Abstract
In the cerebellum, the bHLH transcription factors Ptf1a and Atoh1 are expressed in distinct neuroepithelial regions, the ventricular zone (VZ) and the rhombic lip (RL), and are required for producing GABAergic and glutamatergic neurons, respectively. However, it is unclear whether Ptf1a or Atoh1 is sufficient for specifying GABAergic or glutamatergic neuronal fates. To test this, we generated two novel knock-in mouse lines, Ptf1aAtoh1 and Atoh1Ptf1a, that are designed to express Atoh1 and Ptf1a ectopically in the VZ and RL, respectively. In Ptf1aAtoh1 embryos, ectopically Atoh1-expressing VZ cells produced glutamatergic neurons, including granule cells and deep cerebellar nuclei neurons. Correspondingly, in Atoh1Ptf1a animals, ectopically Ptf1a-expressing RL cells produced GABAergic populations, such as Purkinje cells and GABAergic interneurons. Consistent results were also obtained from in utero electroporation of Ptf1a or Atoh1 into embryonic cerebella, suggesting that Ptf1a and Atoh1 are essential and sufficient for GABAergic versus glutamatergic specification in the neuroepithelium. Furthermore, birthdating analyses with BrdU in the knock-in mice or with electroporation studies showed that ectopically produced fate-changed neuronal types were generated at temporal schedules closely simulating those of the wild-type RL and VZ, suggesting that the VZ and RL share common temporal information. Observations of knock-in brains as well as electroporated brains revealed that Ptf1a and Atoh1 mutually negatively regulate their expression, probably contributing to formation of non-overlapping neuroepithelial domains. These findings suggest that Ptf1a and Atoh1 specify spatial identities of cerebellar neuron progenitors in the neuroepithelium, leading to appropriate production of GABAergic and glutamatergic neurons, respectively.
Keywords: cerebellum, development, identity, neuron, spatial, transcription factor
Introduction
The cerebellum presents as a good model to investigate the machinery for neuronal subtype specification, because it contains only ∼10 types of neurons, all of which are well characterized (Chan-Palay et al., 1977; Hatten and Heintz, 1995; Wang and Zoghbi, 2001). Cerebellar neurons are generated from two distinct neuroepithelial zones; the ventricular zone (VZ) and the more dorsally located rhombic lip (RL; Hatten et al., 1997). Genetic fate-mapping studies suggested that the VZ produces all GABAergic inhibitory-type neurons, whereas the RL generates all glutamatergic excitatory neurons (Hoshino et al., 2005; Machold and Fishell, 2005; Wang et al., 2005). Two bHLH proteins, Ptf1a and Atoh1, are exclusively expressed in the VZ and RL, respectively, in the developing cerebellum. Loss of Ptf1a and Atoh1 results in failure of GABAergic and glutamatergic neuron production in the cerebellum, respectively, suggesting their involvement in the generation of corresponding neuronal types (Hoshino et al., 2005; Machold and Fishell, 2005; Wang et al., 2005).
Ptf1a and Atoh1 are longitudinally expressed in the neuroepithelium of the hindbrain [rhombomeres (r) 1–8] in a non-overlapping manner, defining distinct neuroepithelial domains. Additionally, the Atoh1- and Ptf1a-expressing neuroepithelial domains generate excitatory and inhibitory neurons of the cochlear nucleus, respectively, in the middle hindbrain (r2–r5; Fujiyama et al., 2009). In the caudal hindbrain (r6–r8), the Atoh1- and Ptf1a-expressing neuroepithelial domains produce mossy fiber and climbing fiber neurons, respectively (Landsberg et al., 2005; Wang et al., 2005; Yamada et al., 2007; Dun, 2012; Hori and Hoshino, 2012; Hoshino et al., 2012). Furthermore, loss of Atoh1 and Ptf1a results in loss of production of corresponding neurons. These findings suggest that Atoh1 and Ptf1a participate in the specification of distinct neuron subtypes within each subregion (rostral, middle, or caudal) of the hindbrain, raising the hypothesis that these bHLH factors may confer spatial identity of neuroepithelial domains along the dorsoventral axis, enabling each neuroepithelial domain to produce specific types of neurons (Hoshino, 2006, 2012).
To test this, we aimed to clarify whether Atoh1 and Ptf1a are sufficient to produce glutamatergic and GABAergic neurons in the cerebellum, despite the fact that they are required for generating corresponding neurons. First, we generated two lines of knock-in mice in which Atoh1 and Ptf1a are expressed ectopically in the cerebellar VZ and the RL, respectively. This resulted in the ectopic generation of glutamatergic and GABAergic neurons from the VZ and the RL, respectively, suggesting that expression of these bHLH factors is sufficient to change neuronal fates. Transient introduction of Atoh1 and Ptf1a into cerebellar primordia by in utero electroporation confirmed this outcome. Furthermore, birthdating studies revealed that the ectopically produced fate-changed neurons were generated at temporal schedules closely mimicking normal development, suggesting common temporal characteristics of the neuroepithelium between the RL and VZ. In addition, endogenous expression of Ptf1a and Atoh1 was suppressed by ectopic expression of Atoh1 and Ptf1a, respectively, suggesting that these factors mutually suppress each other's expression and further indicating that this machinery may contribute to the formation of non-overlapping neuroepithelial domains, the RL and VZ.
Materials and Methods
Animals.
All animal experiments in this study have been approved by the Animal Care and Use Committee of the National Institute of Neuroscience, National Center of Neurology and Psychiatry (Tokyo, Japan; project 2008005). The Ptf1aCre, Ptf1acbll, Rosa26R (R26R), Tg–Atoh1–Cre, and Atoh1CreERn mouse lines were described previously (Soriano, 1999; Kawaguchi et al., 2002; Hoshino et al., 2005; Fujiyama et al., 2009). The pgk–neo cassette of the Atoh1CreERn mice were removed by crossing with CAG–FLP (Flippase recombinase) mice (Kanki et al., 2006) in which the Cre recombinase was active in the germ line. In this study, this Atoh1CreER line was used as an Atoh1 null allele and therefore renamed Atoh1null.
Ptf1aAtoh1 and Atoh1Ptf1a knock-in mice were generated as follows (Fig. 1; see Fig. 6). The targeting vector for Ptf1aAtoh1 contained the genomic DNA around the Ptf1a gene whose open reading frame (ORF) was replaced with the Atoh1 cDNA and pgk–neo flanked by LoxP sequences (Fig. 1A). The targeting vector for Atoh1Ptf1a included the genomic DNA around the Atoh1 gene whose ORF was replaced with the Ptf1a cDNA and pgk–neo flanked by LoxP sequences (see Fig. 6A). These targeting vectors were electroporated into ES cells in previously described conditions of culture, transfection, and selection of clones. For the Ptf1aAtoh1 or Atoh1Ptf1a allele, a recombinant ES cell clone was injected into BALB/c blastocysts, and resulting chimeric males were bred with C57BL/6 females to obtain germ-line transmission of the mutation. In addition, the Ptf1aAtoh1 strain mice (F1) were crossed with CAG–Cre transgenic mice to remove the pgk–neo cassette. The Ptf1aAtoh1 and Atoh1Ptf1a strains were maintained by crossing into a C57BL/6 background. PCR genotyping was performed on genomic tail DNA. Genotyping of the Ptf1aAtoh1 allele was performed with primers situated in the upper (AGTGACTCTGATGAGGCCAGTTAG) and lower (TTCCGATCATATTCAATAACCCTTA) strands. PCR conditions for the Ptf1aAtoh1 allele were 35 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 3 min. Genotyping of the Atoh1Ptf1a allele was performed with primers situated in the upper (ATCTTTCGACAACATAGAGAACGA) and lower (ATACGACATTTTAGCAGCATATTGG) strands. PCR conditions for the Atoh1Ptf1a allele were 35 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 3 min.
