Abstract
The steep rise in the incidence and prevalence of food allergy (FA) in the last decades have focused attention of environmental mechanisms which act to promote disease, chief amongst which is the microbiome. Recent studies have now established the presence of pathogenic dysbiosis in FA that could be precipitated by a variety of environmental insults, including amongst others antibiotic usage and the mode of delivery, that acts to subvert the immune regulatory response that enforce tolerance to dietary antigens. A key attribute of this dysbiosis is the loss of Clostridial bacterial species that act to promote the formation of food allergen-specific nascent regulatory T cells in the gut. Significantly, different immunoprotective commensal bacteria, including members of the Clostridiales and Bacteroidales orders act to induce the transcription factor RORγt in nascent Treg cells via an upstream MyD88-dependent mechanism to promote tolerance to dietary antigens. Activation of this axis is disrupted by the dysbiosis, and can be restored by treatment with therapeutic microbiota. These findings highlight the potential for novel microbiota-based approaches to the prevention and treatment of the FA epidemic.
Keywords: Bacteroidales, Clostridiales, FOXP3, MyD88, Regulatory T Cells, RORC, RORγt, Subdoligranulum variable
Introduction
The mammalian immune system is tailored to tolerate self-antigens while responding appropriately to foreign antigens such as those of pathogens. However, although dietary antigens are foreign to the immune system, a healthy immune response to innocuous dietary antigens entails a state of immune non-responsiveness, referred to as “oral tolerance”. In food allergy (FA), there is failure to achieve oral tolerance to one or more foods that is associated with the emergence of a pathogenic T-helper 2 (Th2) response directed against food antigens [1]. Although allergic diseases were well recognized in the pre-industrial era, there has been a dramatic increase in their incidence and prevalence in the past decades, rendering them a major public health concern [2]. In particular, the prevalence of FA in recent years has increased worldwide, now affecting up to 8% of children and 3% of adults in affluent societies [3,4]. The sudden emergence of the allergy epidemic has sharply coincided with lifestyle changes associated with modernity, suggesting a possible role for environmental factors in influencing the development of allergies [2].
In 1989 Strachan provided a hypothesis to explain the increased prevalence of allergic diseases by proposing that improved hygiene coupled with dwindling family size reduced the risk of childhood cross-infections that would normally protect against allergic diseases [5]. Later studies have modified the original hygiene hypothesis to suggest a role for an altered microbial flora in predisposing to allergies, reflective of the impact of an affluent life style in restricting exposure and colonization with commensal bacteria [6].
The gastrointestinal tract is a niche for diverse microbial communities that harbors an estimated 1011 bacteria per milliliter of the luminal content in the colon [7]. Early life microbial colonization is largely influenced by the mode of delivery (vaginal versus caesarian-section birth), diet (breast versus formula feeding), antibiotic exposure and probiotic intake, all of which can impact and restructure the infant’s microbial communities [8]. Normally, healthy, diverse communities of commensal bacteria program favorable tolerogenic immune responses that offer effective long-term protection against the development of allergic and inflammatory diseases [9]. In contrast, aberrant colonization leads to dysbiosis and enhances the susceptibility of the host to atopic and inflammatory diseases. In particular, dysbiosis has been implicated as playing a critical initiating role in the pathogenesis of FA and its persistence. Below, we will review studies that have detailed the cellular and molecular circuitries by which the microbiota promotes tolerance to foods, and how dysbiosis disrupts these mechanisms to promote FA.
Role of Dysbiosis in FA.
There is substantial evidence for the role of dysbiosis in the pathogenesis and course of FA in both human FA subjects and in mouse models of FA [10]. Initial studies based on culturing microbes on selective media suggested that in comparison to healthy infants, cow milk FA infants had higher total and anaerobic bacterial counts [11]. However, there was no association of food sensitization and culturable commensal bacteria in 3 European cohorts [12]. However, culture-based techniques are inherently biased in favor of specific subsets of bacteria that grow in selective media, thus sidestepping a large proportion of the gut flora are unculturable.
