by Rrp5 (Fig. 6). Rrp5 binds early in 40S assembly (Vos et al. 2004; Perez-Fernandez et al. 2007; Chaker-Margot et al. 2015) in a conformation where the three carboxy-terminal S1 domains are not engaged with the rRNA near the A2 site. While Rcl1 is also present in these precursors, its access to the A2 cleavage site is blocked, thereby inhibiting separation of 40S and 60S precursors at site A2. This model is consistent with the recent structures of early 40S precursors, which show Rcl1 (as well as Utp24)—but not near the A2 cleavage site (Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017). Indeed, ITS1 (containing the A2 cleavage site) is not resolved in the structures, suggesting it is flexible (and thereby not near Rcl1, or Utp24, which are in the core of the molecule). As a result, A2 cleavage is blocked in these molecules (Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017). Continued transcription of 25S rRNA eventually creates the binding site for the early 60S AFs Noc1/Noc2, which bind within 25S rRNA domain I (Chen et al. 2017). These may be repositioned to domain I from previous locations, or newly recruited to the pre-40S. Once localized to pre-rRNA, Noc1/Noc2 binds S1 domain 7 of Rrp5, thereby changing the Rrp5-RNA binding mode, allowing for repositioning of Rcl1 or A2 cleavage site (or both). In this new conformation, rRNA can be cleaved at the A2 cleavage site. Thus, the binding of Noc1/Noc2 to D1 and Rrp5 is a checkpoint required for 40S maturation.
FIGURE 6.
Rrp5 links 40S and 60S maturation in response to early steps of 60S assembly. Early during ribosome assembly Rrp5 is in conformation that is incompatible with A2 cleavage. This conformation is induced by S1 domain 7 (in red) and blocks access of Rcl1 to the A2 cleavage site. (Note that Rcl1 is nevertheless a component of these assembly intermediates.) After transcription of domain 1 of 25S, Noc1/Noc2 are recruited (or repositioned from within pre-40S) and bind S1 domain 7 of Rrp5, thereby releasing its inhibitory effect and inducing changes in the Rrp5•RNA interaction. This allows Rcl1 to bind at the A2 site, thereby promoting A2 cleavage either directly or indirectly. Afterwards Rok1 releases Rrp5 from pre-40S subunits (Khoshnevis et al. 2016).
The data herein, as well as our and other's previous data (Horn et al. 2011; Wells et al. 2016) demonstrate that recombinant Rcl1 can cleave an RNA at the A2 cleavage site. Nonetheless, the significance of this result has been questioned because the protein lacks a nuclease signature, and it is unclear how it would carry out its catalytic activity (Tanaka et al. 2011). Furthermore, A2 cleavage activity has also been shown for Utp24 (Wells et al. 2016). Thus, this question has remained controversial, and it is certainly possible that the observed nuclease activity is an in vitro artifact due to the evolution of Rcl1 from Rtc1, a cyclic phosphatase. Nonetheless, it is important to note that the model for temporal regulation of A2 cleavage proposed here is independent of the exact nature of the A2 nuclease. In vivo and in vitro data show that productive RNA binding by Rrp5 (such that S1-domains 10–12 are engaged) near the A2 cleavage site is dependent on Noc1/2, to relieve an inhibitory interaction that arises from S1 domain 7. In addition, even if the resulting cleavage is artifactual, the data also demonstrate that access of Rcl1 to rRNA mimics depends on Noc1/2 in vitro, and genetics link Rcl1 to Noc1/2 in vivo. While we would like to suggest that this Noc1/Noc2-dependent switch in the RNA binding mode regulates A2 cleavage via Rcl1 activity, it might similarly be possible that Rcl1 simply supports the activity of another nuclease, Utp24, or a yet-to-be discovered protein, by binding in an Noc1/Noc2/Rrp5-dependent manner near the A2 cleavage site to carry out whatever its essential role in A2 cleavage is (Billy et al. 2000; Horn et al. 2011).
Regardless of the identity of the A2 nuclease, this model suggests that Rrp5 regulates A2 cleavage, an essential step in 40S ribosome maturation, in response to transcription and successful assembly of Noc1/Noc2, two early 60S AFs, onto pre-60S rRNA. This model explains the surprising previous observations (Osheim et al. 2004; Koš and Tollervey 2010) that A2 cleavage is delayed by about 40 sec until about 1000–1500 nt comprising domain I of 25S rRNA have been transcribed: Their transcription is necessary to provide the binding site for Noc1/Noc2. This observation also rationalizes why the cotranscription of 18S, 5.8S, and 25S rRNAs is conserved, as complete separation of the 40S and 60S assembly pathways would otherwise allow for evolution away from this genomic organization.
