Abstract
Small RNA (sRNA) regulators promote efficient responses to stress, but the mechanisms for prioritizing target mRNA regulation remain poorly understood. This study examines mechanisms underlying hierarchical regulation by the sRNA SgrS, found in enteric bacteria and produced under conditions of metabolic stress. SgrS post-transcriptionally coordinates a nine-gene regulon to restore growth and homeostasis. An in vivo reporter system quantified SgrS-dependent regulation of target genes and established that SgrS exhibits a clear target preference. Regulation of some targets is efficient even at low SgrS levels, whereas higher SgrS concentrations are required to regulate other targets. In vivo and in vitro analyses revealed that RNA structure and the number and position of base pairing sites relative to the start of translation impact the efficiency of regulation of SgrS targets. The RNA chaperone Hfq uses distinct modes of binding to different SgrS mRNA targets, which differentially influences positive and negative regulation. The RNA degradosome plays a larger role in regulation of some SgrS targets compared to others. Collectively, our results suggest that sRNA selection of target mRNAs and regulatory hierarchy are influenced by several molecular features and that the combination of these features precisely tunes the efficiency of regulation of multi-target sRNA regulons.
Keywords: sugars; stress, physiological; RNase E; Hfq; base pairing; degradosome
ABBREVIATED SUMMARY
Bacterial small RNAs (sRNAs) are important regulators of gene expression in response to environmental stresses. SgrS sRNA controls several genes to alleviate growth-inhibitory effects related to intracellular accumulation of phosphosugars. Here we uncover that regulation of target mRNAs is prioritized by SgrS via a combination of molecular mechanisms, including binding site multiplicity, sRNA-mRNA binding affinity, RNase E-dependent degradation and Hfq interactions, collectively leading to stress recovery.
INTRODUCTION
Bacteria live in diverse niches, often encountering rapidly changing and stressful environments. Bacterial stress responses can mitigate the negative effects of stress on cell structure and function. Stress responses are usually coordinated by regulators, either RNAs or proteins, that alter expression of a regulon comprised of multiple genes. Coordinated control of the regulon prepares the cell to survive or adapt to the stress (Fang et al., 2016). Proteins control expression of target regulons by binding to DNA sequences and modulating the frequency of transcription initiation, whereas RNAs often modulate gene expression post-transcriptionally. A prevalent type of RNA regulator in bacteria is referred to simply as small RNA (sRNA). The sRNAs are often produced in response to a particular stress, and regulate target mRNAs through base pairing interactions that modify mRNA translation or stability (Georg and Hess, 2011; Storz et al., 2011). Hundreds of sRNAs have been identified in diverse bacteria (Carroll et al., 2016; Koo and Lathem, 2012; Zhang et al., 2003). While the majority of sRNAs have not been characterized, many studies suggest that sRNA regulatory networks are as extensive and complex as those controlled by proteins (Salvail and Masse, 2012; Sharma et al., 2011).
A large body of work has illuminated base pairing-dependent molecular mechanisms of post-transcriptional regulation by sRNAs (Bobrovskyy and Vanderpool, 2013; Bobrovskyy et al., 2015; De Lay et al., 2013; Kavita et al., 2018; Papenfort and Vanderpool, 2015). The sRNA SgrS (sugar-phosphate stress sRNA) has been an important model for discovery of both negative and positive mechanisms of target mRNA regulation. SgrS is induced in response to metabolic stress associated with disruption of glycolytic flux and intracellular accumulation of sugar phosphates (also referred to as glucose-phosphate stress) (Vanderpool and Gottesman, 2004; Vanderpool and Gottesman, 2007; Richards et al., 2013). SgrS regulates at least 9 genes and promotes recovery from glucose-phosphate stress. SgrS-dependent repression of mRNAs encoding sugar transporters (ptsG, manXYZ) (Rice et al., 2012; Rice and Vanderpool, 2011; Vanderpool and Gottesman, 2004) reduces uptake of sugars to prevent further sugar-phosphate accumulation. Activation of a sugar phosphatase (yigL) mRNA promotes dephosphorylation and efflux of accumulated sugars (Papenfort et al., 2013), and repression of other mRNAs is hypothesized to reroute metabolism to promote recovery from stress (Bobrovskyy and Vanderpool, 2016). Each target of SgrS is regulated by a distinct molecular mechanism. How different mechanisms of regulation yield effects of variable magnitude with respect to mRNA stability and translation is an open question.
Temporally-ordered and hierarchical patterns of gene regulation carried out by protein transcription factors have been characterized in many systems (Chevance and Hughes, 2008; Syed et al., 2009; Tonner et al., 2015; Yu and Gerstein, 2006). These regulatory patterns allow cells to respond efficiently to environmental signals by prioritizing induction or repression of products needed to respond to those signals. Protein regulators can establish a hierarchy of regulation based on their affinities for binding sites in the operator regions of different target genes. As the concentration of active regulator increases, genes are sequentially regulated based on binding site affinity (Gao and Stock, 2015). There is growing evidence that sRNAs also regulate their target genes hierarchically (Feng et al., 2015; Levine et al., 2007). However, the mechanisms involved in establishing and maintaining prioritized regulation of sRNA targets are not known.
We hypothesize that the temporal progression of target regulation by SgrS is optimized to promote efficient recovery from glucose-phosphate stress. To test this hypothesis, we first defined the efficiency of SgrS regulation of each target and found that SgrS indeed prioritizes regulation of some targets over others. We examined the factors that determine regulatory efficiency, including the arrangement and strength of SgrS target binding sites and the roles of other factors like RNase E and Hfq. Detailed characterization of a specific SgrS-mRNA target interaction revealed cooperative binding of SgrS to two sites and a requirement for both binding sites for maximal SgrS-dependent regulation. Quantifying SgrS-dependent regulation in strains producing different Hfq variants revealed that the mode of Hfq interaction with SgrS and targets varies between negatively- and positively-regulated targets, and between stronger and weaker targets, implying that Hfq plays a major role in determining efficiency of sRNA-dependent regulation. Collectively, our results uphold the hypothesis that sRNAs regulate expression of genes in their target regulons hierarchically, and that this is influenced by features of each sRNA-mRNA pair, different molecular mechanisms of regulation, and the role of accessory factors that precisely determine the regulatory priority for each target.
RESULTS
SgrS-target mRNA interactions have a range of predicted stabilities.
The thermodynamic stability of sRNA-target mRNA interactions likely influences the regulatory outcome. SgrS regulates genes post-transcriptionally by base pairing with target binding sites on mRNAs (Bobrovskyy and Vanderpool, 2016), which can be located in coding or non-coding regions of the mRNA. The thermodynamic stability of sRNA-mRNA duplexes for SgrS and its target mRNAs ptsG, manX, purR, asd and yigL were predicted in silico. The IntaRNA (Mann et al., 2017) hybridization energy value accounts for the energy required to unfold each partner RNA to make binding sites accessible for base pairing, and the energy of hybridization of the partners (Table 1, Folded ΔG). We used full-length SgrS (+1 to +227) for the sRNA input, and 5’ regions of target mRNAs: ptsG (+1 to +180), manX (+1 to +180), purR (+1 to +300), asd (+1 to +180) and yigL (+1 to +258, where +1 is the processed end of the ‘pldB-yigL transcript) (Papenfort et al., 2013). SgrS interactions with ptsG and yigL mRNAs were predicted to be the most thermodynamically favorable (Table 1, Folded ΔG). Intermediate stabilities were predicted for SgrS base pairing with asdI (the primary SgrS binding site on asd, near the translation initiation region (Bobrovskyy and Vanderpool, 2016)) and purR. SgrS interactions with manX and asdII (the secondary SgrS binding site located in the coding region (Bobrovskyy and Vanderpool, 2016)) were predicted to be the weakest (Table 1, Folded ΔG).
Table 1.
In silico predicted thermodynamic stability of sRNA-mRNA duplexes for SgrS and its target mRNAs.
| Target mRNA |
mRNA positions shown1 |
Base-pairing interactions (Top SgrS sRNA 3′→5′, bottom mRNA 5′→3′)2 |
Folded ΔG3 (kcal/mol) |
Unfolded ΔG4 (kcal/mol) |
|---|---|---|---|---|
|
ptsG |
+73 → +106 |
AAAUGUGGUUAUGAGUCAGUGUGUACUACGUCCG AAAGCACCCAUACUCAGGAGCACUCUCAAUUAUG |
−16.7 | −27.6 |
|
yigL |
+93 → +127 |
AAAUGUGGUUA-UGAGUCAGUGUGUACUACGUCCG AGGACGCAAUGCGCUCAGUCGCGCUCCACGCCAUC |
−15.5 | −20.9 |
|
asdI |
+17 → +50 |
AAAUGUGGUUAUGAGUCAGUGUGUA-CUACGUCCG CCGGCACAUUUAUACAG-CACACAUCUUUGCAGGA |
−10.5 | −21.4 |
|
purR |
+193 → +225 |
AAAUGUGGUUAUGAGUCAGUGUGUACUACGUCCG UUUCCACUACAACUGUGUCACACGUGAU-CAACA |
−10.1 | −20.1 |
|
manX |
+119 → +154 |
AAAUGUGGUUAUGA-GUCAGUGUGUACUACGUCCG CCAUUGCUAUUGUUAUAGGCACACAUGGUUGGGCU |
−5.5 | −15.5 |
|
asdII |
+98 → +130 |
AAAUGUGGUUAUGAGUCAGU-GUGUACUACGUCCG GUCGGCUCCGUUCUCAUGCAACGCAUGGU-UGAAG |
−1.1 | −14.6 |
mRNA positions are relative to transcriptional start (+1) for all targets, except yigL, where +1 is relative to a processing site within pldB-yigL mRNA
Sequence in red is conserved among SgrS orthologs, sequence in blue is predicted RBS, sequence in green is start codon. Bars represent G:C and A:U base interactions and dots represent G:U interactions
Predicted energy of interaction (energy score of the interaction in kcal/mol = sum of the energy of the hybridization and the unfolding energy for sRNA and mRNA) for full-length SgrS (+1 to +227) with ptsG (+1 to +180), manX (+1 to +180), purR (+1 to +300), asd (+1 to +180) and yigL (+1 to +258 relative to pldB-yigL processing site)
Minimum free energy of interaction for full-length SgrS (+1 to +227) with ptsG (+1 to +180), manX (+1 to +180), purR (+1 to +300), asd (+1 to +180) and yigL (+1 to +258 relative to pldB-yigL processing site)
Note: In silico should be italicized.