Subsequent studies employing the 16S rRNA gene sequencing have provided a clearer perspective on the impact of dysbiosis in FA.
Among a cohort of 166 infants participating in the Canadian Health Infant Longitudinal Development (CHILD) study found that reduced microbial diversity early in life (3 months of age) were associated with increased likelihood of food-sensitization later in life (12 months of age). The reduction in microbial diversity was accompanied by diminished Bacteroidaceae species and an enriched Enterobacteriaceae species [13]. Every quartile increase in microbial diversity or richness reduced the risk of sensitization to food by 55%, and conversely reduced Bacteroidaceae/Enterobacteriaceae ratio enhanced the risk of sensitization to food by two-fold [13].
Another study from a cohort of 226 milk-allergic infants examined the gut microbiome composition at 3–6 months of age and corelated the persistence and resolution of milk allergy at 8 yrs. Infants enriched with Firmicutes and Clostridia early in life tended to resolve their milk allergy later in life. However, the gut microbiome composition at older age (8yrs) were similar between patients who persisted milk allergy in comparison to those whose milk allergy resolved, thus suggesting that early immune maturation in the presence of certain commensals may harbor long-term effects later in life [14].
The role of dysbiosis in eczema and its relationship with FA has not been sufficiently addressed. In one study, an analysis of Infants with eczema by 18 months showed increased bacterial genera of Enterobacteriaceae and Parabacteroides species in the first 26 weeks, as well as decreased lactate-utilizing bacteria producing butyrate, including Eubacterium and Anaerostipes taxa, with increased lactate and decreased butyrate levels [15]. Collectively, studies on the role of dysbiosis in human FA have remained correlative, raising the question of whether the observed changes in fecal flora had any impact on disease pathogenesis.
Direct evidence for a role for dysbiosis in the pathogenesis of FA was first described in studies that employed a FA-prone strain of mice, the Il4raF709 mice, which harbor a gain-of-function mutation in the Interleukin −4 receptor-alpha chain (IL-4Rα) [16]. Acquisition of FA in the Il4raF709 mice were associated with a gut microbiota signature that was distinct form that of similarly treated but FA-resistant wild-type (WT) mice [17]. Importantly, fecal microbiota transfer (FMT) from the Il4raF709 to WT germ-free (GF) mice transmitted heightened disease susceptibility to FA. The Il4raF709 microbiota-reconstituted GF mice had increased ovalbumin (OVA)-specific IgE responses and symptoms consistent with anaphylaxis upon oral OVA sensitization and challenge. Interestingly, enforcing oral tolerance in Il4raF709 mice by means of therapy with OVA-specific regulatory T cells (Tregs) suppressed allergen sensitization and restructured the pathogenic gut microbiota. In contrast to the pathogenic role for the dysbiotic bacteria in FA, the gut microbiota of healthy infants is largely protective. Transfer of healthy infant microbiome characterized by a dominance of Bifidobacterium and Bacteroides into wild type GF mice suppressed β-lactoglobulin specific IgG1, mast cell degranulation and anaphylaxis upon β-lactoglobulin challenge [18].
Precision microbiota therapies in FA.
Building on these aforementioned studies, two new reports further our understanding on the impact of gut flora in influencing the development of FA. Feehley et.al performed FMT from four healthy infants and four cow’s milk allergic infants in to GF mice [19]. Similar to the previous findings, they found that FMT from healthy infants suppressed the development of allergic sensitization to β-lactoglobulin. Of note they also demonstrated that the GF mice that received the FMT from milk-allergic subjects failed to suppress the development of FA. In line with these studies we have also observed that FA subjects manifest a dysbiotic flora that fail to suppress the development of FA in GF Il4raF709 mice, whereas those of heathy infants are protective even in the context of a genetically FA prone host. Moreover, these two reports separately identify FA protective bacteria that are impacted by dysbiosis. By analyzing the fecal microbiota of GF mice colonized with healthy versus cow milk allergic human infant fecal flora, Feehley and colleagues identified Anaerostipes caccae to be the most closely matching bacteria to operational taxonomix units (OTUs) that were abundant in colonized GF mice, and were also associated with a differentially expressed epithelial transcriptional signature. Monocolonization of GF mice with A. caccae protected against the induction of FA by suppressing antigen specific allergic responses and symptoms of anaphylaxis [19].
