Abstract
Gut cell losses contribute to overall feed efficiency due to the energy requirement for cell replenishment. Intestinal epithelial cells are sloughed into the intestinal lumen as digesta passes through the gastrointestinal tract, where cells are degraded by endonucleases. This leads to fragmented DNA being present in faeces, which may be an indicator of gut cell loss. Therefore, measuring host faecal DNA content could have potential as a non-invasive marker of gut cell loss and result in a novel technique for the assessment of how different feed ingredients impact upon gut health. Faecal calprotectin (CALP) is a marker of intestinal inflammation. This was a pilot study designed to test a methodology for extracting and quantifying DNA from pig faeces, and to assess whether any differences in host faecal DNA and CALP could be detected. An additional aim was to determine whether any differences in the above measures were related to the pig performance response to dietary yeast-enriched protein concentrate (YPC). Newly weaned (∼26.5 days of age) Large White × Landrace × Pietrain piglets (8.37 kg ±1.10, n = 180) were assigned to one of four treatment groups (nine replicates of five pigs), differing in dietary YPC content: 0% (control), 2.5%, 5% and 7.5% (w/w). Pooled faecal samples were collected on days 14 and 28 of the 36-day trial. Deoxyribonucleic acid was extracted and quantitative PCR was used to assess DNA composition. Pig genomic DNA was detected using primers specific for the pig cytochrome b (CYTB) gene, and bacterial DNA was detected using universal 16S primers. A pig CALP ELISA was used to assess gut inflammation. Dietary YPC significantly reduced feed conversion ratio (FCR) from weaning to day 14 (P<0.001), but not from day 14 to day 28 (P = 0.220). Pig faecal CYTB DNA content was significantly (P = 0.008) reduced in YPC-treated pigs, with no effect of time, whereas total faecal bacterial DNA content was unaffected by diet or time (P>0.05). Faecal CALP levels were significantly higher at day 14 compared with day 28, but there was no effect of YPC inclusion and no relationship with FCR. In conclusion, YPC reduced faecal CYTB DNA content and this correlated positively with FCR, but was unrelated to gut inflammation, suggesting that it could be a non-invasive marker of gut cell loss. However, further validation experiments by an independent method are required to verify the origin of pig faecal CYTB DNA as being from sloughed intestinal epithelial cells.
Keywords: gut, swine, faeces, sloughing, cytochrome b
Implications
The host DNA in faeces presumably originates from gut cell losses. This development of a non-invasive assessment method is potentially a novel indirect measurement of gut cell losses, although further validation experiments are required. A yeast-inclusion diet significantly reduced pig DNA in faeces and therefore may reduce gut cell losses. Turnover of gut cells contributes to feed efficiency; therefore, assessment of host DNA in faeces could potentially, following further methodology development, form the basis of a procedure to indirectly assess gut health and feed conversion ratio.
Introduction
Global food security is driving demand for increased feed efficiency in livestock production, resulting from an ever-growing human population (Gill, et al., 2010). One important component of feed efficiency that is often ignored is gut turnover, which relates to the continual proliferation, differentiation, movement and subsequent sloughing of epithelial cells lining the gastrointestinal tract. The rate at which this occurs can be very high (Bregendahl et al., 2004), requires both energy and nutrients for replenishment of cells (Baskin and Taegtmeyer, 2011), and therefore influences the feed efficiency. Consequently, accurate measurements of gut turnover are important when attempting to establish ways to improve feed efficiency. Currently all methods to measure gut turnover are invasive or expensive involving the use of stable isotope-labelled amino acids (Buttery, 1981) or deuterium-labelled water and expensive detection systems (Gasier et al., 2010). Hence a simple non-invasive method of measuring gut cell losses could potentially be extremely useful to understand growth efficiency and diet suitability.
Epithelial cells lining the small intestine are rapidly turned over and lost into the small intestine lumen where they are degraded by endonucleases, leading to DNA debris being present in faeces (Creamer, et al., 1961; Loretz, et al., 2006). Most literatures regarding DNA extraction from faeces in humans and animals relate to (i) determination of the microbial population in the gut (Ariefdjohan et al., 2010); (ii) use in human diagnostics of particular cancers (Roperch et al., 2015); (iii) species confirmation in wildlife research (Ramón-Laca et al., 2015); or (iv) use in forensics (Boonseub et al., 2012). In all situations, the extraction of sufficient DNA of a suitable quality is the primary concern since faecal material contains inhibitors, such as bile salts, complex polysaccharides and haemoglobin degradation products (McOrist et al., 2002), that can impair analysis methods (e.g. PCR). Previous studies have assessed human faecal DNA content as a marker of gut inflammation due to the expected increase in gut cell loss as a result of disturbance to the gut homeostasis (Vincent, et al., 2015); however, to the best of our knowledge, the methodology has not been assessed in production animals. Our overall hypothesis is that the concentration of animal DNA present in faeces will be directly proportional to the number of cells lost from the gut. Therefore, the aim of this study was to use DNA extraction and PCR methodology to detect animal (host) DNA in faecal samples as a potential novel, simple, non-invasive measure of gut cell losses. However, faecal DNA will also include DNA from a range of other sources including bacteria and feed ingredients, meaning a host species-specific gene target is required. Mitochondrial DNA gene targets have previously been used as host-specific markers in studies assessing faecal contamination of water sources (Martellini et al., 2005). Another advantage of using mitochondrial gene targets is the relatively high copy number in cells, including the sloughed epithelial cells present in faecal material (Caldwell, et al., 2011). DNA extracted from faeces is likely to be highly degraded, so choosing a high copy number gene should increase the likelihood of detection. Pig cytochrome b (CYTB) was therefore chosen as the gene for assessing host DNA content in pig faeces in the present study.
