Abstract
Exposure to air pollution is associated with enhanced risk of developing asthma, notably in the presence of genetic risk factors. Interaction analyses have shown that both outdoor and indoor air pollution interact with genetic variability to increase the incidence of asthma. In this review, we summarize recent progress in candidate gene-based studies, as well as genome-wide gene-air pollution interaction studies. Advances in epigenetics have provided evidence for DNA methylation as a mediator in gene-air pollution interactions. Emerging strategies for study design and statistical analyses may improve power in future studies. Improved air pollution exposure assessment methods and asthma endo-typing can also be expected to increase the ability to detect biologically driven gene-air pollution interaction effects.
Background
Asthma is a highly heterogeneous disease for which a large number of both genetic and environmental risk factors have been identified[1,2]. Twin studies have allowed estimates of asthma heritability ranging from 0.35 to 0.8[3]. It has, however, become clear that the asthma-associated genetic variants identified so far collectively account for a very small amount of the estimated asthma heritability[4]. One of several possible explanations for this is that some genetic variants only confer risk in individuals exposed to certain environmental factors, and these interactions between genes and environmental risk factors need to be taken into account to fully understand the impact of genetics on asthma risk (Figure 1). Exposure to ambient air pollution (AAP) emitted by vehicles and industrial sources has been associated with asthma risk in both children and adults[5–8]. AAP is a complex mix of particulate matter (PM) and gaseous components, of which PM < 2.5 μm in diameter (PM2.5), PM < 10 μm in diameter (PM10), nitrogen dioxide (NO2), sulphur dioxide (SO2), and ozone (O3) are among the most studied. Exposure to AAP is known to initiate complex signaling pathways involving oxidative stress and inflammation, resulting in an increased risk of respiratory disease[9]. In addition to outdoor sources of air pollution, exposure to air pollution may also occur indoors, and the most commonly studied source of indoor air pollution is environmental tobacco smoke (ETS). Similar to AAP, exposure to ETS can provoke oxidative stress and inflammation, and is well known to be associated with asthma development in both children and adults[10].
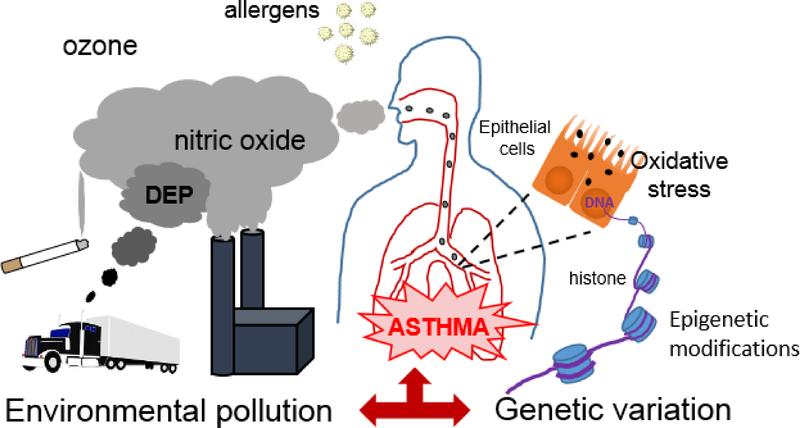
Figure 1.
Summary of the processes leading to gene-air pollution interaction effects in asthma. Inhaled particulate and gaseous components and allergens activate inflammatory and oxidative stress-related pathways in the airways. Genetic variation may result in allele-specific air pollution-related epigenetic modifications of genes involved in the development of asthma.
The study of gene by environment (GxE) interactions has the potential to greatly increase our understanding of the etiology underlying the development of different asthma phenotypes, and to identify vulnerable groups who are at increased risk of asthma. The complex statistical tests, however, involved require large sample sizes and very high-quality data on environmental exposures, confounders, and outcomes. So far, the discovery of novel GxE interactions is lagging behind expectations[11]. In this review, we summarize the most recent studies of gene-air pollution interactions. Among the vast number of studies in this field, we limit ourselves to studies of interactions between genes and AAP, TRAP, and ETS exposure, although other types of air pollution can also contribute to respiratory health risks. This review is not meant to be comprehensive, but to provide an update on recent studies of interest. In addition, the role of epigenetic changes in mediating GxE interactions will be explored, and finally novel strategies and future approaches in this field will be discussed.