Figure 1.
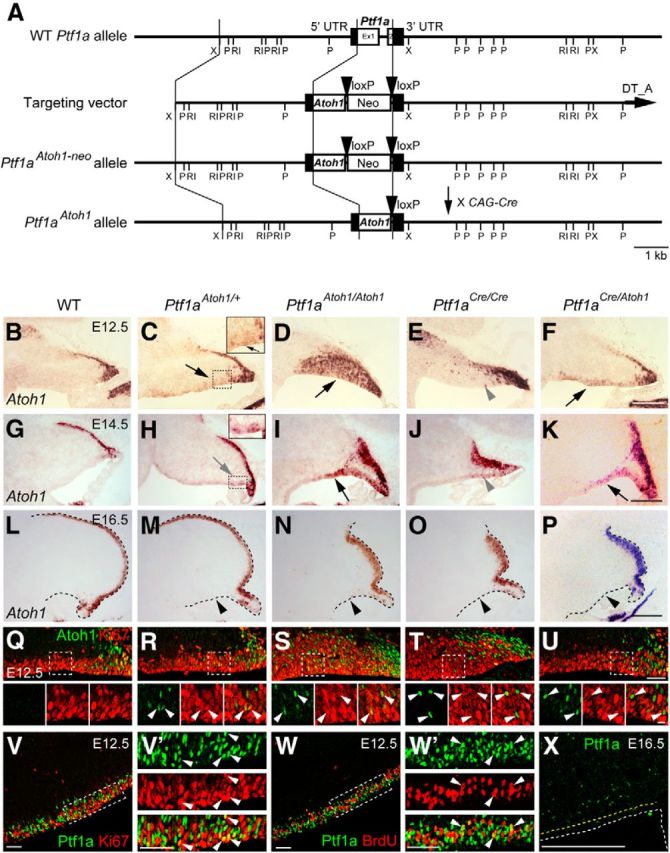
Expression of Atoh1 and Ptf1a in the cerebellar primordium at embryonic stages. A, Generation of Ptf1aAtoh1 knock-in mouse line. Diagram showing Ptf1a wild-type allele, targeting construct, Ptf1aAtoh1-neo, and Ptf1aAtoh1 knock-in alleles. The pgk–neo cassette of the Ptf1aAtoh1-neo mice was removed by crossing with CAG–Cre mice. Boxes in wild-type Ptf1a allele represent noncoding (black) and coding (white) Ptf1a exon sequences. Black arrowheads represent loxP sequences. Ex, Exon; Neo, neomycin-resistance gene under the control of the pgk promoter; P, PstI; RI, EcoRI; X, XbaI. B–X, Sagittal sections of cerebella of indicated genotypes at indicated embryonic stages. Top is dorsal, and left is rostral. B–P, Atoh1 transcripts (in situ hybridization) visualized by in situ hybridization probe that detects both endogenous and exogenous Atoh1 transcripts. Black and gray arrows and gray arrowheads indicate cells that expressed Atoh1 ectopically in the VZ. Black arrowheads indicate the disappearance of ectopic Atoh1 expression in the VZ at later stages. Q–U, Expression of Atoh1 and Ki67 in the VZ at E12.5. Arrowheads indicate colocalization of Atoh1 and Ki67. V–W′, Expression of Ptf1a and markers (Ki67 or BrdU) is visualized by immunostaining. BrdU was administered 30 min before fixation. V′, W′, High magnification of boxed regions in V and W, respectively. Arrowheads indicate coexpression of Ptf1a and markers in the VZ. X, Ptf1a expression in the cerebellar primordium at E16.5. Yellow dotted lines indicate the edges of the VZ. WT, Wild-type. Scale bars: B–P, X, 200 μm; Q–W′, 50 μm; small panels at bottom in Q–U, 10 μm.
Figure 6.
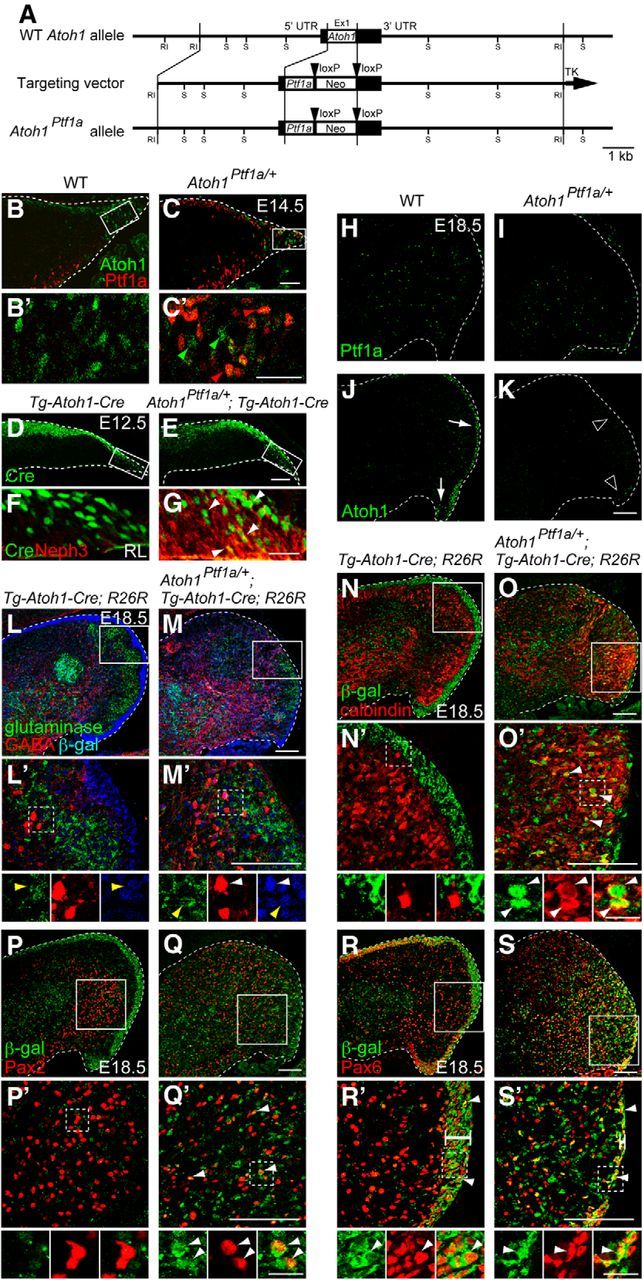
Lineage tracing analysis of cells produced from the RL that ectopically expresses Ptf1a. A, Generation of Atoh1Ptf1a knock-in mouse line. Diagram showing Atoh1 wild-type allele, targeting construct, and Atoh1Ptf1a mutant alleles. Boxes in wild-type Atoh1 allele represent noncoding (black) and coding (white) Atoh1 exon sequences. Black arrowheads represent loxP sequences. Ex, Exon; Neo, neomycin-resistance gene under the control of the pgk promoter; RI, EcoRI; S, SacI. B–K, Localization of Ptf1a, Atoh1, Cre, and Neph3 proteins in the cerebellar primordium of embryos of indicated genotypes at E14.5 (B–C′), E12.5 (D–G), and E18.5 (H–K). C′, Green and red arrowheads show Atoh1 and Ptf1a expression in the RL, respectively. F, G, Cre and Neph3 expression in the rectangular regions in D and E, respectively. White arrowheads in G show colocalization of Cre and Neph3 in the RL. L–M′, Triple immunostaining with β-gal (blue), GABA (red), and glutaminase (green). Yellow and white arrowheads indicate colocalization of β-gal with glutaminase and GABA, respectively. N–S′, Double immunostaining with β-gal (green) and cell type-specific markers (red), such as calbindin (N–O′), Pax2 (P–Q′), and Pax6 (R–S′). B′, C′, L′–S′, High-magnification pictures of boxed regions in B, C, and L–S, respectively. Small panels below L′–S′ are single colored or merged pictures corresponding to the rectangular regions in L′–S′. White bars in R′ and S′ indicate the width of the EGL. All are sagittal sections. Top is dorsal, and left is rostral. WT, Wild-type. Scale bars: B–E, 50 μm; B′, C′, F, G, 25 μm; H–S′, 100 μm; high-magnification views in L′–S′, 20 μm.
