Abstract
Two major flaviviruses, dengue virus (DENV) and Zika virus (ZIKV), cause severe health and economic burdens worldwide. Recently, genome-wide screenings have uncovered the importance of the Hrd1 ubiquitin ligase-mediated endoplasmic reticulum (ER)-associated degradation (ERAD) pathway for flavivirus replication in host cells. Here we report the identification of the compound Bardoxolone methyl (CDDO-me) as a potent inhibitor of the Hrd1 ubiquitin ligase-mediated ERAD, which possesses a broad-spectrum antiviral activity against both DENV and ZIKV. Cellular thermal shift assay (CETSA) suggested that CDDO-me binds to grp94, a key component of the Hrd1 pathway, at a low nanomolar concentration, whereas interaction was not detected with its paralog Hsp90. CDDO-me and the grp94 inhibitor PU-WS13 substantially suppressed DENV2 replication and the cytopathic effects caused by DENV and ZIKV infection. The antiviral activities of both compounds were demonstrated for all four DENV serotypes and four ZIKV strains in multiple human cell lines. This study defines grp94 as a crucial host factor for flavivirus replication and identified CDDO-me as a potent small molecule inhibitor of flavivirus infection. Inhibition of grp94 may contribute to the antiviral activity of CDDO-me. Further investigation of grp94 inhibitors may lead to a new class of broad-spectrum anti-flaviviral medications.
Keywords: ERAD, CDDO-me, Dengue virus, Zika virus, grp94, Antiviral
Introduction
Dengue virus (DENV) and Zika virus (ZIKV) (family Flaviviridae, genus Flavivirus) are pathogens borne by Aedes mosquitoes and disseminated worldwide. There are approximately 390 million cases of human DENV infections each year and the infections cause a range of symptoms from mild fever to dengue hemorrhagic fever with a mortality rate of 2–5% (Bhatt et al., 2013; Morra et al., 2018; Schaffner and Mathis, 2014; Horstick et al., 2014). ZIKV infection, which typically is asymptomatic or only causes mild symptoms, became a global health emergency in recent years due to its unprecedented high prevalence in several regions and its association with severe neurological complications including catastrophic microcephaly in newborns and Guillain-Barre syndrome in adults (Solomon and Mallewa, 2001; Pyke et al., 2014; Tappe et al., 2014; Rothan et al., 2019).
Currently there is no approved vaccine for ZIKV. The recently approved vaccine for DENV has regional and age based restrictions due to the limitation of vaccine efficacy and the potential deadly side effects (Hueston et al., 2017; Castanha et al., 2017; Dejnirattisai et al., 2016; Rothan et al., 2018). Moreover, epidemics of ZIKV infections have mostly occurred in the DENV endemic areas and there is evidence to show co-infection of these two flaviviruses, which makes vaccine development even more complicated (Shan et al., 2018; Dejnirattisai et al., 2016; Rothan et al., 2018). There are currently also no approved antiviral drugs specific for treatment or prophylaxis of either DENV or ZIKV infection. Traditionally antivirals are developed to target viral pathogens directly and specifically. However, antivirals that target host cell components that are essential for viral infection or replication represent an alternative approach (Plummer et al., 2015; Barrows et al., 2016; Boldescu et al., 2017; Scaturro et al., 2018). Host factor-targeted antivirals would address not only two limitations associated with vaccines: 1) evasion of immunity caused by viral mutations (Schein et al., 2005) (Chiappelli et al., 2014; Maillard et al., 2014; Silveira et al., 2016; Chang et al., 2016; Sulczewski et al., 2018), and 2) DENV and ZIKV co-infection (Shan et al., 2018; Dejnirattisai et al., 2016; Rothan et al., 2018), as these flaviviruses exhibit the same lifecycle progression and require similar host factors (Gerold et al., 2017; Wang and Zhang, 2017; Puschnik et al., 2017). Thus, targeting the shared host factors would have a broad-spectrum of anti-flavivirus activity in co-infected patients (Boldescu et al., 2017).
Flaviviruses use the endoplasmic reticulum (ER) for viral proteins production and new virion assembly (Romero-Brey and Bartenschlager, 2016). Recently, genome-scale RNAi and CRISPR/Cas9 screenings have identified many host factors that are required for DENV, West Nile Virus (WNV), and ZIKV replication (Krishnan et al., 2008; Mairiang et al., 2013; Ma et al., 2015; Zhang et al., 2016; Marceau et al., 2016; Marceau et al., 2016; Boldescu et al., 2017; Scaturro et al., 2018), including proteins in the Hrd1 complex (Krishnan et al., 2008; Mairiang et al., 2013; Ma et al., 2015; Scaturro et al., 2018). The Hrd1 complex mediates a protein quality control mechanism in the ER by which misfolded proteins are dislocated from the ER lumen to the cytosol for degradation by the proteasome, a process known as ER-associated degradation or ERAD (Vembar and Brodsky, 2008). Although how the Hrd1 complex is involved in flavivirus replication is currently not understood, its essential role in flaviviral replication suggests that it is an attractive target for developing broad spectrum anti-flaviviral agents. In this study, we identified a small molecule CDDO-me that inhibits ER-to-cytosol protein dislocation and has broad-spectrum anti-flaviviral activities in vitro. Furthermore, we identified grp94 (Christianson et al., 2008), an ER luminal chaperone and a critical component of the Hrd1 ubiquitin ligase complex, as a potential new target for CDDO-me. CDDO-me and the grp94 inhibitor PU-WS13 (Patel et al., 2013) exhibited potent antiviral activities against both DENV and ZIKV replications at low nanomolar concentrations and protected human cells from viral cytopathic effects.
Materials and Methods
Viruses, cells, antibodies, and other materials
All viruses were obtained from Emerging Infections Research Resources Repository (BEI Resources, Manassas, VA, USA). Monkey plasmas neutralizing DENV or ZIKV were provided by Dr. Gregory Gromowski, Viral Diseases Branch, Walter Reed Army Institute of Research. Aedes albopictus mosquito C6/36 cells (ATCC CRL-1660), African green monkey kidney epithelial Vero cells (CCL-81), human embryonic kidney HEK-293 cells (CRL-1573), SH-SY5Y cells (CRL-2266), U-251 MG cells (HBT-17), and HeLa cells (CCL-2) were obtained from ATCC (Manassas, VA, USA). Huh-7 cells were provided by Dr. Hongbing Wang, University of Maryland School of Pharmacy. All cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) as growth medium or 2% FBS as maintenance medium. Bortezomib (BTZ), CDDO-me, and PU-WS13 were purchased from Sigma-Aldrich (St. Louis, MO, USA), Adooq Biosciences (Irvin, CA, USA), EMD Chemicals (San Diego, CA, USA), respectively. The sources for antibodies are: Anti-grp94 antibody (Affinity Bioreagents, Golden, CO, USA), anti-OS9 antibody (Thermo Fisher Scientific, Waltham, MA, USA), Anti-Hsp70 and anti-Tubulin antibodies (Enzo Life Sciences, Farmingdale, NY, USA), anti-Hsp90 and anti-BiP antibodies ( BD Biosciences, San Jose, CA, USA), Anti-DENV2 E protein and anti-DENV2 NS3 protein antibodies (GeneTex, San Antonio, TX, USA).
