Abstract
Metabolic syndrome has become a global health challenge and was recently reported to be positively correlated with increased sucrose consumption. Mechanistic analyses of excess sucrose-induced progression of metabolic syndrome have been focused mainly on abnormal hepatic lipogenesis, and the exact contribution of excess sucrose to metabolic disorders remains controversial. Considering that carbohydrate and lipid metabolisms exhibit clear circadian rhythms, here we investigated the possible contribution of diurnal oscillations to responses of hepatic lipid metabolism to excess sucrose. We found that excess sucrose dose-dependently promotes fatty liver and hyperlipidemia in in rats fed a high-sucrose diet (HSD). We observed that excess sucrose enhances the oscillation amplitudes of the expression of clock genes along with the levels of hepatic lipid and carbohydrate metabolism-related mRNAs that increase lipogenesis. We did not observe similar changes in the levels of the transcription factors regulating the expression of these genes. This suggested that the excess sucrose-induced, circadian rhythm–dependent amplification of lipogenesis is post-transcriptionally regulated via the stability of metabolic gene transcripts. Of note, our findings also provide evidence that fructose causes some of the HSD-induced, circadian rhythm–dependent alterations in lipogenic gene expression. Our discovery of HSD-induced circadian rhythm–dependent alterations in lipogenesis at the post-transcriptional level may inform future studies investigating the complex relationships among sucrose uptake, circadian rhythm, and metabolic enzyme expression. Our findings could contribute to the design of chrono-nutritional interventions to prevent or manage the development of fatty liver and hyperlipidemia in sucrose-induced metabolic syndrome.
Keywords: metabolic syndrome, fatty acid metabolism, circadian rhythm, dyslipidemia, lipogenesis, excess sucrose intake, nonalcoholic fatty liver diseases, lipid metabolic disorders, high-sucrose diet (HSD), gene regulation
Introduction
Metabolic syndrome has become a global health problem, representing an increased risk for developing type-2 diabetes mellitus or cardiovascular diseases (1). Although no internationally recognized clinical diagnosis standard has been established for metabolic syndrome (2), it is identical for several symptoms, such as hyperlipidemia, fatty liver, obesity, and insulin resistance (2). Lifestyle changes, such as a lack of exercise, excess energy consumption from processed foods, and increased stress, promote metabolic syndrome prevalence (1). Intervention on diets, especially a high-fat diet, has been focused on controlling metabolic syndrome development, whereas the threat of added sugars to public health has also been recognized (3). Distinct from a high-fat diet accelerating metabolic diseases via obesity, excess sucrose contributes to noncommunicable diseases independently of excess energy intake (4). Since the first critics of sucrose in the 1960s, the related mechanisms have been investigated in both animals and humans (5–8), most of which focused on increased hepatic lipogenesis induced by the composition of fructose. Fructose has been considered to provide a rapid substrate flux for lipogenesis, partially due to its being less controlled by rate-limiting enzymes than glucose (6, 8, 9). Recent biochemistry textbooks also state that the rapid substrate influx from fructose into the lipogenesis pathway is the main reason for fatty liver induced by excess fructose (10, 11). It does not appear that the rapid substrate influx can explain accumulation of lipids in blood and liver. Despite accumulating evidence implicating sucrose in metabolic syndrome-associated symptoms (3), the mechanisms underlying such associations remain controversial because several contrasting phenotypes have been found in animal and human studies, as reviewed by Macdonald (5). Moreover, on the molecular level, several animal studies generated conflicting results with regard to whether the expression of sucrose- or fructose-induced enzymes involved in lipogenesis increased or was unaffected (12–15). Thus, we performed this study to examine whether other unknown factors may explain the differing results.
Circadian rhythms can play important roles in metabolism homeostasis in mammals (16). In contrast to the suprachiasmatic nucleus located in the hypothalamus that mainly receives the regulation by light signals, cell-autonomous circadian oscillations in peripheral tissues (including the liver, adipose tissue, and gastrointestinal tract) are sensitive to feeding and diet (17). For instance, feeding-regulated insulin can function as a synchronizer for the liver clock (18). Numerous metabolites and gene expression oscillations can increase energy efficiency and prevent futile cycles (19). Previously, we reported that an effective intervention strategy, time-restricted feeding, can alleviate metabolic disorders induced by excess sucrose (20). Although several questions remain regarding this feeding regimen (19), time-restricted feeding has become a practical method for ameliorating diet-induced metabolic disorders (21–23). These findings suggest the possibility of circadian rhythm–dependent metabolic responses to various diets, and to investigate the responses of lipid metabolism to a high-sucrose diet (HSD),3 we studied rhythmic changes in the expression patterns of clock genes and lipogenic genes. Here, we found that genes involved in hepatic lipid and carbohydrate metabolism in rats fed an HSD showed circadian rhythm–dependent amplification rather than persistent induction. Unexpectedly, this response occurred post-transcriptionally instead of at the transcription level, which would indicate a novel mechanism mediating high sucrose–induced fatty liver.
Results
Excess sucrose promoted fatty liver and hyperlipidemia in a dose-dependent manner in rats
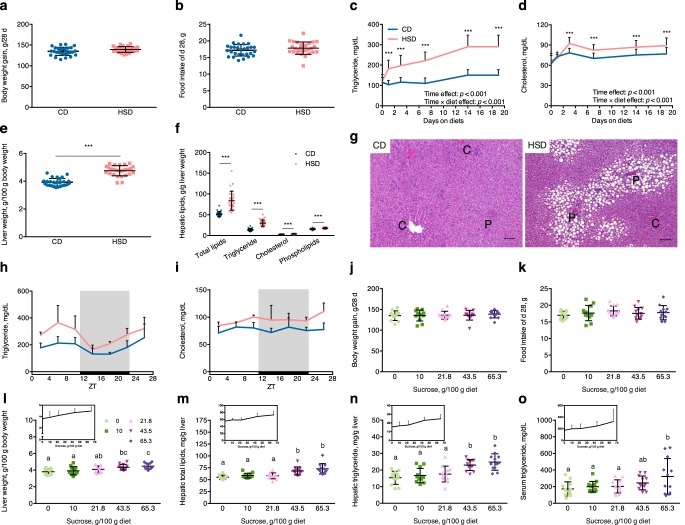
To confirm the pathogenetic effects of excess sucrose, rats were fed with diets containing either starch (control starch diet (CD)) or sucrose (HSD) as a sole carbohydrate source, for 4 weeks. In agreement with our previous study (20), rats showed no significant differences in body weight gain (Fig. 1a) or food intake (Fig. 1b). Fasting plasma triglyceride and cholesterol concentrations significantly increased in HSD rats (Fig. 1, c and d). The results of two-way repeated measurement analysis of variance (ANOVA) revealed different variation patterns over days on these two different diets, which showed a more robust elevation according to the days on diets in HSD group. This result showed that the HSD induced hyperlipidemia in rats. Liver weights also significantly increased in HSD rats, with obvious hepatic lipid accumulation (Fig. 1, e–g). Hepatic lipid-fraction analysis showed that the triglyceride level was most strongly induced by HSD and that cholesterol and phospholipids were also robustly induced (Fig. 1f). Analysis of liver histology revealed that hepatic macrovesicular steatosis occurred in rats fed an HSD (Fig. 1g). Substantial formation of lipid droplets were observed in periportal regions, which was consistent with sucrose-induced periportal steatosis found previously with metabolic zonation of the liver (24). Nonfasting serum triglyceride levels on the final experimental day displayed diurnal rhythmic variations (JTK_CYLCE analysis: CD, p = 0.0034; HSD, p = 0.0014), with ∼1.5-fold increased amplitude in HSD rats (Fig. 1h and Table S2). Cholesterol levels were not rhythmic during the day (p = 1) but were induced by the HSD (Fig. 1i and Table S2).
