Abstract
Members of the diazaquinomycin class of natural products have shown potent and selective activity against Mycobacterium tuberculosis. However, poor aqueous solubility has prevented extensive studies in animal models thus far. Our long-term goal is to harness knowledge regarding diazaquinomycin biosynthesis towards the generation of derivatives for structure-activity relationship studies. We have previously sequenced the genomes of two diazaquinomycin-producing, actinomycete bacteria and identified putative daq biosynthetic gene clusters. Here we report the heterologous expression of the daq gene cluster from the marine Streptomyces sp. F001 in S. coelicolor M1152. In addition to serving as functional proof for gene cluster assignment, the heterologous expression system reported here is expected to facilitate investigations aimed at elucidating diazaquinomycin biosynthesis.
Keywords: Tuberculosis, natural product, biosynthesis, antibiotic, Streptomyces
Introduction
Diazaquinomycins (DAQs) are diazaanthraquinone natural products produced by Actinobacteria of the genera Streptomyces and Micromonospora [6–10]. Their potent and selective antituberculosis activity [8] has attracted the attention of the scientific community, as exemplified by the recent report of optimized total synthesis routes [12,11]. The reported minimum inhibitory concentration (MIC) is in the 0.1 μM range which is on par with approved antituberculosis drugs. Moreover, potency was maintained against strains resistant to currently approved drugs. Most remarkably, DAQ A appears to be selective toward Mycobacterium tuberculosis, showing at least 40-fold increase in MIC against other gram-positive and gram-negative bacteria tested and even against other mycobacteria, the exception being Mycobacterium bovis [8]. However, DAQs suffer from poor aqueous solubility which has prevented translation of their in vitro activity to animal models.
Streptomyces and Micromonospora produce DAQ analogs with different alkyl substitutions at the side rings, suggesting that the respective Daq enzymes may have distinct substrate preferences [1]. Enzymes that catalyze biosynthesis of DAQ’s side rings could be exploited to introduce functional groups leading to increased solubility. We are interested in uncovering the molecular basis for the different analogs produced by Micromonospora and Streptomyces with the ultimate goal of generating structural derivatives via biosynthetic methods. Towards that end, we have recently sequenced the genomes of Lake Michigan-derived Micromonospora sp. B006 [2] and marine-derived Streptomyces sp. F001 and identified daq biosynthetic gene clusters in both strains [1]. We have also developed reverse genetics for Micromonospora sp. B006 [2]. However, genetic engineering of Streptomyces sp. F001 proved to be challenging. Here we report the heterologous expression of the daq gene cluster from Streptomyces sp. F001 (Fig. 1) in S. coelicolor M1152, not only setting the stage for in vivo functional investigations of Streptomyces daq genes, but also providing an alternative route for generating DAQ derivatives.
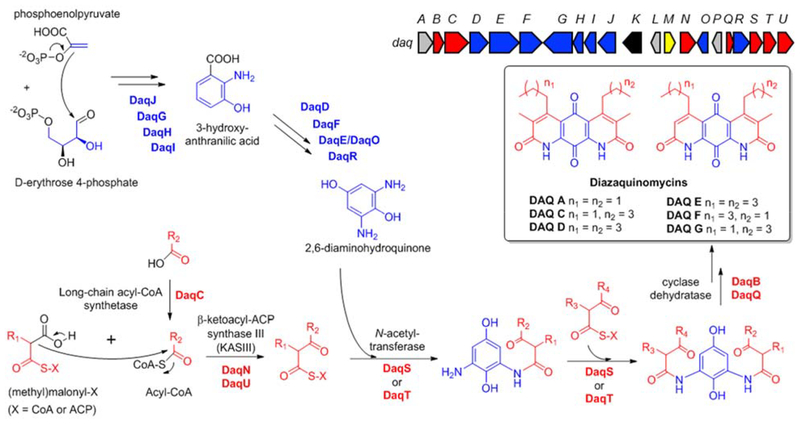
Fig. 1.