Deletion or depletion of other early 60S AFs, including Noc1, Noc2, Pwp1, Rlp7, Nop12, Cic1, and Erb1, or the neighboring ribosomal protein Rpl8 also affects Rcl1-dependent A2 cleavage (Dunbar et al. 2000; Milkereit et al. 2001; Pestov et al. 2001; Gadal et al. 2002; Granneman et al. 2011; Jakovljevic et al. 2012; Talkish et al. 2014), although these are not strictly required for this process. Large-scale yeast two-hybrid experiments indicate interactions between a subset of these proteins and Rrp5 (Tarassov et al. 2008; McCann et al. 2015). Furthermore, the DEAD-box protein Has1 is also required for 40S and 60S assembly (Emery et al. 2004; Rocak et al. 2005; Dembowski et al. 2013), and directly interacts with Rrp5 (Khoshnevis et al. 2016), suggesting it might also play a role in this process. Future experiments will be required to determine whether and how these proteins might modulate the Rrp5•RNA complex and temporally regulate A2 cleavage.
MATERIALS AND METHODS
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S1. Yeast strains were generated using standard recombination techniques and verified by western blotting and/or colony PCR. Plasmids used in this study are listed in Supplemental Table S2.
RNA design and nomenclature
All rRNA fragments we use are designed to start and end at defined secondary structure elements, taking care to preserve such structural elements, to ensure that the resulting RNAs are stably folded into biologically relevant structures. Native gels, and DMS probing were used to validate thee RNAs as previously described (Lamanna and Karbstein 2009; Young and Karbstein 2011). These RNAs are named according to the sequence or structural features at the beginning and end. Thus, H45-5.8S starts at the beginning of H45 and ends at the end of 5.8S. The numbering of these RNAs in the context of the 35S pre-rRNA is as follows. H45-5.8S: 2476-2989; H44-A2: 2340-2712; H44-ITS1: 2340-2861.
Protein expression and purification
All proteins were expressed in E. coli Rosetta2 (DE3) cells (Novagen). Cells were grown at 37°C to OD600 of 0.6 in 2 × YT media supplemented with the appropriate antibiotic and then transferred to 18°C. Protein expression was induced by addition of 0.3 mM or 1 mM IPTG for pGEX-6-P3 or pET23/pSV272 harboring cells, respectively, and cultures were harvested after 18 h.
Rcl1, Rrp5_FL and Rrp5_S10-12T, Rrp5_S5-12T, Rrp5_ S6-12T, Rrp5_S7-12T, and Rrp5_S8-12T were purified as previously described (Horn et al. 2011; Young and Karbstein 2011; Khoshnevis et al. 2016).
MBP-Noc1/Noc2 were coexpressed and purified using amylose resin (GE Healthcare) in MBP-binding buffer (200 mM NaCl, 50 mM HEPES/NaOH [pH 7.5], 5% glycerol). Protein was eluted in MBP-binding buffer supplemented with 20 mM maltose. The complex was further purified using a MonoQ ion exchange column (GE Healthcare) equilibrated with MBP-binding buffer. The protein was eluted with a salt gradient (1 M NaCl, 30 mM HEPES/NaOH, 10% glycerol and 2 mM βME) over 15 column volumes. The protein was further polished using a Superdex S-200 gel filtration column (GE Healthcare) equilibrated in 200 mM NaCl, 20 mM HEPES/NaOH, 5% glycerol and 1 mM DTT.
Utp24 WT and D138N were expressed as His-MBP-tagged fusion proteins and purified on Ni-NTA resin in 250 mM NaCl, 50 mM Tris/HCl (pH 7.5), 25 mM imidazole, 5% glycerol and 1 mM MnCl2. The protein was eluted with 250 mM imidazole, followed by overnight TEV cleavage during dialysis (150 mM NaCl, 30 mM Tris/HCl [pH 7.6], 5% glycerol and 1 mM MnCl2). The protein was further purified over MonoS and MonoQ ion exchange columns, and then stored in 250 mM NaCl, 50 mM Tris/HCl (pH 7.5), 25 mM imidazole, 5% glycerol and 1 mM MnCl2.
