Abstract
Advances in tandem mass spectrometry (MS/MS) acquisition rate have steadily led to increased performance in shotgun proteomics experiments. To that end, contemporary mass spectrometers are outfitted with multiple analyzers allowing for the simultaneous collection of survey (MS1) and MS/MS spectra. In the latest generation Orbitrap hybrid, MS/MS scans can be acquired at a high rate using the dual cell linear ion trap analyzer, all while the next precursor is being dissociated in a collision cell and a MS1 scan is occurring in the Orbitrap. Often overlooked in these experiments is that the ion trap scan duration is highly variable and dependent upon precursor mass. Here, we examine the use of various static mass-to-charge ratio scan ranges for ion trap MS/MS acquisition and determine performance relative to conventional dynamic mass-to-charge ratio range scanning. We demonstrate that a fixed mass-to-charge ratio scan range can generate 12% more MS/MS scans and more unique peptide identifications as compared to the standard dynamic approach, respectively.
Graphical Abstract
Tandem mass spectrometry (MS) is an essential technology for comprehensive and rapid characterization of proteomes.1–6 In data-dependent shotgun proteomics, complex mixtures of peptides are chromatographically separated and individually sequenced using tandem MS.7 A major limitation, of this approach, however, is the speed at which tandem spectra (MS/MS) are collected.8 Historically ion trap mass analyzers have acquired MS/MS scans at rates ranging between 1 and 20 Hz, too slow to identify every peptide observed by the MS system.9–11 Recent improvements in ion trap mass analyzers permit much higher MS/MS scan rates, providing acquisition speeds that considerably improve sampling depth.11–14 Still, many peptidic features remain unsampled, underscoring the need for even faster MS/MS acquisition.
In the latest generation of Orbitrap hybrid systems, these faster scanning ion trap mass analyzers must also work in concert with other devices, i.e., quadrupole mass filter, Orbitrap analyzer, and a collision cell.11 Considerable effort has been expended to synergize the various ion processing activities so that maximum parallelization is achieved.15–17 In other words, ion injection, MS1 mass analysis, ion dissociation, and MS/MS scanning are coordinated so that each device is operating at or near 100% duty cycle. In practice this can be difficult to achieve as each disparate step has optimal conditions, particularly MS/MS data acquisition since the mass-to-charge ratio range scanned defines the time required to collect each scan. Given peptides have masses that range from less than 900 to greater than 2 000, MS/MS data acquisition times can fluctuate over 2-fold from scan-to-scan. Given this, we wondered whether static mass-to-charge ratio ranges, coupled with optimal parameters, could increase both the rate of MS/MS data acquisition but also the number of identified peptides over a single-shot LC−MS experiment.
To test this hypothesis, we examined whether the time and parallelization benefits achieved by using a smaller, static mass-to-charge ratio scanning range would outperform the current approach. To do this we considered several parameters including optimal MS/MS mass-to-charge ratio range, scan rate, and injection time. We tested these parameters using a complex mixture of tryptic peptides derived from cultured human cells. Indeed, with a fixed scan range from 175 to 1 075 m/z, we generated more MS/MS scans (12% increase) and more unique peptide identifications (45 000 vs 43 000) as compared to the standard auto approach. These data evince an improvement in overall system parallelization and suggest careful consideration of instrument parameters lead to further increases in performance.
MATERIALS AND METHODS
LC−MS/MS.
Sample digestion conditions are reported in the Supporting Information. Human cell line K562 tryptic peptides were resuspended in 0.2% formic acid at a concentration of 1 μg/μL. All separations were performed with a Thermo Dionex Ultimate 3000 RSLC-nanoliquid chromatography instrument, using an in-house fabricated heater set at 50°C to aid with controlling the column pressure.18 LC−MS/MS analysis was performed with 1 μg of peptides, injected onto a reversed phase nanoultra-high-performance column. Separation was performed on a 30 cm, 75−360 μM inner-outer diameter, PicoFrit nanospray column (New Objective, Woburn, MA) that was packed with 130 A pore size, 1.7 μm particle size Bridged Ethylene-Hybrid C18 (Waters, Milford, MA) as previously described.19 The sample was loaded with 100% A (0.2% formic acid), gradient elution increasing %B over 51 min with an additional 9 min of re-equilibration times in 100% A, for a total of 60 min LC−MS/MS analysis.