This was a pilot study designed to explore whether host DNA content in faeces could be extracted and quantified and whether host DNA content in faeces was related to animal performance measures that might be influenced by gut cell sloughing. Inclusion of yeast extract in livestock diets had previously been shown to affect gut morphology, by increasing gut maturation, and also significantly improved animal performance (Muthusamy et al., 2011). Therefore, inclusion of yeast-enriched protein concentrate (YPC) was used as a dietary treatment in this pilot study to test the hypothesis, since a significant reduction in gut cell sloughing was expected in YPC-treated animals, which should then be detected using the CYTB DNA assay.
Faecal calprotectin (CALP) levels have previously been used as a marker of gut inflammation in pigs (Lallès and Fagerhol, 2005). As previously mentioned, studies in humans have assessed human DNA content in faeces as a marker of bowel inflammation (Vincent, et al., 2015). Since gut turnover might be influenced by gut inflammation, we also assessed the CALP levels in pig faeces along with any relationships between faecal CALP, CYTB DNA and animal performance, particularly feed conversion ratio (FCR).
Material and methods
Animals
The feeding study was carried out at Harper Adams University and was approved by the internal ethical review committee (Approval number: 00583940-STAFF-1-201607). The study did not use any ‘regulated procedures’ and was therefore not carried out under Home Office Licence. Newly weaned Large White × Landrace × Pietrain piglets (8.37 ± 1.10 kg, n = 180) were assigned to one of four dietary treatment groups (n = 9 pens per treatment, 5 animals per pen). The piglets were in single sex pens, with both males and females used in the study, but balanced across the treatment groups. There were five pens of males and four pens of females on each treatment. The pens were 1.5 m² with plastic slatted floors, in line with the commercial Assured Food Standard requirement of 0.3 m² per animal up to 30 kg in body weight. The diets differed in YPC (AB Agri) content, as follows: 0% (control), 2.5%, 5% and 7.5% (w/w) (Table 1). Diets were formulated by AB Agri Limited. All diets were formulated to be equivalent in net energy and standard ileal digestible lysine, methionine, threonine, tryptophan and valine and were balanced using potato protein and wheat. Diet 1 was offered from weaning (∼26.5 days) to day 14 of the trial, and diet 2 was offered from day 14 to day 28 (Table 1). No therapeutic zinc oxide or dietary antibiotics were used. Animals were weighed at the beginning of the trial (at weaning), on day 14 and day 28. Feed intake was determined by offering a specified weight of feed, measuring the amount of feed weighed back at the end of each week and using these values to calculate feed eaten. FCR was calculated as feed intake divided by animal body weight gain and provided as an average for each experimental pen. A single fresh faecal sample was taken immediately after defecation from a minimum of three pigs from each pen at weighing and these samples were then pooled for the pen. The pooled sample was approximately 40 g in weight. Weighing of the pigs was typically carried out at midday. The YPC product is a maize-based protein concentrate with a high content (30%) of inactive yeast from the bioethanol production industry. Its nutritional value is shown in Table 2.
Table 1.
Composition of pig diets
| % Ingredient unless otherwise specified | Diet 1 (wean to day 14) | Diet 2 (day 14 to day 28) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control 0% YPC | 2.5% YPC | 5.0% YPC | 7.5% YPC | Control 0% YPC | 2.5% YPC | 5.0% YPC | 7.5% YPC | |
| Raw wheat | 38.3 | 37.4 | 35.6 | 33.9 | 53.8 | 52.0 | 49.8 | 47.6 |
| Micronised barley | 15.0 | 15.0 | 15.0 | 15.0 | 15.0 | 15.0 | 15.0 | 15.0 |
| Micronised oats | 5.0 | 5.0 | 5.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| HiPro soya | 10.0 | 10.0 | 10.0 | 10.0 | 15.0 | 15.0 | 15.0 | 15.0 |
| Soya oil | 5.1 | 5.3 | 5.5 | 5.7 | 1.1 | 1.3 | 1.6 | 1.8 |