Gene-air pollution interaction in asthma risk
As is the case for genetic association studies, investigations of GxE interactions involve either hypothesis-driven studies of candidate genes or agnostic genome-wide scans. The statistical analyses are more challenging, however, because of the inherently low power of tests that include interaction terms. Crucial to the success of all GxE interaction studies is accurate and valid exposure assessment. A further complication is that the underlying biological mechanism of the response to individual pollutants may differ between low- and high-range exposures. The most common method for exposure assessment in recent gene-AAP interaction studies is land use regression (LUR), which usually combines sampling of pollutants at stationary locations with regression modeling that takes into account environmental characteristics and other predictor variables to obtain estimates for unmonitored locations, e.g. residential addresses. Other methods include air dispersion models, remote sensing by satellites, and traffic-related air pollution (TRAP) proxy measurement such as distance to major highways. The advantages and limitations of different air pollution assessment methods have been reviewed elsewhere[12,13]. Air pollution is a global problem that is especially severe in developing countries in Asia, Africa and South America. Local specificities regarding the nature and severity of air pollutants as well as the ancestry of the population will likely results in the identification of different genes and pathways from cohorts of European origin. Studies from these developing countries are therefore of high importance.
Studies of GxE interactions in highly admixed populations presents additional difficulties. Different ancestral populations often has differing allele frequencies, which may obscure the genetic component. Furthermore, the genetic component may be confounded by environmental covariates, and this can inflate Type I errors. Admixture, the mixing of genetically diverse populations, can, however also be leveraged to detect interaction effects. Localizing disease genes using an admixed population is called admixture mapping. In the study of GxE interaction, admixture mapping may provide clues to the relative contribution and the effect size of genetic factors contributing to the differential risk. It can also track environmental influences such as socioeconomic status, access to health care, and socio-cultural factors that influence complex diseases including asthma[14]. For example, if an excess of global African (or European) ancestry is noted across the entire genome in the affected group relative to the control group but there is no significant rise in local ancestry at a particular locus, this may point to a stronger role for non-genetic or socio-environmental factors (e.g., access to health care, diet, or lifestyle). These insights are critical to developing public health policies and interventions to reduce the asthma burden due to environmental factors, and to improve clinical outcomes for diseases with a biological basis through ancestry-specific, personalized, drug therapies, and genetic screening.
Candidate gene approaches
Focused studies of individual candidate genes selected based on biological plausibility has so far been the main approach for the analysis of GxE interactions, and most studies of interactions between genes and air pollution have focused on genes involved in oxidative stress and inflammatory responses.
Members of the family of glutathione S-transferases (GST) are among the best studied candidate genes in gene by air pollution interactions. Two recent systematic reviews have summarized the evidence for interactions between GSTs, including GSTM1, GSTP1, and GSTT1, and TRAP exposure and ETS, respectively[15,16]. A majority of studies of GSTP1 found that this gene interacted with one or several TRAP components to modify the risk of allergic airway disease, but not all of the findings were consistent with regard to direction of the effects[15]. The heterogeneity of the studies with regard to age of exposure, TRAP exposure assessment method, geographic area, and outcome definition precluded meta-analysis, and may explain inconsistencies in the results. With regard to indoor air pollution, mainly ETS[16], found that 15 of 22 studies supported an interaction effect between indoor air pollution and GSTs. Similarly to TRAP exposure, findings were not consistent with regard to risk alleles and specific exposures, and, again, meta-analysis of the studies was not possible because of variability in exposure age and definitions, as well as outcome measures.
The chromosomal region 17p11 has been implicated in an early genome-wide linkage screen of asthma and bronchial hyper-responsiveness (BHR) in the presence of a gene x early life ETS exposure interaction[17]. More recently[18], conducted a fine-scale mapping of this region to identify specific SNPs interacting with ETS to modify the risk of BHR. A single SNP mapping to the ciliary gene Dynein Axonemal Heavy Chain 9 (DNAH9) interacted significantly with ETS.