Another algorithm, RNAhybrid (Kruger and Rehmsmeier, 2006), predicts the minimum free energy of SgrS hybridization with targets assuming that both RNA partners are unfolded and accessible for intermolecular interactions. The predicted thermodynamic stabilities of SgrS-mRNA interactions predicted by this method were (from highest to lowest): ptsG, asdI, yigL, purR, manX and asdII (Table 1, Unfolded ΔG). If sRNA-mediated regulatory outcomes are determined only by properties inherent to the RNAs themselves, then simplistically, interactions with the highest thermodynamic stability should result in faster and more efficient regulation than less favorable interactions.
sRNA-dependent regulation in cells is almost certainly influenced by both thermodynamic and kinetic controls. Indeed, previous work demonstrated that SgrS-dependent regulation of ptsG and manXYZ is influenced by the kinetics of target search, with slower formation of SgrS-manXYZ mRNA complexes compared to SgrS-ptsG mRNA complexes (Fei et al., 2015). Predicted ΔG values, even those generated by algorithms like IntaRNA that take into account unfolding of individual RNAs prior to hybridization, do not take into account the role of interactions with multiple sRNA binding sites (e.g., asd and manXYZ mRNAs, which contain multiple SgrS binding sites (Bobrovskyy and Vanderpool, 2016; Rice et al., 2012)) or the role of Hfq in promoting RNA structural changes. Co-transcriptional sRNA-mRNA interactions may also occur, and these would generate structures that would change as transcription elongation proceeds. The remaining experiments in this study use tools that allowed us to measure and compare the efficiency of regulation and to dissect the impact of features of sRNA-mRNA interactions and the roles of protein cofactors in determining the outcome of sRNA-mediated regulation.
SgrS differentially regulates targets at the level of translation.
To assess the efficiency of SgrS-mediated regulation of targets at the mRNA level, we first performed RT-qPCR to measure changes in RNA levels of select target mRNAs after SgrS expression was induced by the addition of α-methyl D-glucopyranoside (αMG). In the wild-type strain, SgrS rapidly accumulated, whereas it was not detected in the control ΔsgrS strain (Fig. 1A). As shown in previous studies (Papenfort et al., 2013; Rice and Vanderpool, 2011; Vanderpool and Gottesman, 2004), upon SgrS induction (Fig. 1A), target transcripts decreased (ptsG) or increased (yigL) in the wild-type strain, but not in the ΔsgrS strain (Fig. 1B, C). Notably, basal expression levels of the target genes differ and certain target mRNA (e.g., purR, asd) levels were too low to reliably quantify, preventing a broad comparison of regulatory efficiency across targets using RT-qPCR. Additionally, some targets are regulated at the level of translation, with only modest effects on target mRNA stability, as we previously observed for asd mRNA (Bobrovskyy and Vanderpool, 2016). These obstacles called for an alternative approach to study and compare the efficiency of regulation among different targets of SgrS and to get at the molecular mechanisms determining regulatory outcomes.
Figure 1. Relative RNA levels of SgrS, ptsG and yigL mRNAs under glucose-phosphate stress conditions.
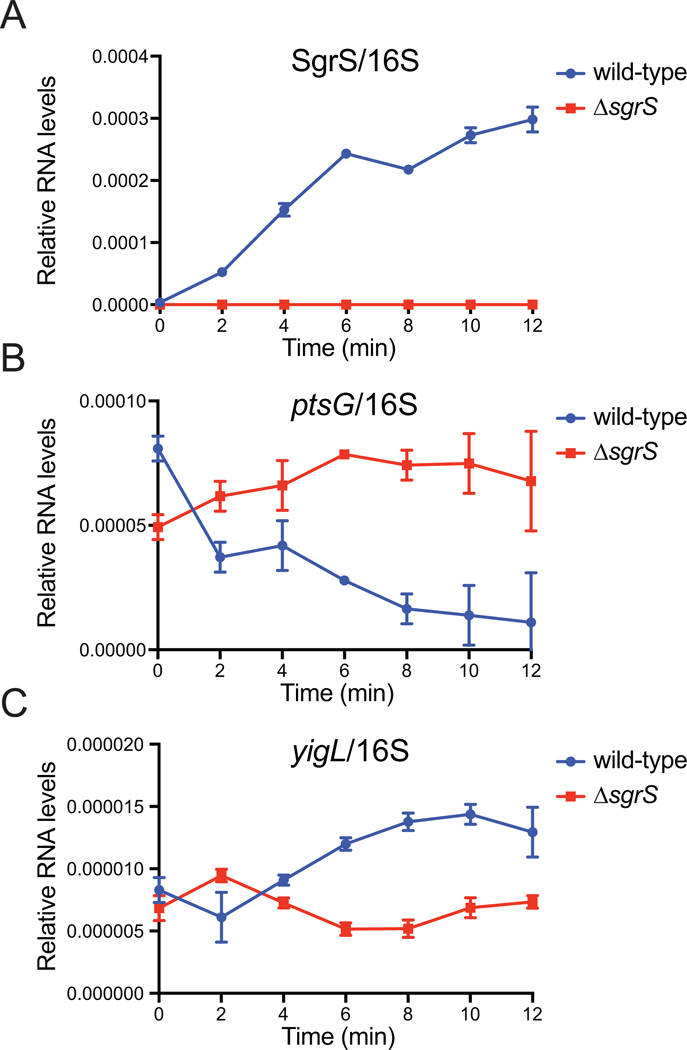
RT-qPCR measuring relative levels of A) SgrS sRNA, B) ptsG mRNA and C) yigL mRNA in the wild-type and ΔsgrS strains after induction of glucose-phosphate stress with 1% αMG. Transcript levels were normalized to rrsA, encoding 16S rRNA in E. coli, and plotted as a function of time.
To this end, we used a two-plasmid system (Fig. 2A) to modulate expression of SgrS and target translational fusions, allowing for finer control of their stoichiometry. Target transcript fragments fused to superfolder gfp (sfgfp) were chosen because they contain experimentally-confirmed SgrS binding sites. To quantify translational regulation by SgrS and facilitate comparisons of regulatory efficiency among targets, we analyzed the data as described previously (Levine et al., 2007). Activity of reporter fusions was measured by monitoring GFP fluorescence over time. By plotting the GFP fluorescence (RFU) as a function of growth (OD600) for target-sfgfp fusions in the absence of SgrS, we defined “basal activity” as the slope of the curve (RFU/OD600) at different IPTG inducer concentrations (Fig. S1A). While the absolute values for basal activity differ among different fusions, all fusions responded to induction by IPTG in a dose-dependent manner (Fig. S1G). Similar plots (RFU/OD600) were generated for each fusion in the presence of SgrS. We define “regulated activity” as the slope of the curve (RFU/OD600) under conditions where both the fusion and SgrS are induced (Fig. S1B–F). This method of quantifying fusion activity accounts for the fact that fluorescence levels are not directly proportional to inducer concentrations ((Levine et al., 2007) and Fig. S1A–F). As levels of SgrS increase, clear patterns of repression or induction are seen for all target fusions (Figs. S1A–L). An advantage of this experimental system is that it allows us to monitor regulation across a range of concentrations of both target and sRNA.
Figure 2. Efficiency of target regulation by SgrS.
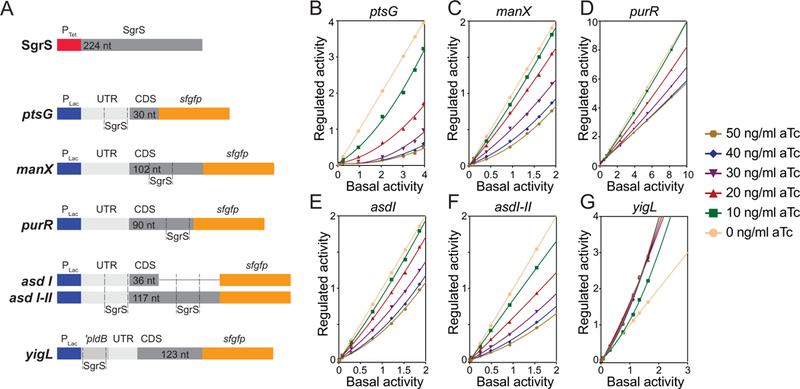
A) Representation of genetic constructs in two compatible plasmids used to study target regulation by SgrS. One plasmid contains full-length SgrS under the control of the aTc-inducible Ptet promoter. A second plasmid contains a Plac promoter and the relevant region encoding each SgrS target (including the SgrS binding site) translationally fused to a superfolder gfp (sfgfp) reporter gene. B-G) Regulated activity plotted as a function of basal activity (see text for description) for ptsG (B), manX (C), purR (D), asdI (E), asdI-II (F) and yigL (G) fused to sfgfp reporter gene. Without SgrS-mediated regulation, line has a slope =1. The plots with slopes <1 indicate repression of ptsG (B), manX (C), purR (D), asdI (E) and asdI-II (F) by SgrS. The plot with a slope of >1 is indicative of activation of yigL (G).
To define the efficiency of regulation of each target, we plotted regulated activity as a function of basal activity for ptsG, manX, asdI, asdI-II, purR, and yigL. (The two different asd fusions contain one [asdI] or both [asdI-II] previously discovered SgrS binding sites (Bobrovskyy and Vanderpool, 2016).) In the absence of SgrS-mediated regulation (0 ng/mL aTc, Fig. 2B–G), a line with a slope of 1 is seen for all targets (Fig. 2B–G). Slopes less than 1 indicate that a fusion is repressed by SgrS, as seen for ptsG (Fig. 2B), manX (Fig. 2C), purR (Fig. 2D), asdI (Fig. 2E) and asdI-II (Fig. 2F) reporter fusions. Slopes greater than 1 are indicative of activation by SgrS, as seen for yigL (Fig. 2G). The regulation was responsive to SgrS levels over the range of SgrS inducer (aTc) concentrations for most targets (Fig. 2B–G). In contrast, for yigL, the magnitude of activation did not increase beyond a maximal level obtained at 20 ng/mL of inducer (Fig. 2G). While the basis for this difference is unclear, it is likely to reflect the inherently different molecular mechanisms of regulation, i.e., mRNA stabilization for yigL and translational repression for other targets.
We then compared regulatory efficiency of targets at different levels of SgrS induction. At the two lowest levels of SgrS induction (10 ng/mL and 20 ng/mL aTc), ptsG, asdI-II and yigL showed substantial repression and activation, respectively (Fig. 3A, B). In contrast, manX, asdI and purR fusions yielded curves whose slopes remained at ~1, indicating no regulation at lower levels of SgrS. Our interpretation is that ptsG, asdI-II and yigL are high-priority targets of SgrS, as they are regulated preferentially when SgrS levels are low. With increasing SgrS levels (30–50 ng/ml aTc), regulation of “weaker” targets manX, asdI and purR became apparent (Fig. 3C–E). As SgrS levels increased, ptsG repression became more efficient up to a maximal repression at 40 ng/mL of aTc, and it remained the most strongly repressed target at all levels of SgrS. Collectively these data suggest that SgrS targets are preferentially regulated in the following order: 1/2) ptsG and yigL, 3) asd, 4) manX, 5) purR (Fig. 3A–E).
Figure 3. Regulatory hierarchy established by SgrS.
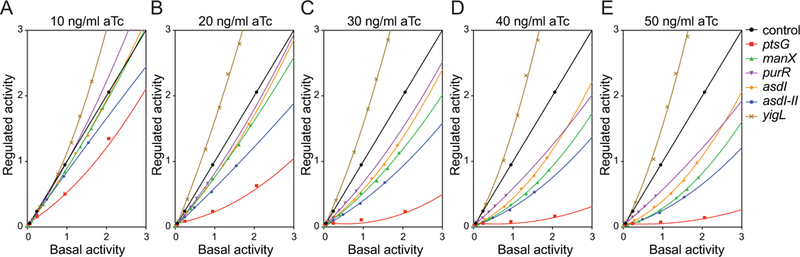
Regulated activity was plotted as a function of basal activity for ptsG, manX, purR, asdI, asdI-II and yigL fusions to sfgfp. Lack of SgrS regulation is indicated by a line with a slope =1. The plots with slopes <1 indicate repression (ptsG, manX, purR, asdI and asdI-II) by SgrS. The plot with slope >1 indicates activation (yigL). Target fusion activity was monitored at different levels of SgrS induction by aTc at concentrations of: A) 10 ng/ml, B) 20 ng/ml, C) 30 ng/ml, D) 40 ng/ml, E) 50 ng/ml.