In a separate approach, by analyzing differentially expressed OTUs in the fecal microbiota of healthy control (HC) infants versus those with FA at different age groups ranging from 1 to 30 months of age, we identified dysbiotic changes involving up to 77 OTUs in the microbiota of FA infants. Interestingly, most of these changes were maintained when milk allergic subjects were excluded, indicating that the dysbiosis is a common feature across subjects with different FA. The dysbiosis in FA was determined to be pathogenic, evidenced by the failure fecal flora of FA infants to protect GF Il4raF709 mice from developing FA, whereas those of HC infants did. Similarly, the fecal flora of FA-resistant wild-type mice but not that of the FA-prone Il4raF709 mice protected against FA when transplanted into GF Il4raF709 mice. Thus, dysbiosis is an essential feature of FA across mammalian species and plays a critical role in disease pathogenesis.
Of the OTUs affected by the dysbiosis, several were commensals of the of the order Clostridiales that were decreased in the FA infants at one or more age group. In particular, we identified one Clostridiales species, Subdoligranulum variabile, to be underrepresented in the FA infants age 1 year and older irrespective of the food allergens they are reactive to, suggesting that deficiency may act as a switch to promote FA [20]. Importantly, treatment of conventional (bacteria sufficient) Il4raF709 with S. variabile protected against their development of FA by suppressing antigen specific allergic responses and attributes of anaphylaxis [20].
Together, these reports extend our understanding of the impact microbial flora in regulating immune response to food antigens, they also raise an important translational question of whether the gut microbiota can be harnessed in treating and preventing FA.
Host gene-microbiota interactions mediating immune tolerance in FA.
Commensals play an essential role in mediating immune tolerance at the mucosal interface by promoting a number of immunoregulatory mechanisms, most notably the generation of regulatory T (Treg) cells [21–23]. In the gut, nascent induced Treg specific for dietary antigens are derived upon the interaction of naïve CD4+ T cells with antigen-presenting classical dendritic cells (cDC) that express the surface marker CD103, MHCIIhi, and CCR7hi, consistent with a migratory cDC phenotype [24]. The mechanisms by which commensals promote tolerance in FA have been explored in a series of studies. Commensals of the order Clostridiales and Bacteroidales drive the differentiation of induced Treg (iTreg) cells expressing the transcription factor RORγt, which have been implicated in maintaining mucosal tolerance in the gut [25,26]. In particular, induction of RORγt+ Treg cells in the draining lymph nodes of the small intestine appears critical to establish tolerance to luminal antigens in the gut [27]. However, different outcomes have be described as a consequence of Treg cell-specific deletion of floxed Rorc allele, encoding Rorγt, using a Foxp3-driven Cre recombinase [25,26,28,29]. Sefik et al have reported dysregulated Th17 cell responses in the Foxp3CreRorcΔ/Δ mice in models of inflammatory bowel disease [25]. In contrast, Ohnmacht et al found that deletion of Rorc in Treg cells resulted in dysregulated Th2 responses [26]. Several lines of evidence indicated that deficiency of RORγt+ Treg cells directly contributes to the pathogenesis of FA. First, Both FA human subjects and Il4raF709 mice had decreased RORγt+ Treg cells in circulation, and the latter also in the mesenteric lymph nodes and the small intestinal lamina propria. Instead, there is the emergence of Th2 cell-like reprogrammed Treg cells that fail to suppress allergen-specific T cell responses [30,31], and which contribute to disease pathogenesis by secreting IL-4 to promote allergen-specific IgE responses and mast cell expansion [30]. Furthermore, the dysbiotic flora of FA infants ineffectively induced RORγt+ Treg cells when transplanted in GF Il4raF709 mice as compared to those of HC infants. Significantly, we found that therapy with small consortia of Clostridiales and Bacteroidales species, or monobacterial therapy with S. variabile, protected against the development of FA in the Il4raF709 mice and suppressed established disease by inducing RORγt Treg cells [20]. Deletion of Rorc in Treg cells in otherwise FA-resistant wild-type mice precipitated their susceptibility to FA, while its deletion in Il4raF709 mice abrogated the efficacy of the consortia in protecting these mice against FA. Collectively, these results established a causal relationship between the induction by the commensal microbiota of protective RORγt+ Treg cells and protection against FA, and also established the failure of this mechanism as a critical link in the development of FA in human infants.