Live-cell imaging and quantification of drGFP
Live-cell imaging of protein dislocation was performed using the previously developed drGFP assay (Zhong and Fang, 2012). Briefly, drGFP HeLa cells reporting NHK dislocation (NHK-drGFP cells) were seeded in chamber slides (177402 Lab-Tak). After overnight culture, the cells were washed once with PBS and then treated with BTZ (1 µM) alone or BTZ (1 µM) together with CDDO-me (1 µM). Live cell images were acquired immediately after addition of the inhibitors and then every 30 min under a 40x objective lens mounted on a Nikon TRE fluorescence microscope equipped with a high-sensitivity CCD camera (QuantEM 512SC, Photometrics, Tucson, AZ), environment control units and an Autofocus module.
Cellular Thermal Shift Assay (CETSA)
This assay was performed as previously described (Martinez Molina et al., 2013; Martinez Molina and Nordlund, 2016). In brief, cells were trypsinized and washed with PBS. The cell lysates were diluted with appropriate buffer and divided into aliquots that were treated with CDDO-me (1 µM). After 30 min incubation at room temperature, the respective lysates were divided into smaller (50 µL) aliquots and heated individually at different temperatures for 3 min (Veriti Thermal Cycler, Applied Biosystems, Foster City, CA, USA) followed by cooling for 3 min at room temperature. The heated lysates were centrifuged at 20,000 × g for 20 min at 4°C to separate the soluble fractions from precipitates. The supernatants were transferred to new tubes and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting.
Immunoblotting analysis
A previously reported protocol was used (Zhong and Fang, 2012). Briefly, equal amounts of protein samples were separated by SDS-PAGE and transferred onto PVDF membrane. Non-specific binding sites were blocked with 5% milk for 1 hr at room temperature followed by incubation with the specific primary antibody. Blots were incubated with the HRP-conjugated secondary antibody (1:5000) for 1 hr. ECL Immunoblotting Substrate (Thermo Scientific Pierce) was used to detect membrane-bound antibodies. Immunoblots were imaged by Fluorchem M System (ProteinSimple, San Jose, CA, USA).
Virus infection and titration
Huh-7 cells were seeded at 3 × 105 cell/well in 12-well plates for 24 hrs using DMEM media containing 2% FBS. Then, the cells were infected with either DENV2 or ZIKV at various multiplicity of infection and treated with the test compounds. The culture supernatant was collected at 48 hrs post-infection, and plaque assay was used to measure virus titer as we previously described (Rothan et al., 2014; Rothan et al., 2016). In brief, a 10-fold serial dilution of culture supernatant collected from virus-infected cells was added to fresh Vero cells grown in 6-well plates (5 × 105 cells) and incubated for 1 hr at 37°C. The cells were overlaid with DMEM (maintenance medium) containing 0.5% agarose. Viral plaques were stained with crystal violet dye after a 5-day incubation. Virus titers were calculated according to the following formula: Titer (pfu/ml) = number of plaques/volume of the diluted virus added to the well × dilution factor of the virus used to infect the well in which the plaques were counted.
Immunostaining
Huh-7 cells were seeded on a glass coverslip in 12-well plate and infected with DENV2 at a multiplicity of infection (MOI) of 0.1. Cells were then treated with 100 nM each compound and after 48 hrs fixed using 4% paraformaldehyde for 1 hr at 4°C. Viral particles were stained using anti-flavivirus group antigen-antibody (clone D1–4G2–4-15, EMD Millipore) at 1:1,000 in blocking buffer for 1 hr, washed three times and incubated with goat anti-mouse immunoglobulin G (IgG) Alexa 488 (Life Technologies) and DAPI (Insitus) for 30 min. After three washes with PBS, cells were visualized using confocal microscopy. Virus infectivity was analyzed by selecting five random fields of view for each condition. ImageJ software was used for fluorescence dye quantification, and the percentage of infected cells was calculated by dividing the number of Alexa-488-positive cells by the number of DAPI-positive cells.
Cell viability assay
Huh-7 cells (1 × 104 cells per well of a 96-well plate) were treated with increasing concentrations (25, 50, 100, 200 and 400 nM) of the compound for 72 hrs. The cytotoxic effects of the test compounds were determined based on MTT (3-(4,5-dimethylthiazol 2-yl)-2,5-diphenyltetrazolium bromide). The percent cell viability was calculated as follows: 100% - (Absorbance of treated cells / Absorbance of untreated cells) × 100%. For calculation of the adherent to apoptotic cell percentage, the apoptotic cells were collected from the supernatant, and the adherent cells were trypsinized, and both cells were counted using TC™ Automated Cell Counter (Bio-Rad, Portland, ME, USA).
Real-time cell proliferation assay
Effect of CDDO-me treatment or antibody neutralization on the proliferation of ZIKV-infected cells was assessed using IncuCyte Live-Cell Analysis System (Essen BioScience, Ann Arbor, MI). Vero cells were seeded in 96-well plates and allowed to grow to 10–30% confluence. To infect cells, growth media was removed and 250 pfu of virus was added to each assay well for a MOI of approximately 0.02. Virus and cells were incubated together for 1 hr before the removal of viral supernatant followed by the addition of maintenance media containing the respective treatments in duplicate. Proliferation was measured through quantitative kinetic processing metrics derived from images acquired from each well every 3 hours and presented as a percentage of culture confluence over time. The wells were imaged over 90 hours (DENV infected cells) or 110 hours (ZIKV infected cells).
Real-Time RT-PCR
Huh-7 cells were plated in 24-well plates (in triplicate for each condition), infected at a MOI of 0.1, and treated with 100 nM CDDO-me or PU-WS13 and 50 µg/ml Heparin as an inhibitor for virus entry and 10 µM of 2’CMA as a viral polymerase inhibitor. Viral RNA was harvested using the QIAamp viral RNA extraction mini kit (Qiagen, USA) at 48 post-infection (hpi). Real-time quantitative reverse transcription and PCR (qRT-PCR) was performed and analyzed on a CFX96™ Real Time System (Bio-Rad (Portland, ME, USA) using DENV2 forward primer 5’ACATCTCAAGTGCAGGCTGA3’, and reverse primer 5’GTCTCCGAATGGAGGTTCTG3. Viral RNA levels were normalized to GAPDH RNA levels.