Structural analyses of SgrS-asd mRNA interactions.
Our data thus far indicate that SgrS regulates mRNA targets with varying degrees of efficiency (Fig. 3A–E). To further understand the features that influence the efficiency of target regulation, we analyzed SgrS-asd mRNA interactions in more detail. Previous work demonstrated that SgrS binding site I encompasses nts +31 to +49 and site II encompasses nts +110 to +127 ((Bobrovskyy and Vanderpool, 2016), Fig. 4A). The predicted energy of SgrS binding to each site on unfolded asd mRNA is −18 kcal/mol for site I and −7.4 kcal/mol for site II (Table 1, Unfolded ΔG). When energy required for unfolding the asd mRNA to make the SgrS binding sites accessible is considered, interaction of SgrS with asd site I has a predicted ΔG of −10.5 kcal/mol, while SgrS pairing with site II has a predicted ΔG of −1.1 kcal/mol (Table 1, Folded ΔG). These predictions suggest that SgrS interaction with site II is less favorable, particularly in the context of the longer structured asd mRNA.
Figure 4. Secondary structure of 5’ end of asd.
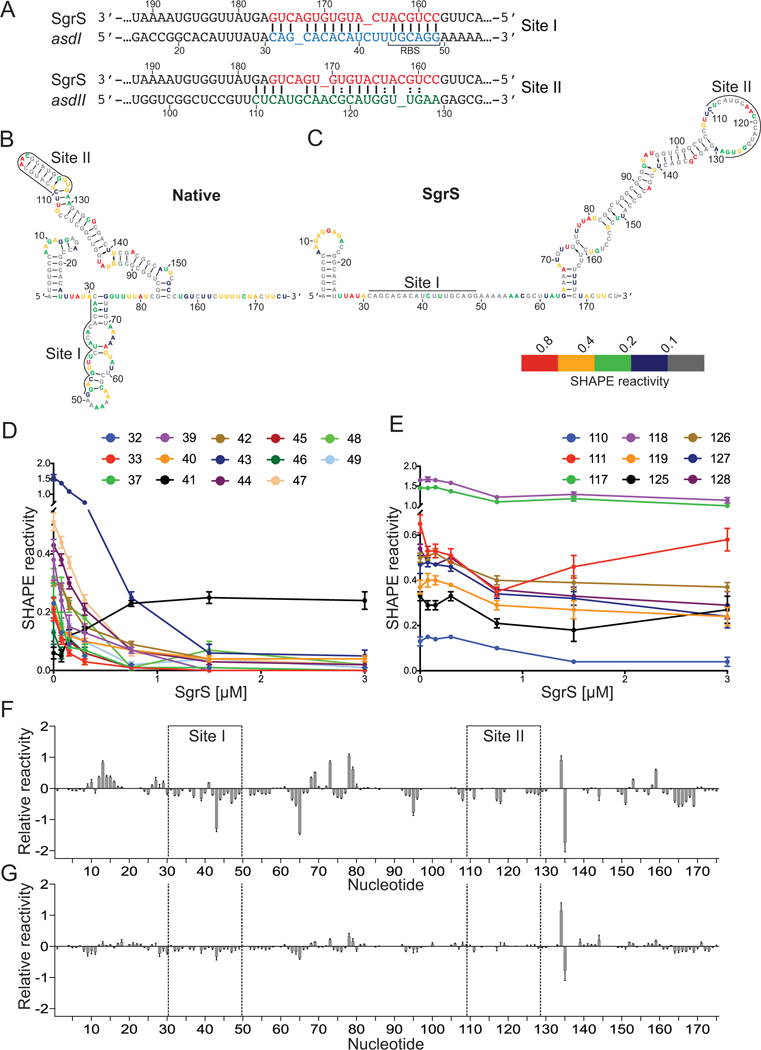
A) Diagram showing base-pairing interactions of SgrS with binding sites I and II of asd mRNA. B,C) Native structure of the asdI-II RNA alone or in the presence of SgrS was probed with NMIA and the modified RNA was analyzed by primer extension inhibition. SHAPE reactivity (difference between the frequency of primer extension products at each nucleotide in +NMIA vs. -NMIA samples) was then used as a parameter in the Vienna RNAprobing WebServer (Lorenz et al., 2016) to predict the secondary structure of the asdI-II RNA. The SHAPE reactivity of asdI-II RNA alone (B) or in the presence of 5-fold excess SgrS (C) is mapped to the predicted secondary structures. Colors indicate SHAPE reactivity as follows: red, highly reactive (≥ 0.8); gold, reactive (0.4–0.79); green, moderately reactive (0.2–0.39); blue, minimally reactive (0.1–0.19); grey, unreactive (< 0.01). D-E) SHAPE reactivity as a function of SgrS concentration for asd binding site I (D) and site II (E). Only nucleotides with a significant (≥ 0.1) change in reactivity are shown. Error bars denote SEM, n = 9. F-G) Relative SHAPE reactivity (difference in the SHAPE reactivity in the presence of SgrS vs. the absence of SgrS) of the asdI-II RNA in the presence of wild-type (F) or mutant SgrS (G). Error bars denote SEM, n = 9 (WT), 6 (MT). The asdI-II RNA nucleotides are numbered below the X-axis and the SgrS binding sites are indicated.
We investigated the structure of asdI-II with selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE), in which accessible nts are modified by N-methylisotoic anhydride (NMIA), while nts constrained in helices are not reactive. In the absence of SgrS, the sequence encompassing the asd ribosome binding site (+44 to +50) is predicted to be located within a structured loop (+36 to +69) on top of a short helix (+31 to +35 pairing with +70 to +74) (Fig. 4B, Fig. S2A). The nts in site I (+31 to +49, Fig. 4A) are located on the 5’ side of the helix. Most of the nts in this structure are reactive, which is indicative of a flexible conformation that is accessible for ribosome binding or base pairing with the seed sequence of SgrS (Fig. 4B). The seed interaction with SgrS is likely to promote opening of the structure. Downstream of the site I structure is a highly structured second helix (+83 to +155) that contains site II in the apical region (+110 to +129) (Fig. 4B, Fig. S2A). Site II is sequestered in a helix and would not be accessible to base pair with SgrS (Fig. 4B).
We next used SHAPE to probe changes in the asdI-II structure in the presence of SgrS (Fig. 4C). The reactivity of site I nts +31 to +49 decreased as the concentration of SgrS increased (Fig. 4D), with the exception of nt +41 which is not predicted to base pair with SgrS (Fig. 4A). The SHAPE reactivity plateaued between 5- and 10-fold excess SgrS (Fig. S2B–G), which suggests that binding to site I was saturated. This is consistent with a strong base-paring interaction between SgrS and site I. In contrast, the reactivity of the site II nts +110 to +129 decreased more slowly and to a lesser extent (Fig. 4E), consistent with a weaker interaction. Fewer site II nts showed changes in SHAPE reactivity upon addition of SgrS; this is likely to be due to the highly structured nature of site II in the absence of SgrS.
The reactivity of nts outside of the SgrS binding sites also changed in the presence of SgrS (Fig. 4F). In contrast, when a mutant SgrS that is not predicted to bind to asdI-II was used, minimal changes in SHAPE reactivity of asdI-II mRNA were observed, which suggests that the changes in the presence of wild-type SgrS are due to the specific interactions between SgrS and asdI-II mRNA and not to the presence of additional RNA in the system (Fig. 4G, Fig. S2I–J). These results indicate that SgrS binding changes the overall structure of the asd RNA. A secondary structure predicted using the SHAPE data suggests that these changes are limited to opening the SgrS binding sites and extending the site II helix (Fig. 4C). We note an important caveat to these analyses. The structure prediction algorithms were not designed to account for intermolecular interactions, so this analysis may not be able to capture the in vivo relevant structure of asd mRNA in complex with SgrS. Nonetheless, SHAPE data are consistent with other analyses in demonstrating binding of SgrS to asd mRNA, prominently at site I and to a lesser extent at site II.
Optimal repression by SgrS involves both pairing sites within asd mRNA.
To further investigate the role of the two SgrS pairing sites on asd in determining the efficiency of regulation, we used a single molecule imaging and analysis platform (Fei et al., 2015) to follow the abundance of SgrS and asd RNAs. Our platform uses stochastic optical reconstruction microscopy (STORM) coupled with single-molecule fluorescence in situ hybridization (smFISH) and allowed us to monitor SgrS regulation of asd-lacZ mRNA variants asdI, asdII, and asdI-II (Fig. 5A). In these experiments, bacteria were grown in the presence of L-arabinose to induce expression of chromosomal asd-lacZ mRNAs, and glucose-phosphate stress was induced for 10 min by the addition of 1% α-methyl D-glucopyranoside (αMG). 3D super-resolution images show asd-lacZ mRNAs (Fig. 5B–D, green) and SgrS (Fig. 5B–D, red), as projected on 2D planes, with cells outlined. Numbers of asd-lacZ mRNAs and SgrS sRNAs were counted and represented as “copy number per cell” in histograms, with average copy number per cell indicated above the histogram (Fig. 5B–D). SgrS reduced the copy number of asdI-lacZ RNA by 3-fold (Fig. 5B, and S3A, B). In contrast, the copy number of asdII-lacZ RNAs was not strongly affected by high SgrS levels after αMG treatment (Fig. 5C and Fig. S3C, D). Copy numbers of asdI-II-lacZ RNA were reduced by ~8-fold after 10 min of SgrS induction (Fig. 5D and Fig. S3E,F). These data demonstrate that both binding sites on asd mRNA participate in efficient SgrS-dependent regulation of mRNA stability.
Figure 5. STORM imaging of SgrS regulation of asd variants.
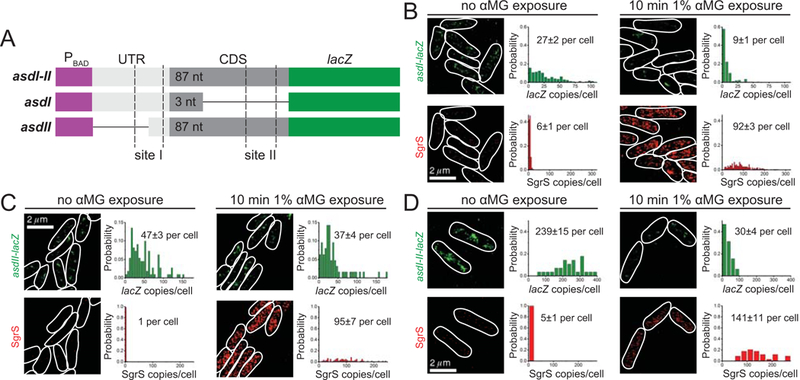
A) Illustration of asdI-II, asdI and asdII translationally fused to lacZ reporter with SgrS binding sites I and II marked. B-D) 2D projection of 3D super-resolution images of SgrS and lacZ mRNA for asdI-lacZ (B), asdII-lacZ (C), and asdI-II-lacZ (D) labeled by smFISH, before and after 10 min induction with 1% αMG. Probability distributions of RNA copy numbers in individual cells for 30–250 cells are plotted next to the representative images.