Semi-agnostic screens can be a useful way to explore many candidate genes simultaneously with far smaller multiple testing penalties than in genome-wide scans. This approach was taken in a study involving children recruited from either highly polluted urban areas or less polluted rural regions in the Czech Republic in order to assess the interactions between ambient Benzo[a] pyrene (B[a]P) and 621 SNPs in genes involved in oxidative stress response, DNA repair, xenobiotics metabolism, and inflammation[19]. The large 8-fold difference in median B[a]P between rural and urban areas during the period of investigation may have increased the power of the analysis, which found that the immune response genes CTLA4 and STAT4 and the cytochrome P450 gene CYP2E1 interacted with high levels of B[a]P to increase the risk of pediatric asthma.
Genome-wide environment interaction (GWEI) studies
Although hypothesis-driven studies of candidate genes are useful, they are limited to genes already identified by the existing knowledge about the etiology of asthma. A more comprehensive and unbiased approach is to scan the entire genome for G×E effects.. These genome-wide GxE scans are very challenging because of the large samples sizes required and the multiple testing corrections involved. Most GWEI studies adopt a step-wise analytical approach, with an initial screen for marginal genetic effects or a test that models G-E associations in controls and cases combined[20]. SNPs selected in the first step are included in a formal interaction test.
Gref[21] performed a meta-analysis of genome-wide gene-NO2 interaction results from three European birth cohorts, followed by validation by look-up analysis of the SNPs with the lowest interaction P values in two North American cohorts. LUR was used for NO2 exposure assessment. The discovery primary model identified 186 SNPs with a P < 1 × 10−4, which were selected for validation a validation cohort. Eight SNPs located in or near B4GALT5, MOCOS, ADCY2, and DLG2 had nominally significant interaction effects (P < 0.05), although the direction of the effect was only consistent for the G-protein-coupled receptor associated enzyme encoded by ADCY2. This study illustrates the advantages of larger sample sizes with meta-analysis over several cohorts, but also the drawbacks that can lower the power of the analysis, such as study heterogeneity involving, for example, differences in timing and methodology of exposure assessment and different outcome definitions.
In a Dutch GWEI study of ETS in adults with FEV1 as the outcome, 45 SNP-by-ETS interaction were identified after multiple testing correction[22], and two of these were replicated in at least one of two replication cohorts. Pathway analysis identified one significantly enriched pathway involving apoptosis. In addition, the p38 MAPK pathway and the tumor necrosis factor pathway were suggestively enriched.
The role of epigenetics in air pollution-gene interactions
Due to recent advances in epigenetics, we are beginning to gain an understanding of possible mechanisms underlying air pollution-gene interactions. The vast majority of asthma-associated SNPs are located in non-coding regions of genes[23], and many of these SNPs overlap with CpG dinucleotides that are potential targets of methylation. Epigenetic methylation of DNA occurs at CpG sites in promoter regions as well as enhancer regions in introns and elsewhere. Both global and specific methylation changes are associated with transcriptional dysregulation leading to asthma (Figure 2)[24,25].
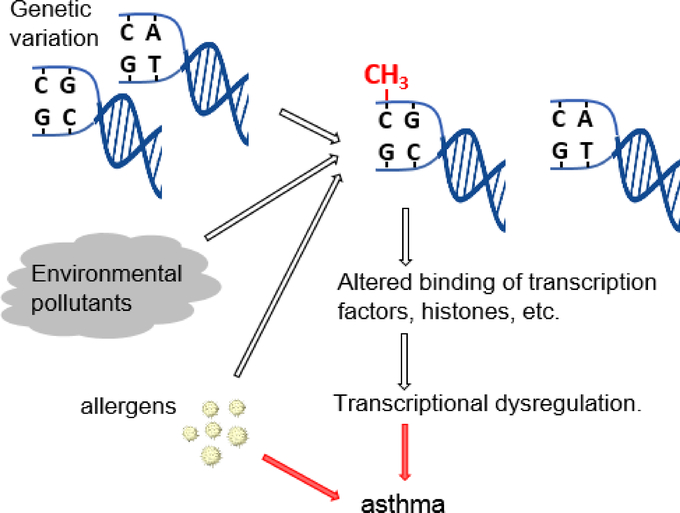
Figure 2.
Schematic representation of a suggested biological mechanism for gene-air pollution interactions involving allele-specific DNA methylation of genetic loci as a result of exposure to air pollution.