Figure 1.
HSD induced fatty liver and hyperlipidemia in a dose-dependent manner. a, total body weight gain during 28 days in rats fed a CD or HSD (p = 0.057; n = 28/group). b, food intake during the final experimental day (d 28) (p = 0.178; n = 28/group). c and d, 4-h fasting plasma triglyceride and cholesterol variations (***, p < 0.001; n = 28/group); time effect and time × diet effect, as shown by the results of two-way repeated measurement ANOVA. e, liver weights after 4 weeks of a CD or HSD (***, p < 0.001; n = 28/group). f, hepatic total lipids were measured, and the proportions of triglyceride, cholesterol, and phospholipids were determined after rats were administered a CD or HSD for 4 weeks (***, p < 0.001; n = 28/group). g, representative pictures of H&E-stained liver sections. Scale bar, 100 μm. The letters C and P indicate the central vein and portal vein, respectively. h and i, diurnal variations of serum (harvested when the rats were sacrificed) triglyceride and cholesterol concentrations were analyzed with JTK_CYCLE (shown in Table S2; n = 4/group at each time point). j, total body weight gain during 28 days in rats fed with diets containing different doses of sucrose (p = 0.948; n = 12/group). k, food intake on the final experimental day (d 28) (p = 0.538; n = 12/group). l, liver weights (p < 0.001; n = 12/group). m–o, hepatic total lipids (p < 0.001), triglyceride levels (p < 0.001), and serum triglyceride concentrations (p = 0.029) after rats were fed for 4 weeks with diets containing different doses of sucrose (n = 12/group); line charts over bars indicate linear variations, according to the sucrose dose. Statistical comparison was performed with Student's t test between two groups, and one-way ANOVA followed by Duncan's multiple-range test was performed among five groups. Different letters over each bar indicate significant differences, as determined by Duncan's multiple-range tests. Error bars, S.D.
To determine whether the induction of lipid accumulation by the HSD correlated with dose of sucrose, rats were fed diets containing different ratios of starch and sucrose as carbohydrate sources (described under “Experimental procedures”). Although the sucrose doses in the diets did not influence body weight gain (Fig. 1j) or food intake (Fig. 1k) in rats during the 4-week experiment, liver weights increased linearly with greater sucrose doses. And a significant difference from the CD group appeared at a dose of 43.5 g sucrose per 100 g of diet (Fig. 1l). Hepatic total lipids and triglycerides were also significantly increased at a dose of 43.5 g of sucrose/100 g of diet (Fig. 1, m and n), whereas a significant difference in serum triglyceride levels was found at a higher dose of 65.3 g of sucrose/100 g of diet (Fig. 1o). Despite these significant differences, approximately linear corrections between sucrose doses and lipid accumulation in the liver and blood were observed.
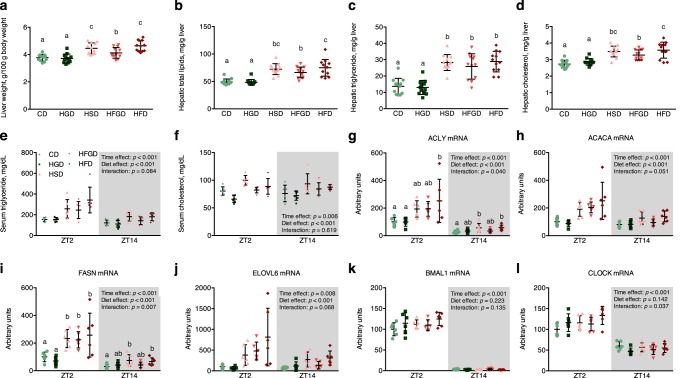
Considering that the monosaccharide fructose has been speculated to be responsible for most excess sucrose-related complications (25), we investigated the role of fructose in HSD-induced fatty liver and hyperlipidemia. Rats were fed a CD, high-glucose diet (HGD), HSD, high-fructose and -glucose diet (HFGD; 1:1 ratio by weight), or high-fructose diet (HFD) ad libitum for 4 weeks. The liver weights increased significantly in the HSD, HFGD, and HFD groups when compared with the CD group, but not in the HGD group (Fig. 2a). Similar variations were found in hepatic lipids, including total lipids, triglycerides, and cholesterol (Fig. 2, b–d). These changes clearly occurred in a fructose-dependent manner. Although fructose dose dependence was not observed (as 2-fold more fructose was present in the HFD than in HSD and HFGD), the liver weights and hepatic lipids did not double. Serum triglycerides and cholesterol were also increased by diets containing fructose (HSD, HFGD, and HFD), and triglycerides, but not cholesterol, showed larger increases at Zeitgeber time 2 (ZT2; ZT0 is defined as the point when the lights were turned on), which was similar to the results shown in Fig. 1, h and i (Fig. 2, e and f).
Figure 2.
The sucrose-induced, circadian rhythm–dependent amplification of lipogenesis was confirmed to be fructose-dependent. a, liver weights in rats after being fed an experimental diet for 4 weeks, which contained or lacked fructose (p < 0.001; n = 12/group). b–d, hepatic total lipids (p < 0.001), triglyceride levels (p < 0.001), and cholesterol levels (p < 0.001) after a 4-week treatment (n = 12/group). e and f, serum triglyceride and cholesterol concentrations at ZT2 and ZT14 after a 4-week treatment (n = 6/group at either time point). g–l, the mRNA levels of several fatty acid synthesis genes and clock genes at ZT2 and ZT14 (n = 6/group at either time point). One-way ANOVA followed by Duncan's multiple-range test was performed among five groups. Two-way ANOVA was performed to determine the interaction effect of time and diet. When a significant interaction effect was observed, Duncan's multiple-range post hoc test was performed for either time point. Different letters over bars indicate significant differences, as determined by Duncan's multiple-range test. Error bars, S.D.
Excess sucrose caused circadian rhythm–dependent amplification of hepatic lipogenic gene expression
To investigate how excess sucrose affects lipid metabolism by influencing the circadian system, we analyzed oscillations in the expression levels of genes that play important roles in regulating metabolism, including clock genes, transcription factor genes, lipid– and carbohydrate metabolism–related genes, and amino acid metabolism–related genes. All rhythmic analysis results pertaining to rhythmic assessment (p values), phases, and amplitudes are shown in Table S2.
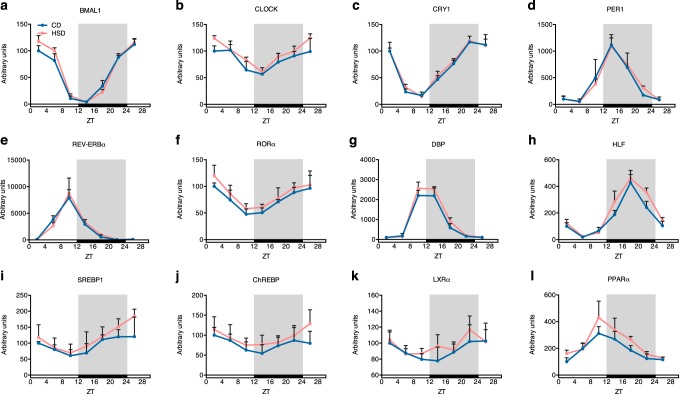
Cell-autonomous circadian oscillations consist of molecular transcription feedback loops that further mediate circadian rhythm-controlled gene expression (16). BMAL1 (also known as ARNTL1; aryl hydrocarbon receptor nuclear translocator-like 1) and CLOCK (circadian locomotor output cycles kaput) proteins form heterodimers that function as core activators in the circadian transcription feedback loop (16). The mRNA levels of BMAL1 and CLOCK showed clear diurnal oscillations with slightly shifted phases (2 h) induced by the HSD, although the amplitudes did not change (0.5 < -fold change < 1.5) between groups (Fig. 3 (a and b) and Table S2) (20). Downstream repressor genes that are activated by BMAL1/CLOCK heterodimers, such as CRYs (cryptochrome 1 and 2), PERs (period 1 and 2), and REV-ERBs (also known as NR1D1 and NR1D2; nuclear receptor subfamily 1, group D, member 1 and 2) (26), also displayed rhythmic expression, but the HSD rarely altered the phases and amplitudes (Fig. 3 (c–e), Fig. S1, and Table S2) (20).