Peptides eluting from the LC were analyzed on an Orbitrap Fusion Lumos Tribrid platform with Instrument Control Software, version 3.1.2412.25. All MS analyses used a 120 000 resolving power survey scan with a cycle time set at 0.6 s. The MS scan maximum injection time was set to 50 ms and automatic gain control (AGC) of 1 × 106. Advanced peak determination was turned on and charge states 2–5 were included. Dynamic exclusion filter temporarily excluded precursor mass with an MS/MS scan for 10 s and the Monoisotopic Precursor Selection Mode was set to Peptide. Followed by the most intense precursor, all MS/MS experiments were performed in the linear ion trap. The MS/MS parameters common across analyses included a 0.7 m/z isolation in the quadrupole, 25% HCD collision, and AGC set at 2 × 104. For the MS/MS analyses with a scan range 100–2000 m/z collected at the “turbo” scan rate, the maximum ion injection was set to 25 ms. For the optimized MS/MS scan ranges experiments, the mass-to-charge ratio ranges and maximum ion injections are summarized in Table 1.
Table 1.
Summary of Ranges and Optimal MS/MS Ion Injection Timesa
| MS/MS | optimal ion injection times (ms) | |||
|---|---|---|---|---|
| scan range (m/z) | scan width (m/z) | “turbo” | “rapid” | “normal” |
| 200–900 | 700 | 10 | 15.5 | 26 |
| 175–1075 | 900 | 12 | 18.5 | 32 |
| 125–1125 | 1000 | 13 | 20 | 35 |
| 125–1225 | 1100 | 13.5 | 21.5 | 38 |
| 125–1425 | 1300 | 15 | 27.5 | 44 |
Five mass-to-charge ratio ranges were subjected to LC−MS/MS analysis each with their respective optimal injection times set according to the ion trap scan rate.
Data Analysis.
Thermo Scientific.raw data files were converted into a .dta file (DTA Generator v1.1.6260). The optimization of MS/MS mass-to-charge ratio range was performed by the postacquisition processing of one 60 min tryptic K562 LC MS/MS experiment. The software used to perform postacquisition truncation of mass-to-charge ratio range was written in-house and developed in C# with .NETFramework 4.5. For each truncated spectrum, a .dta file was generated containing a list of precursors each associated with a companion list of product ion mass-to-charge ratio and intensities. The number of unique peptides, product ions, and number of MS/MS scans were determined using COMPASS software suite.20 See the Supporting Information for more details.
MS/MS Acquisition Rate.
The MS/MS acquisition rate (Hz) was calculated by counting the number of MS/MS scans collected in ambient ion trap conditions over a period of 15 s. Ion trap scan rates “turbo”, “rapid”, and “normal” were compared at 20 000 resolving power with a 4-s cycle. The MS/MS maximum ion injection was set to 1 ms with a 0.7 Da isolation window. Note, MS scans were removed, and survey time was accounted for to calculate the estimated MS/MS acquisition rate.
MS/MS Maximum Ion Injection Time.
The optimal MS/MS maximum ion injection time was determined by monitoring MS/MS acquisition rate for 12 s in ambient ion trap conditions. The maximum ion injection was increased by 1 ms for a scan width (mass-to-charge ratio) until MS/MS acquisition rate dropped, indicating MS/MS analysis is out of parallelization. The greatest maximum ion injection while maintaining parallelization was determined for scan widths between 200 m/z and 1900 m/z.
RESULTS AND DISCUSSION
Optimal Mass-to-Charge Ratio Range.