Several recent studies have strengthened the evidence for associations between DNA methylation and air pollution exposure in utero, in childhood, and in adults[26]. In a meta-analysis of cord blood methylation data from epigenome-wide scans in four European and North American studies, significant associations were found between maternal NO2 exposure and methylation of CpGs in catalase and thyroid peroxidase in newborns[27]. Interestingly, in a controlled human cross-over study, sequential exposure to aeroallergens and diesel exhaust particles (DEP) was associated with greater changes in methylation patterns in the bronchial epithelium than either exposure alone[28].
Furthermore, methylation changes are also known to be associated with asthma[29–31]. In a recent study, asthma was shown to be associated with methylation of Forkhead Box P3 (FOXP3) and IL10, and FOXP3 methylation was associated with NO2, CO, and PM2.5 exposures, respectively[32]. Methylation at a single site in the Tet Methylcytosine Dioxygenase 1 (TET1) promoter was associated with both TRAP exposure and asthma in African-American children[33].
The GWEI study by Gref[21] mentioned above illustrates the usefulness of integrating epigenetics and gene expression analysis with GxE studies. SNPs identified in the GxE analyses were evaluated for expression quantitative trait locus effects and for gene expression and methylation changes in peripheral blood, and it was found that SNPs in ADCY2 was associated with ADCY2 gene expression in peripheral blood. Furthermore, methylation changes were found for SNPs in ADCY2, DLG2, and MOCOS.
There are so far few studies of the effect of air pollution on DNA methylation in the context of allergic responses using cell based or animal models. Exposure of human bronchial epithelial cells to DEP or house dust mites both results in elevated expression TET1 and DNMT1, which are involved in DNA methylation[34]. Prenatal exposure to ETS in mice was shown to result in enhanced inflammatory responses to later house dust mite exposure, as well as methylation changes in IL4, IL13, IFNγ, and FOXP3[35]. These results in part mirror an earlier study, in which increased inflammatory responses following co-exposure to Aspergillus fumigatus and DEP in mice was associated with a greater level of methylation at CpG sites in the IFNγ promoter, whereas DEP exposure alone had no effect[36].
Further use of animal models and in vitro systems with relevant cell types will likely be useful tools for exploring the role of asthma loci as epigenetic regulators and have the potential to contribute a great deal to the understanding of the mechanisms behind GxE interactions.
Future directions
New analytical strategies
Low power due to multiple-testing penalties remains a substantial challenge in GWEI studues. To address this problem, strategies that have been developed recently to increase samples sizes and to further improve analytical techniques. One way to increase power in GWEI studies is to reduce multiple testing penalties by consolidating genetic variation loci into biologically defined sets, such as sets of variants mapping to a particular gene, a genomic region, or a biological pathway, and test the interaction between the set as a whole and the exposure of interest[37–39]. A possible strategy to leverage existing biological information is to assign weight to genetic loci based on transcriptomics and epigenomics annotation, locations of eQTLs and regulatory regions, and/or biological pathway analysis. Biologically assigned weights of loci can be used as input in genome-wide GxE screens, which may result in greater power and more functionally relevant results. This strategy may be combined with a two-step approach including an initial screen of loci followed by GxE analysis to further increase power (Figure 3).
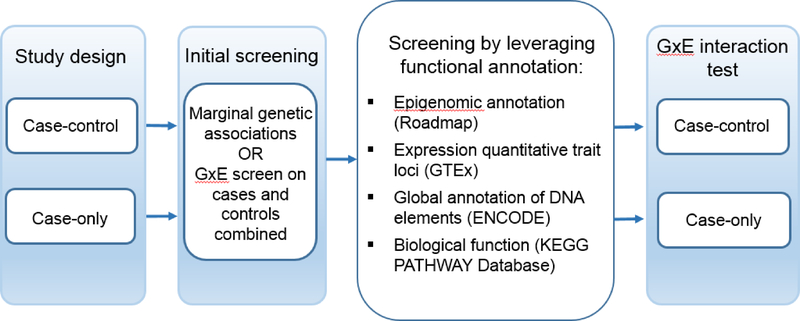
Figure 3.