SgrS binding affinity for target mRNAs in vitro.
We next examined whether in vitro SgrS-target binding affinity correlates with the predicted thermodynamic stability of SgrS-mRNA interactions or the differential post-transcriptional regulation of targets. Electrophoretic mobility shift assays (EMSAs) were performed to measure binding of SgrS to mRNA targets, including asd variants asdI (+1 to +110), asdII (+71 to +310) and asdI-II (+1 to +240) (Fig. 6A, B, C) as well as ptsG, manX, purR and yigL (Fig. 6D, E). The first set of mobility shift assays were performed using a conventional protocol where the target mRNAs and SgrS were mixed together and denatured before adding binding buffer. In these assays, SgrS bound to asdI with a KD of 0.15 μM (Fig. 6A, B), and interaction with asdII did not result in a quantifiable shift at the tested maximum target RNA concentration (Fig. 6A). SgrS bound to asdI-II with a KD of 0.07 μM, an interaction stronger than with asdI (Fig. 6C). We observed two distinct shifts for SgrS-asdI-II, corresponding to binding of either one or two SgrS sRNAs to a single asdI-II RNA (Fig. 6A). SgrS bound to other targets with varying affinities: SgrS-ptsG mRNA KD = 0.11 μM (Fig. 6D, E), SgrS-manX mRNA KD = 19.7 μM (Fig. 6D, F). SgrS binding to purR and yigL mRNAs was substantially weaker and we could not determine KD values for SgrS interaction with purR and yigL at the tested RNA concentrations (Fig. 6D). These results showed that the in vitro binding affinity of SgrS with different targets (Fig. 6A–F) is only partially consistent with the predicted thermodynamic stability of sRNA-mRNA duplexes (Table 1) and is not consistent with the observed efficiency of regulation in vivo (Fig. 3A–E).
Figure 6. SgrS binding to target mRNAs under denatured conditions in vitro.
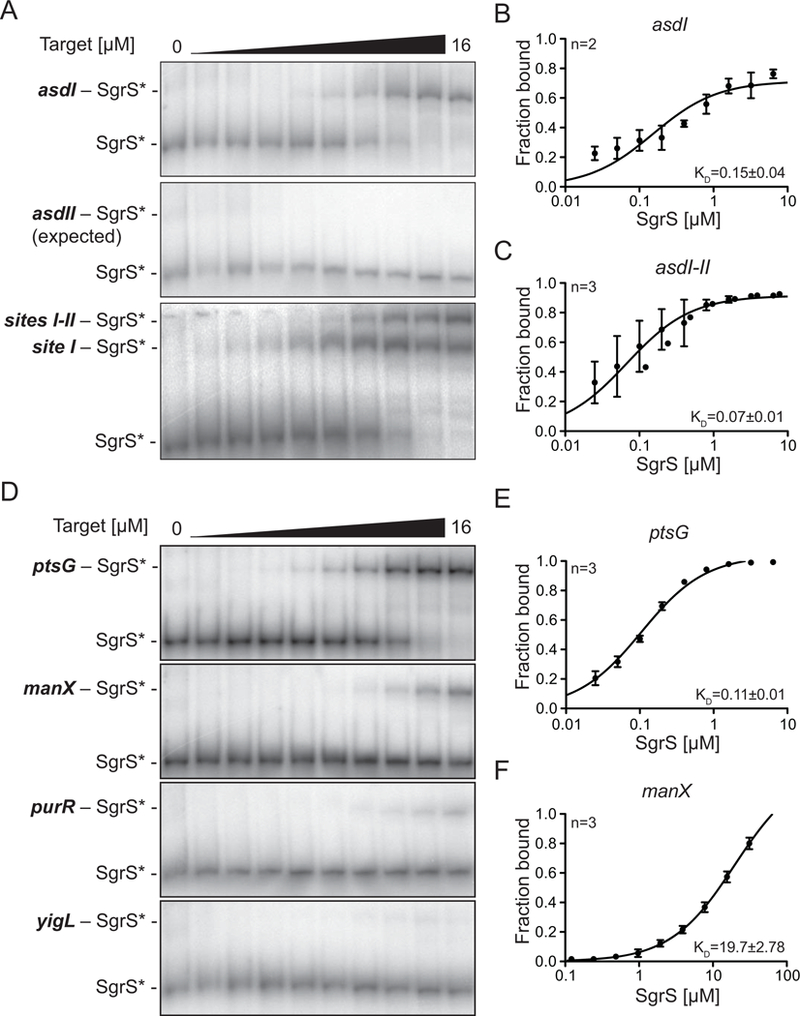
SgrS was labeled with 32P at the 5’ end and incubated with unlabeled target transcripts at final concentrations of 0 μM - 16 μM. A) EMSAs were performed after denaturing at 90 ºC and incubating full-length SgrS (+1 to +227) with its target transcripts asdI (+1 to +110), asdII (+71 to +310) and asdI-II (+1 to +240). Band densities were measured for replicate experiments (n, top left) and plotted to determine dissociation constant (KD, bottom right) values for SgrS complex with B) asdI and C) asdI-II. D) EMSAs were performed after denaturing at 90 ºC and incubating 32P-labeled SgrS with ptsG (+1 to +240), manX (+1 to +240), purR (+1 to +230) and yigL (−191 to +50 relative to ATG translation start of yigL). Band densities were measured and plotted to determine KD for SgrS complex with E) ptsG and F) manX.
We next investigated how transcript secondary structures affect in vitro binding affinity by conducting EMSAs without the denaturation step (Fig. 7A). In these experiments, in vitro transcribed RNAs were mixed together in binding buffer under annealing conditions without prior denaturation. This modified protocol yielded a set of dissociation constants (KD) ~10-fold higher than the values obtained with the conventional protocol. SgrS bound to ptsG with the lowest KD of 1.4 μM, followed by asdI-II with a KD of 25.5 μM (Fig. 7B). Interestingly, in these experiments we observed a single shifted band for the asdI-II RNA instead of two distinct shifts observed in the mobility shift assay using denatured RNAs (Fig. 6A). This result is consistent with structural analyses suggesting that site II is located in a more highly structured region of asd mRNA (Fig. 4B) and may not be accessible for binding SgrS in vitro when the RNAs are not denatured prior to annealing. SgrS bound to two other targets, manX and yigL, with lower affinities: SgrS-manX mRNA KD = 153.8 μM, SgrS-yigL mRNA KD = 234.7 μM (Fig. 7B). These binding affinities are more consistent with the predicted thermodynamic stabilities of SgrS-mRNA duplexes (Table 1), and except for yigL mRNA, are consistent with the observed regulatory hierarchy in vivo.
Figure 7. SgrS binding to target mRNAs under native conditions in vitro.
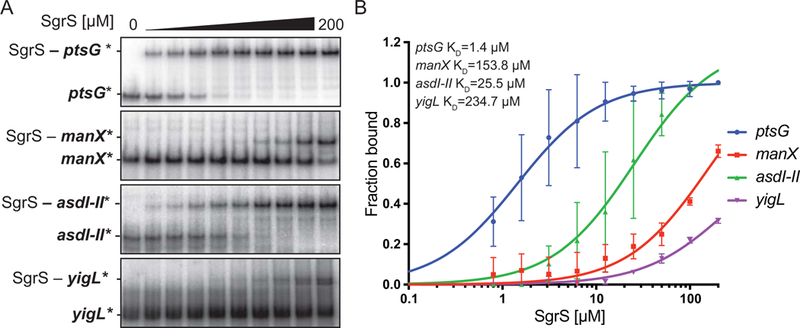
In vitro transcribed ptsG, manX, asdI-II and yigL were labeled with 32P at the 5’ end and incubated with unlabeled SgrS at final concentrations of 0 μM - 200 μM. A) EMSAs were performed after incubating full-length SgrS (+1 to +227) with its target transcripts ptsG (+1 to +240), manX (+1 to +240), asdI-II (+1 to +240) and yigL (−191 to +50 relative to ATG translation start of yigL). B) Band densities were measured for replicate experiments (n=3) and plotted to determine dissociation constant (KD, top left) values.
Role of Hfq in SgrS-dependent target regulation.
The RNA chaperone Hfq mediates sRNA-dependent regulation by interacting with both sRNA and mRNA partners and modulating their stability, structure, and annealing properties. Recent work by Schu et al. (Schu et al., 2015), described at least two distinct modes of interaction between sRNAs and Hfq and defined the consequences of each for sRNA stability and interaction with specific target mRNA sequences. Hfq is a donut-shaped hexamer with three main binding surfaces, the proximal and distal faces of the donut, and the rim of the donut (Link et al., 2009; Mikulecky et al., 2004; Sauer et al., 2012; Schumacher et al., 2002). Different modes of binding to each of these surfaces of Hfq define two major classes of sRNAs. Class I sRNAs bind to residues on the proximal and rim surfaces of Hfq, while Class I sRNA targets are mRNAs with ARN (A=adenine, R=any purine, N=any nucleotide) motifs and are bound on Hfq’s distal face. Class II sRNAs bind to residues on both proximal and distal faces of Hfq, leaving rim residues for target mRNA binding (Schu et al., 2015). While most sRNAs fall clearly into one or the other of these two classes, SgrS has characteristics of both Class I and Class II (Schu et al., 2015; Zhang et al., 2013).
We postulated that SgrS might use different Hfq binding modes for different target mRNAs, and that this could impact the efficiency of regulation. To test this, we constructed host strains with three different chromosomal hfq mutations, yielding proteins Hfq Q8A (proximal face mutant), Hfq Y25D (distal face mutant), and Hfq R16A (rim mutant) (Schu et al., 2015; Zhang et al., 2013). We introduced target reporter and sgrS plasmids into these strains and measured basal activity (Fig. S4A, C, E, G) and regulated activity (Fig. S4B, D, F, H) when SgrS was expressed (Fig. S4I–M). In the parent strain with wild-type Hfq, the regulatory hierarchy for repressed targets as described above was apparent – ptsG was the most stringently regulated, followed by manX, asdI, and purR (Fig. 8A). The single activated target, yigL, was also maximally regulated under these conditions in the strain with wild-type Hfq (Fig. 8A). Mutation of the proximal face residue in the Hfq Q8A strain resulted in loss of regulation of all targets (Fig. 8B). This was expected, as SgrS levels were shown to be very low in this mutant background due to reduced SgrS stability (Schu et al., 2015). The distal face mutation, Y25D, does not reduce SgrS levels; in fact, SgrS is more abundant in this strain background (Schu et al., 2015). However, Hfq Y25D is defective for binding ARN motifs of Class I mRNA targets (Zhang et al., 2013), so we expected that if SgrS regulates some targets by a Class I-like mechanism, regulation of those targets would be affected in the Y25D mutant. Interestingly, this mutation only affected negative regulation by SgrS. Repression of ptsG, asd, purR and manX was very impaired or lost altogether in the Hfq Y25D background, whereas SgrS could still activate yigL in this strain (Fig. 8C). This pattern of regulation is consistent with SgrS acting as a Class I-like regulator for the repressed targets and a Class II-like regulator for the activated target, yigL. We note that regulation of manX and purR fusions by SgrS appears to be reversed in the Hfq Y25D mutant – instead of being repressed, these targets are slightly activated in this background (Fig. 8C). We showed previously that these two targets are repressed by a non-canonical mechanism involving SgrS and Hfq. Unlike ptsG and asd, where SgrS base pairing interactions overlap the Shine-Dalgarno region and directly inhibit ribosome binding, SgrS binds to purR and manX mRNAs at sites in the coding region, far from the site of translation initiation (Bobrovskyy and Vanderpool, 2016; Azam and Vanderpool, 2018). For these targets, SgrS acts as a guide to direct Hfq to bind at a site near the Shine-Dalgarno and inhibit binding of ribosomes (Azam and Vanderpool, 2018; Bobrovskyy and Vanderpool, 2016). These results implicate the distal face of Hfq in binding and repression of purR and manX and suggest that in the absence of Hfq binding to these mRNAs, SgrS pairing in the coding region has a modest stabilizing effect.