Antibodies and immunohistochemistry.
Primary antibodies used in this study were anti-Pax6 (1:300; rabbit; Covance), Tbr1 (1:1000; rabbit; Millipore Bioscience Research Reagents), Tbr2 (1:1000; rabbit; Millipore Bioscience Research Reagents), GABA (1:500; rabbit; Sigma), calbindin (1:500; rabbit; Millipore Bioscience Research Reagents), Pax2 (1:200; rabbit; Invitrogen), glutaminase (1:200; guinea pig; a kind gift from Dr. Watanabe, Hokkaido University, Hokkaido, Japan), Atoh1 (1:50,000; guinea pig; a kind gift from Dr. Y. Sasai, RIKEN CDB, Japan), Corl2 (1:500; rabbit), Neph3 (1:250; hamster; kind gifts from Dr. Y. Ono, KAN Research Institute, Kobe, Japan), GFP (1:50; rat; a kind gift from Dr. A. Imura, Foundation for Biomedical Research and Innovation, Kobe, Japan), GFP (1:2000; rabbit; Invitrogen), β-galactosidase (β-gal; 1:800; goat; Biogenesis), β-gal (1:1000; chick; Abcam), Ki67 (1:500; rat; eBioscience), BrdU (1:200; rat; Serotec), and Cre (1:1000; rabbit; Novagen). Anti-Ptf1a and Atoh1 antibodies were generated as follows. The fragment of mouse Ptf1a full-length (Full: 1–324 aa) or N-terminal (NT: 1–145 aa) or mouse Atoh1 full-length (Full; 1–351 aa) or N-terminal (NT: 1–154 aa) was inserted into pMAL-c2 (New England Laboratories) or pGEX-4T-2 (GE Healthcare), respectively. Maltose-binding protein (MBP) and glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 (DE3) and purified according to the instructions of the manufacturer. Polyclonal rabbit anti-Ptf1a or anti-Atoh1 antibody was prepared against MBP–Ptf1a–Full or GST–Atoh1–Full as an antigen and then purified by use of GST–Ptf1a–NT or MBP–Atoh1–NT, respectively. These antibodies detect no signals in the Ptf1a null (Ptf1aCre/Cre) or Atoh1 null (Atoh1null/null) mutants.
Immunohistochemistry was performed as follows. Embryos were fixed in 4% paraformaldehyde (PFA) in PBS at 4°C, 2–3 h, equilibrated in 10, 20, and 30% sucrose in PBS progressively at 4°C, embedded in OCT compound (Sakura Finetek), and frozen at −20°C. Sections were made by cryostat (Leica) at 12–14 μm and blocked in 5% normal donkey or goat serum and 0.1% Triton X-100/PBS (PBST) for 1 h. Then, these sections were incubated with primary antibodies diluted in 0.1% PBST containing 5% normal donkey or goat serum overnight at 4°C, washed with PBS, and incubated with secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 568, Alexa Fluor 594, or Alexa Fluor 647 (1:400; Invitrogen) and TOPRO-3 (1:2000; Invitrogen) in 0.1% PBST for 1.5 h. In Figure 1V–W′, the number of Ptf1a and Ki67 (n = 18) or BrdU (n = 35) colocalized cells in the VZ was measured using MetaMorph software (Molecular Devices). In Figure 4J–L, the number of Tbr2-positive cells in the cerebellar primordium was also measured using the MetaMorph software (n = 6). In Figure 6B–C′, the number of Atoh1- or Ptf1a-positive cells in the RL was measured (n = 16–18). Student's t test (two-tailed) was performed to calculate p values.
Figure 4.
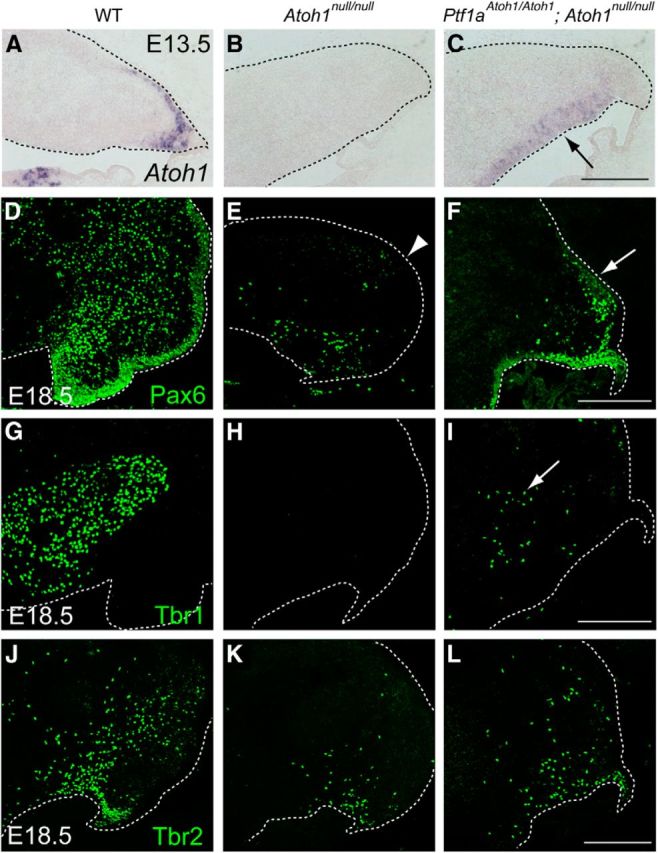
Identity of cells produced in the cerebellum that expresses Atoh1 only in the VZ but not in the RL. A–C, Expression of all (endogenous + exogenous) Atoh1 transcripts in the cerebellum at E13.5. Black arrow in C indicates ectopic expression of Atoh1 in the VZ. D–L, Immunostaining with cell type-specific markers, such as Pax6 (D–F), Tbr1 (G–I), and Tbr2 (J–L), to the E18.5 cerebella. White dotted lines indicate the edge of the cerebellar primordium. All are sagittal sections. Top is dorsal, and left is rostral. WT, Wild-type. Scale bars, 200 μm.