Proposed strategy for incorporating existing biological information into the analysis of gene-air pollution interactions. The approach can be combined an initial screening step, and the study design can be case-control or case only.
Another approach to increase power in GxE studies by increasing the total sample size is the use of cohort consortia. As mentioned under “Gene-air pollution interaction in asthma risk”, however, AAP is a complex mix of particulate and gaseous components, and the composition can vary from region to region depending on the sources of emission. This may complicate the interpretation of results from consortium-based studies. In addition, confounding factors, such as diet, socio-economic status, and ancestry may differ from region to region and from cohort to cohort, and every effort needs to be made to control for these factors.
Improved air pollution exposure assessment and phenotyping methods
Critical to the success of GxE studies, as well as replication and meta-analyses of previous findings, is the refinement and harmonization of air pollution exposure assessment. Although recently developed low-cost monitors do not yet provide the accuracy and sensitivity of scientific-grade instruments, their low cost means that greater density of measurements can be obtained for longer periods of time[40,41]. Furthermore, various types of mobile vehicle-based monitoring stations have been developed for measurements of fine-scale spatial variations that can be used for LUR estimates[42–44].
New approaches for dynamic exposure assessment have also been developed to take into account the spatial mobility of study subjects. This may involve incorporating GPS to obtain time-weighted exposures at multiple locations, and cell phones provide a convenient means of retrieving location data. Portable personal monitors have the advantage of providing direct exposure values at given time points. However, developing accurate and truly wearable personal monitors of air pollutants represents a technological challenge[45], and so far, there are few studies incorporating their use.
As mentioned above, the age of assessed exposure varies greatly for both AAP and ETS studies, and better consistency between studies would help facilitate replication and meta-analyses of results. Animal models could be of help in formulating hypotheses regarding critical windows of exposure.
Finally, as mentioned above, asthma is a highly heterogeneous disease, and progress is being made in identifying clinically, biologically, and genetically defined phenotypes and endotypes that may have different etiologies and risk factors[46,47]. More precise outcome definitions in the study of asthma GxE interactions may increase the power in the analyses and produce more meaningful results.
Conclusions
Although recent gene-air pollution interactions studies have incorporated some promising strategies, low power is still a major challenge and continues to impede the identification and replication of interacting genetic loci. Improvements in air pollution exposure assessment and more precise asthma phenotype definitions may help increase power in analyses and should be a priority. New analytical strategies such as set-based analysis should be applicable in this area and may help in exploring interactions between air pollution and candidate genes or pathways. Integrating transcriptomics and epigenetics with genome-wide GxE scans can provide supporting evidence for interactions. Progress has been made in examining the role of epigenetics in mediating interaction effects, and animal models should provide further insight into how DNA methylation and other epigenetic mechanisms are involved in gene-air pollution interactions.
Highlights.
Pollution-driven DNA methylation may mediate gene-environment interactions.
Studies of gene-air pollution interaction in asthma are still limited by low power.
New approaches in design, analysis and exposure assessment should improve power.
Improved pheno- and endotyping of asthma will result in more meaningful results.
Acknowledgements
We kindly thank Angela Sadler for editorial assistance.
Funding
This work was supported by the National Institutes of Health [U19AI70235, R01HL132344].