Figure 8. Efficiency of target regulation by SgrS in the strains expressing Hfq variants.
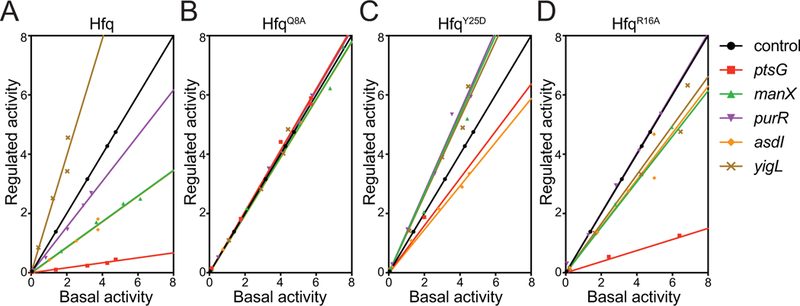
Regulated activity plotted as a function of basal activity (see text for description) for ptsG, manX, purR, asdI, and yigL fused to sfgfp reporter gene. Regulatory efficiency of target-sfgfp fusions was monitored at SgrS induction by 50 ng/ml aTc in A) wild-type, B) HfqQ8A, C) HfqY25D and D) HfqR16A strains. The plots with slopes =1 indicate lack of SgrS regulation, <1 indicate repression and >1 indicate activation by SgrS.
Results in the Hfq R16A rim mutant strain also implicate different SgrS binding modes for different mRNA targets. This mutation abrogated SgrS-dependent positive regulation of yigL, which again is consistent with a Class II-like mode of action because Class II mRNA targets bind to Hfq rim sites (Schu et al., 2015). Repression of ptsG in the Hfq R16A background was nearly as efficient as in the Hfq wild-type strain (Fig. 8D), which is consistent with Class I-like regulation. Repression of other targets was much less efficient than in wild-type, suggesting that something about the mode of Hfq binding differs between ptsG, the most strongly regulated target, and the other weakly repressed targets.
Altogether, these results are consistent with the observations in previous work (Schu et al., 2015; Zhang et al., 2013) that SgrS has Hfq binding and mRNA regulatory properties that are intermediate between Class I and Class II sRNAs. In general, SgrS behaves more like a Class I regulator for repressed targets like ptsG and more like a Class II regulator for the activated target, yigL. The loss of negative regulation but not positive regulation by SgrS in the Y25D background and the differential effect of the R16A rim mutation on negatively regulated targets indicates that Hfq plays distinct roles in regulation of each target.
Effects of degradosome defect on target regulation by SgrS in vivo.
We next investigated how the RNA degradosome impacts SgrS-dependent regulation of different mRNA targets. For sRNAs that repress target mRNA translation by interfering with ribosome binding, degradation is often not strictly required for regulation (Maki et al., 2008). However, translational repression of an mRNA often leads to faster mRNA turnover by a “passive” process owing to increased accessibility of nuclease-sensitive sites on ribosome-free mRNA. In other cases, sRNA-mRNA duplexes seem to more actively recruit RNase E to specific cleavage sites, leading to more rapid degradation of an sRNA-mRNA duplex (Wagner, 2009; Lalaouna et al., 2013; Bandyra et al., 2012). We reasoned that the mechanism of sRNA-dependent degradation of mRNAs and whether it occurs by active or passive mechanisms could influence the efficiency of regulation, even for mRNA targets that are primarily regulated at the level of translation initiation. To test this, we monitored regulatory efficiency in an rne701 mutant strain. The rne701 allele encodes a truncated form of RNase E that is unable to assemble into the degradosome because it lacks the C-terminal scaffold region of the protein (Morita et al., 2004), and a number of studies have shown that degradosome assembly is required for SgrS-dependent mRNA decay (Fei et al., 2015; Morita et al., 2006; Rice et al., 2012; Rice and Vanderpool, 2011). We introduced the sgrS and target reporter plasmids into the rne701 mutant strain to probe the effect of the degradosome on the efficiency of regulation by SgrS. We determined the basal activity (Fig. S5A) and regulated activity (Fig. S5B–F) at specific induction levels of the reporter (IPTG) and SgrS (aTc). All the reporter fusions exhibited dose-dependent response to increasing inducer concentrations (Fig. S5A–F).
To determine the efficiency of regulation, we plotted regulated activity as a function of basal activity for ptsG, manX, purR, asdI, asdI-II, and yigL translational sfgfp fusions at different levels of SgrS induction (Fig. S5G–L). The efficiency of regulation of all targets was slightly reduced in the rne701 background. In the wild-type strain, maximal regulation of all targets was achieved at ~40 ng/mL aTc (SgrS inducer, Fig. 3D). In the rne701 strain, maximal regulation was not reached until ~80 ng/mL of SgrS inducer (Fig. 9D). In addition to the global reduction in SgrS regulation efficiency, we observed a pronounced defect in ptsG regulation in the rne701 mutant strain (Fig. 9A–E). In contrast with the wild-type strain, where repression of ptsG was more efficient than all other repressed targets at all SgrS induction levels (Fig. 3A–E), in the rne701 strain, regulation of ptsG by SgrS was less efficient compared to other targets at lower SgrS induction levels (20–60 ng/mL aTc, Fig. 9A–C). Impaired ptsG regulation in the rne701 mutant strain was overcome by increasing SgrS expression levels (80–100 ng/mL aTc, Fig. 9D, E). It is worth noting that SgrS regulated asdI-II more efficiently compared to asdI in both wild-type (Fig. 3A–E) and the rne701 (Fig. 9A–E) backgrounds. Therefore, the second binding site on asd mRNA enhances the efficiency of SgrS-mediated regulation independent of the degradosome.
Figure 9. Efficiency of target regulation by SgrS in the RNase E mutant strain.
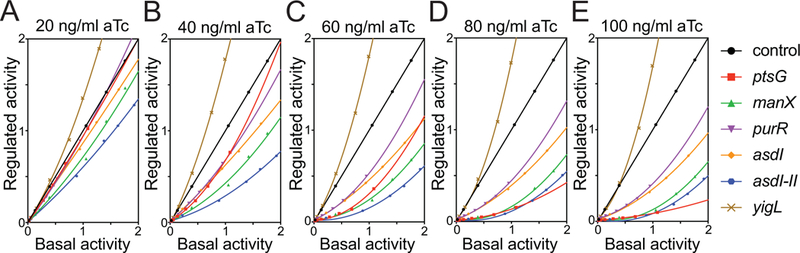
Regulatory efficiency of all the target-sfgfp fusions was monitored at different levels of SgrS induction by aTc: G) 20 ng/ml, H) 40 ng/ml, I) 60 ng/ml, J) 80 ng/ml, K) 100 ng/ml. The plots with slopes =1 indicate lack of SgrS regulation, <1 indicate repression and >1 indicate activation by SgrS.
DISCUSSION
In this study, we set out to define the hierarchy of regulation by a model bacterial sRNA. SgrS is a good model for this study because it has a modestly-sized regulon, and the mechanisms of regulation of several targets have been characterized in detail (Bobrovskyy and Vanderpool, 2016; Kawamoto et al., 2006; Rice et al., 2012; Papenfort et al., 2013). Since one of our goals in this study was to directly compare the regulatory efficiency of SgrS on its various targets, we set up an experimental system that allowed us to manipulate both SgrS and mRNA target levels (Fig. 2A). Our results demonstrate a clear pattern of prioritized regulation of mRNA targets (Fig. 2B–G, Fig. 3A–E) that was consistent across a wide range of RNA expression levels. Two targets in particular, ptsG and yigL, were “high-priority” targets that were efficiently regulated even at low levels of SgrS. Other targets, manX, purR, and asd, were less impacted by SgrS and were regulated only when SgrS was produced at higher levels. When cells experience glucose-phosphate stress, their first response is to induce SgrS to block sugar uptake via repression of ptsG-encoded glucose transporter (EIICBGlc), and to promote dephosphorylation and efflux of phosphosugars by activating yigL-encoded sugar phosphatase (Fig. 10). During prolonged stress, SgrS levels continue to increase (Vanderpool and Gottesman, 2004; Balasubramanian and Vanderpool, 2013), allowing for regulation of lower priority targets manX, asd and purR, encoding a variety of metabolic functions that help reroute cellular metabolism and allow cells to use other carbon sources. This pattern of prioritized regulation is established at a molecular level by a complex interplay of factors. The RNase E degradosome enhances efficiency of SgrS regulation of ptsG mRNA specifically. The binding affinity between each target mRNA and SgrS as well as the number and position of SgrS binding sites on each target also influence the efficiency of regulation. The RNA chaperone Hfq uses different modes of binding to repressed and activated targets of SgrS, thereby influencing competition among targets for SgrS and Hfq binding. Collectively, these molecular mechanisms drive hierarchical regulation of the SgrS regulon (Fig. 10).
Figure 10. Model for SgrS target prioritization during glucose-phosphate stress.