| YPC | 0.0 | 2.5 | 5.0 | 7.5 | 0.0 | 2.5 | 5.0 | 7.5 |
| AlphaSoy 530 | 7.5 | 7.5 | 7.5 | 7.5 | 5 | 5 | 5 | 5 |
| Vitamin and mineral premix | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Whey powder | 10.4 | 10.4 | 10.4 | 10.4 | 3.5 | 3.5 | 3.5 | 3.5 |
| Potato protein | 4.0 | 2.0 | 1.0 | 0.0 | 2.0 | 1.0 | 0.5 | 0.0 |
| L-Lysine HCl | 0.57 | 0.66 | 0.68 | 0.70 | 0.59 | 0.61 | 0.60 | 0.59 |
| DL-Methionine | 0.27 | 0.29 | 0.28 | 0.28 | 0.26 | 0.26 | 0.25 | 0.24 |
| L-Threonine | 0.27 | 0.31 | 0.31 | 0.31 | 0.28 | 0.28 | 0.26 | 0.25 |
| L-Tryptophan | 0.06 | 0.07 | 0.07 | 0.08 | 0.05 | 0.05 | 0.05 | 0.05 |
| L-Valine | 0.15 | 0.20 | 0.21 | 0.22 | 0.14 | 0.15 | 0.14 | 0.13 |
| Limestone flour | 0.32 | 0.38 | 0.44 | 0.50 | 0.08 | 0.14 | 0.19 | 0.25 |
| Dicalcium phosphate | 1.87 | 1.86 | 1.84 | 1.82 | 2.04 | 2.02 | 2.00 | 1.98 |
| Salt | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | 0.03 | 0.04 | 0.04 |
| Sodium biocarbonate | 0.58 | 0.61 | 0.60 | 0.60 | 0.62 | 0.61 | 0.58 | 0.55 |
| Calculated nutritive values | ||||||||
| Protein | 19.7 | 19.5 | 19.9 | 20.2 | 19.3 | 19.7 | 20.3 | 20.9 |
| Digestible energy (MJ/kg) | 15.3 | 15.2 | 15.2 | 15.2 | 14.1 | 14.2 | 14.2 | 14.2 |
| Oil | 7.6 | 7.7 | 7.9 | 8.2 | 3.3 | 3.6 | 3.8 | 4.1 |
| Fibre | 2.2 | 2.3 | 2.4 | 2.5 | 2.4 | 2.5 | 2.6 | 2.7 |
| Calcium | 0.77 | 0.79 | 0.81 | 0.83 | 0.77 | 0.79 | 0.80 | 0.82 |
| Digestible phosphorus | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 |
| Lysine a | 1.35 | 1.35 | 1.35 | 1.35 | 1.28 | 1.28 | 1.28 | 1.28 |
| Methionine a | 0.54 | 0.54 | 0.54 | 0.54 | 0.51 | 0.51 | 0.51 | 0.51 |
| Methionine + Cysteine a | 0.81 | 0.81 | 0.81 | 0.82 | 0.78 | 0.79 | 0.79 | 0.80 |
| Threonine a | 0.90 | 0.90 | 0.90 | 0.90 | 0.86 | 0.86 | 0.86 | 0.86 |
| Isoleucine a | 0.72 | 0.68 | 0.67 | 0.66 | 0.69 | 0.67 | 0.68 | 0.69 |
| Tyrosine a | 0.57 | 0.53 | 0.52 | 0.52 | 0.54 | 0.54 | 0.55 | 0.57 |
| Valine a | 0.95 | 0.95 | 0.95 | 0.95 | 0.90 | 0.90 | 0.90 | 0.90 |
YPC=yeast-enriched protein concentrate.
Amino acid levels are expressed as standardised ileal digestible content.
Table 2.
Nutrient composition of YPC fed to the pigs
| Ingredient | YPC |
|---|---|
| CP (%) | 49.0 |
| DM (%) | 95.0 |
| Gross energy (MJ/kg) | 20.2 |
| Ether extract (%) | 3.80 |
| Crude fibre (%) | 4.50 |
| Ash (%) | 5.0 |
| Calcium (%) | 0.02 |
| Phosphorus (%) | 0.92 |
| NDF | 2.9 |
| ADF | 8.5 |
| Starch | 3.0 |
| Sugars | 0.5 |
YPC = yeast-enriched protein concentrate.
DNA extraction
Genomic DNA was extracted from pig faeces using a modified phenol chloroform extraction method, followed by ethanol precipitation. All centrifugation steps were at 16 000×g and room temperature. Faeces were not dried prior to weighing. Briefly, 30 mg of frozen faeces was homogenised in 600 µl Nuclei Lysis Solution (Promega) using a hand-held homogeniser, the solution centrifuged for 2 min and the resulting supernatant used for a standard phenol : chloroform : isoamyl alcohol (25 : 24 : 1) (Sigma) extraction followed by an overnight precipitation with 3 M sodium acetate (0.1 × sample volume) and 100% (v/v) ethanol (2.5 × (sample + sodium acetate)). The DNA was re-suspended in 100 µl of water (DNase and RNase free). Following re-suspension, 25 µl of DNA was incubated with 0.5 µl of RNase A (Promega) at 37°C for 15 min to remove RNA contaminants. After incubation, 200 µl of water (DNase and RNase free) was added to increase the volume and a further two-step phenol : chloroform : isoamyl alcohol (25 : 24 : 1) (Sigma) clean-up was carried out with a final overnight precipitation with 3 M sodium acetate (0.1 × sample volume) and 100% (v/v) ethanol (2.5 × (sample + sodium acetate)). Full method is presented in Supplementary Material S1. The DNA was re-suspended in a final volume of 10 µl. Deoxyribonucleic acid concentration (ng/µl) and DNA purity (260 nm/280 nm) were measured using the NanoDrop™ 2000 (Thermo Scientific) spectrophotometer. DNA yield was calculated as nanogram per milligram of faecal material. Extracted DNA samples were stored at –20˚C prior to analysis.