Footnotes
Declarations of Interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burbank AJ, Sood AK, Kesic MJ, Peden DB, Hernandez ML: Environmental determinants of allergy and asthma in early life. J Allergy Clin Immunol 2017, 140:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta J, Johansson E, Bernstein JA, Chakraborty R, Khurana Hershey GK, Rothenberg ME, Mersha TB: Resolving the etiology of atopic disorders by using genetic analysis of racial ancestry. J Allergy Clin Immunol 2016, 138:676–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Los H, Koppelman GH, Postma DS: The importance of genetic influences in asthma. Eur Respir J 1999, 14:1210–1227. [DOI] [PubMed] [Google Scholar]
- 4.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson W, et al. : A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010, 363:1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khreis H, Kelly C, Tate J, Parslow R, Lucas K, Nieuwenhuijsen M: Exposure to traffic-related air pollution and risk of development of childhood asthma: A systematic review and meta-analysis. Environ Int 2017, 100:1–31. [DOI] [PubMed] [Google Scholar]
- 6.Buteau S, Doucet M, Tetreault LF, Gamache P, Fournier M, Brand A, Kosatsky T, Smargiassi A: A population-based birth cohort study of the association between childhood-onset asthma and exposure to industrial air pollutant emissions. Environ Int 2018, 121:23–30. [DOI] [PubMed] [Google Scholar]
- 7.Hehua Z, Qing C, Shanyan G, Qijun W, Yuhong Z: The impact of prenatal exposure to air pollution on childhood wheezing and asthma: A systematic review. Environ Res 2017, 159:519–530. [DOI] [PubMed] [Google Scholar]
- 8.Ai S, Qian ZM, Guo Y, Yang Y, Rolling CA, Liu E, Wu F, Lin H: Long-term exposure to ambient fine particles associated with asthma: A cross-sectional study among older adults in six low- and middle-income countries. Environ Res 2019, 168:141–145. [DOI] [PubMed] [Google Scholar]
- 9.Huang SK, Zhang Q, Qiu Z, Chung KF: Mechanistic impact of outdoor air pollution on asthma and allergic diseases. J Thorac Dis 2015, 7:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.: In The Health Consequences of Involuntary Exposure to Tobacco Smoke: A Report of the Surgeon General. Edited by; 2006. Publications and Reports of the Surgeon General; [PubMed] [Google Scholar]
- 11.Kraft P, Aschard H: Finding the missing gene-environment interactions. Eur J Epidemiol 2015, 30:353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khreis H, Nieuwenhuijsen MJ: Traffic-Related Air Pollution and Childhood Asthma: Recent Advances and Remaining Gaps in the Exposure Assessment Methods. Int J Environ Res Public Health 2017, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoek G: Methods for Assessing Long-Term Exposures to Outdoor Air Pollutants. Curr Environ Health Rep 2017, 4:450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mersha TB: Mapping asthma-associated variants in admixed populations. Front Genet 2015, 6:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowatte G, Lodge CJ, Perret JL, Matheson MC, Dharmage SC: Interactions of GST Polymorphisms in Air Pollution Exposure and Respiratory Diseases and Allergies. Curr Allergy Asthma Rep 2016, 16:85. [DOI] [PubMed] [Google Scholar]
- 16.Dai X, Bowatte G, Lowe AJ, Matheson MC, Gurrin LC, Burgess JA, Dharmage SC, Lodge CJ: Do Glutathione S-Transferase Genes Modify the Link between Indoor Air Pollution and Asthma, Allergies, and Lung Function? A Systematic Review. Curr Allergy Asthma Rep 2018, 18:20. [DOI] [PubMed] [Google Scholar]
- 17.Dizier MH, Bouzigon E, Guilloud-Bataille M, Siroux V, Lemainque A, Boland A, Lathrop M, Demenais F: Evidence for gene x smoking exposure interactions in a genome-wide linkage screen of asthma and bronchial hyper-responsiveness in EGEA families. Eur J Hum Genet 2007, 15:810–815. [DOI] [PubMed] [Google Scholar]
- 18.Dizier MH, Nadif R, Margaritte-Jeannin P, Barton SJ, Sarnowski C, Gagne-Ouellet V, Brossard M, Lavielle N, Just J, Lathrop M, et al. : Interaction between the DNAH9 gene and early smoke exposure in bronchial hyperresponsiveness. Eur Respir J 2016, 47:1072–1081. [DOI] [PubMed] [Google Scholar]
- 19.Choi H, Tabashidze N, Rossner P Jr., Dostal M, Pastorkova A, Kong SW, Gmuender H, Sram RJ: Altered vulnerability to asthma at various levels of ambient Benzo[a]Pyrene by CTLA4, STAT4 and CYP2E1 polymorphisms. Environ Pollut 2017, 231:1134–1144.* This GxE study utilizes a varient of the candidate gene approach by simultaneously analyzing SNPs for 97 genes involved in oxidative stress responses, DNA repair, and inflammation. The multiple testing penalty was considerably smaller than for true genome-wide scans.