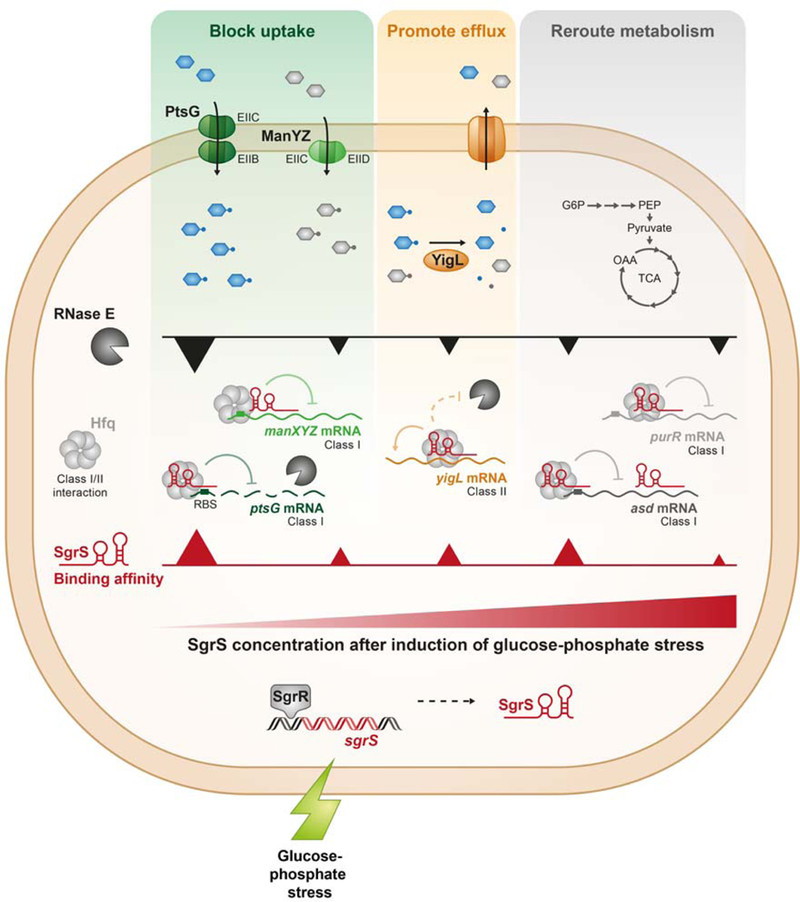
Glucose or the analogs αMG and 2-deoxyglucose are phosphorylated during transport through the phosphotransferase system proteins EIICBGlc (PtsG) or EIICDMan (ManYZ). If sugar-phosphates are not metabolized, the glucose-phosphate stress response is triggered, and the transcription factor SgrR becomes active and promotes sgrS transcription. The RNA chaperone Hfq promotes SgrS-mediated translational repression of ptsG and manXYZ mRNAs, reducing synthesis of sugar transporters. SgrS stabilizes yigL mRNA, promoting sugar phosphatase (YigL) synthesis. SgrS-mediated repression of asd, purR, folE and adiY likely reroutes metabolism to restore homeostasis during stress recovery. The hypothetical sequence of regulatory events following stress induction is represented from left to right as SgrS levels increase over time. When SgrS concentrations are low, only the highest priority targets are regulated. When stress persists and concentrations of SgrS increase, lower priority targets are regulated.
Hfq is a conserved RNA chaperone that facilitates target mRNA regulation by many sRNAs. Different sRNAs can bind to distinct sites on Hfq and this impacts their regulation of particular targets (Schu et al., 2015; Santiago-Frangos et al., 2016). Our experiments in three Hfq mutant backgrounds underscore this point. While all the repressed targets had patterns of regulation most consistent with Class I-like sRNA-mRNA interactions, the only target activated by SgrS, yigL mRNA, behaved in a more Class II-like fashion (Fig. 8 and 10). EMSAs were conducted to measure dissociation constants for SgrS and its target mRNAs. Binding affinities of SgrS for ptsG (KD 1.4 μM), asd (KD 25.5 μM) and manX (KD 153. μM) mRNAs (Fig. 7), regulated in Class I-like manner, correlate with the observed translational efficiency of regulation (Fig. 3A–E). The dissociation constant for SgrS pairing with yigL mRNA, a Class II-like interaction, is higher (KD 234.7 μM) than all of the Class-I repressed targets, yet yigL is regulated efficiently even at low SgrS concentrations. Since yigL mRNA is the only known activated target of SgrS and it uses a distinct mode of binding to Hfq, lack of competition with repressed targets for Hfq binding may promote more efficient regulation of yigL at lower SgrS levels. The mode of Hfq participation in regulation clearly impacts the regulatory outcome – for yigL, making the regulation more efficient than expected based on in vitro behavior in the absence of Hfq (Fig. 10).
Several lines of evidence implicate mRNA structure and sRNA-dependent structural rearrangements in determining regulatory efficiency. When in vitro transcribed RNAs were denatured, mixed and then annealed (Fig. 6), the measured KDs for individual SgrS-target mRNA complexes did not match what was expected based on observed in vivo regulation. When folded RNAs were not denatured prior to annealing (Fig. 7), the KDs were at least an order of magnitude higher than in the other experiment, and they more closely matched the expectations based on the measured in vivo hierarchy. These results imply that some SgrS-target interactions involve structural elements in one or both partners that impact base pairing and ultimately the regulatory outcome. Our experiments with asd mRNA and the differential accessibility of the two different binding sites (Fig. 4) illustrates this point. To understand how specific molecular features of SgrS-mRNA interactions and cofactors influence regulation efficiency, we investigated the role of multiple sRNA binding sites on a single mRNA target (asd mRNA, Figs. 4, 5). SgrS binding site I overlaps the asd ribosome binding site and binding site II is ~60 nt downstream in the asd coding sequence ((Bobrovskyy and Vanderpool, 2016) and Fig. 4A, 5A). EMSAs using denatured and annealed RNAs demonstrated SgrS pairing at site I (in the absence of site II), but no detectable pairing at site II in the absence of site I (Fig. 6A, B). Binding of SgrS to asdI-II mRNA (containing both sites) appeared to be cooperative (Fig. 6A, C). Structural analyses of asd mRNA in the absence and presence of SgrS demonstrated that SgrS indeed pairs preferentially at site I over site II and induces substantial structural rearrangement in the mRNA (Fig. 4, Fig. S2). Consistent with in vitro analyses, in vivo quantification of SgrS-dependent regulation of asd mRNA showed that site I is important, but sites I and II together promote the most efficient regulation (Figs. 2, 3, 5, Fig. S3). These results, along with those of prior studies from our lab (Bobrovskyy and Vanderpool, 2016; Rice et al., 2012; Rice and Vanderpool, 2011), suggest that the number and position of sRNA binding sites on certain mRNA targets plays an important role in the regulatory outcome in vivo.
In many cases, sRNA-mediated regulation of translation is thought to indirectly affect mRNA stability by making untranslated mRNA more susceptible to degradation by RNase E. There are also examples of sRNA regulation, including SgrS regulation of yigL (Papenfort et al., 2013), where modulation of mRNA stability is translation-independent. Truncation of RNase E (encoded by rne), removing the C-terminal scaffold for degradosome assembly, often prevents sRNA-mediated degradation of mRNA targets (Fei et al., 2015; Morita et al., 2006; Rice and Vanderpool, 2011). Two different mechanisms of sRNA-mediated mRNA decay have been proposed. Translational repression leading to mRNAs unoccupied by ribosomes can inherently leave mRNAs susceptible to nucleases, including RNase E, by a process that has been referred to as passive nucleolytic repression (Lalaouna et al., 2013; Wagner, 2009). In contrast, some sRNA-mRNA duplexes seem to actively recruit RNase E to promote specific mRNA cleavage (Bandyra et al., 2012). If an sRNA’s primary effect on a given mRNA target is modulation of translation leading to passive degradation, we might expect that loss of the degradosome would not change the overall pattern of regulation, although it might alter the speed or magnitude of the regulatory effect. On the other hand, if an sRNA-mRNA interaction actively recruits the degradosome as an essential mechanism for regulation, then loss of degradosome function would be expected to substantially impair regulation. Our results show that compared to the wild-type strain (Fig. 3A–E) higher concentrations of SgrS are required to achieve target regulation in the rne701 degradosome mutant but the regulatory hierarchy is mostly preserved (Fig. 9). These results suggest that for most targets, SgrS repression of translation leads to passive RNase E-mediated degradation. A notable exception is ptsG. In the wild-type background, ptsG is the most efficiently-regulated target (Fig. 3A–E). In the rne701 host, ptsG is weakly regulated at lower levels of SgrS induction (Fig. 9A–C) and only efficiently regulated at the highest levels of SgrS (Fig. 9D–E). This result suggests that ptsG may be unique among the repressed targets of SgrS in its requirement for RNase E-dependent degradation for its efficient regulation. This is consistent with the observation that ptsG mRNA levels decrease at least 10-fold whereas other targets exhibit a modest 2-fold decrease in mRNA levels upon SgrS expression (Bobrovskyy and Vanderpool, 2016). Our recent study quantifying SgrS-dependent mRNA degradation at single molecule resolution indicated that ptsG mRNA exhibits faster degradation kinetics than manXYZ mRNA (Fei et al., 2015), which could enhance the efficiency of regulation in a wild-type but not rne701 mutant background where translational regulation and mRNA degradation are uncoupled.
In summary, we established target prioritization within SgrS regulon and demonstrated that multiple molecular events contribute to target prioritization, such that ptsG is the primary and most efficiently repressed target. Hfq-defined SgrS interaction with ptsG mRNA, along with other repressed targets manX, asd and purR, have features of Class I sRNA regulation. Uniquely, SgrS-stabilized yigL mRNA is regulated in a Class II-like fashion. Importantly, our findings indicate that sRNA-mRNA affinity is a strong determinant of regulatory outcome, highlighted by the cooperative binding of SgrS to two sites on asd, which significantly improves its repression. Furthermore, we demonstrate that RNase E degradation is essential in determining target priority for Class I, negatively regulated SgrS targets, exemplified by robust and highly efficient repression of ptsG (Fig. 10). It is worth noting that while these conclusions are supported by in vitro and in vivo results, each method used has caveats. For example, while target reporter fusions contain all sRNA binding site determinants previously identified as important for regulation, reporter target mRNAs may not share certain important structural features with native mRNAs. Future work using proteomics approaches to directly and simultaneously quantify SgrS-dependent changes in target protein levels will extend this work and shed more light on these questions.
Beyond molecular features of sRNA-mRNA interactions, defining regulatory hierarchy for sRNA regulons can provide insights relevant for bacterial physiology. The vast majority of sRNA regulons remain undefined, and thus many sRNA functions are unknown. For novel sRNAs, distinguishing high-priority from weaker targets may provide crucial clues to the predominant role of the sRNA in cell physiology. For SgrS, the regulatory hierarchy we have defined here is perfectly consistent with growth studies that demonstrate the primary importance of SgrS regulation of sugar transport and efflux under glucose-phosphate stress conditions (Sun and Vanderpool, 2011). While in this system, regulatory efficiency is correlated with physiological outcomes, it is also possible that in some cases, weakly regulated targets could play important roles in regulatory networks. Nevertheless, hierarchical regulation of multi-target regulons by sRNAs is likely one aspect that evolved to promote rapid and efficient responses to environmental signals that would provide cells with a competitive growth advantage in their specific niche. It remains a key challenge in sRNA research to develop tools to more rapidly and quantitatively characterize sRNA regulons. Progress in this area will drive better understanding of the functions of the hundreds of uncharacterized sRNAs in diverse bacteria.
EXPERIMENTAL PROCEDURES
Strain and plasmid construction.
Strains and plasmids used in this study are listed in Table S1. Primers used in this study are described in Table S2. All strains used in this study are derivatives of E. coli K-12 strain MG1655. Oligonucleotide primers and 5’ biotinylated probes used in this study are listed in Table S2 and were purchased from Integrated DNA Technologies. Chromosomal alleles were moved between strains by P1 vir transduction (Miller, 1972) and inserted using λ Red recombination (Datsenko and Wanner, 2000; Yu et al., 2000).