Results
Bardoxolone methyl (CDDO-me) is an Hrd1-mediated protein dislocation inhibitor
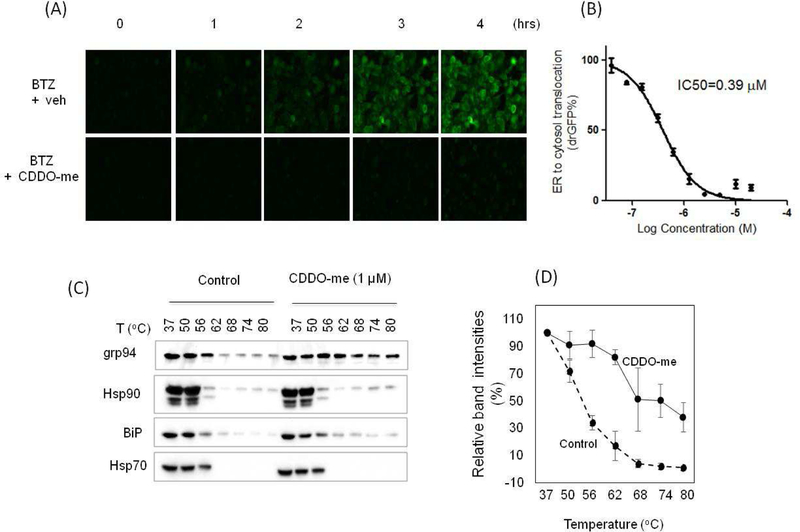
CDDO-me is a noncytotoxic and multifunctional synthetic triterpenoid that has potential applications for various diseases (Wang et al., 2014). Its multifunctionalities could be attributed to its ability to inhibit multiple targets in cells, including Keap1 (Wang et al., 2014; Wong et al., 2016), IKKβ (Ahmad et al., 2006), Arp2/3 (To et al., 2010), USP7 (Qin et al., 2016), Hsp90 (Qin et al., 2015), and Lon protease (Bernstein et al., 2012). In addition, CDDO-me also induces ER stress in cells (Zou et al., 2008; Jeong et al., 2015), indicating a buildup of misfolded proteins in the ER, but the underlying mechanism is not known. To address this question, we used the previously established drGFP assay (Zhong and Fang, 2012) to determine the effects of CDDO-me on ER-to-cytosol protein dislocation in live cells. A time-lapse imaging of mutant α−1-antitrypsin protein (NHK) dislocation in HeLa cells revealed that treatment with the proteasome inhibitor Bortezomib (BTZ) to block the degradation of dislocated NHK caused a time-dependent increase in GFP fluorescence (drGFP), an indicator of NHK dislocation (Fig. 1A, upper panel). The increase in NHK dislocation was significantly diminished when CDDO-me was applied along with BTZ (Fig. 1A, lower panel). A dose response experiment showed that CDDO-me inhibited NHK dislocation with an IC50 of 0.39 µM (Fig. 1B). CDDO-me did not affect the expression of S11-NHK and S1–10 (Fig. S1), suggesting that the CDDO-me-induced decreases in drGFP is due to its inhibition of NHK dislocation.
Figure 1. CDDO-me is a dislocation inhibitor that may target grp94.
(A) Time-lapse imaging of mutant α−1-antitrypsin protein (NHK) dislocation by drGFP in HeLa cells revealed that CDDO-me is an inhibitor of protein dislocation. Cells were treated with 1 µM bortezomib (BTZ) + 1 µM CDDO-me or DMSO (veh) and dislocation was monitored over 4 hrs (B) CDDO-me inhibited ER to cytosol dislocation of NHK at an IC50 of 0.39 µM. (C) Examining CDDO-me target engagement by CETSA. (D) Densitometric analysis of grp94 band intensities in CETSA were quantified and expressed as mean +/− SEM, n=3.
grp94 is a potential molecular target for CDDO-me
As a dislocation inhibitor, we expect that CDDO-me targets one of the key regulators in the dislocation pathway. We, therefore, used the cellular thermal shift assay (CETSA) to identify its potential target(s). CETSA is a well-established assay for detecting interaction between a small molecule and protein, based on the biophysical principle of ligand-induced thermal stabilization of a target protein (Martinez Molina et al., 2013; Martinez Molina and Nordlund, 2016). Ligand-stabilized protein target can be detected in soluble cellular fraction by immunoblotting or mass spectrometry (Martinez Molina et al., 2013; Martinez Molina and Nordlund, 2016). Using CETSA, we found that CDDO-me increased the thermal stability of grp94 compared to the vehicle control (Fig. 1C and 1D) while other related chaperones, including Hsp90, Hsp70, and BiP/grp78, showed little to no changes under the same condition as revealed by immunoblotting (Fig. 1C). Previous studies have shown that knockdown of grp94 expression inhibited degradation of NHK, indicating an important role of grp94 in the ERAD pathway (Christianson et al., 2008). Thus, CDDO-me may target grp94 to inhibit dislocation.
grp94 is essential for virus replication
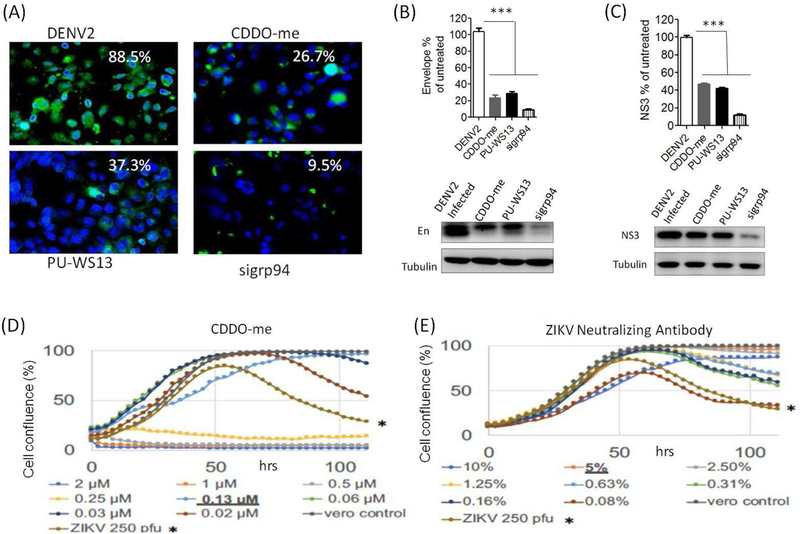
Several proteins in the Hrd1 pathway are required for Dengue and/or Zika replication (Krishnan et al., 2008), but it is not known whether grp94 is required. Therefore, we determined the effects of grp94 knockdown on DENV2 replication in Huh-7 cells. The results showed that grp94 knockdown markedly inhibited DENV2 replication as indicated by decreased viral particle production and viral protein (Envelope and NS3) synthesis (Fig. 2A–C). Interestingly, CDDO-me treatment almost recapitulated the antiviral results by grp94 knockdown (Fig. 2A–C), which support the possibility that CDDO-me is an inhibitor of grp94. However, CDDO-me is known to have multiple molecular targets in cells, including Keap1 (Wang et al., 2014; Wong et al., 2016), IKKβ (Ahmad et al., 2006), Arp2/3 (To et al., 2010), USP7 (Qin et al., 2016), Hsp90 (Qin et al., 2015), and Lon protease (Bernstein et al., 2012). Therefore, CDDO-me can simultaneously affect different cellular functions and may inhibit DENV replication by multiple mechanisms. Indeed, an additive effect of CDDO-me treatment and siRNA knockdown of grp94 or its binding partner OS9 on the decrease in DENV E protein production (Fig. S2), suggesting that mechanisms in addition to grp94 inhibition were involved in the antiviral activity of CDDO-me.