MBP-Noc1/Noc2 and Rrp5 or Rcl1 interaction studies
Five micromolars of FL Rrp5, Rrp5 fragments or Rcl1 were mixed with 3 µM MBP-Noc1/Noc2 (or MBP) in 150 mM NaCl and 20 mM HEPES/NaOH (pH 7.5), 5% glycerol, and preincubated on ice for 15 min before addition of 25 µL of equilibrated amylose resin (New England BioLabs). The mixture was incubated for 30 min at 4°C, flow-through was collected, resin washed and eluted with binding buffer supplemented with 20 mM maltose.
His-Rrp5 and Rcl1 interaction studies
Five micromolars of Rcl1 was mixed with 3 µM His-Rrp5 in 150 mM NaCl and 20 mM HEPES/NaOH (pH 7.5), 5% glycerol and 60 mM imidazole, and preincubated on ice for 15 min before addition of 25 µL of equilibrated Ni-NTA resin (New England BioLabs). The mixture was incubated for 30 min at 4°C, flow-through was collected, resin washed and eluted with binding buffer supplemented with 250 mM imidazole.
In vitro RNA•protein binding assay
rRNA was folded in the presence of 10 mM Mg2+ as described previously (Karbstein and Doudna 2006). Prefolded rRNA and appropriate protein, or protein complexes were incubated together for 20 min in 10 mM Tris, pH 7.5, 100 mM KCl, 10 mM MgCl2 at 30°C before being loaded on a running 6% acrylamide/THEM (Tris, HEPES, EDTA at pH 7.5, MgCl2) gel for 2 h at 4°C. Control experiments where the incubation time was varied to 2 h indicate that equilibrium was achieved. For MBP-Noc1/2 RNA binding experiments, 1 mg/mL of yeast total tRNA was used to compete for nonspecific binding. Protein-bound and unbound fractions were quantified using Quantity One (BioRad), and data were fit to a single binding isotherm using Kaleidagraph (Synergy Software).
RNA cleavage assays
RNA was folded as described previously (Karbstein and Doudna 2006). Rcl1 or Rcl1/Rrp5 fragments were preincubated at 30°C for 10 min in Rcl1 cleavage buffer (100 mM KCl, 50 mM MOPS [pH 7.6], 10 mM MgCl2), before mixing with RNA. For experiments with Noc1/Noc2, all proteins were incubated for 5 min with 0.1 µM prefolded D1 or U3 RNA before the addition of the substrate. Utp24 cleavage experiments were carried out in Utp24 cleavage buffer (100 mM KCl, 50 mM Tris/HCl [pH 7.5], 5 mM MnCl2, 2 mM MgCl2). For Nob1 cleavage experiments, 2 µM Nob1 was mixed with H44-A2 with or without 1 µM Rrp5 in Nob1 cleavage buffer (100 mM KCl, 50 mM Tris/HCl [pH 7.5], 3 mM MnCl2, 2 mM MgCl2). Aliquots were removed and quenched after 0, 5, 10, 15, 30, and 60 min.
Aliquots were removed at the indicated time points and mixed with an equal volume of 2× loading dye supplemented with 2 mg/mL heparin. Samples were boiled for 5 min at 95°C and held on ice before separation on a 6% acrylamide, 50 mm MES (pH 6.2), 8 M urea gel. Gels were dried and then exposed overnight. Cleavage gels were quantified using phosphoimager analysis and graphs fit with Equation 1 to obtain cleavage rate constants.
| (1) |
To obtain inhibition constants, Ki, rRNA cleavage rate constants at different Rrp5 concentrations were measured, plotted against Rrp5 concentration and fit with Equation 2.
| (2) |
Primer extension assays
To map cleavage sites, reverse transcription and sequencing gels were performed as previously described (Lamanna and Karbstein 2009). The sequence of the primer for mapping A0 and A1 cleavage was CGATAACTGATTTAATGAGCCATTCGCAG and the 5.8S probe was used to probe A2 cleavage.