The mechanisms operative in the generation by the commensal microbiota, including the Clostridiales and Bacteroidales consortia, of protective RORγt+ Treg cell proceeded via a Treg cell-specific common upstream pathway involving the Myeloid differentiation primary response 88 (MyD88), an essential signal transducer of several innate immune cytokines (IL-1, IL-18, IL33) and the Toll-like receptor signaling pathways. Deletion of MyD88 in Treg cells abrogated the protective effect of the consortia, thus establishing a MyD88-RORγt signaling axis operative in nascent Treg cells in the gut that mediates tolerance induction by the commensals in FA [20] (Figure 1). The activation of this axis implies antigen-independent mechanisms (such as TLR activation) that would promote the differentiation of RORγt+ Treg cells specific for commensal bacterial, including those that are potentially pathogenic (pathobionts), as well as dietary antigens. Further studies would be required to delineate the contribution to the RORγt+ Treg cell T cell receptor repertoire of dietary and bacterial antigen specificities.
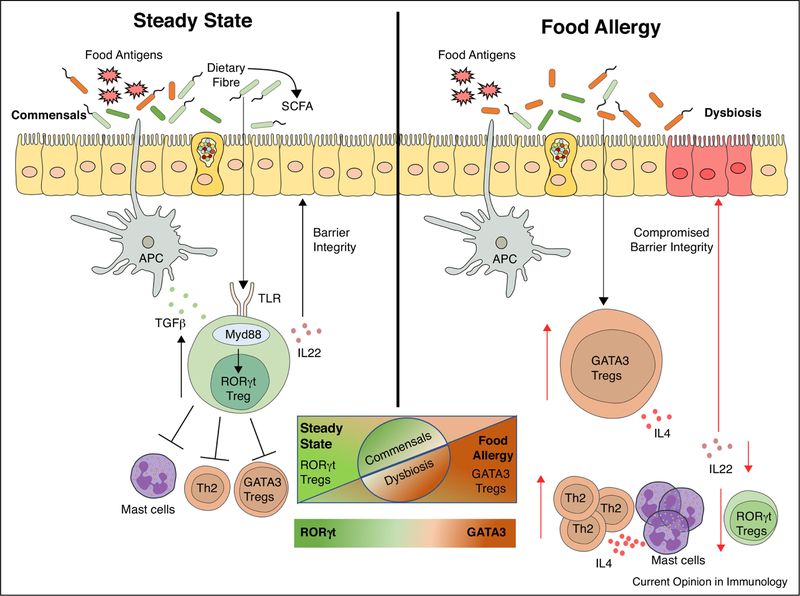
Figure 1.
(Left) Under homeostatic conditions, antigen presenting cells (classical CD103+ dendritic cells) promote the formation of nascent dietary antigen-specific iTreg cells. Further signals delivered by the commensal microbiota via MyD88 in nascent iTreg cells drives the expression of ROR-γt, which may regulate tolerance to dietary antigens by a range of mechanisms, including inhibition of antigen-specific helper T 2 (Th2) cell responses, suppression of pathogenic Th2 cell-like reprogramming of Treg cells and of mast cell activation, and the production of barrier-protective cytokines such as IL-22. (Right) Under conditions of FA, dysbiosis compromises the differentiation of naïve T cells into ROR-γt+ iTreg cells. Instead, there is expansion of iTreg cells with a Th2 cell-like phenotype characterized by increased GATA3 expression and IL-4 secretion. These pathogenic Treg cells are unable to suppress mast cell activation or Th2 cell expansion, leading to a dysregulated FA response with dietary allergen-specific IgE responses and a compromised barrier integrity.