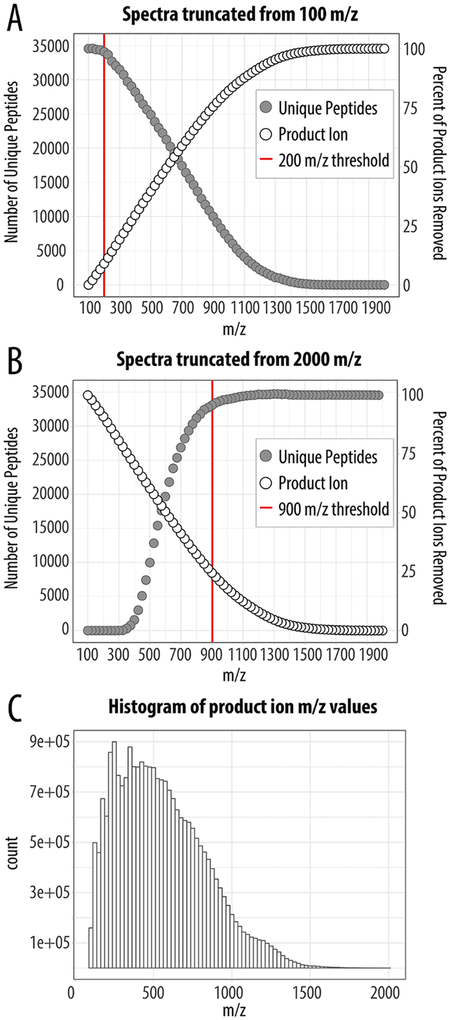
For HCD MS/MS analysis, where dissociation is done external to the ion trap, the m/z range is defined by a user selected low m/z and an automatically calculated high m/z based on precursor mass. For example, a precursor having a mass of 1900 Da would typically have an MS/MS spectrum ranging from 100 to 1910 m/z, taking 50 ms at the fastest scan rate of 38 Hz. In most cases, the low mass is set in the range of 100–200 m/z for HCD MS/MS. Note, isobaric tagged experiments would require a scan range that starts around 100 m/z; however, high-resolution mass analysis is typically preferred when using isobaric tags. This strategy ensures all theoretical product ions could be measured. However, to our knowledge, it is currently unclear if using a full scan range is required for bottom-up proteomics experiments. To address this question, we first generated a complex mixture of tryptic peptides by digestion of a mammalian cell whole proteome (human K562). Next, we collected a nanoLC–MS/MS data set using a static MS/MS scan range of 100–2000 m/z and “turbo” ion trap scan rate. In this experiment, we collected 84 702 MS/MS scans and proceeded to generate 76 versions of each MS/MS differing in scan range. Specifically, we iteratively removed 25 m/z in silico, starting from the low end of the mass range (i.e., 100–2 000 m/z, 125–2 000 m/z, 150–2 000 m/z…1 975–2 000 m/z (Figure 1A)). MS/MS spectra were grouped by mass range, searched against a database of human tryptic peptides, and the resulting identifications filtered to a 1% FDR. Figure 1A plots the number of unique peptides identified for each mass-to-charge ratio truncation point. The full scan range (100–2 000 m/z) yielded 34 579 unique peptide identifications which precipitously decline when the range is truncated above 200 m/z, suggesting there is minimal opportunity for optimizing the starting mass-to-charge ratio point. Not surprisingly, few peptides are identified when the range starts at, or above, ~1 300 m/z.
Figure 1.
Effects of truncating the MS/MS mass-to-charge ratio scan range with “turbo” ion trap scan rate. Both the number of uniquely identified peptides and the percentage of product ion loss was determined by either truncating from the low-end (A) or high-end (B) of the MS/MS 100–2 000 m/z scan range. The distribution of product ions produced is densely populated between 100 and 1300 m/z (C). Note the scarcity of product ions above 1 500 m/z, suggesting this mass-to-charge ratio region may not be necessary.
Having established the effect of truncating the low-end of the mass-to-charge ratio range, we next shifted our focus to the upper end. Similarly, we generated another 76 versions of each MS/MS spectrum, but with the truncation now starting at the upper-end while leaving the low-end fixed at 100 m/z (i.e., 100–2 000 m/z, 100–1 975 m/z, 100–1 950 m/z…100–125 m/z, Figure 1B). Unlike the low range truncation, we determined that no consequential impact was induced until about 900 m/z. That is, we can remove the entire product ion region from 900 to 2 000 m/z with only a loss of 5% of the IDs. We also examined the loss of product ions as a function of the various m/z truncations (Figure 1A,B). Removal of product ions from 100–200 m/z appears to have very little impact on the overall number of identifications. This is not surprising as this range would primarily be occupied by b1 or y1; y1 is not specific, as tryptic peptides end in R or K, and b1 is rarely observed.21,22 Figure 1C plots the distribution for over 24 million product ions that were measured for the 100–2 000 m/z scan. These data suggest the MS/MS scan duration can likely be reduced substantially by reducing the upper mass-to-charge ratio scan range. Additionally, only 8% of product ions have mass-to-charge ratio values above 1 300 and their removal is largely inconsequential (Figure 1C). We conclude product ions having mass-to-charge ratio values ranging from 200–1 300, however, are both abundant and useful for spectral identification. This can be observed by the steep loss in both product ions and identifications when this region is removed (from either the upper or lower end). Indeed, 95% of peptides are still identified when only product ions from the range of 200–1 300 m/z are utilized.
Optimal Maximum Injection Times.