Diazaquinomycins produced by Streptomyces spp. (box), the daq biosynthetic gene cluster (block arrows), and diazaquinomycin biosynthesis hypothesis [1]. Genes are color-coded based on proposed function: biosynthesis of DAQ core (blue), biosynthesis of DAQ side rings (red), regulation (yellow), transposase (black), unknown (grey)
Material and Methods
General Experimental Procedures
All chemicals were acquired from Sigma-Aldrich, VWR, and Fisher Scientific. Solvents were of HPLC grade or higher. Restriction enzymes were purchased from New England Biolabs. Oligonucleotide primers were synthesized by Sigma-Aldrich. Molecular procedures were carried out according to the manufacturer’s instructions (New England Biolabs, Fisher Scientific, Qiagen, Sigma-Aldrich, Zymo Research) unless otherwise indicated.
Strains and Cultivation Conditions
Streptomyces sp. F001 was routinely cultivated on A1 medium (36.9 g instant ocean sea salt (Marineland®, 10 g starch, 4 g yeast extract, 2 g peptone, 1 g calcium carbonate, 100 mg potassium bromide, and 40 mg iron sulfate per liter deionized water, and 20 g agar per liter for solid medium) at 30 °C. To isolate genomic DNA, Streptomyces sp. F001 was cultivated in liquid A1 medium for seven days at 30 °C and 200 rpm. A defrosted frozen stock of the wild-type strain was used for inoculation. Frozen stocks were prepared with mycelium from three to five-day old TSB (3% tryptic soy broth) liquid cultures by adding glycerol to 20% [v/v] final concentration followed by storage at −80 °C.
Streptomyces coelicolor M1152 was routinely cultivated on mannitol soya flour (MS) agar medium (2% mannitol, 2% soya flour per 1 L tap water, and 20 g agar) [4]. For triparental conjugation, 10 mM MgCl2 (final concentration) was added to the MS agar medium. For genomic DNA isolation and the preparation of frozen stocks, the S. coelicolor M1152 exconjugants were grown in TSB broth for four days at 30 °C and 200 rpm. These TSB cultures were inoculated with a loopful of cell material from four-day old, pure cultures that had been grown on MS agar.
E. coli strains were cultivated in LB medium supplemented with the appropriate antibiotics. The following antibiotics were used as selection markers for E. coli and Streptomyces when appropriate: apramycin (final concentration: 50 μg/mL), kanamycin (50 μg/mL), chloramphenicol (25 μg/mL), and nalidixic acid (25 μg/mL).
Construction of plasmid pJB038EL
Primer pairs oJB207/oJB208 and oJB209/oJB210 (Table 1) were used to amplify the daq BGC from genomic DNA of Streptomyces sp. F001 (GenBank accession code for the genome sequence: QZWF00000000) via PCR. Genomic DNA was isolated from strain F001 using the Blood & Cell Culture DNA Midi Kit (Qiagen).
Table 1.
Oligonucleotide primers used in this study
| Name | Sequence (5′to 3′) |
|---|---|
| oJB207 | TTT GAC GCC TCC CAT GGTATA AATAGT GGC CTC GCT AAC CCC ATA GTC CAC C |
| oJB208 | CCT CCT TCG TAT GAG GTG ACA GC |
| oJB209 | CAC CTC AGC TGT CAC CTC ATA CG |
| oJB210 | G4G G4C GTT CCT TATATG TAG CTT TCG ACA ACC CGC AGC AAC TAC CAG С |
| oJB222 | GGT CCT CGGAAG GAC GCA TCA TGA AGG CTG CCT АСА TCG AGC |
| oJB223 | ACC GGG TCC GTC AAG TCG |
| oJB225 | GGT CCT CGGAAG GAC GCA TCA TGG TCA AAG TGG CCA TCA TCT TC |
| oJB226 | GTG TTC CGT GCC GTT TCC |
Overhangs are shown in italics. Note that the overhangs for primers oJB222 and oJB225 used to amplify daqA and daqP, respectively, are not necessary for the purposes of this study (these primers were designed for another experiment that required overhangs)
The PCR reactions (50 μL) to amplify the two ~12-kb fragments of the daq BGC (Fig. 2a) consisted of 0.2 mM of each dNTP, 1x Q5 High GC Enhancer, 0.5 μM of each primer, and 0.02 U μL−1 Q5 High-Fidelity DNA Polymerase (New England Biolabs) in Q5 reaction buffer supplied with the enzyme. The following thermal cycling conditions were used: 30 s at 98 °C; 35 cycles of 98 °C for 10 s, 70 °C for 20 s, and 72 °C for 375 s; and a terminal hold for 7 min at 72 °C. The vector pCAP03-acc(3)IV [13] was linearized with NdeI and XhoI. The 10.5-kb vector backbone was gel purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research). The two PCR-amplified, 12.3-kb daq BGC fragments (0.06 pmol; 240 ng) and the linearized pCAP03-acc(3)IV vector (0.03 pmol; 205 ng) were assembled using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs) to generate plasmid pJB038EL. The reaction was incubated for 1 h at 50 °C and then transferred into NEB 10-beta competent E. coli cells (New England Biolabs). Obtained clones were confirmed by restriction digest. Sanger sequencing of the obtained plasmid indicated that there was one PCR mutation at position 1,152 of gene daqD which turned out to be synonymous (CAC → CAT, both coding for histidine).