Quantitative PCR
To assess the DNA composition, SYBR Green (Roche) quantitative PCR methodology was used. Primers for detecting host (pig) DNA were designed specifically to the pig CYTB gene (Accession no. AY920909.1) [forward primer 5′-TTCATAGGCTACGTCCTGCC-3′ and reverse primer 5′-TCGTGTGAGGGTTGCTTTGT-3′], with a predicted amplicon length of 150 bp. For the detection of bacterial 16S DNA, published universal 16S primers (Mieszkin et al., 2009) were used with a predicted amplicon length of 142 bp. Polymerase chain reaction was performed on 384-well plates using a LightCycler® 480 (Roche) instrument. Each well contained 7.5 µl of SYBR Green (Roche) Mastermix, 1.6 µl of water (DNase and RNase free), 0.45 µl of each primer (10 µM, Forward and Reverse), and 5 µl of either the PCR standard, sample, or water (DNase and RNase free) as a negative control. The PCR protocol used was as follows: pre-incubation at 95˚C for 5 min, followed by 45 amplification cycles (95˚C for 10 s, 60˚C for 15 s, 72˚C for 15 s), a single melt cycle (95˚C for 5 s followed by 65˚C for 1 min) and then a cooling cycle of 40˚C for 10 s. A melt cycle was carried out to confirm a single product was produced in the PCR reaction. A pool of extracted faecal DNA samples diluted to 10 ng/µl was used in a fourfold dilution series to create standard curves for both CYTB and bacterial 16S DNA. Individual pig faecal DNA samples were then diluted 1 in 8 and tested in triplicate against the standard curve. Negative template controls (DNase and RNase free water) were also tested in triplicate at two points on the plate. Intra-assay CV for CYTB DNA assay, which takes into account the DNA extraction, is 0.66%. Intra-assay CV for the bacterial 16S DNA assay is 0.93%. The assays are carried out on the same day with all samples analysed on the same plate and therefore there is no reported inter-assay CV. Polymerase chain reaction products were verified by Sanger Sequencing (BioScience, Nottingham, UK).
Pig Calprotectin ELISA
Calprotectin content in faeces was assessed using the Pig Calprotectin (CALP) ELISA kit (Cusabio, China) according to manufacturer’s instructions, using a FLUOstar® Omega platereader (BMG LABTECH Ltd, UK) to measure absorbance at 450 nm. Intra-assay CV is <8% and inter-assay CV is <10%. The protocol was optimised for use with faecal samples, whereby samples were diluted 1 : 5 with sample diluent rather than the suggested 1 : 500 for plasma or serum samples. Pig faecal samples were tested in duplicate, requiring two plates, and each plate contained a standard curve also in duplicate. The standard curve was created by generating a four-parameter logistic (4-PL) curve-fit using online software (elisaanalysis.com). The standard curve was then used to calculate CALP concentration values in the faecal samples and the resulting value was multiplied by the dilution factor (x5) to give a final faecal CALP concentration, as per the manufacturer’s instructions.
Statistical analysis of results
All data were analysed using Genstat Statistical Software (17th edition), with significance at P<0.05. One-way ANOVA was used to assess the effects of diet on animal performance (BW, daily feed intake, daily live weight gain and FCR). Two-way (factors: diet × day of trial, blocking for pen) ANOVA were used to assess whether there were any effects on the resulting DNA concentration (ng/µl), DNA quality (260 nm/280 nm), DNA yield (ng/mg faecal material) and DNA contents (CYTB, 16S and CYTB : 16S ratio). Three-way ANOVA (factors: diet × day of trial × plate, blocking for pen) was used to analyse the faecal pig CALP levels. Where significant differences were observed, post hoc Bonferroni tests were then performed. The relationships between pig faecal CYTB DNA content and FCR and pig faecal CALP and FCR were assessed by Pearson’s correlation for both time points separately.
Results
Effects of Yeast Protein Concentrate on pig performance
There were no observable health issues to report and there were no significant treatment differences in daily intake or growth rate (Table 3). Pigs fed diets containing 2.5%, 5% or 7.5% YPC had significantly reduced FCR recorded from wean to day 14 (P<0.001) and over the entire 28-day trial (wean-day 28, P = 0.019) for 5% and 7.5% YPC inclusion compared to animals on the control diet (Table 3). However, there was no effect (P = 0.220) of YPC on FCR from day 14 to day 28 (Table 3).
Table 3.