Northern analysis of rRNA processing
Cells were grown in the presence of glucose, with or without the addition of 50 mM LiCl, to OD ∼ 0.8 and total RNA was isolated using the hot phenol method. rRNA processing intermediates were analyzed either by reverse transcription (Lamanna and Karbstein 2009) or by northern blotting using the following probes: 35S, 23S and 27SA2: probe 003 (between A2 and A3), 27S and 7S: probe D (between A3 and 5.8S), 20S: probe B (between D and A2), 18S, 5.8S and 25S: probes G, 5.8S and Y, respectively (within the mature rRNA of interest).
Limited proteolysis
Endoproteinase Glu-C (Sigma) was incubated with Rrp5_S6-12T or an equimolar mixture of MBP-Noc1/Noc2 and Rrp5_S6-12T in a ratio of 1:100 (w/w) in a buffer containing 100 mM KCl and 50 mM Tris (pH 7.5) at 25°C. In all experiments, the protein concentrations were set at 1 µM. At various timepoints the reaction was stopped by the addition of SDS loading dye and samples were analyzed by SDS-PAGE on 4%–20% precast Mini-Protean TGX gels (Bio-Rad). In order to detect changes in the proteolytic pattern of individual proteins, samples were analyzed by western blotting using anti-Rrp5 antibodies raised by Josman LLC against recombinant full-length protein.
Rrp5 release assay
A total of 0.7 µM prefolded H45-5.8S RNA, 0.5 µM Rrp5_S8-12T, and Rrp5_S1-7, Rrp5_S1-5 or MBP-Noc1/Noc2 were mixed in 100 mM KCl, 50 mM Tris/HCl, 10 mM MgCl2. Mixtures were incubated for 30 min at 25°C prior to separation on a native 6% acrylamide/bis-acrylamide/THEM gel. The position of Rrp5_S8-12T on the gel was detected by western blotting using anti-Rrp5 antibody raised against Rrp5_S10-12T.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Philip Milkereit for sharing sequence information about the Noc1-1 allele, S. Pincus and Z. A. Lee for assistance in cloning, A. Corbett for providing laboratory space for S.K., and members of the Karbstein laboratory for comments on the paper. M.D.D. was partially supported by a High School Intern Fellowship from the BallenIsles Charities Foundation, Inc. This work is supported by National Institutes of Health (NIH) grant R01-GM086451 and Howard Hughes Medical Institute (HHMI) Faculty Scholars grant 55108536 to K.K.
Author contributions: S.K., X.L., and M.D.D. performed the experiments and contributed unique reagents. S.K., X.L., and K.K. designed and analyzed the experiments. S.K. and K.K. wrote the paper.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.071225.119.
REFERENCES
- Arcus VL, Bäckbro K, Roos A, Daniel EL, Baker EN. 2004. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem 279: 16471–16478. 10.1074/jbc.M313833200 [DOI] [PubMed] [Google Scholar]
- Barandun J, Chaker-Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S. 2017. The complete structure of the small-subunit processome. Nat Struct Mol Biol 24: 944–953. 10.1038/nsmb.3472 [DOI] [PubMed] [Google Scholar]
- Billy E, Wegierski T, Nasr F, Filipowicz W. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J 19: 2115–2126. 10.1093/emboj/19.9.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. 2006. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci 103: 9464–9469. 10.1073/pnas.0603673103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. 2015. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22: 920–923. 10.1038/nsmb.3111 [DOI] [PubMed] [Google Scholar]
- Chen W, Xie Z, Yang F, Ye K. 2017. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res 45: 6837–6847. 10.1093/nar/gkx254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kellner N, Berninghausen O, Hurt E, Beckmann R. 2017. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat Struct Mol Biol 24: 954–964. 10.1038/nsmb.3476 [DOI] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M Jr, Stevens SW. 2006. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 26: 523–534. 10.1128/MCB.26.2.523-534.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, Woolford JL Jr. 2015. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem 84: 93–129. 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A, Al Kadri Y, Pérébaskine N, Monfoulet C, Henry Y, Henras AK, Fribourg S. 2014. Crucial role of the Rcl1p-Bms1p interaction for yeast pre-ribosomal RNA processing. Nucleic Acids Res 42: 10161–10172. 10.1093/nar/gku682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski JA, Kuo B, Woolford JL Jr. 2013. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res 41: 7889–7904. 10.1093/nar/gkt545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Stevens A, Tollervey D. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J 16: 7184–7195. 10.1093/emboj/16.23.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Dragon F, Lee SJ, Baserga SJ. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc Natl Acad Sci 97: 13027–13032. 10.1073/pnas.97.24.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol 52: 141–158. 10.1111/j.1365-2958.2003.03973.x [DOI] [PubMed] [Google Scholar]