Figure 3.
Clock genes and transcription factors involved in lipid metabolism showed little changes in mRNA levels in rats fed an HSD. a–h, diurnal mRNA level variations of several important clock genes in maintaining metabolism hemostasis (n = 4/group at each time point). i–l, diurnal mRNA level variations of key transcription factors involved in lipid metabolism (n = 4/group at each time point). Rhythmic analysis was performed with JTK_CYCLE, and the results are shown in Table S2. Error bars, S.D.
The HSD did not affect the mRNA expression levels of several other rhythmically expressed genes, including RORα (retinoic acid–related orphan receptor α), DBP (D site of albumin promoter-binding protein), HLF (hepatic leukemia factor), DECs (deleted in esophageal cancer 1 and 2), TEF (thyrotroph embryonic factor), and E4BP4 (also known as NFIL3; nuclear factor, interleukin 3–regulated), whose products all play important roles in regulating carbohydrate and lipid metabolism homeostasis (26) (Fig. 3 (f–h), Fig. S1, and Table S2). There are important transcription factors controlling the expression of carbohydrate and lipid metabolism–related genes downstream of the core clock transcriptional feedback loop, such as SREBP1 (sterol regulatory element–binding protein 1) and ChREBP (carbohydrate-responsive element–binding protein). These transcription factors are well-known for their roles in regulating expression of rate-limiting enzymes in lipid and carbohydrate metabolism (27, 28). Their mRNA levels showed a diurnal rhythm, but little difference was found between the CD and HSD groups (Fig. 3 (e and f) and Table S2). LXRα (liver X receptor α) and PPARα (peroxisome proliferator–activated receptor α), which are transcription factors that connected the clock genes with lipid metabolism (29, 30), displayed circadian rhythms at the mRNA levels but were not affected by the HSD (Fig. 3 (k and l) and Table S2).
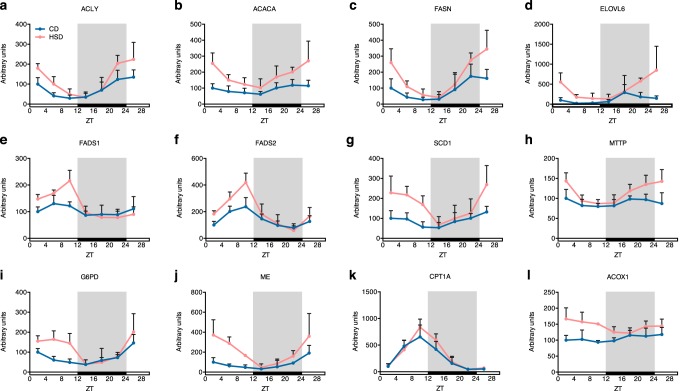
Although core clock genes and transcription factors were not significantly changed by the HSD, the enzymes that directly participate in lipid metabolism showed different rhythmic expression patterns between the CD and HSD groups. ACLY (ATP citrate lyase), ACACA (acetyl-CoA carboxylase α), and FASN (fatty acid synthase) contribute to de novo lipogenesis from citrate, and the amplitudes of their mRNA oscillations all increased by approximately twice (amplitude -fold change: 2.04, 1.81, and 1.99, respectively), with slightly shifted phases (2 h), in rats fed an HSD (Fig. 4 (a–c) and Table S2). Fatty acid elongase ELOVL6 (elongation of very long chain fatty acids protein 6) and desaturase FADS1 (fatty acid desaturase 1), FADS2, and SCD1 (stearoyl-CoA desaturase 1) also showed larger mRNA expression amplitudes (-fold change: 4.44, 2.12, 1.71, and 2.19, respectively) in rats of the HSD group (Fig. 4 (d–g) and Table S2). In contrast, MTTP (microsomal triglyceride transfer protein), which plays a central role in the assembly of lipoproteins, such as very low-density lipoprotein, was not rhythmically expressed in the CD group (p > 0.05). However, the HSD induced de novo diurnal oscillations in its mRNA levels (Fig. 4h and Table S2).
Figure 4.
The HSD increased the oscillation amplitudes of mRNA levels of key enzymes in lipid metabolism. Diurnal mRNA levels of enzymes contributing to fatty acid synthesis (a–c), elongation (d), desaturation (e–g), lipoprotein assembly (h), fatty acid oxidation (i and j), and NADPH synthesis for lipogenesis (k and l) are shown (n = 4/group at each time point). Rhythmic analysis was performed with JTK_CYCLE, and the results are shown in Table S2. Error bars, S.D.
Lipogenesis requires NADPH as an energy source. The major source of NADPH in animal cells is the pentose phosphate pathway (PPP), and we found that expression of G6PD (glucose-6-phosphate dehydrogenase), which encodes the rate-limiting enzyme mediating the reaction step of NADPH production in the PPP (31), showed a slightly shifted phase (2 h) and a largely increased amplitude (-fold change: 2.84) (Fig. 4i and Table S2). These variations were similar to those found in the enzymes that catalyze fatty acid synthesis, including ACLY, ACACA, and FASN. Another reaction step producing NADPH in the PPP is regulated by PGD (phosphogluconate dehydrogenase), which showed little difference in mRNA levels between two groups (Fig. S2a and Table S2). A third source of NADPH production mediated by the NADP+-dependent ME (malic enzyme) also displayed mRNA levels with an ∼4-fold increased amplitude of mRNA levels in the HSD group (Fig. 4j and Table S2).
Finally, we also studied whether HSD affected fatty acid oxidation. CPT1A (carnitine palmitoyltransferase 1A) is the essential enzyme of the long-chain fatty acid shuttle system that helps long-chain fatty acids enter the mitochondria to become oxidized. Our results showed similar mRNA oscillation patterns between the CD and HSD groups (Fig. 4k and Table S2). However, the expression of ACOX1 (acyl-CoA oxidase 1), whose gene product catalyzes the first step of peroxisomal very long-chain fatty acid oxidation, was induced de novo rhythmicity by the HSD (Fig. 4l and Table S2). Overall, the results described above indicated that the HSD induced circadian rhythm–dependent mRNA expression changes in lipid metabolism–related genes, including shifted phases, increased amplitudes, or de novo rhythmic oscillations; these changed patterns could mostly be attributable to increases in the light phase, rather than the dark phase. These circadian rhythm–dependent changes could have contributed to the large increase in lipogenesis found in rats fed an HSD, leading to the promotion of fatty liver and hyperlipidemia.