The effect of dietary YPC on pig performance
| Animal performance | YPC inclusion level (%) | SED | P-value | |||
|---|---|---|---|---|---|---|
| 0 | 2.5 | 5.0 | 7.0 | |||
| Average BW at weaning (kg) | 8.38 | 8.33 | 8.38 | 8.37 | 0.025 | 0.175 |
| Average BW at day 14 (kg) | 12.30 | 12.91 | 13.03 | 12.79 | 0.291 | 0.081 |
| Average BW at day 28 (kg) | 21.95 | 21.66 | 22.68 | 21.84 | 0.666 | 0.457 |
| DFI wean to day 14 (g/pig per day) (diet 1) | 340 | 339 | 338 | 317 | 19.0 | 0.561 |
| DFI day 14 to day 28 (g/pig per day) (diet 2) | 830 | 776 | 823 | 791 | 41.0 | 0.542 |
| DFI wean to day 28 (g/pig per day) (treatment diets) | 580 | 557 | 580 | 554 | 27.0 | 0.585 |
| DLWG wean to day 14 (g/pig per day) (diet 1) | 280 | 327 | 332 | 316 | 21.1 | 0.076 |
| DLWG day 14 to day 28 (g/pig per day) (diet 2) | 687 | 625 | 689 | 646 | 36.4 | 0.240 |
| DLWG wean to day 28 (g/pig per day) (treatment diets) | 483 | 476 | 511 | 481 | 24.6 | 0.510 |
| FCR wean to day 14 (diet 1) | 1.24a | 1.04b | 1.02b | 1.01b | 0.048 | <0.001 |
| FCR day 14 to day 28 (diet 2) | 1.21 | 1.25 | 1.20 | 1.23 | 0.025 | 0.220 |
| FCR wean to day 28 (treatment diets) | 1.21a | 1.17ab | 1.14b | 1.15b | 0.023 | 0.019 |
DFI = daily feed intake; DLWG = daily live weight gain; FCR = feed conversion ratio (feed intake/weight gain); YPC = yeast-enriched protein concentrate; SED = standard error of the differences of the means.
Means with different superscript letters in the same row differ significantly.
DNA extraction
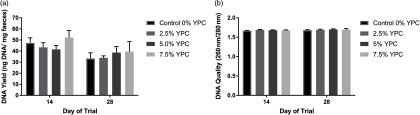
DNA yield (ng/mg faecal material) was significantly (P = 0.016) higher at day 14 compared with day 28 (Figure 1a). However, there was no effect of dietary treatment (P = 0.551), nor any interaction (P = 0.739) between day and dietary treatment (Figure 1a). When analysing DNA quality (260 nm/280 nm) there were no significant (P>0.05) interactions or effects of either diet or day of the trial (Figure 1b).
Figure 1.
The effect of an YPC-inclusion diet and when the pig faeces were collected on (a) DNA yield (ng/mg faecal material) and (b) the DNA quality (260 nm/280 nm). Mean values + Standard Error of the Mean, n = 9 for all groups. SEDs for YPC × day interaction=: (a) 7.60 and (b) 0.021. DNA yield was greater at day 14 compared with day 28 (P = 0.016). YPC=yeast-enriched protein concentrate; SED=standard error of the differences of the means.
Effects of Yeast Protein Concentrate on faecal host and bacterial DNA composition and calprotectin concentration
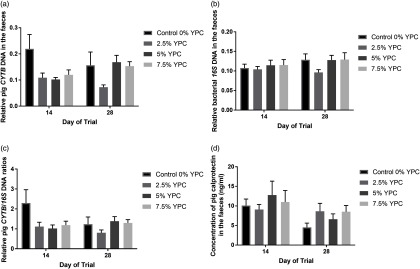
There was no day × YPC interaction (P = 0.222) nor any effect of day (P = 0.997) on pig CYTB DNA in the faeces, but there was a significant (P = 0.008) effect of YPC diet (Figure 2a). Bonferroni post hoc analysis showed that faeces from animals on the control diet (0% YPC) contained significantly (P<0.01) more pig CYTB DNA than those on the 2.5% YPC diet (Figure 2a and Table 4). Interestingly, even though there was no day × YPC interaction, all three YPC diets appeared to reduce faecal pig CYTB DNA contents at day 14, but only 2.5% YPC did so at day 28 (Figure 2a).
Figure 2.
The effect of an YPC-inclusion diet and when the faeces were collected on faecal (a) pig CYTB DNA content, (b) bacterial 16S DNA content, (c) pig CYTB DNA normalised to bacterial 16S DNA content and (d) pig CALP concentrations (ng/ml). Mean values + SEM, n = 9 for all groups. SEDs for YPC × day interaction=: (a) 0.045, (b) 0.018, (c) 0.445 and (d) 0.600. There were significant effects of diet on both CYTB DNA content (P = 0.008) and CYTB normalised to 16S DNA (P = 0.045), but not 16S DNA content and no significant diet × day interactions or effects of day (P>0.05). See Table 4 for post hoc Bonferroni analyses for effects of diet on CYTB DNA content and CYTB normalised to 16S DNA. Pig CALP concentration was greater at day 14 compared with day 28 (P = 0.009). YPC=yeast-enriched protein concentrate; CYTB=cytochrome b; SED=standard error of the differences of the means; CALP=calprotectin.
Table 4.
The effect of diet on mean faecal host DNA content in pig faecal samples collected on days 14 and 28 combined
| Faecal DNA component | Mean faecal DNA content in dietary group | SED | P-value for effect of diet (ANOVA) | |||
|---|---|---|---|---|---|---|
| Control (0% YPC) | 2.5% YPC | 5% YPC | 7.5% YPC | |||
| CYTB DNA | 0.188 A | 0.091 B | 0.135 AB | 0.136 AB | 0.0259 | 0.008 |
| CYTB DNA normalised for 16S DNA | 1.77 a | 0.96 b | 1.20 ab | 1.24 ab | 0.278 | 0.045 |
CYTB=cytochrome b; YPC=yeast-enriched protein concentrate; SED=standard error of the differences of the means.