Figure 2. Pharmacologic and genetic inhibition of grp94 reduced virus replication and protected cells from death.
Infection with DENV2 were performed at MOI of 0.1 and the cells were treated with 100 nM of the test compounds for 48 hrs. (A) grp94 knockdown, CDDO-me, and PU-SW13 reduce virus particle production in Huh-7 cells. (B and C) grp94 knockdown and treatment with CDDO-me or PU-WS13 reduced the expression levels of DENV2 envelope and NS3 proteins in Huh-7 cells. (D and E) ZIKV infected Vero cells were treated with different dilutions of CDDO-me or neutralizing antibody. Cell confluence was monitored over 110 hours. * denotes the cytopathic effects caused by infection with 250 pfu ZIKV (MOI of approximately 0.02). CDDO-me at 0.13 µM protected ZIKV-infected Vero cells with the effects comparable to that with 5% ZIKV-neutralizing antibody. Densitometric analysis of the band intensities were quantified and expressed as mean +/− SEM, n=3. One-Way ANOVA with Bonferroni’s post test. *** p<0.001.
To further determine the involvement of grp94 in viral replication, we assessed the effects of PU-WS13, a well characterized specific inhibitor of grp94 (Patel et al., 2013), on DENV2 replication. Indeed, treatment of DENV2-infected Huh-7 cells with PU-WS13 also reduced DENV2 particles and viral protein expression as seen in grp94 knockdown and CDDO-me treated cells (Fig. 2A–C). In addition, a real-time cell viability assay showed that CDDO-me protected Vero cells against ZIKV and DENV infection. The cell viability assay is a dye-free method that allows for drug-induced cytotoxicity and drug-induced changes in proliferation to be observed concurrently for a nuanced approach to drug toxicity screening. After the initial growth curve, virus-induced cell death due to viral infection is observed as a decrease in confluence while a maintenance of confluence is indicative of viral protection. The results demonstrated the antiviral effect with the CDDO-me treatment, in a dose-dependent manner comparable to that by virus neutralizing antibody treatment. CDDO-me protected Vero cells against ZIKV infection with efficacy that was comparable to 5% ZIKV-neutralizing plasma (Fig. 2D, E). CDDO-me showed a significant inhibitory effect on proliferation kinetics at higher drug concentrations (0.25 µM – 2 µM), but did not show a significant effect on proliferation at low concentrations. Protection against ZIKV infection was seen at low CDDO-me concentrations including 0.03 µM, 0.06 µM and 0.13 µM. Weak or partial protection was seen at 0.02 µM. Similar antiviral effect was also seen against DENV infection (Fig. S3).
PU-WS13 and CDDO-me exhibited a broad spectrum of anti-DENV and anti-ZIKV activity
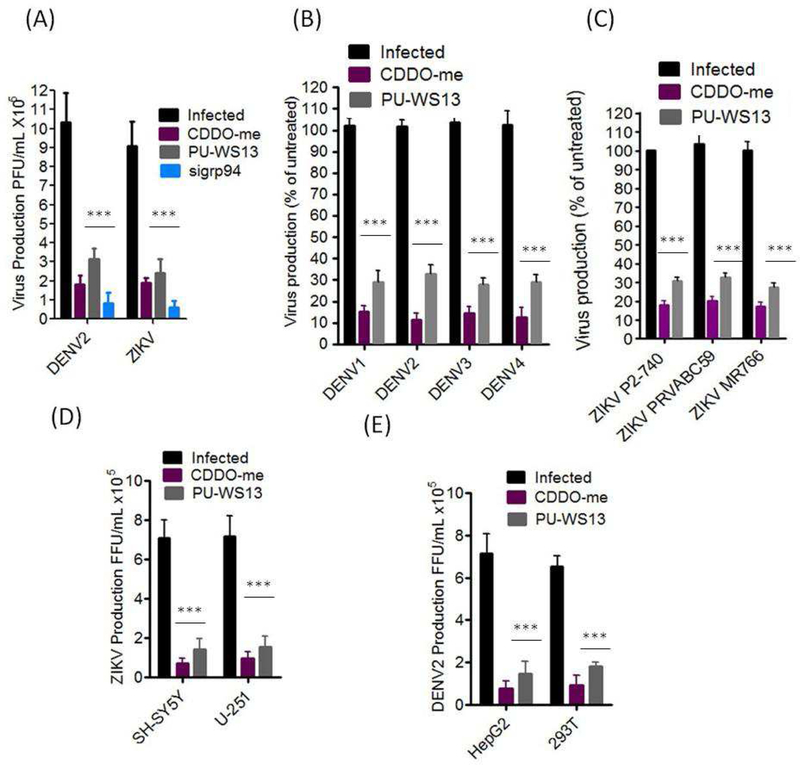
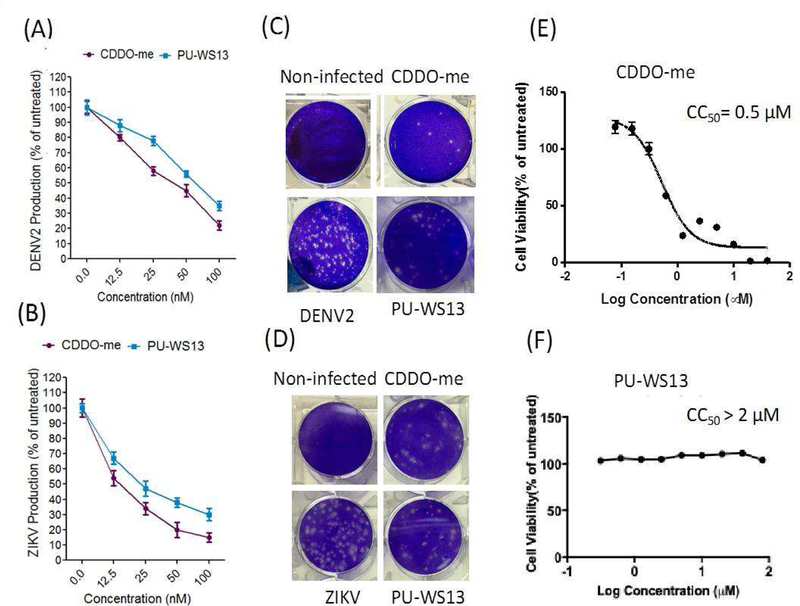
We showed that the knockdown of grp94 or treatment with either CDDO-me or PU-WS13 led to significant reduction in viral titers of DENV2 and ZIKV in Huh-7 cells (Fig. 3A). These anti-viral activities of CDDO-me and PU-SW13 were translated across multiple DENV serotypes and ZIKV strains (Fig. 3B, C), and also across several human cell lines (Fig. 3D, E). These findings indicate that grp94 inhibitors have a broad spectrum of anti-DENV and ZIKV activity and may be considered as drug candidates to combat DENV and ZIKV infections and co-infections. Next, we determined the potencies of the antiviral activities of CDDO-me and PU-WS13 (Fig. 4). We infected Huh-7 cells with DENV or ZIKV and treated the cells with increasing concentrations of the compounds. Viral replication was measured 48 hours post-infection (hpi) (Fig. 4A, B). A significant reduction in viral replication was observed at 100 nM, and the half-maximal effective concentrations (EC50) were 17 nM for CDDO-me and 27 nM for PU-WS13 against DENV infection (Fig. 4A) and 15 nM for CDDO-me and 25 nM for PU-WS13 against ZIKV inhibition, respectively (Fig. 4B). Furthermore, we observed a significant reduction in viral plaque formation of DENV- or ZIKV-infected Huh-7 cells (Fig. 4C, D).