- Eppens NA, Rensen S, Granneman S, Raué HA, Venema J. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5: 779–793. 10.1017/S1355838299990313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E. 2002. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J Cell Biol 157: 941–951. 10.1083/jcb.200111039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL Jr. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210. 10.1101/gad.228825.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019. 10.1038/emboj.2011.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, Hamperl S, Ohmayer U, Rachel R, Jacob A, et al. 2013. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res 41: 1191–1210. 10.1093/nar/gks1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn DM, Mason SL, Karbstein K. 2011. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J Biol Chem 286: 34082–34087. 10.1074/jbc.M111.268649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. 10.1261/rna.032540.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Doudna JA. 2006. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J Mol Biol 356: 432–443. 10.1016/j.jmb.2005.11.052 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Askenasy I, Johnson MC, Dattolo MD, Young-Erdos CL, Stroupe ME, Karbstein K. 2016. The DEAD-box protein Rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from Pre-40S ribosomes to allow for 60S maturation. PLoS Biol 14: e1002480 10.1371/journal.pbio.1002480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koš M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820. 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K. 2009. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci 106: 14259–14264. 10.1073/pnas.0905403106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. 2005. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 25: 9269–9282. 10.1128/MCB.25.21.9269-9282.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe A, French SL, Dudnakova T, de Lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D. 2013. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol Cell 52: 707–719. 10.1016/j.molcel.2013.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 2008. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res 36: 4988–4999. 10.1093/nar/gkn469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. 2006. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol 26: 513–522. 10.1128/MCB.26.2.513-522.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Wilbur JS, So M, Brennan RG. 2006. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J Biol Chem 281: 37942–37951. 10.1074/jbc.M605198200 [DOI] [PubMed] [Google Scholar]
- McCann KL, Charette JM, Vincent NG, Baserga SJ. 2015. A protein interaction map of the LSU processome. Genes Dev 29: 862–875. 10.1101/gad.256370.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509. 10.1016/S0092-8674(01)00358-0 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954. 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27: 5414–5429. 10.1128/MCB.00380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Stockelman MG, Strezoska Z, Lau LF. 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res 29: 3621–3630. 10.1093/nar/29.17.3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Emery B, Tanner NK, Linder P. 2005. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res 33: 999–1009. 10.1093/nar/gki244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. 2017. Molecular architecture of the 90S small subunit pre-ribosome. Elife 6: e22086 10.7554/eLife.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL Jr. 2014. Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 34: 1863–1877. 10.1128/MCB.01322-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Smith P, Shuman S. 2011. Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis. RNA 17: 595–602. 10.1261/rna.2571811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. 2008. An in vivo map of the yeast protein interactome. Science 320: 1465–1470. 10.1126/science.1153878 [DOI] [PubMed] [Google Scholar]
- Torchet C, Hermann-Le Denmat S. 2000. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA 6: 1498–1508. 10.1017/S1355838200000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C, Jacq C, Hermann-Le Denmat S. 1998. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA 4: 1636–1652. 10.1017/S1355838298981511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. 1996. RRP5 is required for formation of both 18S and 5.85 rRNA in yeast. EMBO J 15: 5701–5714. 10.1002/j.1460-2075.1996.tb00954.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos HR, Bax R, Faber AW, Vos JC, Raue HA. 2004. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res 32: 5827–5833. 10.1093/nar/gkh904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, Watkins NJ, Schneider C. 2016. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res 44: 5399–5409. 10.1093/nar/gkw213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CL, Karbstein K. 2011. The roles of S1 RNA-binding domains in Rrp5's interactions with pre-rRNA. RNA 17: 512–521. 10.1261/rna.2458811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C, Karbstein K. 2012. Analysis of cofactor effects on RNA helicases. Methods Enzymol 511: 213–237. 10.1016/B978-0-12-396546-2.00010-3 [DOI] [PubMed] [Google Scholar]
- Young CL, Khoshnevis S, Karbstein K. 2013. Cofactor-dependent specificity of a DEAD-box protein. Proc Natl Acad Sci 110: E2668–E2676. 10.1073/pnas.1302577110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