- 20.Murcray CE, Lewinger JP, Gauderman WJ: Gene-environment interaction in genome-wide association studies. Am J Epidemiol 2009, 169:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gref A, Merid SK, Gruzieva O, Ballereau S, Becker A, Bellander T, Bergstrom A, Bosse Y, Bottai M, Chan-Yeung M, et al. : Genome-Wide Interaction Analysis of Air Pollution Exposure and Childhood Asthma with Functional Follow-up. Am J Respir Crit Care Med 2017, 195:1373–1383.** This consortium-based GWEI study of gene-NO2 interactions in asthma risk incorporates epigenetics and gene expression analysis to find supporting functional evidence for GxE findings. It exemplifies several promising strategies as well as challenges for GWEI studies.
- 22.de Jong K, Vonk JM, Imboden M, Lahousse L, Hofman A, Brusselle GG, Probst-Hensch NM, Postma DS, Boezen HM: Genes and pathways underlying susceptibility to impaired lung function in the context of environmental tobacco smoke exposure. Respir Res 2017, 18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerasimova A, Chavez L, Li B, Seumois G, Greenbaum J, Rao A, Vijayanand P, Peters B: Predicting cell types and genetic variations contributing to disease by combining GWAS and epigenetic data. PLoS One 2013, 8:e54359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesselbach K, Kim GJ, Flemming S, Haupl T, Bonin M, Dornhof R, Gunther S, Merfort I, Humar M: Disease relevant modifications of the methylome and transcriptome by particulate matter (PM2.5) from biomass combustion. Epigenetics 2017, 12:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reese SE, Xu CJ, den Dekker HT, Lee MK, Sikdar S, Ruiz-Arenas C, Merid SK, Rezwan FI, Page CM, Ullemar V, et al. : Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alfano R, Herceg Z, Nawrot TS, Chadeau-Hyam M, Ghantous A, Plusquin M: The Impact of Air Pollution on Our Epigenome: How Far Is the Evidence? (A Systematic Review). Curr Environ Health Rep 2018, 5:544–578. [DOI] [PubMed] [Google Scholar]
- 27.Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, Ballereau S, Bellander T, Bousquet J, Bustamante M, et al. : Epigenome-Wide Meta-Analysis of Methylation in Children Related to Prenatal NO2 Air Pollution Exposure. Environ Health Perspect 2017, 125:104–110.* In this study, the effect of prenatal air pollution exposure on DNA methylation was evaluated using a epigenome-wide approach as wellas a literature-based multi-candidate approach for antioxidant and anti-inflammatory genes.
- 28.Clifford RL, Jones MJ, MacIssac JL, McEwen LM, Goodman SJ, Mostafavi S, Kobor MS, Carlsten C: Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. Journal of Allergy and Clinical Immunology 2016, S0091-6749(16)30273-1.** The combined effect of allergens and diesel exhaust particles was explored in a controlled human cross-over study. The synergistic effect on DNA methylation was shown to be highly associated with the timing of the two exposures.
- 29.Brunst KJ, Leung YK, Ryan PH, Khurana Hershey GK, Levin L, Ji H, Lemasters GK, Ho SM: Forkhead box protein 3 (FOXP3) hypermethylation is associated with diesel exhaust exposure and risk for childhood asthma. J Allergy Clin Immunol 2013, 131:592–594 e591–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Biagini Myers JM, Brandt EB, Brokamp C, Ryan PH, Khurana Hershey GK: Air pollution, epigenetics, and asthma. Allergy Asthma Clin Immunol 2016, 12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu CJ, Soderhall C, Bustamante M, Baiz N, Gruzieva O, Gehring U, Mason D, Chatzi L, Basterrechea M, Llop S, et al. : DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med 2018, 6:379–388. [DOI] [PubMed] [Google Scholar]
- 32.Prunicki M, Stell L, Dinakarpandian D, de Planell-Saguer M, Lucas RW, Hammond SK, Balmes JR, Zhou X, Paglino T, Sabatti C, et al. : Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin Epigenetics 2018, 10:2.* In this study,methylation of IL10 and FOXP3 was associated with the pollutants NO2, CO, and PM2.5, as well as with asthma. Furthermore, IL10 methylation was associated with altered IL10 expression levels, and FOXP3 methylation was associated with altered levels of regulatory T cells.