The geometry of the tribrid Orbitrap system allows for parallel ion accumulation/fragmentation and product ion m/z analysis. As such, the instrument duty cycle approaches unity when the timing of these two events are synced. First, we wondered what the maximal ion trap MS/MS acquisition rate could be for three ion trap scan rates: 33 m/z per millisecond “normal”; 67 m/z per millisecond “rapid”; 125 m/z per millisecond “turbo” (Figure 2A). Note this experiment capped ion injection time at 1 ms so that the MS/MS acquisition rate can be assessed. As expected, increasing scan range decreased acquisition rate. At the narrowest mass-to-charge ratio range scanned (100 m/z), MS/MS acquisition rates up to 71 Hz were achieved. Highlighting the importance of our work to identify optimal mass-to-charge ratio ranges, the widest m/z range scanned (1900 m/z) roughly halved the acquisition rate (35 Hz). For a more practical mass-to-charge ratio range of 1 000, the highest performance is 47, 35, and 27 Hz for the respective scan rates (Figure 2A).
Figure 2.
Determining the optimal maximum ion injection time in context of ion trap scan rate and mass-to-charge ratio range. (A) MS/MS acquisition rates were calculated at specific mass-to-charge ratio scan widths at various ion trap scan rates (i.e., turbo, rapid, and normal). (B) The optimal maximum ion injection was determined by monitoring the MS/MS acquisition rate. For a 1 000 m/z scan range, the optimal maximum injection times were 13, 20, and 36 ms for turbo, rapid, and normal ion trap scan rates, respectively. (C) We expanded this analysis to mass-to-charge ratio scan widths between 200 m/z and 1900 m/z.
We next determined the ideal maximum ion injection time, i.e., to balance the best signal while maintaining parallelization, in the context of ion trap scan rate and mass-to-charge ratio range. For each scan width, hereafter defined as the number of mass-to-charge ratio units scanned without regard to start and stop point and scan rate tested, we determined the greatest maximum ion injection time that could be set while still maintaining full parallelization. Accordingly, we monitored the MS/MS acquisition rates while iteratively increasing the maximum ion injection times (1 ms steps) for a scan width of 1 000 m/z (Figure 2B). When the ion accumulation time exceeds the MS/MS scan duration and associated overhead, the MS/MS acquisition rate drops. For example, at the highest scan rate, MS/MS scans are collected at a rate of 46 Hz when the maximum ion injection time of 13 ms or less; however, at longer ion injection times the parallelization breaks resulting in a reduced MS/MS acquisition rate. We expanded this analysis for scan widths from 200 m/z to 1 900 m/z at all three scan rates (Figure 2C). These data now provide a framework to expedite selection of maximum injection time for any desired ion trap scan rate and width.
Benefits of Optimized Parameters.
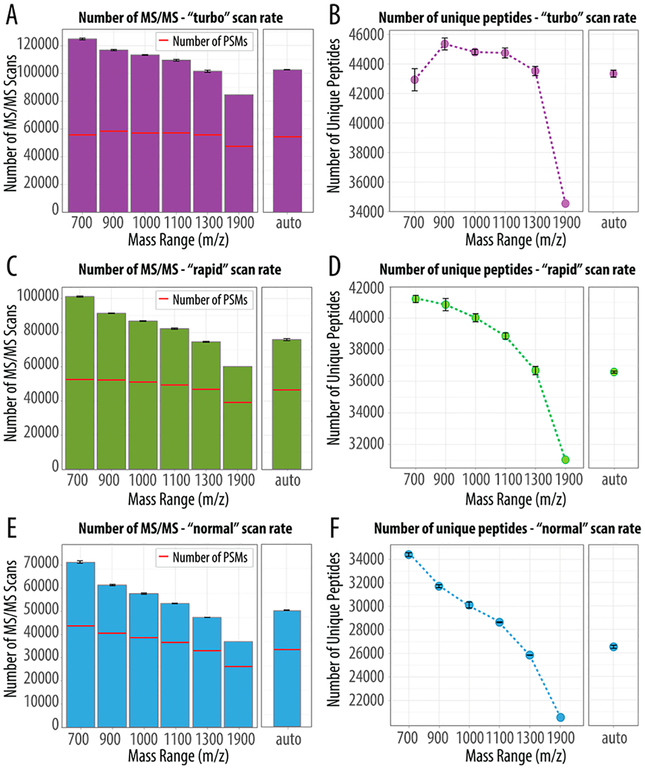
Next, we sought to determine whether these optimized parameters could be leveraged to improve shotgun proteome performance. We prepared a complex mixture of tryptic peptides by extracting and digesting proteins from a cultured human cell line. These peptides were loaded onto a capillary LC column and gradient eluted into the MS over 60 min using the optimized maximum ion injection times for selected scan ranges and speeds (Table 1). The specific scan ranges tested were 200–900, 175–1075, 125–1125, 125–1225, and 125–1425 m/z. The select scan ranges were determined as previously described in the truncation experiment, taking into consideration “normal”, “rapid”, and “turbo” ion trap scan rates (Figure S1). Further we employed an additional range termed “auto”, where the MS automatically determines the mass-to-charge ratio range. Note this function is an existing default option on this commercial instrument system that uses precursor mass to calculate a mass-to-charge ratio range.