Figure 3. Broad-spectrum antiviral activity of PU-WS13 and CDDO-me against different DENV serotypes and ZIKV strains.
(A) Knockdown of grp94 or treatment with either CDDO-me or PU-SW13 led to a significant reduction in viral titers of DENV2 and ZIKV in Huh-7 cells. (B and C) grp94 inhibitors significantly reduced the virus titers of different DENV serotypes and ZIKV strains. (D and E) CDDO-me and PU-WS13 inhibited ZIKV and DENV2 replication in different permissive cell lines. All virus infections were performed at MOI of 0.1 and the cells were treated with 100 nM of the indicated compound for 48 hrs. One-Way ANOVA with Bonferroni’s post test. *** p<0.001.
Figure 4. PU-WS13 and CDDO-me showed dose-dependent antiviral activity and inhibited DENV2 and ZIKV in Huh-7 cells.
(A and B) Huh-7 cells were infected with DENV or ZIKV (MOI, 0.1) and treated cells with increasing concentrations of the indicated compounds and viral replication was measured 48 hrs post-infection. The percentages of DENV2 and ZIKV production were remarkably reduced with increasing concentrations of the test compounds (C and D) The test compounds at 100 nM exhibited remarkable reduction in virus plaque formation at 48 hrs post-infection. (E and F) Huh-7 cell viabilities were measured 72 hrs after CDDO-me or PU-WS13 treatment.
Because CDDO-me leads to cell-cycle arrest, reduces proliferation, and apoptosis in a wide variety of human cancer cells (Chintharlapalli et al., 2005; Konopleva et al., 2002; Gao et al., 2007; Samudio et al., 2008), we determined the effects of increasing concentrations of CDDO-me and PU-WS13 on Huh-7 proliferation. The half-maximum cytotoxic concentration (CC50) in Huh-7 cells was 0.5 µM for CDDO-me and more than 2 µM for PU-WS13 (Fig. 2D, E), indicating that the antiviral effects could not be explained by reduced proliferation. The therapeutic indices (CC50/EC50) for CDDO-me and PU-WS13 in Huh-7 cells were ~29.4 and 80, respectively. This finding suggests that the antiviral activities of the grp94 inhibitors is due to inhibition of DENV and ZIKV replication and not due to a general cytotoxic effects.
PU-WS13 and CDDO-me inhibited virus replication at a late stage of the virus life cycle
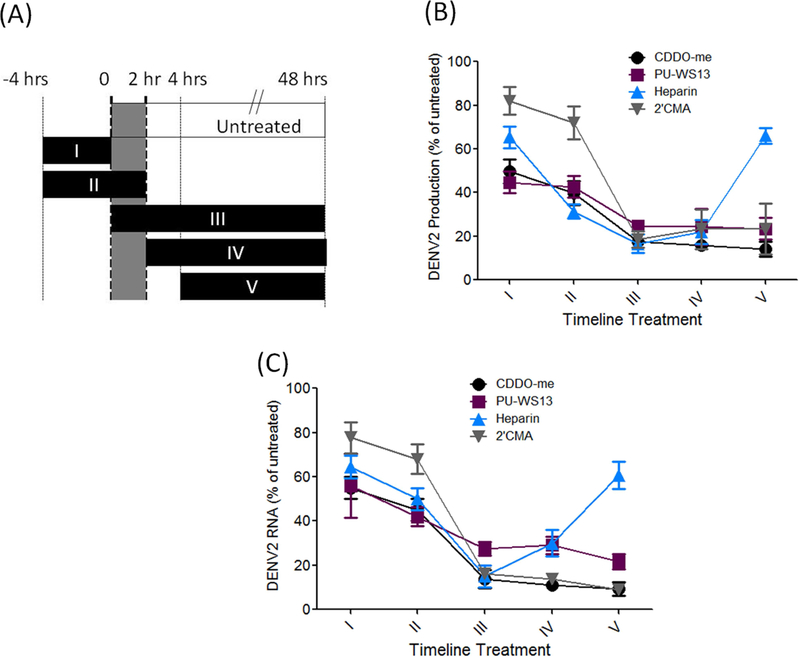
To dissect which steps in the viral life-cycle require grp94 in human cells, we performed a time-of-addition experiment using CDDO-me, PU-WS13, the NS5 polymerase inhibitor 2’C-methyladenosine (2’CMA,) and the entry inhibitor heparin (Fig. 5A). The activity of grp94 inhibitors on DENV replication was comparable to 2’CMA. The grp94 inhibitors and 2’CMA suppressed viral production (Fig. 5B) and RNA synthesis (Fig. 5C) in human cells. As expected, heparin only blocked viral production and viral RNA replication when added before or concurrently with DENV infection. In contrast, 2’CMA was effective only when added concurrently with or after infection. Both CDDO-me and PU-WS13 considerably inhibited DENV production and viral RNA replication at the last three treatment regimes, suggesting grp94 is required for post-entry steps of the viral life cycle (Fig. 5B, C), which is inhibited by CDDO-me and PU-WS13.
Figure 5. PU-WS13 and CDDO-me interfered with the late stage of virus life cycle.