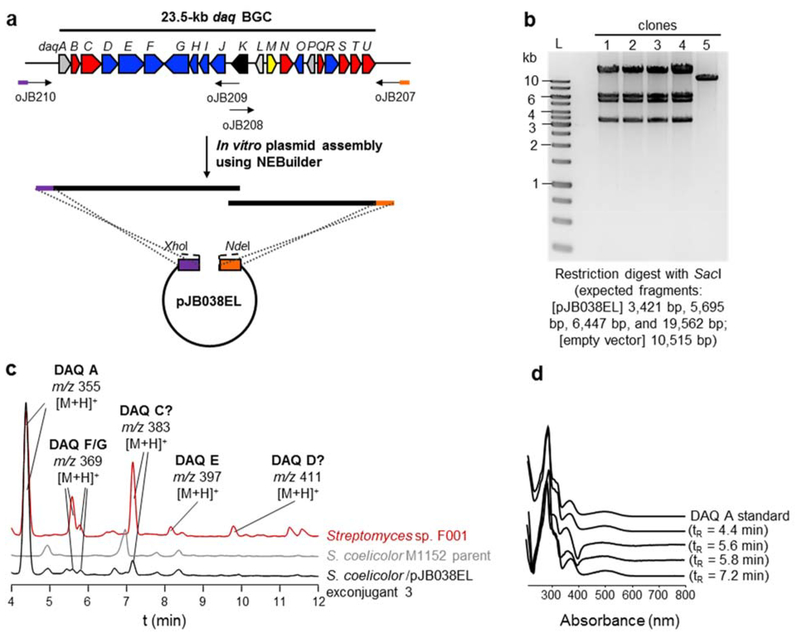
Fig. 2.
Cloning and heterologous expression of the daq biosynthetic gene cluster. (a) Schematic representation of the in vitro cloning strategy. The daq BGC was PCR-amplified in two fragments of ~12 kb and assembled in vitro using NEBuilder. (b) Restriction digest and gel electrophoresis of plasmids isolated from five different clones. Clones #1–4 were confirmed as positive, and the plasmid was named pJB038EL. L, DNA molecular ladder. (c) HPLC analysis of culture extracts with detection at 280 nm. The extract from the S. coelicolor / pJB038EL exconjugant is ten-fold more concentrated than the extract from the F001 wild-type strain. DAQ analogs produced are indicated. (d) Overlaid UV- Vis spectra of DAQ peaks shown in panel c and a DAQ A standard
Plasmid pJB038EL was transferred into S. coelicolor M1152 by triparental conjugation [4] from E. coli ET12567/pUB307 and E. coli ET12567. Exconjugants were streaked on MS solid medium containing kanamycin and nalidixic acid till pure cultures were obtained. Exconjugants were confirmed by PCR using primer pairs oJB222/oJB223 to amplify daqA and oJB225/226 to amplify daqP, respectively (Table 1). The 25 μL PCR reactions consisted of 0.2 mM of each dNTP, 1x Q5 High GC enhancer, 0.5 μM of each primer, and 0.02 U/μL Q5 High-Fidelity DNA Polymerase (New England Biolabs) in Q5 reaction buffer supplied with the enzyme. Thermocycling parameters were initial denaturation at 98 °C for 30 s; amplification: 30 cycles (98 °C for 10 s, 71 °C/68 °C for 20 s, 72 °C for 30 s); and terminal hold at 72 °C for 2 min.