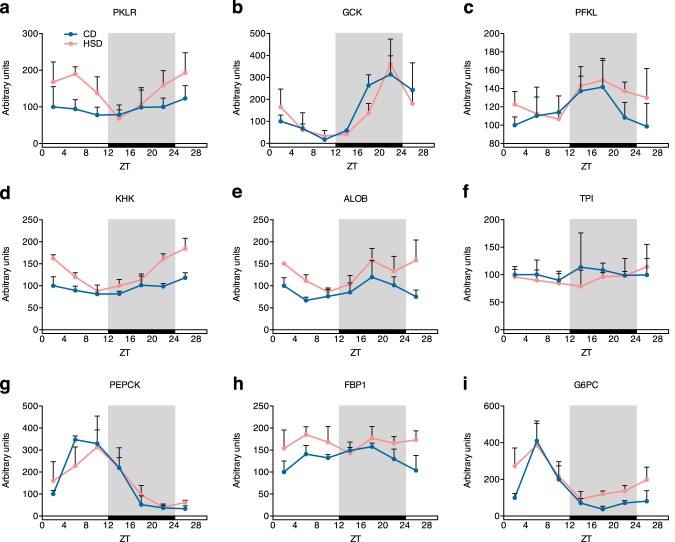
Because de novo lipogenesis was induced to display circadian rhythm–dependent amplifications by the HSD, we further investigated carbohydrate catabolism, which supplies substrates for lipogenesis. The enzyme in the catabolic pathway important for both fructose and glucose catabolism, PKLR (pyruvate kinase, liver, and red blood cell type), showed rhythmic expression in the HSD group, but not in the CD group, and an obvious increase in mRNA amplitude (Fig. 5a and Table S2). Rate-limiting enzymes of the glycolysis pathway, such as GCK (glucokinase) and PFKL (phosphofructokinase, liver type), showed oscillatory mRNA levels, but the oscillations were not largely changed by the HSD (Fig. 5 (b and c) and Table S2). However, an enzyme that regulates fructolysis, KHK (ketohexokinase), showed similar variations in rats fed an HSD, displaying de novo rhythmic expression and approximately a 5-fold increase in the mRNA amplitude (Fig. 5d and Table S2). ALOB (aldolase B) also showed an increased mRNA amplitude and a 4-h delayed phase in the HSD group, whereas TPI (triosephosphate isomerase) expression did not change between the groups (Fig. 5 (e and f) and Table S2). In contrast, the HSD barely affected the mRNA expression levels of genes whose products mediate gluconeogenesis, such as PEPCK (phosphoenolpyruvate kinase), FBP1 (fructose-1,6-bisphosphatase), and G6PC (glucose-6-phosphatase, catalytic subunit) (Fig. 5 (g–i) and Table S2). Amino acid and ammonia metabolism-related genes, such as GOT1 (aspartate aminotransferase), GPT1 (also known as ALAT, alanine aminotransferase), ASNS (asparagine synthetase), CPS1 (carbamoyl phosphate synthetase), and ASS1 (argininosuccinate synthetase), were barely affected by the HSD (Fig. S2 (b–f) and Table S2). Only ASNS had a higher amplitude in the HSD group (Fig. S2d and Table S2). These results indicate that HSD induced large circadian rhythm–dependent increases of mRNA amplitudes of lipid metabolism– and fructolysis–related genes, which further enhanced the de novo lipogenesis.
Figure 5.
Several enzymes involved in carbohydrate metabolism also showed increased oscillation amplitudes of mRNA levels, induced by the HSD. Diurnal mRNA levels of enzymes regulating glycolysis (a–c), fructolysis (d–f), and gluconeogenesis (g–i) are shown (n = 4/group at each time point). Rhythmic analysis was performed with JTK_CYCLE, and the results are shown in Table S2. Error bars, S.D.
Excess sucrose–induced circadian rhythm–dependent amplifications were regulated at the post-transcriptional level
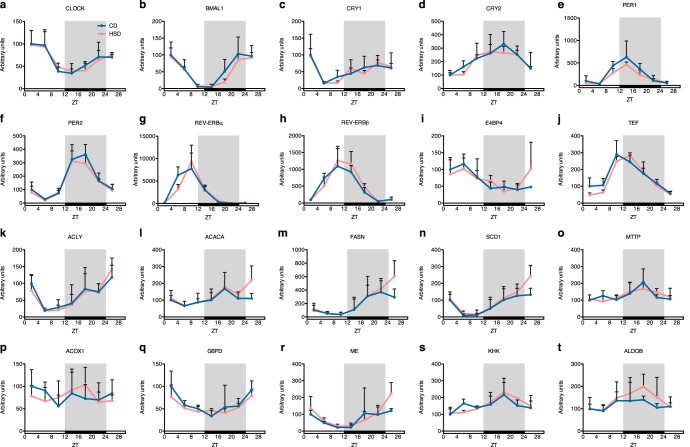
Next, we investigated whether the HSD-induced circadian rhythm–dependent amplification of genes involved in lipid and carbohydrate metabolisms at the transcriptional level. We estimated the gene transcription rates by measuring the nascent mRNA levels (32). The expression of clock genes is known to be regulated transcriptionally. As expected, the transcription rates of clock genes were not changed by the HSD (Fig. 6, a–j), as their mRNA levels showed no differences (Fig. 3). Surprisingly, although the mRNA amplitudes were enriched for lipid metabolism- and fructolysis-related genes (Figs. 4 and 5), we found that the transcription rates of the genes showed little changes between both groups (Fig. 6, k–t). These results indicate that the HSD-induced, circadian rhythm–dependent expansion of mRNA amplitudes was not mediated through transcriptional regulations. The results suggest that some sucrose-derived metabolites or molecules modulate the circadian rhythm–dependent mRNA stability.
Figure 6.
Transcription rates of lipogenic genes displayed different oscillation patterns compared with those of mRNA levels with HSD treatment. Nascent mRNA levels of important clock genes (a–j), and genes involved in lipid metabolism (k–t) are shown (n = 4/group at each time point). Error bars, S.D.
Circadian rhythm–dependent inductions occurred in a fructose-dependent manner
Lipogenic gene mRNA levels showed the largest fluctuation between ZT2 and ZT14 (Fig. 3). To investigate how excess sucrose induced circadian rhythm–dependent amplifications of lipogenic gene expression, we measured the mRNA levels at these two time points to estimate the amplitudes of lipogenic gene oscillations in rats fed different types of sugars. The mRNA levels of lipogenic genes, such as ACLY, ACACA, FASN, and ELOVL6, displayed higher levels in HSD, HFGD, and HFD groups but did not increase in the HGD group (Fig. 2, g–j). Similar to the findings presented in Fig. 4, the increases were more obvious at ZT2, indicating that the amplitudes of lipogenesis gene mRNA levels expanded. In contrast, BMAL1 and CLOCK showed clear day-night expression differences, but the fluctuations were not affected by diets (Fig. 6, k and l). These results suggest that fructose caused the HSD-induced, circadian rhythm–dependent amplifications.