An hfq::cat-sacB was introduced into strain JH111 from the donor strain SA1408 by P1 transduction to create strain XM102. Primers SA035 and SA036 were used to amplify hfq from strains DJS2411, DJS2420, SA1410 to obtain hfq fragments with desired mutations Q8A, Y25D and R16A respectively. The three PCR products were then individually recombined into XM102 strain by λ Red recombination to create strains XM106, XM107, and XM108 (Table S1).
Translational reporter fusion alleles PBAD-asdI-II-lacZ (MBP151F/MBP193R primers), PBAD-asdI-lacZ (MBP151F/MBP151R primers) and PBAD-asdII-lacZ (MBP193F/MBP193R primers) were constructed by PCR amplification of desired fragments using primers containing homologies to PBAD and lacZ. Similarly, transcriptional fusions PBAD-asdI-II-lacZ (MBP151F/MBP206R3 primers) and PBAD-asdI-lacZ (MBP151F/MBP206R1 primers) were generated by PCR amplification using a forward primer with homology to PBAD and reverse primers containing lacZ RBS and lacZ homology. PCR products were then recombined into PM1205 strain using λ Red homologous recombination.
Construction of plasmids Ptet0–1-sgrS (pZAMB1), Plac0–1-ptsG-sfgfp (pZEMB8) and Plac0–1-purR-sfgfp (pZEMB25) was described previously (Bobrovskyy and Vanderpool, 2016). Plasmids with translational reporter fusions Plac0–1-manX-sfgfp (pZEMB10), Plac0–1-yigL-sfgfp (pZEMB15), Plac0–1-asdI-sfgfp (pZEMB26) and Plac0–1-asdI-II-sfgfp (pZEMB27) were constructed by PCR amplification of manX (+1 to +217), yigL (−191 to +123 relative to ATG translation start of yigL; −191 corresponds to the processing site within pldB-yigL polycistronic RNA), asdI (+1 to +97) and asdI-II (+1 to +178) from E. coli MG1655 chromosomal DNA using primer pairs MBP2F/ MBP2R34, MBP16F2/ MBP16R41, MBP92F/MBP92R1 and MBP92F/MBP92R2 respectively. Amplified DNA fragments were digested with KpnI and EcoRI restriction endonucleases and ligated into pZEMB8 vector.
Media and reagents.
Bacteria were cultured in Luria-Bertani (LB) broth medium or LB agar plates (Miller, 1972) at 37°C, unless stated otherwise. Bacteria were grown in MOPS (morpholine-propanesulfonic acid) rich defined medium (Teknova) with 0.2% fructose for reporter fluorescence assays. When necessary, media were supplemented with antibiotics at the following concentrations: 100 μg/ml ampicillin (Amp), 25 μg/ml chloramphenicol (Cm), 25 μg/ml kanamycin (Kan) and 50 μg/ml spectinomycin (Spec). Isopropyl β-D-1-thiogalactopyranoside (IPTG) was used at concentrations of 0.1–1.5 mM for induction of Plac0–1 promoter, anhydrotetracycline (aTc) was used at 0–50 ng/ml for induction of Ptet0–1 promoter and L-arabinose was used at 0.000002%−0.2% for induction of PBAD promoter, unless otherwise noted. To induce glucose-phosphate stress, 0.5% α-methylglucoside (αMG) was added to the growth medium.
Quantitative real time polymerase chain reaction (RT-qPCR)
Escherichia coli strains DJ480 and CV104 were cultured in MOPS medium supplemented with 0.2% glucose to OD600 ~ 0.2. αMG was added to a final concentration of 1% and 1 ml aliquots were collected at 2, 4, 6, 8, 10 and 12 minutes, and frozen with liquid nitrogen. Cells were pelleted by centrifugation at 7,000 × g for 1 minute at 4. 100 μl of 1 mg/ml lysozyme in TE buffer (10 mM Tris-HCl and 1 mM EDTA) was used to resuspend the cell pellet and mixed by vortexing for 10 seconds at room temperature and then incubated at room temperature for 5 minutes while vortexing gently. Total RNA was extracted from the lysozyme-treated cells using a RNeasy Mini kit (Qiagen). 2.5 μg of RNA was used to remove genomic DNA contamination using TURBO DNA-free™ Kit (Ambion). Following that, 100 ng of DNase-treated total RNA was used to generate cDNA in a reverse transcriptase PCR reaction primed with a mixture of oligo(dT) and random hexamer primers using iScript™ cDNA Synthesis Kit (Bio-Rad). 1/5th of the reaction mixture was subsequently used to prepare samples according to SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) specifications and qPCR was performed using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) to measure relative RNA levels using gene-specific primers (Table S2). The CFX Manager™ Software (Bio-Rad) was used for relative quantification and a housekeeping gene rrsA encoding 16S RNA in E. coli was used to normalize the RNA levels.
Reporter fluorescence assay.
Bacterial strains were cultured overnight in MOPS rich medium supplemented with 0.2% fructose, Amp and Cm, and diluted 1:100 into fresh medium with appropriate inducers (IPTG, aTc) in 48 well plates. Relative fluorescence units (RFU) and optical density (OD600) were measured over time. “Regulated activity” was calculated by plotting RFU over OD600 and determining the slopes of linear regression plots for each IPTG concentration in exponentially growing cells in the presence of aTc to induce SgrS expression. “Basal activity” was calculated by plotting RFU over OD600 and determining the slopes of linear regression plots for each IPTG concentration in exponentially growing cells in the absence of aTc.
In vitro transcription and radiolabeling.
Template DNA for in vitro transcription was generated by PCR using gene-specific oligonucleotides containing the T7 RNA polymerase promoter sequence. The following oligonucleotides were used to generate specific template DNA: MBP84F/MBP213R–ptsG (+1 to +240), O-JH218/MBP214R–manX (+1 to +240), MBP56F/MBP215R–asdI-II (+1 to +240), MBP56F/MBP222R–asdI (+1 to +110), MBP226F/MBP226R–asdII (+71 to +310), MBP65F/MBP174R–purR (+1 to +230), MBP216F/MBP216R–yigL (−191 to +50 relative to ATG translation start of yigL) and O-JH219/O-JH119 were used to generate full-length sgrS template DNA. In vitro transcription of DNA templates was performed according to specifications of the MEGAscript T7 Kit (Ambion). In vitro transcribed RNA was 5’-end labeled with radioisotope 32P using the KinaseMax Kit (Ambion), according to the manufecturer’s instructions. Samples were purified by passing through Illustra ProbeQuant G-50 Micro Columns (GE Healthcare) followed by extraction with phenol-chloroform:isoamyl alcohol (Ambion), and labeled RNA was precipitated with ethanol:3M NaAc (30:1).
RNA-RNA gel electrophoretic mobility shift assay.
Specific (0–16 μM) concentrations of unlabeled target RNA were mixed with 0.02 pmol of 5’-end 32P-labeled SgrS. Samples were denatured at 95°C for 1 min, placed on ice for 5 min, and incubated at 37°C for 30 min in 1x binding buffer (20 mM Tris-HCL [pH 8.0], 1mM DTT, 1 mM MgCl2, 20 mM KCl, 10 mM Na2HPO4 [pH 8.0]) (Morita et al., 2012). For native conformation EMSAs, 0–200 μM concentrations of unlabeled SgrS and 0.02 pmol of 5’-end 32P-labeled ptsG, manX, asdI-II and yigL RNAs were mixed, placed on ice for 5 min and incubated at 37°C for 30 min in 1x binding buffer. Non-denaturing loading buffer was added and samples were resolved for 6 h at 40 V on native 5.6% PAGE. Gels were dried, exposed to phosphor screen that was imaged using Fluorescent Image Analyzer FLA-3000 (Fujifilm) and band intensities of bound and unbound RNA quantitated. Data was plotted using Prism software, and non-linear regression was used to fit the data and calculated KD values.
Protein-RNA gel electrophoretic mobility shift assay.
0.02 pmol of 5’-end labeled mRNA was denatured at 95°C for 1 min, and placed on ice for 5 min. Different concentrations of purified Hfq protein (His-tagged) were added. Samples were incubated at 37°C for 30 min in 1x binding buffer. Non-denaturing loading buffer was added and samples were resolved for 90 min at 20 mA on native 4.0% PAGE (Morita et al., 2012). Gels were dried, exposed to phosphor screen that was imaged using Fluorescent Image Analyzer FLA-3000 (Fujifilm) and band intensities of bound and unbound RNA quantitated. Data was plotted using Prism software, and non-linear regression was used to fit the data and calculated KD values.
SHAPE.
The asdI-II RNA (0.15 μM) and SgrS RNA (0.075 μM, 0.15 μM, 0.30 μM, 0.75 μM, 1.5 μM, or 3.0 μM) were folded separately as in (Wilkinson et al., 2006) using a modified SHAPE buffer (100 mM HEPES [pH 8.0], 2 mM MgCl2, 40 mM NaCl). For each SgrS concentration, the SgrS RNA or the equivalent volume of 0.5X TE was added to the asdI-II RNA and the samples were incubated at 37°C for 30 min. The RNAs were modified with N-methylisatoic anhydride (NMIA, 6.5 mM; Sigma-Aldrich) and collected by ethanol precipitation as in (Wilkinson et al., 2006). Parallel primer extension inhibition and sequencing reactions were performed using fluorescently labeled primers complementary to the 3’ end of the asdI-II RNA (5′-AGATCAAAGGCATCCTGAAG, 22.5 nM; Applied Biosystems, ThermoFisher Scientific) as in (Mortimer and Weeks, 2009) with minor modifications. Prior to primer binding, the RNAs were denatured and snap cooled and the reactions were carried out for 20 min at 52˚C, followed by 5 min at 65˚C. The cDNAs were analyzed on a 3730 DNA Analyzer (Applied Biosystems, Inc.). The data were processed and SHAPE reactivity (difference between the frequency of primer extension products at each nucleotide in +NMIA vs. -NMIA samples) was derived using the QuShape software (Karabiber et al., 2013). Data for each nucleotide were averaged with statistical outliers removed and normalized using the 2–8% rule (McGinnis et al., 2009). Relative reactivity was calculated by subtracting normalized SHAPE reactivity in the absence of the SgrS RNA from reactivity in the presence of the WT or MT SgrS RNA.
Single-molecule fluorescence in situ hybridization (smFISH).
The asdI-lacZ (MB170), asdII-lacZ (MB183) and asdI-II-lacZ (MB171) strains were grown overnight at 37ºC with shaking at 250 rpm in LB Broth Miller (EMD) containing Kan and Spec. The next day, the overnight cultures were diluted 100-fold into MOPS EZ rich defined medium (Teknova) with 0.2% (w/w) sodium succinate, 0.02% glycerol and L-arabinose (0.01% for the asdI-lacZ and asdII-lacZ strains, 0.002% for asdI-II-lacZ) and were allowed to grow at 37ºC till the OD600 reached 0.15–0.25. αMG was added to the culture to a desired concentration to introduce sugar phosphate stress and induce SgrS sRNA expression. After 10 min of induction, the cells were fixed by mixing with formaldehyde (Fisher Scientific) at a final concentration of 4%.