Fermentation and Metabolite Analysis
For diazaquinomycin production, 50 mL TSB liquid medium was inoculated with 200 μL of a frozen stock of Streptomyces sp. F001, S. coelicolor M1152 parent strain and exconjugants, respectively. The seed cultures were incubated for three days at 30 °C and 200 rpm. Subsequently, 4 ml of these seed cultures were used to inoculate 100 ml A1 liquid medium with (for strain F001) and without instant ocean sea salt (for S. coelicolor strains) in 250 mL Erlenmeyer flasks (50 mL per flask). The cultures were incubated at 30 °C and 200 rpm. After seven days, 5% Diaion HP-20 resin (Alfa Aesar) was added to the production cultures. The HP-20 resin was washed prior to use by soaking in methanol and then rinsing thoroughly with deionized water. The production cultures were incubated for another 24 h at 30 °C and 200 rpm. Subsequently, the cultures were harvested by centrifugation. The supernatant was decanted, and the cell/resin pellet was extracted three times with 40 ml methanol for HPLC analysis. The extracts were dried in vacuo.
Subsequently, the extracts were dissolved in 4 mL MeOH:H2O (1:1), of which 1 mL was fractionated using Extract Clean C18 cartridges (Agela Technologies) with a MeOH:H2O gradient to produce two fractions (1 mL each). Fractions were eluted with the following mix of MeOH:H2O, 50:50, and 100:0. The fraction eluting at 100:0 was dried in vacuo and dissolved in 250 μL MeOH for HPLC and LC-MS analyses. The Streptomyces sp. F001 fraction was diluted with MeOH 1:10 before analysis.
HPLC analysis was performed on an Agilent 1260 Infinity system equipped with a Kinetex ® C18 column (150 × 4.6 mm, 5 μm particle size, 100 Å pore size, Phenomenex®). Solvent A was 0.1% [v/v] trifluoroacetic acid (TFA) in water, and solvent B was acetonitrile. The solvent gradient was: initial hold at 50% B for 2 min, linear gradient from 50% to 100% B within 10 min, and hold for 5 min, at a flow rate of 1 mL/min. The detection wavelength range was 200 – 600 nm; chromatograms were extracted at λ= 280 nm. A DAQ A calibration curve was generated by performing serial dilutions of DAQ A standard dissolved in methanol. The area under the curve (y axis) obtained during HPLC analysis was plotted against the amount in μg (x axis). The obtained equation (y = 4.6019x + 120.94, R2 = 0.9999) from five data points was used to calculate the theoretical titers of DAQ A.
Low resolution MS analyses were performed on a Finnigan LCQ Advantage MAX Mass spectrometer system (Thermo Electron Corporation) in positive mode and a Hewlett Packard series 1050 HPLC, equipped with a Kinetex® 5 μm C18 100 Å-column (150 × 4.6 nm, 5 μm particle size, Phenomenex®), at a flow rate of 1 mL/min. Solvent A was 0.02% [v/v] formic acid in water, and solvent B was 0.02% [v/v] formic acid in acetonitrile. The gradient was: initial hold at 50% B for 2 min, linear gradient from 50% to 100% B within 10 min, and held for 5 min. The detection wavelength range was 280 nm. The detection mass range was 200 Da to 2,000 Da (positive mode).
High resolution MS analyses were performed on a Bruker impact II quadrupole time-of-flight (Q-TOF) mass spectrometer (Thermo Electron Corporation) in positive mode and a Shimadzu Nexera X2 UHPLC, equipped with a Kinetex C18 column (50 × 2.1 nm, 1.7 μm particle size, 100 Å pore size, Phenomenex), using aqueous acetonitrile with 0.1% [v/v] formic acid at a flow rate of 0.5 mL/min. The linear gradient used was 50% to 100% acetonitrile for 5 min. The detection mass range was 50 Da to 1,500 Da.
Results and Discussion
Cloning of the daq biosynthetic gene cluster from marine Streptomyces sp. F001
Several in vivo and in vitro cloning methods for large DNA fragments such as complete BGCs have been described. A commonly used in vivo method is Transformation-Associated Recombination (TAR) cloning in yeast [5]. TAR cloning takes advantage of the inherent and efficient homologous recombination system of yeast. It is, however, a laborious method. Although we have used TAR in the past to clone a ~30-kb BGC [3], our first attempt to clone the smaller, 23.5-kb daq BGC using TAR failed. Therefore, we decided to test an in vitro method instead and chose NEBuilder due to its technical simplicity, speed, and commercial availability.