Huh-7 cells were plated in 24-well plates (in triplicate for each condition), infected at a MOI of 0.1, and treated with 100 nM PU-WS13 and 50 µg/ml Heparin as an inhibitor for virus entry and 10 µM 2’CMA. (A) Experimental timeline to assess viral stage at which compounds have effects. 4 hrs before infection, 0–2 hrs during infection, 4 hrs post infection and 48 hrs post infection. (B) PU-WS13 or CDDO-me was applied to determine their effects on the virus life cycle. The compounds inhibited DENV production measured by plaque formation assay at post-entry stages, similar to NS5 polymerase inhibitor, 2’C-methyladenosine (2’CMA). (C) Viral RNA levels measured by qRT-PCR were remarkably decreased after applying the test compounds at the post-entry stages of virus life cycle.
Discussion
In this study, we identified CDDO-me as novel dislocation inhibitor with a broad-spectrum anti-DENV and anti-ZIKV activity. CDDO-me may exert these effects by targeting the ER luminal chaperone grp94, which is strongly supported by the fact that PU-WS13, a characterized grp94 inhibitor that is structurally unrelated to CDDO-me, phenocopied the antiviral activities of CDDO-me. The present study also identified grp94 as a novel host factor for DENV and ZIKV, suggesting that small molecule inhibitors of grp94 may be developed as broad-spectrum anti-DENV and anti-ZIKV drugs. While ER dislocation is a critical cellular function for both infected and non-infected cells, previous studies have shown that dislocation inhibitors can be tolerated in vivo. For CDDO-me, it entered clinical developments for the treatment of chronic kidney disease associated with type 2 diabetes but failed in phase III clinical trial due to heart-related adverse effects (Masullo et al., 2016). In contrast to the chronic kidney condition, DENV and ZIKV cause acute infections, and therefore do not require long-term treatment. Thus, CDDO-me has a great potential to be developed as an anti-DENV and anti-ZIKV drug since it has passed phase I and phase II trials (Masullo et al., 2016). In addition, inhibitors of three other key dislocation regulators, BiP, p97/VCP and Npl4, are under preclinical development as anti-cancer drugs all of which are well tolerated in mice (Anderson et al., 2015; Cerezo et al., 2016; Skrott et al., 2017). Therefore, inhibition of dislocation is a novel and promising strategy to develop broad-spectrum anti-flaviviral drugs.
Although several molecular targets have been reported for CDDO-me, including Hsp90 (Qin et al., 2015), a cytosolic counterpart of grp94, inhibition of grp94 is likely to contribute to the anti-viral effects of CDDO-me, since grp94 knockdown and treatment with PU-WS13, a grp94 inhibitor structurally unrelated to CDDO-me, both potently inhibited DENV and ZIKV replication. A mechanistic understanding of how grp94 regulates DENV and ZIKV replication is not available, however, our results indicate that grp94 is required for post-entry steps of the viral life cycle (Fig. 5). We speculate that grp94 may play a quality control role for viral proteins. Normally, the grp94 chaperone binds to OS9 lectin to deliver misfolded protein to the Hrd1 complex that in turn forwards misfolded protein to the proteasome for degradation (Christianson et al., 2008; Seidler et al., 2014). Therefore, CDDO-me may interrupt the function of grp94 that is necessary for misfolded viral protein degradation by ERAD. Accumulation of misfolded viral proteins would interfere with viral replication. In support for this possibility, previous studies have reported that flaviviruses exploit host cell protein machinery to translate its genomic RNA to viral proteins, fold viral proteins, and alter proteostasis (Valk et al., 1999; Perera and Kuhn, 2008; Mayer, 2005; Nagy and Pogany, 2011). The impact of grp94 on flavivirus replication could also be on the protein folding function of grp94. grp94 is not only an essential chaperone for ERAD, it also facilitates protein folding in the ER (Vembar and Brodsky, 2008; Marzec et al., 2012; Hebert and Molinari, 2007). Flavivirus replication requires host cell chaperones, including the newly identified grp94, for appropriate folding of viral multi-domain proteins (Valk et al., 1999; Perera and Kuhn, 2008; Mayer, 2005; Nagy and Pogany, 2011).
Previous studies have shown an essential role of grp94 in promoting replications of other viruses, such as retrovirus (Valk et al., 1999), herpes simplex type virus 1 (HSV-1) (Ramakrishnan et al., 1995), HCV (Lee et al., 2008) and HBV (Lim et al., 2005). The retention of secreted HCV E2 envelope proteins in the ER frequently induces grp94 expression to attenuate ER stress and inhibit cell apoptosis (Liberman et al., 1999). Knockdown of grp94 during HCV infection inhibits cell apoptosis which in turn promotes virus replication (Mayer, 2005). Interestingly, the data presented in this study revealed that in the context of DENV2 and ZIKV infection, silencing of grp94 has the opposite effect, and inhibits replication. CDDO-me and a previously identified grp94 inhibitor, PU-WS13, both showed significant anti-DENV2 and anti-ZIKV activity.
In conclusion, our study for the first time identified the importance of grp94 in flavivirus replication. We have demonstrated that PU-WS13 and CDDO-me have a broad spectrum of anti-flavivirus activity, and inhibition of grp94 may contribute to the antiviral activity of CDDO-me. However, CDDO-me has multiple molecular targets, it is entirely possible that CDDO-me inhibits DENV and ZIKV replication via targeting multiple proteins in addition to grp94. Further studies are required to define the mechanism of grp94 action in DENV and ZIKV replication and test the anti-DENV and ZIKV activity of grp94 inhibitors in animal models.
Supplementary Material
Highlights.
CDDO-me inhibits ER-to-cytosol protein dislocation potentially by targeting the ER luminal chaperone grp94.
grp94 is a novel host factor required for DENV and ZIKV replication.
The specific grp94 inhibitor PU-WS13 and CDDO-me exhibit a broad-spectrum anti-DENV and anti-ZIKV activity in vitro.
PU-WS13 and CDDO-me significantly suppressed the cytopathic effects caused by DENV and ZIKV infection.
Acknowledgements
This work was primarily supported by the National Institutes of Health [UO1GM117175_2018 to S. F.], and supported in part by the Intramural Research Program of the NIH, National Center for Advancing Translational Sciences, US Military Infectious Diseases Research Program (MIDRP) [S0622_19_WR to J.H.], and Ministry of Higher Education of Malaysia [FP007–2017A to R. Y.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no conflict of interest.
Publisher's Disclaimer: Disclaimer: The views expressed here are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or U.S. Government. This is the work of U.S. government employees and may not be copyrighted (17 USC 105).