It has been previously established that MyD88 in Treg cells regulates the IgA response to gut content, including commensals and dietary antigens [32,33], which in turns plays an essential role in engendering host-microbiome symbiosis [34]. Consistent with the disruption of the commensal microbiota-Treg cell MyD88-RORγt+ axis in FA, we found that FA infants and mice had decreased secretory IgA binding to gut bacteria and, remarkably, increased IgE binding. These findings suggest that FA allergy entails a broader breakdown in oral tolerance than hitherto appreciated, involving impaired tolerance to both dietary and bacterial antigens, underlied by the disruption of a healthy commensal-host symbiotic interaction with a disease promoting pathogenic dysbiosis.
The nature of the signals delivered by the microbiota to promote tolerance to dietary antigens remains unclear. Short chain fatty acids (SCFA) products of the commensal bacteria, including acetate, propionate and butyrate, have previously been proposed to suppress FA by eliciting protective mucosal Treg cell responses and enhancing intestinal barrier integrity [35–38]. SCFAs bind to G-protein coupled receptors (GPCRs): GPR43 or Free fatty acid receptor 2 (FFAR2), GPR41 (FFAR3) and GPR109A with varying affinities. In one model of FA, treatment with SCFA ameliorated the allergic response, while global deletion of Gpr43 rendered mice susceptible to FA induction [38]. However, in FA Il4raF709 mouse model, the mice were not deficient in SCFA, and treatment with SCFA did not induce RORγt+ Treg cells nor did it protect the mice from developing FA. Furthermore, there was no correlation between the production by bacterial consortia of SCFA and their effectiveness in treating FA in mice [20]. These results suggest that the role of SCFA in the recruitment of Treg cells to the mucosal interface and in the consolidation of their phenotype may be distinct from processes governing the differentiation of Treg cells. Overall, the contribution of SCFA to the pathogenesis of FA requires further investigation.
In addition to targeting genetic circuits in Treg cells, the microbiota may act on other cell types, including epithelial cells, innate lymphoid cells (ILC), dendritic cells and macrophages, to promote tolerance and to reinforce barrier integrity. For example, colonization of GF mice with a Clostridial consortia protects mice from allergic sensitization by inducing IL-22 from type 3 ILC. IL-22 promotes epithelial barrier integrity and thus curtailing the leakage of food antigens into the blood stream [39]. A more recent study demonstrated that the fecal microbiota of healthy infants differentially upregulated a number of genes in epithelial cells, including Fbp1, encoding the gluconeogenic enzyme fructose-bisphosphatase 1, relevant to the maintenance of a healthy flora [19]. In contrast, colonization with flora of cow milk allergic infants down-regulated Tgfbr3 and Ror2, relevant to epithelial cell repair. An integrative analysis of microbe-driven host genetic changes in different cell lineages will allow better understanding of how dysbiosis disrupt oral tolerance and promotes FA.
Finally, it remains unclear why certain bacteria from different orders such as Clostridiales and Bacteroidales have the same effect of protecting against FA. One possibility is that they act synergistically in vivo to activate the MyD88-RORγt axis in nascent Treg cells, possibly by producing distinct metabolites unrelated to SCFA. Another possibility is that the strategy of evolving redundant symbionts may act to ward-off the evolution of some of those bacteria into potentially pathogenic ones (pathobionts) under inflammatory conditions. Whether such pathobionts act to promote FA remains to be established. The future identification of the proximal mechanisms/metabolites by which different bacteria activate the MyD88-RORγt axis may provide further insights into this interesting issue.