Excess sucrose changed hepatic levels of fructose-derived metabolites, amino acids, and phosphate
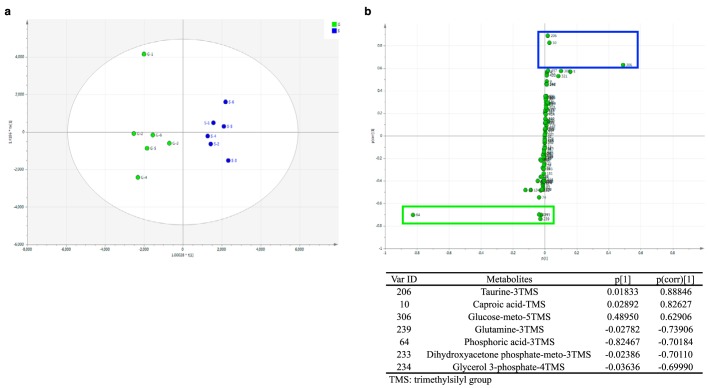
To investigate possible hepatic metabolites that regulate the circadian rhythm–dependent amplifications of lipogenic gene expression at the post-transcriptional level, induced by the HSD, we performed metabolomics analysis between a control glucose diet (CGD) group and an HSD group. Multivariate statistical analysis was used to classify the differences. Orthogonal partial least-squares discriminant analysis (OPLS-DA) revealed separated metabolite clusters in the CGD and HSD groups (Fig. 7a), indicating the production of different primary metabolites in each group. In the corresponding S-plot, the higher absolute p(corr)[1] value represented more significant discrimination between both groups. Variables with an absolute p(corr)[1] value > 0.6 were recognized as significantly changed metabolites and are highlighted with rectangles in Fig. 7b. Hepatic taurine, caproic acid, and glucose levels increased in the HSD group, whereas hepatic glutamine, phosphoric acid, dihydroxyacetone phosphate, and glycerol 3-phosphate levels decreased in the HSD group, when compared with the CGD group (Fig. 7b). Although production of the amino acids taurine and glutamine changed markedly, gene expression of cysteine sulfinate decarboxylase (CSAD) and glutamine synthetase (GS) was not altered by the HSD (Fig. S2, g and h). The decrease of fructose and glucose catabolic metabolites, dihydroxyacetone phosphate, and glycerol 3-phosphate, together with the increase of glucose in the HSD group, suggested a rapid utilization of fructose-derived metabolites by gluconeogenesis or lipogenesis. Moreover, we found decreased phosphate in the HSD group, and this expenditure of Pi would be expected to block ATP synthesis (33).
Figure 7.
Identification of interesting metabolites and profiling by plotting OPLS-DA and corresponding S-plot scores in rats fed a CGD or HSD. a, OPLS-DA score plot for the CGD group (G) and HSD group (S); each point represents a tested liver sample; b, S-plot for the CGD group (p[1] from −1 to 0) and HSD group (p[1] from 0 to 1). Significantly changed metabolites, with an absolute p(corr)[1] value >0.6 between two groups, are circled (green, CGD; blue, HSD) in the figure and noted in the table.
Discussion
The abuse of adding sucrose causes a dramatic increase in sucrose intake from processed, sweet food products. However, excess sucrose is becoming a threat to public health because of its correlation with metabolic syndrome (3). The manifestations of sucrose-induced metabolic syndrome mostly initiate from nonalcoholic fatty liver disease (NAFLD), due to the composition of fructose (34, 35). The pathogenesis of NAFLD can include increased fatty acid influx or impaired lipid clearance, or a combination of both causes in most cases (36). Here, in our 4-week study, an HSD promoted symptoms typical of NAFLD development, with both blood and hepatic lipid accumulation (Fig. 1).
The occurrence of metabolic liver zonation suggests that higher lipogenesis rates are found in the pericentral zone and that hepatocytes in the periportal zone preferentially oxidize fatty acids (37). In patients with NAFLD, hepatic triglycerides were mostly derived from the preferential uptake of circulating free fatty acids (FFAs) by periportal hepatocytes, rather than de novo lipogenesis in pericentral hepatocytes (38). High-fructose diets in rat and mouse NAFLD models were also reported to induce periportal lipid accumulation (24, 36), similar to our results shown in Fig. 1g. The HSD increased the mRNA amplitude of almost all lipid metabolism–related enzymes investigated in this study (Fig. 3). However, we did not find circulating FFA changes in the HSD group (data not shown). Therefore, our findings suggest that periportal lipid deposition in HSD-induced NAFLD could be attributable to new mechanisms that are independent of increased FFA uptake.
Here, we found a novel circadian rhythm–dependent amplification of lipogenic gene expression levels. Concerning the enhanced amplitudes of gene mRNA oscillations (Figs. 4 and 5), transcriptional and post-transcriptional events are taken into consideration. Circadian recruitment of transcription factors to the promoters of target genes, which can activate or repress their transcriptions, comprises oscillations of cellular molecular clocks (16). Transcriptional regulation is a core process of circadian rhythms, so we first hypothesized that an HSD would induce the expression of key transcription factors mediating lipid and carbohydrate metabolism, such as SREBP1c, ChREBP, LXRα, and PPARα. As reported for several studies, the activation of these transcription factors changes in response to an HSD or HFD (9, 34, 39, 40). However, the oscillation patterns of these transcription factors did not vary as obviously as their target genes under an HSD in this study (Fig. 2, i–l). Taking account of possible fluctuating recruitments of transcription factor proteins to the target promoters, we estimated the transcription rates of target genes (i.e. lipid– and carbohydrate metabolism–related genes). Although rhythmic expression of these genes was primarily controlled at the transcriptional level, the HSD did not amplify the circadian oscillations of their transcription rates (Fig. 5). With this, the remaining possibility is post-transcriptional regulation, which could affect the mRNA oscillation patterns. Mechanisms of post-transcriptional regulation, including RNA polyadenylation and mRNA stability regulation, also play central roles in maintaining correct circadian rhythms (41). Hormones and nutrients affect mRNA stability through several distinct mechanisms, such as inducing regulatory proteins binding to poly(A) or 3′-UTR of mRNAs (42). Fructose, but not glucose, has been verified to increase GLUT5 mRNA stability via the cAMP pathway and PABP-interacting protein 2 binding in intestinal Caco-2 cells (43). These findings suggest the possibility that fructose participates in modulating mRNA stability in hepatocytes. As shown in Fig. 7b, the HSD decreased the hepatic phosphate level, which would be accompanied by lower ATP synthesis (33). Factors that require ATP would putatively contribute to regulating mRNA stability via 3′-UTR (44, 45). We compared the temporal expression of putative RNA-binding proteins and miRNAs that possibly bind the mRNAs of lipogenic genes, including ACLY, ACACA, FASN, ELOVL6, SCD1, FADS1, FADS2, G6PD, ME, and MTTP (Table S3). All of these investigated genes showed amplified circadian oscillations of mRNA (Fig. 4). Whereas no common miRNAs were identified that could bind to the mRNAs of lipogenic genes, we found three common RNA-binding proteins, namely EIF4B (eukaryotic translation initiation factor 4B), MBNL1 (muscleblind-like), and PABPC1 (polyadenylate-binding protein 1, cytoplasm), which can bind the mRNA 3′-UTRs of all lipogenic genes investigated in this study (Table S3). PABP has been reported to be involved in fructose-mediated GLUT5 mRNA stability enhancement (43). Moreover, the immediately diminished fructose-derived metabolites, dihydroxyacetone phosphate and glycerol 3-phosphate, should be investigated to determine whether they serve roles in mediating mRNA stability. Thus, despite the upstream regulators, clock genes and transcription factors were not changed by the HSD, and the oscillatory mRNA stability of downstream genes was enhanced by fructose or its metabolites, which promoted the formation of circadian rhythm–dependent amplifications.