References
- Ahmad R, Raina D, Meyer C, Kharbanda S, and Kufe D (2006). Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem 281, 35764–35769. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, and Rolfe M (2015). Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Muñoz G, McGrath EL, Urrabaz-Garza R, Gao J, Wu P, Menon R, Saade G, Fernandez-Salas I, Rossi SL, Vasilakis N, Routh A, Bradrick SS, and Garcia-Blanco MA (2016). A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 20, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein SH, Venkatesh S, Li M, Lee J, Lu B, Hilchey SP, Morse KM, Metcalfe HM, Skalska J, Andreeff M, Brookes PS, and Suzuki CK (2012). The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood 119, 3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, and Hay SI (2013). The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldescu V, Behnam MAM, Vasilakis N, and Klein CD (2017). Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov 16, 565–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanha PMS, Nascimento EJM, Braga C, Cordeiro MT, de Carvalho OV, de Mendonça LR, Azevedo EAN, França RFO, Dhalia R, and Marques ETA (2017). Dengue Virus-Specific Antibodies Enhance Brazilian Zika Virus Infection. J Infect Dis 215, 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M, Lehraiki A, Millet A, Rouaud F, Plaisant M, Jaune E, Botton T, Ronco C, Abbe P, Amdouni H, Passeron T, Hofman V, Mograbi B, Dabert-Gay AS, Debayle D, Alcor D, Rabhi N, Annicotte JS, Héliot L, Gonzalez-Pisfil M, Robert C, Moréra S, Virougoux A, Gual P, Ali MM, Bertolotto C, Hofman P, Ballotti R, Benhida R, and Rocchi S (2016). Compounds Triggering ER Stress Exert Anti-Melanoma Effects and Overcome BRAF Inhibitor Resistance. Cancer Cell 29, 805–819. [DOI] [PubMed] [Google Scholar]
- Chang DC, Hoang LT, Mohamed Naim AN, Dong H, Schreiber MJ, Hibberd ML, Tan MJA, and Shi PY (2016). Evasion of early innate immune response by 2’-O-methylation of dengue genomic RNA. Virology 499, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappelli F, Santos SM, Caldeira Brant XM, Bakhordarian A, Thames AD, Maida CA, Du AM, Jan AL, Nahcivan M, Nguyen MT, and Sama N (2014). Viral immune evasion in dengue: toward evidence-based revisions of clinical practice guidelines. Bioinformation 10, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I, and Safe S (2005). 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor gamma-dependent and -independent pathways. Mol Pharmacol 68, 119–128. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, and Kopito RR (2008). OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 10, 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, and Screaton GR (2016). Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol 17, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Deeb D, Jiang H, Liu Y, Dulchavsky SA, and Gautam SC (2007). Synthetic triterpenoids inhibit growth and induce apoptosis in human glioblastoma and neuroblastoma cells through inhibition of prosurvival Akt, NF-kappaB and Notch1 signaling. J Neurooncol 84, 147–157. [DOI] [PubMed] [Google Scholar]
- Gerold G, Bruening J, Weigel B, and Pietschmann T (2017). Protein Interactions during the Flavivirus and Hepacivirus Life Cycle. Mol Cell Proteomics 16, S75–S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, and Molinari M (2007). In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev 87, 1377–1408. [DOI] [PubMed] [Google Scholar]
- Horstick O, Jaenisch T, Martinez E, Kroeger A, See LLC, Farrar J, and Ranzinger SR (2014). Comparing the usefulness of the 1997 and 2009 WHO dengue case classification: a systematic literature review. Am J Trop Med Hyg 91, 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueston L, Ramirez R, and Mahalingam S (2017). Enhancement of Zika Infection by Dengue Virus-Specific Antibody Is Associated With Low Levels of Antiviral Factors. J Infect Dis 216, 612–614. [DOI] [PubMed] [Google Scholar]
- Jeong SA, Kim IY, Lee AR, Yoon MJ, Cho H, Lee JS, and Choi KS (2015). Ca2+ influx-mediated dilation of the endoplasmic reticulum and c-FLIPL downregulation trigger CDDO-Me-induced apoptosis in breast cancer cells. Oncotarget 6, 21173–21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Tsao T, Ruvolo P, Stiouf I, Estrov Z, Leysath CE, Zhao S, Harris D, Chang S, Jackson CE, Munsell M, Suh N, Gribble G, Honda T, May WS, Sporn MB, and Andreeff M (2002). Novel triterpenoid CDDO-Me is a potent inducer of apoptosis and differentiation in acute myelogenous leukemia. Blood 99, 326–335. [DOI] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, and Fikrig E (2008). RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Song R, Lee MN, Kim CS, Lee H, Kong YY, Kim H, and Jang SK (2008). A molecular chaperone glucose-regulated protein 94 blocks apoptosis induced by virus infection. Hepatology 47, 854–866. [DOI] [PubMed] [Google Scholar]
- Liberman E, Fong YL, Selby MJ, Choo QL, Cousens L, Houghton M, and Yen TS (1999). Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J Virol 73, 3718–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SO, Park SG, Yoo JH, Park YM, Kim HJ, Jang KT, Cho JW, Yoo BC, Jung GH, and Park CK (2005). Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules. World J Gastroenterol 11, 2072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, Manjunath N, and Wu H (2015). A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep 12, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard RA, Liu T, Beasley DW, Barrett AD, Hilser VJ, and Lee JC (2014). Thermodynamic mechanism for the evasion of antibody neutralization in flaviviruses. J Am Chem Soc 136, 10315–10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairiang D, Zhang H, Sodja A, Murali T, Suriyaphol P, Malasit P, Limjindaporn T, and Finley RL (2013). Identification of new protein interactions between dengue fever virus and its hosts, human and mosquito. PLoS One 8, e53535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, and Carette JE (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, and Nordlund P (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D, and Nordlund P (2016). The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In Situ Drug Target Engagement and Mechanistic Biomarker Studies. Annu Rev Pharmacol Toxicol 56, 141–161. [DOI] [PubMed] [Google Scholar]
- Marzec M, Eletto D, and Argon Y (2012). GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim Biophys Acta 1823, 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masullo M, Pizza C, and Piacente S (2016). Oleanane derivatives for pharmaceutical use: a patent review (2000–2016). Expert Opin Ther Pat [DOI] [PubMed] [Google Scholar]
- Mayer MP (2005). Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153, 1–46. [DOI] [PubMed] [Google Scholar]
- Morra ME, Altibi AMA, Iqtadar S, Minh LHN, Elawady SS, Hallab A, Elshafay A, Omer OA, Iraqi A, Adhikari P, Labib JH, Elhusseiny KM, Elgebaly A, Yacoub S, Huong LTM, Hirayama K, and Huy NT (2018). Definitions for warning signs and signs of severe dengue according to the WHO 2009 classification: Systematic review of literature. Rev Med Virol 28, e1979. [DOI] [PubMed] [Google Scholar]