Figure 3 presents a comparison of the total number of MS/MS scans collected, detected peptide spectral matches (PSMs), and the resulting unique identified peptides for each experiment. We evaluated our static mass-to-charge ratio ranges against the “auto” setting and observed that regardless of scan rate we always collected more MS/MS spectra for methods with scan ranges less than 1 100 m/z. Further, these additional scans translated into more PSMs and therefore more identified peptides, except for the 700 m/z range at the highest scan rate (Figure 3A,B). The greatest number of identified peptides was achieved using the 900 m/z range collected at the fastest scan rate with an identification rate of 50% (Figure 3B). As compared to the “auto” setting, this approach resulted in 12% more MS/MS scans and identification of over 2 000 additional unique peptides. As the scan rate is reduced, the benefits of using a shorter static mass-to-charge ratio range are more prominent. For example, panels C and D of Figure 3 summarize the additional MS/MS spectra, PSMs, and peptides identified using the mass range 700 m/z with the rapid scan rate compared to the “auto”. At 700 m/z the identification rate increases to 52% and 61% at rapid and normal scan rates, respectively (Figure 3C,E). In summary, we observe a 15% increase in the number of acquired MS/MS spectra translating into an additional 4 000 unique peptide identifications. Similarly, the 700 m/z range at the slowest scan rate (normal) also resulted in the most substantial increases with 30% more MS/MS spectra acquired (Figure 3E) equating to identification of an additional 7 000 unique peptides (Figure 3F). Note, although the slower scan rates achieve higher identification rates, the fastest scan rates simply collect more MS/MS spectra and, in the end, outperform. Figure S3 depicts the overlap of PSMs for the various mass-to-charge ratio ranges as compared to the 1300 m/z range. These data demonstrate that for the different mass-to-charge ratio ranges there are slight variations in PSMs populations (Figure S3). For example, at the fastest scan rate, the boost of nearly 9 000 unique peptides was identified from the additional 18 000 PSMs detected at the range 1300 m/z versus the widest range, 1900 m/z. In general, as the number of unique peptides increases so does the total number of detected proteins. For instance, in the scan range of 700 m/z (turbo scan rate), we identified 42 930 unique peptide that correlated to 5 146 protein groups, whereas for the scan range of 1900 m/z we identified 34 548 unique peptides corresponding to 3 972 protein groups. That is, in each case we have an average of nine peptides per protein, yet the extra 12% of PSMs collected at 700 m/z scan result in more proteins observed. As such, regardless of scan rate, shotgun proteomics can benefit from short static ranges with optimized maximum injection times. Distribution of peptide confidence scores (i.e., expectation scores) suggest the MS/MS mass-to-charge ratio scan range can be reduced while providing confident peptide identifications (Figure S2). We conclude that limiting the MS/MS scan range shortens scan duration, allows for acquisition of more MS/MS scans, and ultimately results in an increased number of uniquely identified peptides.
Figure 3.
Combining a reduced mass-to-charge ratio scan range with the optimal ion injection time. To benchmark the performance of static mass-to-charge ratio ranges as compared to auto, we selected five optimal mass-to-charge ratio ranges and paired them with the max MS/MS ion injection time summarized in Table 1. Plotted are the number of MS/MS scans, PSMs, and the total number of unique peptides produced following a triplicate analysis of a complex peptide mixture (note the 1 900 m/z range has n = 1) analyzed using either “turbo” (A,B), “rapid” (C,D), or “normal” (E,F).
SUMMARY
In this work we have demonstrated that use of a narrow static mass-to-charge ratio scan range can increase the rate of MS/MS acquisition in an ion trap hybrid mass spectrometer. While mass-to-charge ratio range truncation does lead to data loss, this loss offset by increased parallelization and ultimately produces more peptide identifications as compared to the current dynamic scanning methods. Beyond offering a modest improvement in proteomic depth, these data highlight the complex interplay between multiple analyzers and ion processing that occurs in current hybrid systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National Institutes of Health Grants P41GM108538 (The National Center of Quantitative Biology of Complex Systems) and DE-SC0018409 (Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.9b02979.