BrdU incorporation experiment.
Pregnant mice [embryonic day 10.5 (E10.5), E12.5, and E14.5] were given two 50 mg/kg intraperitoneal injections of BrdU with a 30 min interval. One hour after the first injection, the embryos of pregnant mice (E12.5) were fixed, and frozen sections were subjected to immunostaining with an anti-BrdU antibody as described previously (Kawauchi et al., 2003, 2006; Yamada et al., 2007). For the birthdate analyses, the embryos were fixed at E18.5. Frozen sections were sequentially treated with anti-β-gal antibody and a specific marker (such as anti-Pax6, Tbr1, calbindin, and Pax2 antibodies), incubated with Alexa Fluor 488- or Alexa Fluor 647-conjugated secondary antibodies, refixed with 4% PFA for 20 min, incubated with 2N HCl for 30 min at 37°C, and incubated with anti-BrdU antibody at 4°C overnight, followed by treatment with Alexa Fluor 594-conjugated secondary antibody.
Detection of β-gal using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside.
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining was performed as described previously (Yamada et al., 2007).
In situ hybridization.
The probes used here were Atoh1 (a kind gift from Dr. R. Kageyama, Kyoto University, Kyoto, Japan; Akazawa et al., 1995), Atoh1 3′ untranslated region (UTR; nucleotides 1235–1968, GenBank accession number NM007500) and Atoh1 ORF (nucleotides 327–1184, GenBank accession number NM007500). The antisense probes of Atoh1 3′ UTR and Atoh1 ORF were generated using RT-PCR from total RNA isolated from E11.5 mouse brains with Trizol (Invitrogen). The PCR-amplified DNA fragments were cloned into the pCR–Blunt (Invitrogen). The probes were labeled with digoxigenin–UTP (Roche). In situ hybridization was performed as follows. Cryosections were fixed with 4% PFA in PBS for 10 min and washed with PBS for 10 min. Then the sections were acetylated by incubation in 0.1 m triethanolamine-HCl and 0.25% acetic anhydride for 10 min. After washing twice with PBS for 10 min, the samples were dried. Hybridization was performed with probes in a hybridization solution (50% formamide, 5× SSC, 0.1% SDS, 0.1% N-Lauroylsarcosine (NLS), and 2% blocking reagent) at 60°C for 20 h. After hybridization, the specimens were washed in 5× SSC and 0.1% NLS at 60°C and twice in wash buffer (2× SSC, 50% formamide, and 0.1% NLS) at 60°C for 20 min, followed by RNase treatment in RNase buffer (20 μg/ml RNaseA in 10 mm Tris-HCl, pH 8.0, 0.5 m NaCl, and 1 mm EDTA) at 37°C for 30 min. Then the sections were washed twice with 2× SSC and 0.1% NLS at 37°C for 20 min, twice with 0.2× SSC and 0.1% NLS at 37°C for 20 min, and once with Tris-HCl-buffered saline (TS) 7.5 (0.1 m Tris-HCl, pH 7.5, and 0.15 m NaCl) for 5 min. After treatment with 1% blocking reagent (Roche) in TS7.5 for 1 h, the samples were incubated with anti-DIG alkaline phosphatase conjugate (Roche) diluted 1:2000 with TS7.5 overnight. The sections were washed three times with TS9.5 (0.1 m Tris-HCl, pH 9.5, 0.1 m NaCl, and 50 mm MgCl2). Coloring reactions were performed with NBT/BCIP (Roche) diluted 1:50 with TS9.5 for several hours to overnight and then washed with PBS. The samples were dehydrated and mounted with Entellan (Merck).
Plasmids.
The cDNA of Atoh1 (GenBank accession number NM007500) and Ptf1a (GenBank accession number NM018809) were inserted into pCAG2–IRES–GFP vector to generate pCAG–Atoh1–IRES–EGFP and pCAG–Ptf1a–IRES–EGFP plasmids. As a control, pCAG–EGFP plasmid was used. These plasmids were prepared using the Endo Free plasmid purification kit (Qiagen).
In utero electroporation and quantification.
Pregnant ICR mice were purchased from SLC Japan. In utero electroporation was performed as described previously (Kawauchi et al., 2003, 2006). Briefly, pregnant mice carrying E12.5 embryos were anesthetized. One microliter of plasmid DNA (2 μg/μl) in H2O containing fast green was injected into the fourth ventricles of embryonic brains. Holding the embryo in utero with forceps-type electrode (Nepa Gene), 50 ms of 50 V electronic pulses were delivered five times at intervals of 450 ms with a square electroporator (Nepa Gene). Electroporated brains were cut into 14 μm sagittal sections with a cryostat. Fluorescence images of frozen sections of EGFP-expressing mouse brains were captured by FV1000 (Olympus) or LSM780 (Zeiss) laser scanning confocal microscopes.
The cerebellar RL or VZ of six embryos per pCAG–Ptf1a–IRES–EGFP or pCAG–Atoh1–IRES–EGFP group was analyzed in each experiment. Cells were scored on the basis of staining for GFP. To investigate whether GFP and Ptf1a/Atoh1 signals were detected in the same cells, the z-axis images per 1 μm were acquired. Student's t test (one-tailed) was performed to calculate p values and to determine the results in Figure 10.
Figure 10.