Conclusions and feature directions
The recent advances in FA studies have placed the microbiome at the center stage of disease pathogenesis and thus establish a framework for commensal-host genetic circuit interactions relevant to tolerance induction and its breakdown in FA. Particularly relevant to understanding disease pathogenesis is a better definition of the very early dysbiotic changes conducive to disease onset in infants. It is also unknown at this stage whether the dysbiosis observed in classical IgE-mediated FA in infants is mirrored in adult onset IgE-dependent FA, as well as non-IgE-dependent forms of FA, including eosinophilic esophagitis and food-related allergic colitis. Also, whether allergies to individual foods (or food allergen groups) display distinct albeit overlapping dysbiotic features remains to be determined. More globally, the observation of an aberrant immune response to the microbiota in FA, characterized by decreased IgA and increased IgE binding, raises critical questions regarding its role in promoting disease pathogenesis.
The recent studies by Feehley et.al and Abdel-Gadir et al also highlight the potential of using precision microbiota therapies in FA [19,20]. So far, the use of therapeutic bacterial probiotics as adjuvants to oral immunotherapy has been very limited, and when so employed marred by the lack of clear evidence of synergy [40]. These constraints are related in no small measure to the lack of standardized criteria to choose optimal therapeutic microbiota in FA. The identification of a requisite role for RORγt induction in the protection against FA by microbial therapies now provides one such critical measure. Importantly, the nature of the signals emanating from the commensal bacteria to promote MyD88-dependent differentiation of nascent gut Treg cells into the RORγt+ variety remains unclear. Harnessing microbial small molecule products and/or metabolites capable of replicating protection provided by the intact organism would provide a new class of therapeutics for the treatment of FA and related disorders.
Highlights.
Food allergy (FA) is characterized by pathogenic dysbiosis
The dysbiosis in FA disrupts oral tolerance to dietary antigens by impairing the generation of ROR-γt+ induced regulatory T cells
3)Therapy with Clostridiales and Bacteroidales commensal bacteria activates an MyD88-ROR-γt axis in nascent iTreg cells to restore immune tolerance in FA
Acknowledgements
This work was supported by NIH NIAID grants 1R56AI117983 and 1R01AI126915 to T.A.C., the Bunning Food Allergy Fund the Jasmine and Paul Mashikian Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
T.A.C. is an inventor on published US patent application, 15/801,811, that covers methods and compositions for the prevention and treatment of FA using microbial treatments. T.A.C. and E.S.-V. have pending patent applications related to the use of probiotics in enforcing oral tolerance in FA (62/758,161, and, 62/823,866). T.A.C. is founders of and has equity in Consortia Tx.
Declaration of interests
T.AC. is a co-founder and has equity in Consortia Tx.
References
* of Interest
** of high interest
- *1.Tordesillas L, Berin MC, Sampson HA: Immunology of Food Allergy. Immunity 2017, 47:32–50. [DOI] [PubMed] [Google Scholar]
- 2.Platts-Mills TA: The allergy epidemics: 1870–2010. J Allergy Clin Immunol 2015, 136:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RS, Warren CM, Smith BM, Blumenstock JA, Jiang J, Davis MM, Nadeau KC: The Public Health Impact of Parent-Reported Childhood Food Allergies in the United States. Pediatrics 2018, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RS, Warren CM, Smith BM, Jiang J, Blumenstock JA, Davis MM, Schleimer RP, Nadeau KC: Prevalence and Severity of Food Allergies Among US Adults. JAMA Netw Open 2019, 2:e185630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strachan DP: Hay fever, hygiene, and household size. BMJ 1989, 299:1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills-Karp M, Santeliz J, Karp CL: The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol 2001, 1:69–75. [DOI] [PubMed] [Google Scholar]
- 7.Sender R, Fuchs S, Milo R: Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016, 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamburini S, Shen N, Wu HC, Clemente JC: The microbiome in early life: implications for health outcomes. Nat Med 2016, 22:713–722. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Littman DR: The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535:75–84. [DOI] [PubMed] [Google Scholar]
- 10.Rachid R, Chatila TA: The role of the gut microbiota in food allergy. Curr Opin Pediatr 2016, 28:748–753. [DOI] [PubMed] [Google Scholar]
- 11.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A: Changes in faecal microbiota of infants with cow’s milk protein allergy--a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol 2010, 21:e394–400. [DOI] [PubMed] [Google Scholar]
- 12.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, Perkin MR, Tripodi S, Hesselmar B, Saalman R, et al. : Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 2007, 120:343–350. [DOI] [PubMed] [Google Scholar]