In addition to employing calorie restriction to treat metabolic diseases, time-restricted feeding without changing caloric intake has becoming a prevalent intervention method (20–23). Disruption of molecular circadian rhythms is thought to be a pathogenic manifestation of metabolic syndrome (46–49). However, Chaix et al. (22) recently found that imposed feeding-fasting rhythms achieved from time-restricted feeding prevented metabolic diseases in circadian clock–deficient mice. This benefit may be increased by some preserved hepatic molecular rhythmic oscillations of metabolic processes caused by time-restricted feeding (22). Chaix et al. (22) also indicated that this feeding regimen exerted some previously unknown effects at the post-transcriptional level rather than directly impacting the rhythmic gene transcription (22). Similar to our results, their findings showed desynchronization of downstream metabolic pathways from core circadian clock regulation. It has been well-documented that feeding is an important synchronizer for peripheral circadian clocks and that rhythmic metabolic processes are regulated by circadian clocks (16). Here, the disintegration of the linkage between molecular circadian clock and metabolic pathways reveals that they could be separately synchronized by sucrose. Thus, it is conceivable that fructose or fructose-derived metabolites and the energy status (such as a lowered ATP concentration) could play roles in enhancing the circadian oscillatory amplitudes of lipogenic genes by mediating post-transcriptional events involved in mRNA stability. We also recently reported that time-restricted feeding of an HSD during the active phase of rats suppressed the development of fatty liver and hyperlipidemia (20). As our results showed that increased amplitudes of mRNA oscillations were mainly observed during the inactive phase of rats, excess sucrose may primarily impact mRNA levels during this period. Hence, avoiding sucrose intake during the inactive phase had significant ameliorative effects (20).
In conclusion, we found that excess sucrose intake mediated fatty liver and hyperlipidemia development, possibly via a novel circadian rhythm–dependent induction of lipogenesis, and the monosaccharide fructose played a key role in this mechanism. Our results also indicated that this phenomenon depended on fructose-mediated regulation of lipogenic genes at the post-transcriptional level. In the future, studies on regulation of the proteins and enzyme activity of key glucose and lipid regulatory pathways and verification of the metabolites regulating mRNA stability would help to examine the exact mechanism in greater details. The discovery of such circadian rhythm–dependent amplifications at the post-transcriptional level of metabolic processes should provide a new approach for investigating the complicated network of nutrients, circadian rhythms, and metabolism. In addition, it should help to establish further interventions against sucrose-induced metabolic syndrome, using a chrono-nutritional approach.
Experimental procedures
Animals
All rodent studies were approved by the Animal Research Committee of the Center for Animal Research and Education, Nagoya University (permit numbers 2013052201, 2014080701, and 2017110101) and Oita University (permit number J048002), and the experiments were performed according to the guidelines stipulated by this committee. Wherever possible, efforts were made to minimize animal suffering. Four independent experiments were designed, and 5-week-old male Wistar rats weighing ∼90 g each (Japan SLC, Shizuoka, Japan) were used for each experiment. All animals were independently housed under a 12-h light and 12-h dark cycle (lights on 0800–2000 h) at a temperature of 23 ± 1 °C, with free access to water. After adapting to the housing conditions for 6 days, the rats were randomly assigned to groups with equivalent initial body weights and plasma triglyceride concentrations and fed with an experimental diet for 4 weeks. Body weight and food intake of each rat was measured every day. For circadian oscillation analysis, rats were assigned to two groups (n = 28 rats/group) and fed either a CD or HSD ad libitum for 4 weeks. One hundred grams of the experimental diet consisted of 65.3 g of carbohydrate (only starch or sucrose), 20 g of casein, 5 g of cellulose, 5 g of corn oil, 3.5 g of mineral mixture (AIN93G) (50), 1 g of vitamin mixture (AIN93) (50), and 0.2 g of choline chloride. To analyze sucrose dose dependence, rats were divided into five groups (n = 12 rats/group). The rats in each group were respectively fed ad libitum with diets containing different sucrose/starch ratios (0:65.3, 10:55.3, 21.8:43.5, 43.5:21.8, or 65.3:0 g per 100 g) and the other components described above for 4 weeks. To analyze fructose dependence, rats were assigned to five groups (n = 12 rats/group). They were respectively fed diets with different carbohydrate sources, comprising 65.3 g of starch (CD), glucose (HGD), sucrose (HSD), fructose/glucose at a 1:1 ratio (HFGD), or fructose (HFD) per 100 g, and the other components described above ad libitum for 4 weeks. For metabolomics analysis, rats were assigned to two groups (n = 6 rats/group) fed a CGD or HSD ad libitum for 7 days. One hundred grams of the experimental diet consisted of 56.5 g of carbohydrate (only glucose or sucrose), 25 g of casein, 3 g of cellulose, 10 g of dextrin, 1 g of soybean oil, 3.5 g of mineral mixture (AIN93G) (50), 1 g of vitamin mixture (AIN93) (50), and 0.0014 g of tert-butylhydroquinone. To investigate circadian oscillations, rats were sacrificed by decapitation without anesthesia at ZT2, ZT8, ZT14, ZT18, or ZT22 (n = 4 for each group at each time point) on days 28 and 29. For the latter two experiments, to compare fluctuations occurring between day and night, the rats were sacrificed at ZT2 or ZT14 (n = 12 for each group at each time point). For metabolomics analysis, rats were sacrificed at ZT2. Rats were exposed to light only at the moment of decapitation at ZT14, ZT18, and ZT22 to minimize the influence of light. Livers and epididymal adipose tissues were excised and frozen immediately in liquid nitrogen and stored at −80 °C for further analysis.
Measurement of blood lipids
During the experimental period, blood was sampled at ZT5 after 4-h fasting from the caudal vein of rats, using heparin-coated capillaries. Plasma was prepared by centrifuging blood samples at 1500 × g (4 °C) for 10 min; blood supernatant fraction collected with heparin treatment (as mentioned above) was named as plasma. At the end of each experiment, whole-body blood was collected without heparin treatment when the rats were sacrificed, and serum was obtained by centrifuging blood samples at 1500 × g (4 °C) for 10 min. Plasma and serum triglyceride and cholesterol concentrations were measured using commercial kits (triglyceride E-test and T-cholesterol E-test, respectively; Wako Pure Chemical Industries, Osaka, Japan).
Liver histology and measurement of hepatic lipids
A slice of the left largest liver lobe was fixed in formalin and embedded in paraffin, sectioned into slices ∼3 μm thick, and stained with hematoxylin and eosin (H&E). Liver histology was analyzed with a KEYENCE BZ-X810 microscope and the accompanying software (Keyence, Osaka, Japan).
Hepatic lipids were extracted from the homogenized livers, following the method described by Folch et al. (51). Total lipids in the liver were determined gravimetrically. The extracted hepatic triglycerides, cholesterol, and phospholipids were measured using commercial kits (triglyceride E-test, T-cholesterol E-test, and phospholipid C-test, respectively; Wako Pure Chemical Industries).
Gene expression analysis
Total RNA was isolated from the liver of each rat, according the method of Chomczynski and Sacchi (52). RNA quality was confirmed by microchip electrophoresis with MultiNA (Shimadzu, Kyoto, Japan), which showed the presence of intact 18S and 28S rRNA. cDNA synthesis was performed as described previously (20). The cDNA was used to determine RNA levels by quantitative real-time PCR analysis with the StepOnePlus System (Thermo Fisher Scientific). The transcription rate was estimated by measuring nascent mRNA levels. To measure nascent mRNA, we used primers designed against the first-intron region of each nascent mRNA of interest (32). The APOE mRNA level was not affected by different carbohydrate sources and was used for normalization purposes. Sequences of the primer sets used are shown in Table S1.