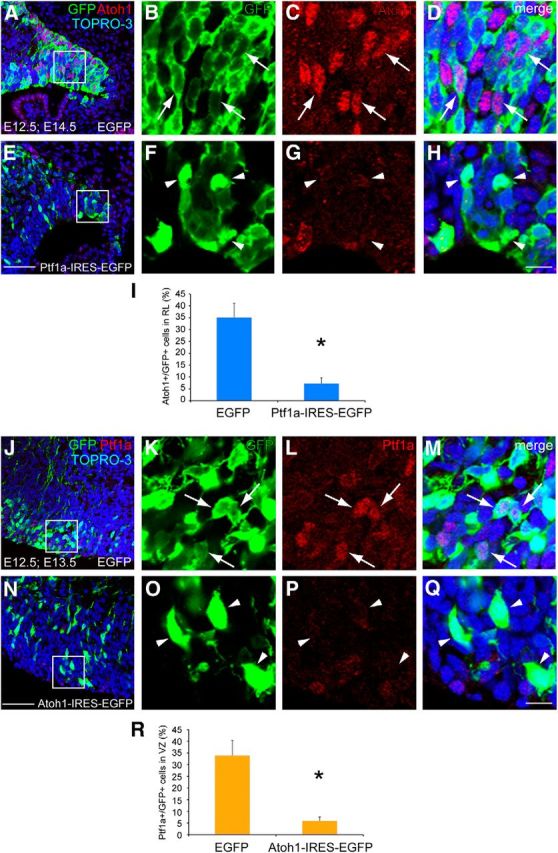
Mutual control of Ptf1a and Atoh1 expression. A–H, Sagittal sections of E14.5 wild-type RL electroporated with control (A–D) or Ptf1a (E–H) at E12.5. GFP (green) and Atoh1 (red) were visualized by immunostaining. Nuclei were also visualized by TOPRO-3 (blue). B–D, F–H, High magnification of boxed region in A and E. I, Ratios of Atoh1-positive cells in the GFP-positive cells in the RL. The ratios are 35.08 ± 5.98 and 7.27 ± 2.41% in control and Ptf1a-electroporated animals, respectively (mean ± SEM). J–Q, Sagittal sections of E13.5 wild-type cerebellar VZ electroporated with control (J–M) or Atoh1 (N–Q) at E12.5. Double immunostaining with GFP (green) and Ptf1a (red) was performed. K–M, O–Q, High magnification of boxed region in J and N. R, Ratios of Ptf1a-positive cells in the GFP-positive cells in the VZ. The ratios are 33.92 ± 6.46 and 5.95 ± 1.64% in control and Atoh1-electroporated animals, respectively (mean ± SEM). Student's t test, *p < 0.01. n = 6. Scale bars: A, E, J, N, 50 μm; B–D, F–H, K–M, O–Q, 25 μm.
Results
Generation of Ptf1aAtoh1 and Atoh1Ptf1a knock-in mice
In the cerebellar primordium, Ptf1a and Atoh1 are expressed in the VZ and RL, respectively. To investigate the roles of Atoh1 and Ptf1a in cerebellar development, we generated two lines of knock-in mice, aiming to ectopically express Ptf1a and Atoh1 in the RL and VZ, respectively. In the Ptf1aAtoh1 line, the ORF of the Ptf1a gene was replaced with a cDNA encoding Atoh1. The pgk–neo cassette was deleted by crossing with a mouse line carrying Cre-recombinase activity in germ-line cells (Fig. 1A). The other is Atoh1Ptf1a in which the ORF of the Atoh1 gene was replaced with a cDNA encoding Ptf1a (see Fig. 6A). Both heterozygous mice (Ptf1aAtoh1/+ and Atoh1Ptf1a/+) were viable and fertile, whereas the homozygous mice (Ptf1aAtoh1/Atoh1 and Atoh1Ptf1a/Ptf1a) died just after birth in a manner similar to Ptf1a and Atoh1 null mice (Ben-Arie et al., 1997; Krapp et al., 1998; Kawaguchi et al., 2002).
Atoh1 expression in the cerebellar primordium
We first examined the expression of Atoh1 transcripts in cerebellar primordium of mice carrying the Ptf1aAtoh1 allele by RNA in situ hybridization with an RNA probe corresponding to the Atoh1 full-length mRNA (Akazawa et al., 1995) using a probe that can detect both endogenous and exogenous Atoh1 transcripts. In the wild-type cerebellar primordium at E12.5, Atoh1 transcripts were observed in the RL and nuclear transitory zone, a proposed transient differentiation zone for cells destined to become glutamatergic deep cerebellar nuclear (DCN) neurons as described previously (Altman and Bayer, 1985; Machold and Fishell, 2005; Fig. 1B). In E14.5 and E16.5 wild-type embryos, Atoh1 was expressed in the RL and the external germinal/granule layer (EGL), as reported previously (Helms et al., 2001; Wang et al., 2005; Fig. 1G,L). In the heterozygous mice (Ptf1aAtoh1/+), Atoh1 expression was observed not only in the RL but also in the VZ at E12.5–E15.5, although the level of Atoh1 transcripts in the VZ was not very strong (Fig. 1C,H, black and gray arrows; data not shown). Interestingly, in the homozygous (Ptf1aAtoh1/Atoh1) mice, the expression of ectopic Atoh1 transcripts in the VZ was very strong (Fig. 1D,I, black arrows).
In the wild-type mice, Ptf1a is expressed in the cerebellar VZ until E15.5 (Fig. 1V,W; see Figs. 6B, 9E; data not shown). Coimmunostaining with Ki67, a marker for proliferating cells, and short-term labeled BrdU confirmed that most of the Ptf1a-expressing cells are mitotic at E12.5 (Fig. 1V′,W′, arrowheads). The expression profile of ectopic Atoh1 in Ptf1aAtoh1/+ and Ptf1aAtoh1/Atoh1 cerebella was basically consistent with that of Ptf1a in the wild type, reflecting the Ptf1a-promoter activity of the Ptf1aAtoh1 allele, as expected. Double-staining with Ki67 confirmed that these cells are mitotic in the VZ (Fig. 1Q–S, arrowheads). However, the ectopic Atoh1 expression appeared weaker at earlier stages (E14.5–E15.5 in Ptf1aAtoh1/+; Fig. 1H, gray arrow; data not shown) than Ptf1a expression in the wild-type mice (at E16.5; Fig. 1X) and disappeared at E16.5 in the VZ of both Ptf1aAtoh1/+ and Ptf1aAtoh1/Atoh1 (Fig. 1M,N, black arrowheads). The relatively low expression of Atoh1 in those cerebella might be caused by our finding that the endogenous Ptf1a promoter activity is partially downregulated by Atoh1, which is assessed in later detailed experiments.
Figure 9.