ΔsgrS and ΔlacZ strains were grown in LB Broth Miller (EMD) at 37ºC with shaking at 250 rpm overnight. The cultures were diluted 100-fold into MOPS EZ rich defined medium (Teknova) with 0.2% glucose and allowed to grow at 37ºC until the OD600 reached 0.2. The cells were then fixed by mixing with formaldehyde at a final concentration of 4%. TK310 cells were grown overnight, similar to the knockout strains. The overnight culture was then diluted 100-fold into MOPS EZ rich defined medium (Teknova) with 0.2% glucose and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Sigma-Aldrich) and allowed to grow at 37ºC for 30 minutes. The cells were then fixed as described above.
The fixation and permeabilization of the cells were done using the methods published previously (So et al., 2011). After fixing with 4% formaldehyde, the cells were incubated at room temperature for 30 min. The cells were then centrifuged at 600 x g for 7 min and the pellets were washed with three times with 1X PBS 3. The cells were then permeabilized with 70% ethanol for 1 h at room temperature and stored at 4ºC before fluorescence in situ hybridization.
The smFISH probes were designed using Stellaris Probe Designer and purchased from Biosearch Technologies (https://www.biosearchtech.com/). The labeling of the probes was performed using equal volumes of each probe. The final volume of sodium bicarbonate was adjusted to 0.1 M by adding 1/9 reaction volume of 1 M sodium bicarbonate (pH = 8.5). The probe solution was mixed with 0.05–0.25 mg of Alexa Fluor 647 or Alexa Fluor 568 succinimidyl ester (Life Technologies) dissolved in 5 μL DMSO. The dye was kept at 20–25 fold in molar excess relative to the probes. After incubation with gentle vortexing in the dark at 37ºC overnight, the reaction was quenched by adding 1/9 reaction volume of 3 M sodium acetate (pH 5). Unconjugated dye was removed by ethanol precipitation followed by P-6 Micro Bio-Spin Column (Bio-Rad).
A previously published protocol (So et al., 2011) was used for the hybridization procedure. 60 μl of permeabilized cells were washed with FISH wash solution (10% formamide in 2X SSC [Saline Sodium Citrate] buffer) and resuspended in 15 μl hybridization buffer (10% dextran sulfate, 1 mg/ml E. coli tRNA, 0.2 mg/ml BSA, 2 mM vanadyl ribonucleoside complexes, 10% formamide in 2X SSC) with probes. Nine probes labeled with Alexa Fluor 647 were used for sRNA SgrS and 24 probes labeled with Alexa Fluor 568 were used for mRNA lacZ. The concentration of the labeled probes for SgrS and lacZ mRNA were 50 nM and 15 nM, respectively. The reactions were incubated in the dark at 3 ºC overnight. The cells were then resuspended in 20X volume FISH wash solution and centrifuged. They were then resuspended in FISH wash solution, incubated for 30 min at 30ºC and centrifuged and this was repeated 3 times. The cells were pelleted after the final washing step and resuspended in 20 μl 4X SSC and stored at 4ºC for imaging. The labeled cells were immobilized on a poly-L-lysine (Sigma-Aldrich) treated 1.0 borosilicate chambered coverglass (Thermo Scientific™ Nunc™ Lab-Tek™). They were then imaged in imaging buffer (50 mM Tris-HCl [pH = 8.0], 10% glucose, 1% β-mercaptoethanol [Sigma-Aldrich], 0.5 mg/ml glucose oxidase [Sigma-Aldrich] and 0.2% catalase [Calbiochem] in 2X SSC).
Single-molecule localization-based super-resolution imaging.
An Olympus IX-71 inverted microscope with a 100X NA 1.4 SaPo oil immersion objective was used for the 3D super-resolution imaging. The lasers used for two-color imaging were Sapphire 568–100 CW CDRH, Coherent (568nm) and DL640–100-AL-O, Crystalaser (647nm) and DL405–025. Crystalaser (405nm) was used for the reactivation of Alexa 647 and Alexa 568 fluorophores. The laser excitation was controlled using mechanical shutters (LS6T2, Uniblitz). A dichroic mirror (Di01-R405/488/561/635, Semrock) was used to reflect the laser lines to the objective. The objective collected the emission signals and then they made their way through an emission filter (FF01–594/730–25, Semrock for Alexa 647 or HQ585/70M 63061, Chroma for Alexa 568) and excitation laser was cleaned up using notch filters (ZET647NF, Chroma, NF01–568/647–25×5.0 and NF01–568U-25, Semrock). Samples were then imaged on a 512×512 Andor EMCCD camera (DV887ECS-BV, Andor Tech). Astigmatism was introduced by placing a cylindrical lens with a focal length of 2 m (SCX-50.8–1000.0-UV-SLMF-520–820, CVI Melles Griot) in the emission path between two relay lenses with focal lengths of 100 mm and 150 mm each, which facilitated 3D imaging. In this setup, each pixel corresponded to 100 nm. We used the CRISP (Continuous Reflective Interface Sample Placement) system (ASI) to keep the z-drift of the setup to a minimum. The image acquisition was controlled using the storm-control software written in Python by Zhuang and coworkers (Rust et al., 2006; Huang et al., 2008) and available at GitHub.
The imaging of the sample began with a DIC image of the sample area. Subsequently two-color super-resolution imaging was performed. 647nm excitation was used first and after image acquisition was completed for Alexa Fluor 647, 568nm excitation was used to image Alexa Fluor 568. 405 nm laser power was increased slowly to compensate for fluorophore bleaching and also to maintain moderate signal density. Imaging was stopped when most of the fluorophores had photobleached and the highest reactivation laser power was reached.
The raw data acquired using the acquisition software were analyzed using the method described in previously published work (Fei et al., 2015), which was a modification of the algorithm published by Zhuang and coworkers (Huang et al., 2008; Rust et al., 2006). The clustering analysis on the localization data was performed using MATLAB codes as described previously (Fei et al., 2015). Background signal was estimated using ΔsgrS and ΔlacZ strains and they were prepared, imaged and analyzed as described above. TK310 cells were prepared, imaged and analyzed in the same way as a low copy lacZ mRNA sample for copy number calculation. The copy number calculation was also performed using MATLAB codes as described previously (Fei et al., 2015).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to extend a special thank you to Erel Levine for providing plasmids. We are grateful to Jennifer Rice, Rich Yemm, Divya Balasubramanian, Chelsea Lloyd, Alisa King, Jessica Kelliher and other current and past members of the Vanderpool lab for strains, plasmids and valuable advice. We thank Prof. James Slauch and members of his lab for fruitful discussions. We appreciate the graphic design assistance of Sandy Pernitzsch from SCIGRAPHIX.
Footnotes
DATA SHARING
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Azam MS, and Vanderpool CK (2018) Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res 46: 2585–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D, and Vanderpool CK (2013) Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J Bacteriol 195: 4620–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, and Luisi BF (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47: 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovskyy M, and Vanderpool CK (2013) Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu Rev Genet 47: 209–232. [DOI] [PubMed] [Google Scholar]
- Bobrovskyy M, and Vanderpool CK (2016) Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol Microbiol 99: 254–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovskyy M, Vanderpool CK, and Richards GR (2015) Small RNAs regulate primary and secondary metabolism in Gram-negative bacteria. Microbiol Spectr 3. [DOI] [PubMed] [Google Scholar]
- Carroll RK, Weiss A, Broach WH, Wiemels RE, Mogen AB, Rice KC, and Shaw LN (2016) Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. MBio 7: e01990–01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, and Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, and Gottesman S (2013) Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288: 7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Frawley ER, Tapscott T, and Vazquez-Torres A (2016) Bacterial stress responses during host infection. Cell Host Microbe 20: 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Singh D, Zhang Q, Park S, Balasubramanian D, Golding I, et al. (2015) RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 347: 1371–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Rutherford ST, Papenfort K, Bagert JD, van Kessel JC, Tirrell DA, et al. (2015) A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell 160: 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, and Stock AM (2015) Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. MBio 6: e00686–00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, and Hess W (2011) Cis-antisense RNA, another level of gene regulation in bacteria. Microbiology and molecular biology reviews : MMBR 75: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, and Zhuang X (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319: 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber F, McGinnis JL, Favorov OV, and Weeks KM (2013) QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavita K, de Mets F, and Gottesman S (2018) New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol 42: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, and Aiba H (2006) Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol 61: 1013–1022. [DOI] [PubMed] [Google Scholar]
- Koo JT, and Lathem WW (2012) Global discovery of small noncoding RNAs in pathogenic Yersinia species. Adv Exp Med Biol 954: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, and Rehmsmeier M (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34: W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Simoneau-Roy M, Lafontaine D, and Masse E (2013) Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta 1829: 742–747. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, and Hwa T (2007) Quantitative characteristics of gene regulation by small RNA. PLoS Biol 5: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, and Brennan RG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 106: 19292–19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Luntzer D, Hofacker IL, Stadler PF, and Wolfinger MT (2016) SHAPE directed RNA folding. Bioinformatics 32: 145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K, Uno K, Morita T, and Aiba H (2008) RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc Natl Acad Sci U S A 105: 10332–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Wright PR, and Backofen R (2017) IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res 45: W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis JL, Duncan CD, and Weeks KM (2009) High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 468: 67–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, and Feig AL (2004) Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH, (1972) Experiments in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, and Aiba H (2004) Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol 54: 1063–1075. [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, and Aiba H (2012) Detection of sRNA-mRNA interactions by electrophoretic mobility shift assay. Methods Mol Biol 905: 235–244. [DOI] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, and Aiba H (2006) Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A 103: 4858–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, and Weeks KM (2009) Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc 4: 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, and Vogel J (2013) Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153: 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, and Vanderpool CK (2015) Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39: 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Balasubramanian D, and Vanderpool CK (2012) Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc Natl Acad Sci U S A 109: E2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, and Vanderpool CK (2011) The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39: 3806–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GR, Patel MV, Lloyd CR, and Vanderpool CK (2013) Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J Bacteriol 195: 4816–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, and Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, and Masse E (2012) Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA 3: 26–36. [DOI] [PubMed] [Google Scholar]
- Santiago-Frangos A, Kavita K, Schu DJ, Gottesman S, and Woodson SA (2016) C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc Natl Acad Sci U S A 113: E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Schmidt S, and Weichenrieder O (2012) Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci U S A 109: 9396–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu DJ, Zhang A, Gottesman S, and Storz G (2015) Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34: 2557–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, and Brennan RG (2002) Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 21: 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, and Vogel J (2011) Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165. [DOI] [PubMed] [Google Scholar]
- So LH, Ghosh A, Zong C, Sepulveda LA, Segev R, and Golding I (2011) General properties of transcriptional time series in Escherichia coli. Nat Genet 43: 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, and Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, and Vanderpool CK (2011) Regulation and function of Escherichia coli sugar efflux transporter A (SetA) during glucose-phosphate stress. J Bacteriol 193: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, et al. (2009) The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol 191: 6555–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonner PD, Pittman AM, Gulli JG, Sharma K, and Schmid AK (2015) A regulatory hierarchy controls the dynamic transcriptional response to extreme oxidative stress in archaea. PLoS Genet 11: e1004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, and Gottesman S (2004) Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076–1089. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, and Gottesman S (2007) The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J Bacteriol 189: 2238–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG (2009) Kill the messenger: bacterial antisense RNA promotes mRNA decay. Nat Struct Mol Biol 16: 804–806. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, and Weeks KM (2006) Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, and Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, and Gerstein M (2006) Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci U S A 103: 14724–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, and Gottesman S (2013) Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425: 3678–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, and Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