Samples differing in lowercase letters indicate significant statistical differences at P<0.05.
Samples differing in uppercase letters indicate significant statistical differences at P<0.01.
In contrast, there were no significant effects of diet (P = 0.348), day (P = 0.205) nor an interaction (P = 0.602) on bacterial 16S DNA content in the faeces (Figure 2b). As there were no differences in bacterial 16S DNA, bacterial DNA was used to normalise the pig CYTB DNA content (Figure 2c). There was still a significant (P = 0.045) effect of YPC diet observed when CYTB DNA was normalised for bacterial 16S DNA content, with animals on the control diet (0% YPC) having significantly (P<0.05) more CYTB/16S DNA in their faeces compared to those on the 2.5% YPC diet (Figure 2c and Table 4), with no effect of day (P = 0.368) nor a day × diet interaction (P = 0.206).
Pig faecal CALP is a marker of inflammation and the levels were significantly higher at day 14 compared to day 28 (P = 0.009), indicating more intestinal inflammation closer to weaning (Figure 2d). However, there was no significant (P = 0.709) effect of diet nor a day × diet interaction (P = 0.391).
Relationship between host DNA, calprotectin and feed conversion ratio
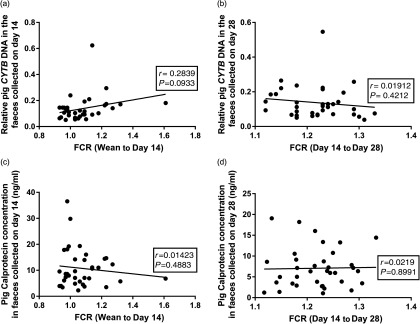
Pearson correlations were used to investigate relationships between the faecal DNA or CALP contents and feed efficiency. There was a trend for a correlation (r = 0.2839, P = 0.093) between the faecal pig CYTB DNA content (at day 14) and the recorded FCR (wean to day 14) over the early stages, where a significant effect of diet on FCR was observed (Figure 3a and Table 3). However, there was no correlation (P = 0.393) at the later phase (day 14 to day 28; Figure 3b), where no effect of diet on FCR was observed (Table 3). Likewise, there were no significant correlations between faecal CALP concentrations and recorded FCR at either day 14 (Figure 3c, P = 0.4883) or day 28 (Figure 3d, P = 0.9667).
Figure 3.
(a) The relationship between the pig CYTB DNA content in faeces at day 14 and recorded FCR (weaning to day 14). (b) The relationship between the pig CYTB DNA content in faeces at day 28 with FCR (day 14 to day 28). (c) The relationship between pig faecal CALP concentration (ng/ml) at day 14 and FCR (weaning to day 14). (d) The relationship between pig faecal CALP (ng/ml) at day 28 and FCR (day 14 to day 28). CYTB=cytochrome b; CALP=calprotectin; FCR=feed conversion ratio.
Discussion
To the best of our knowledge, this is the first (pilot) study to measure host DNA in faeces as a non-invasive marker of gut cell losses, which contribute to feed efficiency in production animals. A yeast-inclusion diet was used to test the hypothesis as a previous study had shown that dietary yeast influenced gut morphology and animal performance (Muthusamy et al., 2011). The current study shows that host CYTB DNA content in faeces may be a novel, simple, non-invasive marker for assessing gut cell losses. DNA yield was significantly greater in samples extracted from day 14 compared to samples extracted on day 28. The reason for this is unclear but it may be due to there being a greater level of feed digestion at day 28 as there being a more mature gut. Faecal DM was not measured and while it is unlikely to impact upon PCR, as measurements are performed on a constant DNA basis, this may account for the difference in DNA yield, although this is purely speculation. The DNA quality was unaffected by either diet or day of the trial the samples were collected. Polymerase chain reaction of both pig CYTB DNA and bacterial 16S DNA was successful, as verified by Sanger Sequencing, illustrating that a modified phenol–chloroform extraction method was suitable for extracting sufficient DNA of a suitable quality from pig faeces. In order for a PCR-based methodology to be used for the evaluation of gut cell losses in faeces, the host gene target needs to be present in sufficient quantities in the faeces to be measurable, as faeces are a heterogeneous material. As pig CYTB was successfully amplified and measured using quantitative PCR, it appears to be suitable as a gene target for the assessment of host DNA in faeces.
The average growth performance of the pigs was as expected for the unit and no undesirable side effects of the treatment were noted. The YPC product contains a maize-derived protein, which is expected to provide a valuable source of amino acids within the diet. However, the product also contains around 30% inactive yeast, from the bioethanol process. This yeast component is expected to provide a functional benefit beyond the protein value. Halder et al. (2011) investigated a low level of inclusion of a similar product in the diets of heat-stressed broilers and found a significant improvement in feed efficiency. Yeast contains substantial amounts of mannan and glucan in the cell wall. These components have been suggested to exert a ‘pre-biotic-like’ mechanism, with varied performance-enhancing effects such as inhibition of pathogenic invasion, modulation of the host immune response and enhancing the host gut morphology, thereby limiting the number of pathogens in the gut lumen (Roto et al., 2015). The main mechanism by which this occurs is thought to involve the mannan and glucan from the yeast cell wall binding to pathogenic bacteria, preventing their attachment to the intestinal epithelia and therefore enabling the host to reserve energy for growth, rather than repair and replenishment of epithelial cells (Roto et al., 2015).