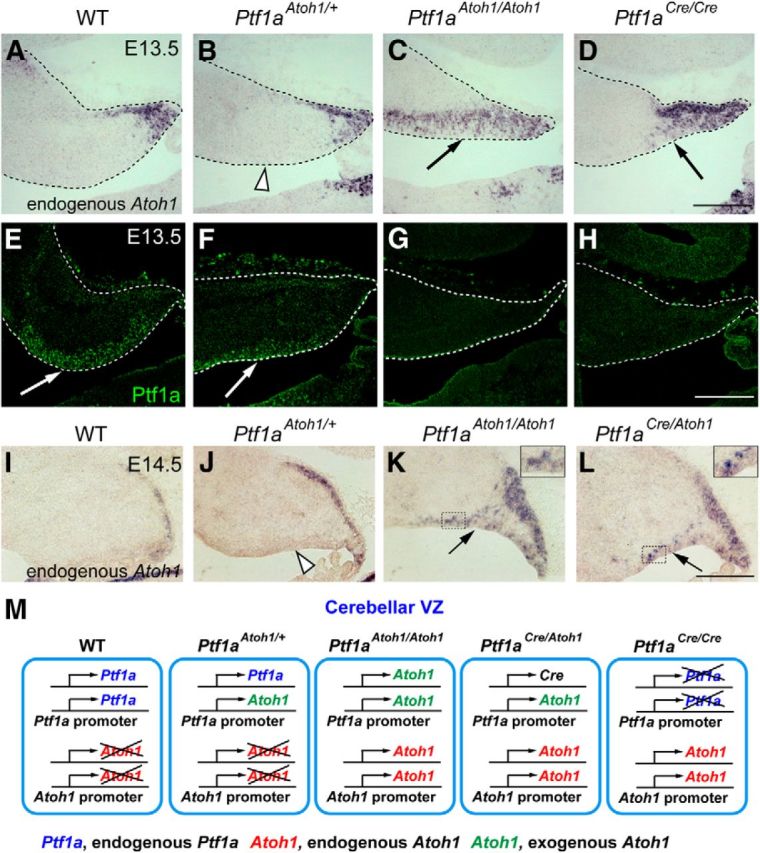
Expression of the endogenous Atoh1 in the cerebellar primordium of the knock-in mice. A–H, Sagittal sections of cerebella of indicated genotypes at E13.5. Localization of endogenous Atoh1 (Atoh1 3′ UTR probes, in situ hybridization, A–D) and Ptf1a (immunofluorescence, E–H) is shown. I–L, Localization of endogenous Atoh1 transcripts in the cerebellar primordium of indicated genotypes at E14.5. Interestingly, ectopic expression of endogenous Atoh1 in the VZ was not detected in Ptf1aAtoh1/+ cerebella (J, white arrow) in contrast to those of Ptf1aCre/Atoh1 and Ptf1aAtoh1/Atoh1 (K, L, black arrows). Insets in K, L, High-magnification pictures of the boxed regions. Scale bars, 200 μm. M, Deduced schematic model for Ptf1a and Atoh1 transcription in cells of the cerebellar VZ. Genotypes are indicated. WT, Wild-type.
In Ptf1aCre/Cre null mutants, we observed ectopic expression of Atoh1 in the VZ at E12.5–E15.5 (Fig. 1E,J, gray arrowheads; data not shown), albeit at very low levels. This may account for previous reports that the cerebellar VZ ectopically produces EGL-like cells in the Ptf1a mutants (Pascual et al., 2007). This ectopic expression of Atoh1 disappears at E16.5 (Fig. 1O, black arrowhead). Similarly, in Ptf1aCre/Atoh1 embryos, ectopic expression of Atoh1 in the VZ was observed at E12.5 and E14.5 (Fig. 1F,K, black arrows) but not at E16.5 (Fig. 1P, black arrowhead), although the expression level of ectopic Atoh1 was weaker than in Ptf1aAtoh1/Atoh1 and stronger than in Ptf1aCre/Cre. Coimmunostaining with Ki67 showed that etopic Atoh1-expressing cells in E12.5 cerebella of Ptf1aCre1Cre and Ptf1aCre/Atoh1 are mainly mitotic (Fig. 1T,U, arrowheads).
Atoh1 can induce glutamatergic neuron production in the VZ
Atoh1 is expressed in the RL and required for the development of the cerebellar glutamatergic neurons, such as glutamatergic DCN neurons, granule cells, and unipolar brush cells (UBCs; Ben-Arie et al., 1997; Machold and Fishell, 2005; Wang et al., 2005; Englund et al., 2006; Fink et al., 2006). However, it is unclear whether Atoh1 expression is sufficient to confer glutamatergic lineage identity in the cerebellum. To test this, we examined whether glutamatergic neurons were produced from VZ that ectopically expressed Atoh1. We used a recombination-based lineage tracing technique to label cells derived from the cerebellar VZ to distinguish them from RL-derived cells. The Ptf1aCre allele was originally generated by replacement of the Ptf1a protein-coding region with that of a Cre recombinase targeted to the nucleus (Kawaguchi et al., 2002). We crossed Ptf1aCre/+ with Gt(ROSA)26Sortm1sor (R26R) mice, which carry a modified lacZ gene driven by a cell type-independent ROSA26 promoter (Soriano, 1999). This allowed us to obtain offspring in which Ptf1a-driven expression of Cre excises a stop cassette upstream of lacZ and activates β-gal expression, resulting in the labeling of not only Ptf1a-expressing VZ cells but also their progeny (Kawaguchi et al., 2002; Hoshino et al., 2005). Detection of β-gal enabled the labeling of cells that were derived from the cerebellar VZ of mice carrying both Ptf1aCre and R26R alleles during development (Fig. 2A–I).
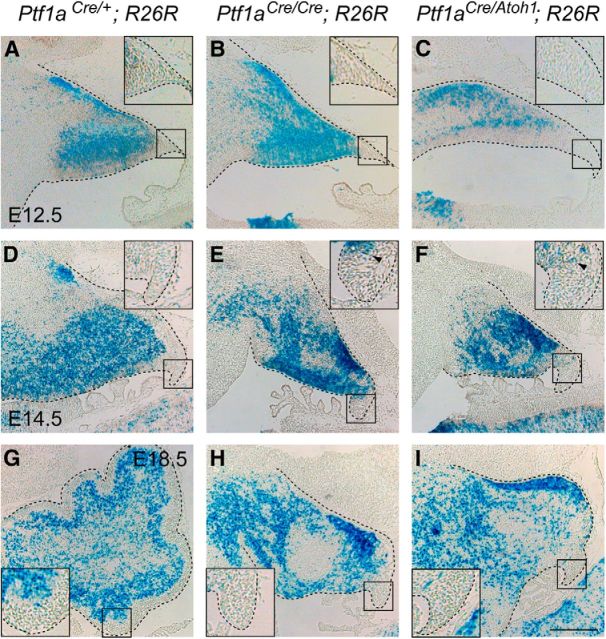
Figure 2.
Dynamics of Ptf1a lineage cells in the cerebellum during embryogenesis. A–I, X-gal-stained sagittal sections of the cerebellar primordia. Insets, High-magnification views of rectangular regions in A–I, respectively. Developmental stages and genotypes are indicated. Some stained cells found at the pial side in A are cells that are supposed to migrate out of the cerebellum (our unpublished data). Scale bar, 200 μm.