- Nagy PD, and Pogany J (2011). The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PD, Yan P, Seidler PM, Patel HJ, Sun W, Yang C, Que NS, Taldone T, Finotti P, Stephani RA, Gewirth DT, and Chiosis G (2013). Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol 9, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R, and Kuhn RJ (2008). Structural proteomics of dengue virus. Curr Opin Microbiol 11, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer E, Buck MD, Sanchez M, Greenbaum JA, Turner J, Grewal R, Klose B, Sampath A, Warfield KL, Peters B, Ramstedt U, and Shresta S (2015). Dengue Virus Evolution under a Host-Targeted Antiviral. J Virol 89, 5592–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschnik AS, Majzoub K, Ooi YS, and Carette JE (2017). A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol 15, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke AT, Daly MT, Cameron JN, Moore PR, Taylor CT, Hewitson GR, Humphreys JL, and Gair R (2014). Imported zika virus infection from the cook islands into australia, 2014. PLoS Curr 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Wang W, Lei H, Luo H, Cai H, Tang C, Wu Y, Wang Y, Jin J, Xiao W, Wang T, Ma C, Xu H, Zhang J, Gao F, and Wu YL (2016). CDDO-Me reveals USP7 as a novel target in ovarian cancer cells. Oncotarget 7, 77096–77109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin DJ, Tang CX, Yang L, Lei H, Wei W, Wang YY, Ma CM, Gao FH, Xu HZ, and Wu YL (2015). Hsp90 Is a Novel Target Molecule of CDDO-Me in Inhibiting Proliferation of Ovarian Cancer Cells. PLoS One 10, e0132337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M, Tugizov S, Pereira L, and Lee AS (1995). Conformation-defective herpes simplex virus 1 glycoprotein B activates the promoter of the grp94 gene that codes for the 94-kD stress protein in the endoplasmic reticulum. DNA Cell Biol 14, 373–384. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I, and Bartenschlager R (2016). Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly. Viruses 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Bahrani H, Abdulrahman AY, Mohamed Z, Teoh TC, Othman S, Rashid NN, Rahman NA, and Yusof R (2016). Mefenamic acid in combination with ribavirin shows significant effects in reducing chikungunya virus infection in vitro and in vivo. Antiviral Res 127, 50–56. [DOI] [PubMed] [Google Scholar]
- Rothan HA, Bahrani H, Shankar EM, Rahman NA, and Yusof R (2014). Inhibitory effects of a peptide-fusion protein (Latarcin-PAP1-Thanatin) against chikungunya virus. Antiviral Res 108, 173–180. [DOI] [PubMed] [Google Scholar]
- Rothan HA, Bidokhti MRM, and Byrareddy SN (2018). Current concerns and perspectives on Zika virus co-infection with arboviruses and HIV. J Autoimmun 89, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Fang S, Mahesh M, and Byrareddy SN (2019). Zika Virus and the Metabolism of Neuronal Cells. Mol Neurobiol 56, 2551–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudio I, Kurinna S, Ruvolo P, Korchin B, Kantarjian H, Beran M, Dunner K, Kondo S, Andreeff M, and Konopleva M (2008). Inhibition of mitochondrial metabolism by methyl-2-cyano-3,12-dioxooleana-1,9-diene-28-oate induces apoptotic or autophagic cell death in chronic myeloid leukemia cells. Mol Cancer Ther 7, 1130–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, and Pichlmair A (2018). An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561, 253–257. [DOI] [PubMed] [Google Scholar]
- Schaffner F, and Mathis A (2014). Dengue and dengue vectors in the WHO European region: past, present, and scenarios for the future. Lancet Infect Dis 14, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Schein CH, Zhou B, and Braun W (2005). Stereophysicochemical variability plots highlight conserved antigenic areas in Flaviviruses. Virol J 2, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler PM, Shinsky SA, Hong F, Li Z, Cosgrove MS, and Gewirth DT (2014). Characterization of the Grp94/OS-9 chaperone-lectin complex. J Mol Biol 426, 3590–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Xie X, and Shi PY (2018). Zika Virus Vaccine: Progress and Challenges. Cell Host Microbe 24, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira GF, Strottmann DM, de Borba L, Mansur DS, Zanchin NI, Bordignon J, and dos Santos CN (2016). Single point mutations in the helicase domain of the NS3 protein enhance dengue virus replicative capacity in human monocyte-derived dendritic cells and circumvent the type I interferon response. Clin Exp Immunol 183, 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, Ozdian T, Bartkova J, Turi Z, Moudry P, Kraus M, Michalova M, Vaclavkova J, Dzubak P, Vrobel I, Pouckova P, Sedlacek J, Miklovicova A, Kutt A, Li J, Mattova J, Driessen C, Dou QP, Olsen J, Hajduch M, Cvek B, Deshaies RJ, and Bartek J (2017). Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T, and Mallewa M (2001). Dengue and other emerging flaviviruses. J Infect 42, 104–115. [DOI] [PubMed] [Google Scholar]
- Sulczewski FB, Liszbinski RB, Romão PRT, and Rodrigues Junior LC (2018). Nanoparticle vaccines against viral infections. Arch Virol 163, 2313–2325. [DOI] [PubMed] [Google Scholar]
- Tappe D, Rissland J, Gabriel M, Emmerich P, Gunther S, Held G, Smola S, and Schmidt-Chanasit J (2014). First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Euro Surveill 19, [DOI] [PubMed] [Google Scholar]
- To C, Shilton BH, and Di Guglielmo GM (2010). Synthetic triterpenoids target the Arp2/3 complex and inhibit branched actin polymerization. J Biol Chem 285, 27944–27957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk PJ, Vankan Y, Joosten M, Jenkins NA, Copeland NG, Löwenberg B, and Delwel R (1999). Retroviral insertions in Evi12, a novel common virus integration site upstream of Tra1/Grp94, frequently coincide with insertions in the gene encoding the peripheral cannabinoid receptor Cnr2. J Virol 73, 3595–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, and Brodsky JL (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9, 944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Zhang P (2017). Recent advances in the identification of the host factors involved in dengue virus replication. Virol Sin 32, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Yang YX, Zhe H, He ZX, and Zhou SF (2014). Bardoxolone methyl (CDDO-Me) as a therapeutic agent: an update on its pharmacokinetic and pharmacodynamic properties. Drug Des Devel Ther 8, 2075–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MH, Bryan HK, Copple IM, Jenkins RE, Chiu PH, Bibby J, Berry NG, Kitteringham NR, Goldring CE, O’Neill PM, and Park BK (2016). Design and Synthesis of Irreversible Analogues of Bardoxolone Methyl for the Identification of Pharmacologically Relevant Targets and Interaction Sites. J Med Chem 59, 2396–2409. [DOI] [PubMed] [Google Scholar]
- Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, and Diamond MS (2016). A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, and Fang S (2012). Live cell imaging of protein dislocation from the endoplasmic reticulum. J Biol Chem 287, 28057–28066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Yue P, Khuri FR, and Sun SY (2008). Coupling of endoplasmic reticulum stress to CDDO-Me-induced up-regulation of death receptor 5 via a CHOP-dependent mechanism involving JNK activation. Cancer Res 68, 7484–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.