Pig CYTB DNA was significantly higher in faeces collected from the pigs on the control diet compared with pigs on the 2.5% YPC-inclusion diet throughout the study, although at day 14, animals on the 5% and 7.5% inclusion level also appeared to have less pig CYTB DNA in their faeces compared to the controls. The reason for this is uncertain but it could be due to gut maturation or microbiome development and the 2.5% YPC inclusion level being optimal for the gut health benefit via a ‘pre-biotic-like’ mechanism previously discussed. Conversely, bacterial 16S DNA content was unaffected by either the diet or the age of the pigs. The fact that the bacterial 16S DNA content remains unchanged suggests it could potentially be used as a normalising control for the analysis of host DNA content in livestock faeces. The level of host DNA in faeces was hypothesised to be directly proportional to the amount of gut cell sloughing. The results of this study indicate that there is significantly more gut cell sloughing in the control fed pigs than the animals on a yeast-inclusion diet. It is therefore possible that the yeast component exerted a protective effect on the gut epithelium as previously described. As these animals are newly weaned pigs, the results suggest that inclusion of YPC in the diet helps the animal cope better with the problems observed when animals are transitioning to solid feed (Fairbrother et al., 2005).
Gut cell losses contribute to overall feed efficiency of the animal due to the high nutrient demand for replenishing the cells lining the small intestine (Bregendahl et al., 2004). There was a trend for a relationship between FCR measured over the first 14 days of the trial and the pig CYTB DNA content in the faeces at day 14, but not at the later phase (day 14 to day 28). In the first 14 days of the study, significant differences in FCR between dietary treatments were observed, and equally no significant differences in FCR were observed at the later phase. These results suggest that the levels of host DNA content in faeces may be a useful method for assessing gut cell losses and their impact on feed efficiency. However, this relies on the assumption that host DNA in faeces originates only from cell sloughing in the gastrointestinal tract, which is yet to be confirmed.
Faecal CALP levels are a recognised measure of intestinal inflammation and have been widely studied in humans (Fagerberg et al., 2005). However, fewer studies have been carried out in animals. Lallès and Fagerhol (2005) determined reference values for faecal CALP levels in pigs of different ages and lactation stage but the present study has assessed dietary intervention of newly weaned piglets. Pig faecal CALP was significantly higher at day 14 of the trial compared with day 28, with no influence of the YPC diet. Previous studies have shown that dietary inclusion of chitosan can reduce faecal CALP and therefore intestinal inflammation in pigs (Xiao, et al., 2014), but the YPC had no effect in the present study. The only differences in pig faecal CALP concentration (ng/ml) were caused by the age of the animal (day 14 > day 28), whereas differences in pig CYTB DNA were caused by diet. This shows that the pig CYTB DNA content is unrelated to the levels of CALP in the faeces, which is a marker of inflammation. Conversely, Varela et al. (2009) found a significant, but weak, correlation between human faecal DNA content and CALP content and hypothesised the weak relationship was due to the different origins of the markers.
In conclusion, dietary YPC positively influenced FCR in newly weaned piglets and reduced faecal CYTB DNA content, suggesting that faecal CYTB DNA content may be a novel non-invasive measure of gut cell losses. However, further work is required to elucidate the origin of the faecal host DNA and determine how sensitive the faecal DNA assay is. These experiments will form the basis for validation studies where the DNA assay will be compared with established methodologies.
Acknowledgements
This work was supported by a PhD studentship for KRS funded by AB Vista and the University of Nottingham and funding from AB Agri Limited and AB Vista. Preliminary results have been published in an abstract form (Slinger et al., 2018).
K. R. Slinger, 0000-0002-7417-165X, A. H. Stewart. https://orcid.org/0000-0003-0309-5343, T. Parr, 0000-0003-2490-9457, J. M. Brameld, 0000-0003-4942-080X
Declaration of interest
There is no conflict of interest.
Ethics statement
Experimental protocol was approved by the Harper Adams University Research Ethics Committee. Ethical approval number: 00583940-STAFF-1-201607.