Actually, Ptf1a-expressing cells in the cerebellar VZ of Ptf1aCre/+;R26R embryos were colabeled with β-gal (Fig. 3A–D, arrowheads). By immunostaining, we also confirmed that mitotic Ptf1a-expressing cells in the E14.5 VZ of Ptf1aCre/+;R26R mice were labeled with β-gal (Fig. 3A–D, arrowheads). Next, by immunostaining with β-gal and several other cell type-specific markers, we examined Ptf1aCre/Atoh1;R26R embryos at E18.5 in which Atoh1 (Fig. 1F,K, arrows) but not Ptf1a (data not shown) had been expressed in the cerebellar VZ at earlier stages and in which cells produced from the VZ were labeled with β-gal (Fig. 2C,F,I). We found that ectopically Atoh1-expressing cells in the E14.5 VZ were labeled with β-gal and Ki67 (Fig. 3E–H, arrowheads), indicating the reliability of the Cre expression from Ptf1aCre allele for this lineage trace experiment. Ptf1acbll is another allele for Ptf1a in which Ptf1a expression is lost in the developing cerebellum (Hoshino et al., 2005). We compared Ptf1aCre/Atoh1;R26R embryos with control Ptf1aCre/+;R26R mice, as well as Ptf1a non-expressing (Ptf1aCre/cbll;R26R) embryos (Fig. 3I–Q′). In the control (Ptf1aCre/+;R26R) mice, most β-gal-positive cells were localized outside the EGL, although a few β-gal-positive cells were found in the EGL (Fig. 3I,I′), as observed previously (Sudarov et al., 2011). However, neither population expressed Pax6 (Fig. 3I,I′), a marker for granule cell lineage (Engelkamp et al., 1999), consistent with our knowledge that the VZ produces only GABAergic, not glutamatergic, neurons (Hoshino et al., 2005, 2006; Fig. 3I,I′). However, in Ptf1aCre/Atoh1;R26R mice, many β-gal-positive cells were found in the EGL and immunoreactive to Pax6 (Fig. 3K,K′, arrowheads). In addition, basically similar results were obtained in Ptf1aCre/cbll;R26R cerebella (Fig. 3J,J′) in which Ptf1a expression was lost and Atoh1 was ectopically expressed in the VZ. Coimmunostaining with β-gal and Zic2, another EGL and granule cell marker (Aruga et al., 2002), further supported these results (data not shown). These findings suggest that the ectopic expression of Atoh1 in the VZ causes production of granule cells from the VZ.
Figure 3.
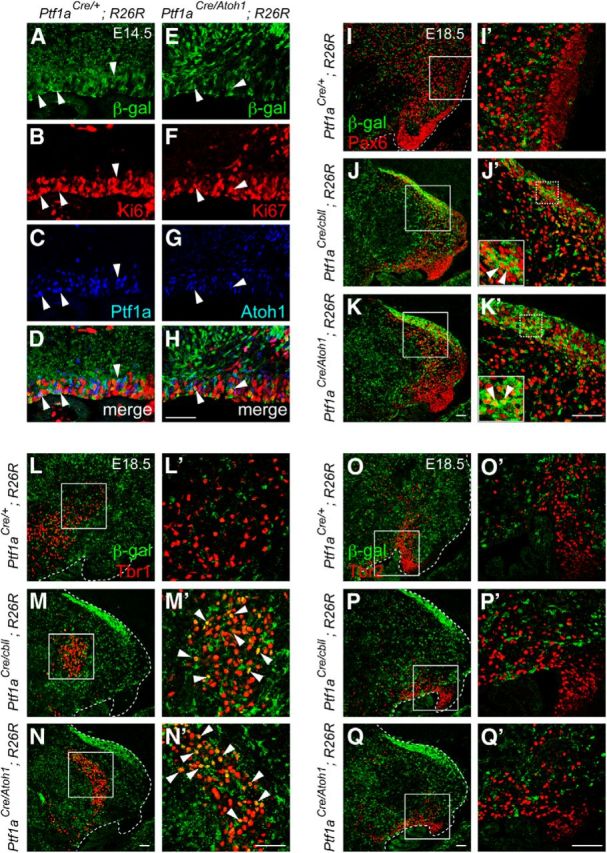
Lineage tracing analysis in the cerebellum of the Ptf1aAtoh1 knock-in mice. A–H, Localization of β-gal (A, E), Ki67 (B, F), and Ptf1a (C) or Atoh1 (G) in the cerebellar VZ of indicated genotypes at E14.5. Arrowheads indicate triple localization of β-gal, Ki67, and Ptf1a or Atoh1. I–Q′, Sagittal sections of indicated genotypes at E18.5. Double immunostaining with β-gal (green) and cell type-specific markers (red), such as Pax6 (I–K′), Tbr1 (L–N′), and Tbr2 (O–Q′) was performed. I′–Q′, High magnification of boxed regions in I–Q, respectively. Arrowheads indicate colocalization of β-gal and a specific marker. Scale bars, 50 μm.
Similarly, we observed that some β-gal-positive cells expressed Tbr1, a marker for glutamatergic DCN neurons (Fink et al., 2006), in the cerebellar primordia of Ptf1aCre/Atoh1;R26R and Ptf1aCre/cbll;R26R embryos (Fig. 3M,M′,N,N′, arrowheads), something never observed in control (Ptf1aCre/+;R26R) mice (Fig. 3L,L′). This indicates that the Atoh1-expressing VZ can generate glutamatergic DCN neurons. Interestingly, there were no β-gal-positive cells that expressed Tbr2, a marker for UBCs (Englund et al., 2006) in any of the examined genotypes (Fig. 3O–Q′), suggesting that the cerebellar VZ of Ptf1aCre/Atoh1 and Ptf1aCre/cbll mice can produce glutamatergic DCN neurons and granule cells but not UBCs.
Occasionally, we observed small numbers of β-gal-positive cells in the RL of Ptf1aCre/Atoh;R26R mice at E14.5 or later but not at E12.5 (Fig. 2C,F,I, arrowheads). However, because these cells were relatively rare (Fig. 2C,F,I), we believe that most β-gal-positive cells were directly derived from the VZ of Ptf1aCre/Atoh;R26R animals.
Using a previously created Atoh1CreERn line (Fujiyama et al., 2009), we generated a new line (Atoh1CreER) by crossing with a mouse line carrying germ-line cell-specific Cre-recombinase activity to delete the pgk–neo cassette (see Materials and Methods). Because we use this line only as a null allele for Atoh1 in this study, we describe this line as Atoh1null in this text. By using this line (Fig. 4B), we investigated whether glutamatergic neurons are produced in the cerebellar primordium of Ptf1aAtoh1/Atoh1;Atoh1null/null mice, in which Atoh1 was expressed only in the VZ but not in the RL or EGL as expected (Fig. 4A–C, black arrow). Because Ptf1a null or Atoh1 null mice die just after birth, we examined wild-type, Atoh1null/null, and Ptf1aAtoh1/Atoh1;Atoh1null/null embryos at E18.5 (Fig. 4D–L).
In the wild-type cerebella at E18.5, we observed many Pax6-, Tbr1-, and Tbr2-expressing cells, which are thought to correspond to immature granule cells, glutamatergic DCN neurons, and UBCs, respectively (Fig. 4D,G,J). In the Atoh1null/null mice, Pax6-positive cells were rarely found in the EGL (Fig. 4E, arrow head) and the RL, although some Pax6-expressing cells are found in the inner region of cerebellar primordium, as reported previously (Flora et al., 2009). In Ptf1aAtoh1/Atoh1;Atoh1null/null mice, we observed Pax6-positive cells in the EGL (Fig. 4F, arrow).