The complete daq BGC including putative promoters from the marine Streptomyces sp. F001 was cloned into pCAP03-acc(3)IV [13] using in vitro NEBuilder assembly (Fig. 2a). The reaction using 0.03 pmol of vector and a two-fold molar ratio of each daq fragment yielded 21 kanamycin-resistant colonies, five of which were analyzed by restriction digest and gel electrophoresis, leading to four positive clones (Fig. 2b). Our results demonstrate that NEBuilder in vitro assembly can be used to construct a plasmid of 35 kb total length, although the user manual states that the largest size previously assembled was 19 kb. Most importantly, by using in vitro assembly, positive clones can be obtained in less than one week. The obtained plasmid was named pJB038EL.
Heterologous expression of the daq BGC in S. coelicolor M1152
Plasmid pJB038EL was transferred into S. coelicolor M1152 by triparental conjugation. The vector contains a ϕC31 integrase and attachment site for site-specific, chromosomal integration. Although the conjugation efficiency (~5 × 1022129) using triparental conjugation was low, we obtained four exconjugants that were confirmed by PCR to contain the daq BGC (Fig. S1).
Comparative metabolite analysis confirms diazaquinomycin production
Two exconjugants, along with the S. coelicolor M1152 parent strain and the native producer Streptomyces sp. F001 were grown in A1 medium for comparative metabolite analysis. Methanol extracts were analyzed by HPLC (Figs. 2c–2d) and LC/MS (Figs. S2–S4). These analyses showed that Streptomyces sp. F001 produced DAQ A and E-G as previously reported [7], in addition to two DAQ analogs with m/z corresponding to DAQ C and D [6], previously reported from a soil Streptomyces sp. (Fig. S2). Whether the compounds produced by F001 are indeed DAQ C and DAQ D remains to be determined.
S. coelicolor M1152 exconjugants harboring the daq BGC produced the major congener found in F001, DAQ A, as evidenced by the UV spectra, molecular ion, and MS fragmentation pattern of the respective peaks and by comparison with an authentic standard (Figs. 2c–2d, S2–S4). The compound with m/z corresponding to DAQ C could also be detected by UV and MS (Figs. 2c–2d and S3–S4). The UV spectrum for the peaks corresponding to DAQ F and G (Figs. 2c–2d) matches that of DAQs (Figs. 2d, S2 and S3). We also detected the corresponding m/z for DAQ F/G (Fig. S4). The minor analog DAQ E could not be unequivocally detected in the heterologous expression system. Although the reason for the different distribution of DAQ analogs observed between the native producer and the heterologous host remains unclear, differential precursor supply may play a role. The theoretical titer of DAQ A calculated for F001 using a DAQ A standard calibration curve was 0.5 mg/L. While the DAQ A theoretical titer using the heterologous host (0.05–0.08 mg/L) was at least six-fold lower compared to the native producer, heterologous expression provides experimental evidence that the genes daqA-daqU are sufficient for DAQ biosynthesis.
Conclusions
Diazaquinomycins are promising antituberculosis natural products [8]. However, their poor aqueous solubility has prevented translation of their in vitro activity to in vivo systems. Streptomyces and Micromonospora strains produce DAQ analogs that bear different alkyl substitutions in the side rings [6–10], suggesting that the corresponding biosynthetic enzymes have distinct substrate preferences [1]. We are interested in elucidating DAQ biosynthesis with the ultimate goal of using biosynthetic methods to generate structural analogs that may address the solubility issue. In order to aid in the functional characterization of daq genes, we have previously developed reverse genetics for Micromonospora sp. B006 [1,2]. However, Streptomyces sp. F001 proved more difficult to engineer genetically. The heterologous expression system reported here sets the stage for investigating the function of daq genes from strain F001. Moreover, heterologous expression also provides experimental evidence that the identified daq cluster [1] is sufficient for DAQ biosynthesis. Finally, heterologous expression may facilitate the future generation of structural derivatives with improved properties.