Software and data repository resources
None of the data were deposited in an official repository.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S1751731119000818.
click here to view supplementary material
References
- Ariefdjohan MW, Savaiano DA and Nakatsu CH 2010. Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutrition Journal 9, 23. doi: 10.1186/1475-2891-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin KK and Taegtmeyer H 2011. Taking pressure off the heart: the ins and outs of atrophic remodelling. Cardiovascular Research 90, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonseub S, Johnston G and Linacre A 2012. Identification of protected avian species using a single feather barb. Journal of Forensic Sciences 57, 1574–1577. [DOI] [PubMed] [Google Scholar]
- Bregendahl K, Liu L, Cant JP, Bayley HS, McBride BW, Milligan LP, Yen J-T and Fan MZ 2004. Fractional protein synthesis rates measured by an intraperitoneal injection of a flooding dose of L-[ring-2H5]phenylalanine in pigs. The Journal of Nutrition 134, 2722–2728. [DOI] [PubMed] [Google Scholar]
- Buttery PJ 1981. Protein turnover in animals. Tropical Animal Production 6, 204–213. [Google Scholar]
- Caldwell JM, Patment P and Villemur R 2011. Mitochondrial DNA as source tracking markers of fecal contamination In Microbial source tracking: methods, applications, and case studies (ed. Hagedorn C, Blanch AR and Harwood VJ), pp. 229–250. Springer, New York City, NY, USA. [Google Scholar]
- Creamer B, Shorter RG and Bamforth J 1961. The turnover and shedding of epithelial cells. Gut 2, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg UL, Lööf L, Myrdal U, Hansson L-O and Finkel Y 2005. Colorectal inflammation is well predicted by fecal calprotectin in children with gastrointestinal symptoms. Journal of Pediatric Gastroenterology and Nutrition 40, 450–455. [DOI] [PubMed] [Google Scholar]
- Fairbrother JM, Nadeau É and Gyles CL 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Animal Health Research Reviews 6, 17–39. [DOI] [PubMed] [Google Scholar]
- Gasier HG, Fluckey JD and Previs SF 2010. The application of 2H2O to measure skeletal muscle protein synthesis. Nutrition & Metabolism 7, 31. doi: 10.1186/1743-7075-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M, Smith P and Wilkinson JM 2010. Mitigating climate change: the role of domestic livestock. Animal 4, 323–333. [DOI] [PubMed] [Google Scholar]
- Halder S, Ghosh TK, Toshiwati and Bedford MR 2011. Effects of yeast (Saccharomyces cerevisiae) and yeast protein concentrate on production of broiler chickens exposed to heat stress and challenged with Salmonella enteritidis . Animal Feed Science and Technology 168, 61–71. [Google Scholar]
- Lallès J-P and Fagerhol MK 2005. Faecal calprotectin: a non invasive marker of inflammation in pigs? ISAH 1, 405–408. [Google Scholar]
- Loretz B, Föger F, Werle M and Bernkop-schnürch A 2006. Oral gene delivery: strategies to improve stability of pDNA towards intestinal digestion. Journal of Drug Targeting 14, 311–319. [DOI] [PubMed] [Google Scholar]
- Martellini A, Payment P and Villemur R 2005. Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Research 39, 541–548. [DOI] [PubMed] [Google Scholar]
- McOrist AL, Jackson M and Bird AR 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. Journal of Microbiological Methods 50, 131–139. [DOI] [PubMed] [Google Scholar]
- Mieszkin S, Furet J-P, Corthier G and Gourmelon M 2009. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Applied and Environmental Microbiology 75, 3045–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy S, Haldar S, Ghosh TK and Bedford MR 2011. Effects of hydrolysed Saccharomyces cerevisiae yeast and yeast cell wall components on live performance, intestinal histo-morphology and humoral immune response of broilers. British Poultry Science 52, 694–703. [DOI] [PubMed] [Google Scholar]
- Ramón-Laca A, Soriano L, Gleeson D and Godoy JA 2015. A simple and effective method for obtaining mammal DNA from faeces. Wildlife Biology 21, 195–203. [Google Scholar]
- Roperch J-P, Benzekri K, Mansour H and Incitti R 2015. Improved amplification efficiency on stool samples by addition of spermidine and its use for non-invasive detection of colorectal cancer. BMC Biotechnology 15, 41. doi: 10.1186/s12896-015-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roto SM, Rubinelli PM and Ricke SC 2015. An introduction to the avian gut microbiota and the effects of yeast-based prebiotic-type compounds as potential feed additives. Frontiers in Veterinary Science 2, 28. doi: 10.3389/fvets.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger KR, Stewart AH, Hall H, Masey O’Neill HV, Bedford MR, Brameld JM and Parr T 2018. The effects of exogenous yeast enriched protein concentrate from the bio-ethanol industry (YPC) on pig cytochrome b (CYTB) genomic DNA content in faeces. Advances in Animal Biosciences 9, 37. [Google Scholar]
- Varela E, Antolín M, Guarner F, Verges R, Giralt J and Malagelada J-R 2009. Faecal DNA and calprotectin as biomarkers of acute intestinal toxicity in patients undergoing pelvic radiotherapy. Alimentary Pharmacology and Therapeutics 30, 175–185. [DOI] [PubMed] [Google Scholar]
- Vincent C, Mehrotra S, Loo VG, Dewar K and Manges AR 2015. Excretion of host DNA in feces is associated with risk of Clostridium difficile infection. Journal of Immunology Research 2015, 246203. doi: 10.1155/2015/246203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Wang Y, Liu G, He J, Qui W, Hu X, Feng Z, Ran M, Nyachoti CM, Kim SW, Tang Z and Yin Y 2014. Effects of Chitosan on intestinal inflammation in weaned pig challenged by enterotoxigenic Escherichia coli. PLoS ONE 9, e104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S1751731119000818.
click here to view supplementary material