- 13.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, Mandhane PJ, Turvey SE, Subbarao P, Becker AB, et al. : Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015, 45:632–643. [DOI] [PubMed] [Google Scholar]
- 14.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, Jones SM, Leung DYM, Sampson H, Sicherer S, et al. : Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016, 138:1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS: Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J Allergy Clin Immunol 2018, 141:1334–1342 e1335. [DOI] [PubMed] [Google Scholar]
- 16.Tachdjian R, Al Khatib S, Schwinglshackl A, Kim HS, Chen A, Blasioli J, Mathias C, Kim HY, Umetsu DT, Oettgen HC, et al. : In vivo regulation of the allergic response by the IL-4 receptor alpha chain immunoreceptor tyrosine-based inhibitory motif. J Allergy Clin Immunol 2010, 125:1128–1136 e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, Desantis T, Warrington J, et al. : A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 2013, 131:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, Labellie C, Nicolis I, Berger B, Mercenier A, et al. : Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol Ecol 2012, 79:192–202. [DOI] [PubMed] [Google Scholar]
- **19.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, Campbell E, Aitoro R, Nocerino R, Paparo L, et al. : Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 2019, 25:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that the microbiota of healthy but not milk allergic subjects protects germ free (GF) mice against FA, an effect that can be reproduced by clonization of GF mice with Anaerostipes caccae.
- **20.Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, Wang L, Li N, Crestani E, Spielman S, et al. : Microbiota Therapy Acts Via a Regulatory T Cell MyD88/RORgt Pathway to Suppress Food Allergy. Nat. Med 2019:(in press). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that FA infants demonstrate pathogenic dysbiosis that interrupts the activation of an MyD88-RORgt axis in nascent gut Treg cells that is necessary for oral tolerance in FA.
- 21.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. : Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK: The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. : Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500:232–236. [DOI] [PubMed] [Google Scholar]
- 24.Esterhazy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D: Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol 2016, 17:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. : MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349:993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. : MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349:989–993. [DOI] [PubMed] [Google Scholar]
- **27.Esterhazy D, Canesso MCC, Mesin L, Muller PA, de Castro TBR, Lockhart A, ElJalby M, Faria AMC, Mucida D: Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that the terminal ileum is particularly important for the generation of RORγt+ Treg cells, relevant to tolerance in FA.
- *28.Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, Littman DR: c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018, 554:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R, Li Z, Mortha A, Merad M, Das A, et al. : Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50:212–224 e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, Rachid R, Chatila TA: Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity 2015, 42:512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Abdel-Gadir A, Schneider L, Casini A, Charbonnier LM, Little SV, Harrington T, Umetsu DT, Rachid R, Chatila TA: Oral immunotherapy with omalizumab reverses the Th2 cell-like programme of regulatory T cells and restores their function. Clin Exp Allergy 2018, 48:825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that oral immunotherapy restores the function of patient allergen-specific regulatory T cells, in part by suppressing their disease-associted pathogenic Th2 cell like phenotype.
- 32.Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G, Bry L, Chatila TA: MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 2015, 43:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, Round JL: MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 2015, 17:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. : Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. : Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504:446–450. [DOI] [PubMed] [Google Scholar]
- 36.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS: The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. : Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504:451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE, Macia L, Mackay CR: Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep 2016, 15:2809–2824. [DOI] [PubMed] [Google Scholar]
- 39.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, et al. : Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014, 111:13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, Licciardi P, Burks W, Donath S: Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J Allergy Clin Immunol 2015, 135:737–744 e738. [DOI] [PubMed] [Google Scholar]