Supplementary Material
Acknowledgments
We thank B. T. Murphy (UIC) for providing Streptomyces sp. F001 and the DAQ A standard used in this study. We also acknowledge Laura Sanchez and Guido Pauli (UIC) for providing access to LC/MS instrumentation. Financial support for this work was provided by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), under grant KL2TR002002 (to ASE), the Fogarty International Center and the National Institute of Allergy and Infectious Diseases under grant D43TW010530–02 (to TAT) and by startup funds from the Department of Medicinal Chemistry and Pharmacognosy and the Center for Biomolecular Sciences, University of Illinois at Chicago (to ASE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Braesel J, Lee J, Arnould B, Murphy BT, Eustáquio AS. (2019) Diazaquinomycin biosynthetic gene clusters from marine and freshwater actinomycetes. J Nat Prod 82:937–946. doi: 10.1021/acs.jnatprod.8b01028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braesel J, Crnkovic CM, Kunstman KJ, Green SJ, Maienschein-Cline M, Orjala J, Murphy BT, Eustáquio AS (2018) Complete genome of Micromonospora sp. strain B006 reveals biosynthetic potential of a Lake Michigan actinomycete. J Nat Prod 81:2057–2068. doi: 10.1021/acs.jnatprod.8b00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braesel J, Eustáquio AS (2019) Heterologous expression of a putative ClpC chaperone gene leads to induction of a host metabolite. J Brazil Chem Soc 30:499–508. doi: 10.21577/0103-5053.20180234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich, England [Google Scholar]
- 5.Kouprina N, Larionov V (2008) Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc 3:371–377. doi: 10.1038/nprot.2008.5 [DOI] [PubMed] [Google Scholar]
- 6.Maskey RP, Grun-Wollny I, Laatsch H (2005) Isolation and structure elucidation of diazaquinomycin C from a terrestrial Streptomyces sp. and confirmation of the akashin structure. Nat Prod Res 19:137–142. doi: 10.1080/14786410410001704741 [DOI] [PubMed] [Google Scholar]
- 7.Mullowney MW, Ó hAinmhire E, Shaikh A, Wei X, Tanouye U, Santarsiero BD, Burdette JE, Murphy BT (2014) Diazaquinomycins E-G, novel diaza-anthracene analogs from a marine-derived Streptomyces sp. Mar Drugs 12:3574–3586. doi: 10.3390/md12063574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullowney MW, Hwang CH, Newsome AG, Wei X, Tanouye U, Wan B, Carlson S, Barranis NJ, Ó hAinmhire E, Chen WL, Krishnamoorthy K, White J, Blair R, Lee H, Burdette JE, Rathod PK, Parish T, Cho S, Franzblau SG, Murphy BT (2015) Diaza-anthracene antibiotics from a freshwater-derived actinomycete with selective antibacterial activity toward Mycobacterium tuberculosis. ACS Infect Dis 1:168–174. doi: 10.1021/acsinfecdis.5b00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omura S, Iwai Y, Hinotozawa K, Tanaka H, Takahashi Y, Nakagawa A (1982) OM-704 A, a new antibiotic active against gram-positive bacteria produced by Streptomyces sp. J Antibiot 35:1425–1429. 10.7164/antibiotics.35.1425 [DOI] [PubMed] [Google Scholar]
- 10.Omura S, Nakagawa A, Aoyama H, Hinotozawa K, Sano H (1983) The structures of diazaquinomycin-A and diazaquinomycin-B new antibiotic metabolites. Tetrahedron Lett 24:3643–3646. doi: 10.1016/S0040-4039(00)88190-3 [DOI] [Google Scholar]
- 11.Prior AM, Sun DQ (2018) Scope and optimization of the double Knorr cyclization: synthesis of novel symmetrical and unsymmetrical tricyclic 1,8-diazaanthraquinones. Synthesis 50:859–871. doi: 10.1055/s-0036-1589134 [DOI] [Google Scholar]
- 12.Prior AM, Sun D (2019) Total synthesis of diazaquinomycins H and J using double Knorr cyclization in the presence of triisopropylsilane. RSC Adv 9:1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang X, Li J, Millan-Aguinaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM, Moore BS (2015) Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem Biol 10:2841–2849. doi: 10.1021/acschembio.5b00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.