Comparison of “normal”, “rapid”, and “turbo” ion trap scan rates with MS/MS spectral truncation; peptide expectation values for optimized parameters; PSMs detected at the selected optimal mass-to-charge ratio ranges; and additional materials and methods (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).De Hoffmann EJ Mass Spectrom. 1996, 31 (2), 129–137. [Google Scholar]
- (2).Domon B; Aebersold R Science 2006, 312 (5771), 212–217. [DOI] [PubMed] [Google Scholar]
- (3).Shevchenko A; Jensen ON; Podtelejnikov AV; Sagliocco F; Wilm M; Vorm O; Mortensen P; Shevchenko A; Boucherie H; Mann M Proc. Natl. Acad. Sci. U. S. A 1996, 93 (25), 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Richards AL; Merrill AE; Coon JJ Curr. Opin. Chem. Biol 2015, 24, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Coon JJ; Syka JEP; Shabanowitz J; Hunt DF BioTechniques 2005, 38 (4), 519–523. [DOI] [PubMed] [Google Scholar]
- (6).Pandey A; Mann M Nature 2000, 405, 837–846. [DOI] [PubMed] [Google Scholar]
- (7).Shishkova E; Hebert AS; Coon JJ Cell Syst. 2016, 3 (4), 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Link AJ; Eng J; Schieltz DM; Carmack E; Mize GJ; Morris DR; Garvik BM; Yates JR Nat. Biotechnol 1999, 17, 676–682. [DOI] [PubMed] [Google Scholar]
- (9).Syka JEP; Marto JA; Bai DL; Horning S; Senko MW; Schwartz JC; Ueberheide B; Garcia B; Busby S; Muratore T; et al. J. Proteome Res 2004, 3 (3), 621–626. [DOI] [PubMed] [Google Scholar]
- (10).Schwartz JC; Senko MW; Syka JE P. J. Am. Soc. Mass Spectrom 2002, 13, 659–669. [DOI] [PubMed] [Google Scholar]
- (11).Senko MW; Remes PM; Canterbury JD; Mathur R; Song Q; Eliuk SM; Mullen C; Earley L; Hardman M; Blethrow JD; et al. Anal. Chem 2013, 85 (24), 11710–11714. [DOI] [PubMed] [Google Scholar]
- (12).Hebert AS; Thöing C; Riley NM; Kwiecien NW; Shiskova E; Huguet R; Cardasis HL; Kuehn A; Eliuk S; Zabrouskov V; et al. Anal. Chem 2018, 90 (3), 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Riley NM; Hebert AS; Coon JJ Cell Syst. 2016, 2 (3), 142–143. [DOI] [PubMed] [Google Scholar]
- (14).Hebert AS; Richards AL; Bailey DJ; Ulbrich A; Coughlin EE; Westphall MS; Coon JJ Mol. Cell. Proteomics 2014, 13 (1), 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Makarov A; Denisov E; Kholomeev A; Balschun W; Lange O; Strupat K; Horning S Anal. Chem 2006, 78 (7), 2113–2120. [DOI] [PubMed] [Google Scholar]
- (16).Eliuk S; Makarov A Annu. Rev. Anal. Chem 2015, 8, 61. [DOI] [PubMed] [Google Scholar]
- (17).Olsen JV; Schwartz JC; Griep-Raming J; Nielsen ML; Damoc E; Denisov E; Lange O; Remes P; Taylor D; Splendore M; et al. Mol. Cell. Proteomics 2009, 8 (12), 2759–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Richards AL; Hebert AS; Ulbrich A; Bailey DJ; Coughlin EE; Westphall MS; Coon JJ Nat. Protoc 2015, 10 (5), 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shishkova E; Hebert AS; Westphall MS; Coon JJ Anal. Chem 2018, 90, 11503–11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wenger CD; Phanstiel DH; Lee MV; Bailey DJ; Coon JJ Proteomics 2011, 11 (6), 1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lau KW; Hart SR; Lynch JA; Wong SC; Hubbard SJ; Gaskell SJ Rapid Commun. Mass Spectrom 2009, 23, 1508–1514. [DOI] [PubMed] [Google Scholar]
- (22).Medzihradszky KF; Chalkley RJ Mass Spectrom. Rev 2015, 34, 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.