- 33.Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, Ryan PH, Khurana Hershey GK, Ji H: Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol 2016, 137:797–805 e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Chen X, Weirauch MT, Zhang X, Burleson JD, Brandt EB, Ji H: Diesel exhaust and house dust mite allergen lead to common changes in the airway methylome and hydroxymethylome. Environ Epigenet 2018, 4:dvy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen S, Jaffar Z, Cole E, Porter V, Ferrini M, Postma B, Pinkerton KE, Yang M, Kim YJ, Montrose L, et al. : Prenatal environmental tobacco smoke exposure increases allergic asthma risk with methylation changes in mice. Environ Mol Mutagen 2017, 58:423–433.* In this study using an allergen-induced murine model, in utero exposure to ETS and subsequent exposure to house and dust mite resulted in greater changes in methylation as well as greater increses in inflammatory markers compared to exposure to either HDM or ETS alone.
- 36.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, Chen LC, Miller RL: Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci 2008, 102:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gauderman WJ, Mukherjee B, Aschard H, Hsu L, Lewinger JP, Patel CJ, Witte JS, Amos C, Tai CG, Conti D, et al. : Update on the State of the Science for Analytical Methods for GeneEnvironment Interactions. Am J Epidemiol 2017, 186:762–770.* An overview of recent advances in analytical methods for gene-environment interaction, including the use of gene sets.
- 38.Jiao S, Peters U, Berndt S, Bezieau S, Brenner H, Campbell PT, Chan AT, Chang-Claude J, Lemire M, Newcomb PA, et al. : Powerful Set-Based Gene-Environment Interaction Testing Framework for Complex Diseases. Genet Epidemiol 2015, 39:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coombes BJ, Biernacka JM: Application of the parametric bootstrap for gene-set analysis of gene-environment interactions. Eur J Hum Genet 2018, 26:1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morawska L, Thai PK, Liu X, Asumadu-Sakyi A, Ayoko G, Bartonova A, Bedini A, Chai F, Christensen B, Dunbabin M, et al. : Applications of low-cost sensing technologies for air quality monitoring and exposure assessment: How far have they gone? Environ Int 2018, 116:286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masiol M, Zikova N, Chalupa DC, Rich DQ, Ferro AR, Hopke PK: Hourly land-use regression models based on low-cost PM monitor data. Environ Res 2018, 167:7–14. [DOI] [PubMed] [Google Scholar]
- 42.Minet L, Liu R, Valois MF, Xu J, Weichenthal S, Hatzopoulou M: Development and Comparison of Air Pollution Exposure Surfaces Derived from On-Road Mobile Monitoring and Short-Term Stationary Sidewalk Measurements. Environ Sci Technol 2018, 52:3512–3519. [DOI] [PubMed] [Google Scholar]
- 43.Simon MC, Patton AP, Naumova EN, Levy JI, Kumar P, Brugge D, Durant JL: Combining Measurements from Mobile Monitoring and a Reference Site To Develop Models of Ambient Ultrafine Particle Number Concentration at Residences. Environ Sci Technol 2018, 52:6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messier KP, Chambliss SE, Gani S, Alvarez R, Brauer M, Choi JJ, Hamburg SP, Kerckhoffs J, LaFranchi B, Lunden MM, et al. : Mapping Air Pollution with Google Street View Cars: Efficient Approaches with Mobile Monitoring and Land Use Regression. Environ Sci Technol 2018. [DOI] [PubMed] [Google Scholar]
- 45.Ueberham M, Schlink U: Wearable sensors for multifactorial personal exposure measurements - A ranking study. Environ Int 2018, 121:130–138. [DOI] [PubMed] [Google Scholar]
- 46.Kuo CS, Pavlidis S, Loza M, Baribaud F, Rowe A, Pandis I, Hoda U, Rossios C, Sousa A, Wilson SJ, et al. : A Transcriptome-driven Analysis of Epithelial Brushings and Bronchial Biopsies to Define Asthma Phenotypes in U-BIOPRED. Am J Respir Crit Care Med 2017, 195:443–455. [DOI] [PubMed] [Google Scholar]
- 47.Yeh YL, Su MW, Chiang BL, Yang YH, Tsai CH, Lee YL: Genetic profiles of transcriptomic clusters of childhood asthma determine specific severe subtype. Clin Exp Allergy 2018, 48:1164–1172. [DOI] [PubMed] [Google Scholar]