Metabolomics analysis
For metabolomics analysis, rats were fed a CGD or HSD for 7 days, and liver extracts were used for metabolomics analysis. Targeted metabolomics analysis was performed by GC-tandem MS (GC-MS/MS) in multiple-reaction monitoring mode. GC-MS/MS analysis was performed on a GCMS-TQ8040 system (Shimazu, Kyoto, Japan) equipped with a DB-5 capillary column (30 m × 0.25-mm inner diameter; film thickness, 1 μm; Agilent, Santa Clara, CA). Each 1.0-μl aliquot of derivatized sample solution was automatically injected in splitless mode into the GLC column using an auto-injector (AOC-20i, Shimazu). During analysis with the GCMS-TQ8040 system, the injector temperature was held at 280 °C, and helium was used as carrier gas at a constant flow rate of 39.0 cm/s. The GLC column temperature was programmed to remain at 100 °C for 4 min and then to rise from 100 to 320 °C at a rate of 10 °C/min and then remain at 320 °C for a further 11 min. The total GLC run time was 37 min. The MS transfer line and ion-source temperatures were 280 and 200 °C, respectively. The ionization voltage was 70 eV. Argon was used as the collision-induced dissociation gas. Metabolite detection was performed using the Smart Metabolites Database version 2 (Shimazu, Kyoto, Japan) following the method of a previous study, with some modifications (53). The 2-isopropylmalic acid contained in the extraction solution was also used to evaluate the stability of our GC-MS/MS analysis system. Peak identification was performed automatically and then confirmed manually based on the specific precursor and product ions and the retention time.
Multivariate statistical analysis
The integral metabolomics data sets were imported into SIMCA version 13.0 (Umetrics, Sweden), and all variables were scaled to Pareto (par) for the OPLS-DA. Significant metabolites were selected using the S-plot method, in which both the intensity and reliability were visualized by statistical significance.
Rhythmicity analysis
Diurnal changes of serum lipids and hepatic gene expression were analyzed using JTK_CYCLE analysis (54), performed on R studio to determine the phases and amplitude in each group. The results of JTK_CYCLE analysis are displayed in Table S2. An adjusted p value of <0.05 was considered as an indicator of rhythmic expression during the assessed period. A phase shift of ±4 h or 0.5 < amplitude -fold change < 1.5 (compared with the control group) was regarded as an obvious experimental difference.
Statistical analysis
The data displayed in all figures represent the mean ± S.D. (or mean + S.D.). Metabolomics analysis and gene expression oscillations were analyzed as described above. Except for metabolomics analysis and JTK_CYCLE analysis, all statistical analyses were performed with IBM SPSS Statistics (version 22) software. Student's t test was used when comparing two groups, and one-way ANOVA followed by Duncan's multiple-range post hoc test was performed when comparing more than two groups. When analyzing plasma parameters, two-way repeated measurement ANOVA was used to determine variance differences over time. Two-way ANOVA was used to assess interactions between diets and time, and Duncan's multiple-range post hoc test was used when a significant interaction effect was observed. p < 0.05 was considered to reflect a statistically significant difference.
Author contributions
S. S. and H. O. conceptualization; S. S., F. H., and H. O. data curation; S. S. formal analysis; S. S., D. K., M. U., S. N., K. S., S. I., S. M., and H. O. investigation; S. S. writing-original draft; S. S., F. H., and H. O. writing-review and editing; H. O. funding acquisition.
Supplementary Material
Acknowledgment
We thank Yasuko Matsuyama for providing technical assistance.
This work was supported by Japan Society for the Promotion of Science (JSPS) Grants 21658052, 25292069, and 16H04922 (to H. O.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1 and S2.
- HSD
- high-sucrose diet
- CD
- control starch diet
- ANOVA
- analysis of variance
- HGD
- high-glucose diet
- HFGD
- high-fructose and -glucose diet
- HFD
- high-fructose diet
- PPP
- pentose phosphate pathway
- CGD
- control glucose diet
- OPLS-DA
- orthogonal partial least-squares discriminant analysis
- NAFLD
- nonalcoholic fatty liver disease
- FFA
- free fatty acid
- UTR
- untranslated region
- H&E
- hematoxylin and eosin.
References
- 1. Huang P. L. (2009) A comprehensive definition for metabolic syndrome. Dis. Model Mech. 2, 231γÇô237 10.1242/dmm.001180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alberti K. G., and Zimmet P. Z. (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 15, 539–553 [DOI] [PubMed] [Google Scholar]
- 3. Lustig R. H., Schmidt L. A., and Brindis C. D. (2012) Public health: the toxic truth about sugar. Nature 482, 27–29 10.1038/482027a [DOI] [PubMed] [Google Scholar]
- 4. Roncal-Jimenez C. A., Lanaspa M. A., Rivard C. J., Nakagawa T., Sanchez-Lozada L. G., Jalal D., Andres-Hernando A., Tanabe K., Madero M., Li N., Cicerchi C., Mc Fann K., Sautin Y. Y., and Johnson R. J. (2011) Sucrose induces fatty liver and pancreatic inflammation in male breeder rats independent of excess energy intake. Metab. Clin. Exp. 60, 1259–1270 10.1016/j.metabol.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macdonald I. A. (2016) A review of recent evidence relating to sugars, insulin resistance and diabetes. Eur. J. Nutr. 55, 17–23 10.1007/s00394-016-1340-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tappy L., Lê K. A., Tran C., and Paquot N. (2010) Fructose and metabolic diseases: new findings, new questions. Nutrition 26, 1044–1049 10.1016/j.nut.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 7. Caliceti C., Calabria D., Roda A., and Cicero A. F. G. (2017) Fructose intake, serum uric acid, and cardiometabolic disorders: a critical review. Nutrients 9, E395 10.3390/nu9040395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun S. Z., and Empie M. W. (2012) Fructose metabolism in humans: what isotopic tracer studies tell us. Nutr. Metab. 9, 89 10.1186/1743-7075-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Softic S., Cohen D. E., and Kahn C. R. (2016) Role of dietary fructose and hepatic de novo lipogenesis in fatty liver disease. Dig. Dis. Sci. 61, 1282–1293 10.1007/s10620-016-4054-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berg J. M., Tymoczko J. L., Gatto G. J. Jr., and Stryer L. (2015) Biochemistry, 8th Ed., pp. 232–234, W. H. Freeman, New York [Google Scholar]
- 11. Rodwell V. W., Bender D., Botham K. M., Kennelly P. J., and Weil P. A. (2015) Harpers Illustrated Biochemistry 30th Edition, pp. 465–466, McGraw-Hill, New York [Google Scholar]
- 12. Zakim D., Pardini R. S., Herman R. H., and Sauberlich H. E. (1967) Mechanism for the differential effects of high carbohydrate diets on lipogenesis in rat liver. Biochim. Biophys. Acta. 144, 242–251 10.1016/0005-2760(67)90154-3 [DOI] [PubMed] [Google Scholar]
- 13. Fitch W. M., and Chaikoff I. L. (1960) Extent and patterns of adaptation of enzyme activities in livers of normal rats fed diets high in glucose and fructose. J. Biol. Chem. 235, 554–557 [PubMed] [Google Scholar]
- 14. Kornacker M. S., and Lowenstein J. M. (1965) Citrate and the conversion of carbohydrate into fat: activities of citrate-clevage enzyme and acetate thiokinase in livers of normal and diabetic rats. Biochem. J. 95, 832–837 10.1042/bj0950832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruckdorfer K. R., Khan I. H., and Yudkin J. (1972) Fatty acid synthetase activity in the liver and adipose tissue of rats fed with various carbohydrates. Biochem. J. 129, 439–446 10.1042/bj1290439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bass J. (2012) Circadian topology of metabolism. Nature 491, 348–356 10.1038/nature11704 [DOI] [PubMed] [Google Scholar]
- 17. Asher G., and Sassone-Corsi P. (2015) Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 10.1016/j.cell.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 18. Yamajuku D., Inagaki T., Haruma T., Okubo S., Kataoka Y., Kobayashi S., Ikegami K., Laurent T., Kojima T., Noutomi K., Hashimoto S., and Oda H. (2012) Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci. Rep. 2, 439 10.1038/srep00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panda S. (2016) Circadian physiology of metabolism. Science 354, 1008–1015 10.1126/science.aah4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun S., Hanzawa F., Umeki M., Ikeda S., Mochizuki S., and Oda H. (2018) Time-restricted feeding suppresses excess sucrose-induced plasma and liver lipid accumulation in rats. PLoS One 13, e0201261 10.1371/journal.pone.0201261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E. A., Gill S., Leblanc M., Chaix A., Joens M., Fitzpatrick J. A. J., Ellisman M. H., and Panda S. (2012) Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 10.1016/j.cmet.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaix A., Lin T., Le H. D., Chang M. W., and Panda S. (2019) Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e4 10.1016/j.cmet.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaix A., Zarrinpar A., Miu P., and Panda S. (2014) Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991–1005 10.1016/j.cmet.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hijmans B. S., Grefhorst A., Oosterveer M. H., and Groen A. K. (2014) Zonation of glucose and fatty acid metabolism in the liver: mechanism and metabolic consequences. Biochimie 96, 121–129 10.1016/j.biochi.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 25. Havel P. J. (2005) Dietary fructose: implications for dysregulation of energy homeostasis and lipid/carbohydrate metabolism. Nutr. Rev. 63, 133–157 10.1111/j.1753-4887.2005.tb00132.x [DOI] [PubMed] [Google Scholar]
- 26. Bass J., and Takahashi J. S. (2010) Circadian integration of metabolism and energetics. Science 330, 1349–1354 10.1126/science.1195027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Lo Sasso G., Moschetta A., and Schibler U. (2009) REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7, e1000181 10.1371/journal.pbio.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iizuka K. (2017) The role of carbohydrate response element binding protein in intestinal and hepatic fructose metabolism. Nutrients 9, E181 10.3390/nu9020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sancar G., and Brunner M. (2014) Circadian clocks and energy metabolism. Cell Mol. Life Sci. 71, 2667–2680 10.1007/s00018-014-1574-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura M. T., Yudell B. E., and Loor J. J. (2014) Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 53, 124–144 10.1016/j.plipres.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 31. Tian W. N., Braunstein L. D., Pang J., Stuhlmeier K. M., Xi Q. C., Tian X., and Stanton R. C. (1998) Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 273, 10609–10617 10.1074/jbc.273.17.10609 [DOI] [PubMed] [Google Scholar]
- 32. Ripperger J. A., and Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 10.1038/ng1738 [DOI] [PubMed] [Google Scholar]
- 33. Karczmar G. S., Kurtz T., Tavares N. J., and Weiner M. W. (1989) Regulation of hepatic inorganic phosphate and ATP in response to fructose loading: an in vivo 31P-NMR study. Biochim. Biophys. Acta. 1012, 121–127 10.1016/0167-4889(89)90084-0 [DOI] [PubMed] [Google Scholar]
- 34. Jegatheesan P., and De Bandt J. P. (2017) Fructose and NAFLD: the multifaceted aspects of fructose metabolism. Nutrients 9, E230 10.3390/nu9030230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedman S. L., Neuschwander-Tetri B. A., Rinella M., and Sanyal A. J. (2018) Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takahashi Y., Soejima Y., and Fukusato T. (2012) Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 18, 2300–2308 10.3748/wjg.v18.i19.2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kietzmann T. (2017) Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 11, 622–630 10.1016/j.redox.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., and Parks E. J. (2005) Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 115, 1343–1351 10.1172/JCI23621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tappy L., and Lê K. A. (2010) Metabolic effects of fructose and the worldwide increase in obesity. Physiol. Rev. 90, 23–46 10.1152/physrev.00019.2009 [DOI] [PubMed] [Google Scholar]
- 40. Bizeau M. E., and Pagliassotti M. J. (2005) Hepatic adaptations to sucrose and fructose. Metab. Clin. Exp. 54, 1189–1201 10.1016/j.metabol.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 41. Bass J., and Lazar M. A. (2016) Circadian time signatures of fitness and disease. Science 354, 994–999 10.1126/science.aah4965 [DOI] [PubMed] [Google Scholar]
- 42. Ross J. (1996) Control of messenger RNA stability in higher eukaryotes. Trends Genet. 12, 171–175 10.1016/0168-9525(96)10016-0 [DOI] [PubMed] [Google Scholar]
- 43. Gouyon F., Onesto C., Dalet V., Pages G., Leturque A., and Brot-Laroche E. (2003) Fructose modulates GLUT5 mRNA stability in differentiated Caco-2 cells: role of cAMP-signalling pathway and PABP (polyadenylated-binding protein)-interacting protein (Paip) 2. Biochem. J. 375, 167–174 10.1042/bj20030661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen H. H., Xu J., Safarpour F., and Stewart A. F. R. (2007) LMO4 mRNA stability is regulated by extracellular ATP in F11 cells. Biochem. Biophys. Res. Commun. 357, 56–61 10.1016/j.bbrc.2007.03.113 [DOI] [PubMed] [Google Scholar]
- 45. Hadadi E., Zhang B., Baidžajevas K., Yusof N., Puan K. J., Ong S. M., Yeap W. H., Rotzschke O., Kiss-Toth E., Wilson H., and Wong S. C. (2016) Differential IL-1β secretion by monocyte subsets is regulated by Hsp27 through modulating mRNA stability. Sci. Rep. 6, 39035 10.1038/srep39035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., and Bass J. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 10.1126/science.1108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamajuku D., Okubo S., Haruma T., Inagaki T., Okuda Y., Kojima T., Noutomi K., Hashimoto S., and Oda H. (2009) Regular feeding plays an important role in cholesterol homeostasis through the liver circadian clock. Circ. Res. 105, 545–548 10.1161/CIRCRESAHA.109.199034 [DOI] [PubMed] [Google Scholar]
- 48. Paschos G. K., Ibrahim S., Song W. L., Kunieda T., Grant G., Reyes T. M., Bradfield C. A., Vaughan C. H., Eiden M., Masoodi M., Griffin J. L., Wang F., Lawson J. A., and Fitzgerald G. A. (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777 10.1038/nm.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimizu H., Hanzawa F., Kim D., Sun S., Laurent T., Umeki M., Ikeda S., Mochizuki S., and Oda H. (2018) Delayed first active-phase meal, a breakfast-skipping model, led to increased body weight and shifted the circadian oscillation of the hepatic clock and lipid metabolism-related genes in rats fed a high-fat diet. PLoS One 13, e0206669 10.1371/journal.pone.0206669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reeves P. G. (1997) Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr. 127, 838S–841S 10.1093/jn/127.5.838S [DOI] [PubMed] [Google Scholar]
- 51. Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 52. Chomczynski P., and Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 10.1006/abio.1987.9999 [DOI] [PubMed] [Google Scholar]
- 53. Nishiumi S., Kobayashi T., Kawana S., Unno Y., Sakai T., Okamoto K., Yamada Y., Sudo K., Yamaji T., Saito Y., Kanemitsu Y., Okita N. T., Saito H., Tsugane S., Azuma T., Ojima N., and Yoshida M. (2017) Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 8, 17115–17126 10.18632/oncotarget.15081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hughes M. E., Hogenesch J. B., and Kornacker K. (2010) JTK_CYCLE: an efficient non-parametric algorithm for detecting rhythmic components in genome-scale datasets. J. Biol. Rhythms 25, 372–380 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.