Although in the Atoh1null/null embryos Tbr1-positive cells (glutamatergic DCN neurons) were not observed (Fig. 4H) as described previously (Wang et al., 2005), some Tbr1-expressing cells were detected in Ptf1aAtoh1/Atoh1;Atoh1null/null cerebella (Fig. 4I, arrow). A previous study showed that the number of Tbr2-positive cells or UBCs was severely reduced in Atoh1 null mutants (Englund et al., 2006), consistent with our results (Fig. 4K). In Ptf1aAtoh1/Atoh1;Atoh1null/null mice, the number of Tbr2-positive cells was significantly reduced compared with wild type (Fig. 4J) and resembled that of Atoh1null/null mice (Fig. 4K,L). Relative cell numbers were 100 ± 11.0% in wild type, 36.5 ± 4.3% in Atoh1null/null, and 31.8 ± 2.1% in Ptf1aAtoh1/Atoh1;Atoh1null/null (mean ± SEM), and the difference between two latter scores was not significant (p > 0.1). Together with our above results (Fig. 3), these data suggest that granule cells and glutamatergic DCN neurons were generated from the ectopic Atoh1-expressing VZ, although UBCs were not.
Molecular genetics-based lineage tracing analyses (Machold and Fishell, 2005; Wang et al., 2005) revealed that each type of glutamatergic neuron in the cerebellum is generated at distinct developmental stages; glutamatergic DCN neurons leave the cerebellar RL at early stages (E10.5–E12.5) and granule cells at middle to late stages (granule cell, E13.5). UBCs are known to emerge at relatively late developmental stages (Englund et al., 2006). However, at late embryonic stages (e.g., E16.5), ectopic Atoh1 expression in the VZ disappears in the knock-in (Ptf1aAtoh1/+, Ptf1aAtoh1/Atoh1, and Ptf1aCre/Atoh1) and Ptf1a null (Ptf1aCre/Cre) mice (Fig. 1M–P, black arrowheads). This may explain the discrepancy in the ability of the VZ to produce DCN neurons/granule cells but not UBCs. The mechanism underlying the earlier disappearance of ectopic Atoh1 in the knock-in mice will be discussed below.
These findings suggest that Atoh1 is sufficient to produce glutamatergic neurons when ectopically expressed in the cerebellar VZ and therefore suggest that Atoh1 has the ability to change the characteristics of neuroepithelial cells from GABAergic neuron progenitors to glutamatergic neuron progenitors. In the wild-type mice, this ability of Atoh1 may confer the capability to generate glutamatergic neurons on neuroepithelial cells of the RL during normal development.
Birthdate analyses of ectopically produced glutamatergic neurons
Previously, it was shown that glutamatergic DCN neurons first leave the RL at approximately E10.5 and then granule cell precursors start to leave the RL at approximately E13.5 during cerebellar development (Machold and Fishell, 2005; Wang et al., 2005). To label cells in S-phase at certain developmental stages, we performed BrdU incorporation studies. BrdU was administered to wild-type and Ptf1aCre/+;R26R embryos at E10.5 and E14.5, and embryos were fixed at E18.5 (Fig. 5A). Immunolabeling with BrdU and cell type-specific markers, such as Tbr1 and Pax6, revealed that cells labeled at E10.5 include glutamatergic DCN neurons but not cells in the granule cell lineage, whereas BrdU-labeled cells at E14.5 contain granule-lineage cells but not glutamatergic DCN neurons, in both Ptf1aCre/+;R26R (Fig. 5B–C′,F–G′, arrowheads and arrows) and wild-type (data not shown) animals. These findings further confirmed that glutamatergic DCN neurons are first generated and then granule-lineage cells emerge during normal cerebellar development.
Figure 5.
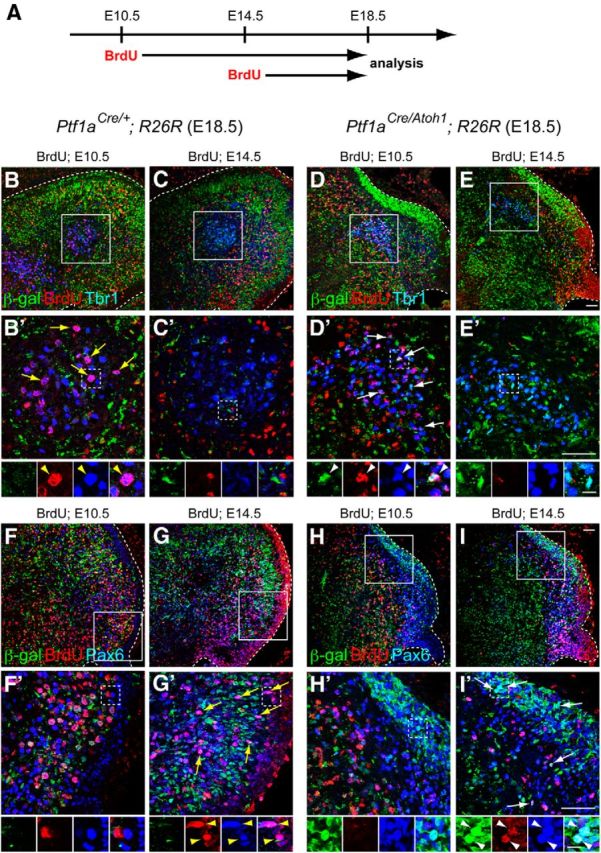
Birthdate analyses of ectopically produced glutamatergic neurons from the VZ of Ptf1aCre/Atoh1;R26R mice. A, Scheme of the experiment. Pregnant mice (E10.5 or E14.5) were given intraperitoneal injections of BrdU, and the embryos (Ptf1aCre/+;R26R and Ptf1aCre/Atoh1;R26R) were fixed at E18.5, followed by immunostaining. B–I, Triple immunolabeling with β-gal (green), BrdU (red), and cell type-specific markers (blue), such as Tbr1 (B–E) and Pax6 (F–I). B′–I′, High-magnification views of rectangular regions in B–I, respectively. Small panels below B′–I′ are single colored or merged pictures corresponding to the rectangular regions in B′–I′. Yellow arrows and arrowheads indicate colocalization of BrdU and the markers. White arrows and arrowheads indicate the triple localization of β-gal, BrdU, and markers. All are sagittal sections. Top is dorsal, and left is rostral. Scale bars: B–I′, 50 μm; small panels below B′–I′, 10 μm.
To investigate the order of generation of ectopically produced glutamatergic neurons from the VZ, BrdU was administered to Ptf1aCre/Atoh1;R26R embryos at E10.5 and E14.5, and embryos were analyzed at E18.5 (Fig. 5A). In this experiment, cells derived from the VZ were labeled with β-gal. We found that glutamatergic DCN neurons generated from the VZ (Tbr1+/β-gal+ cells) were BrdU positive when BrdU was injected at E10.5 (Fig. 5D,D′, arrows and arrowheads) but not at E14.5 (Fig. 5E,E′). In contrast, granule-lineage cells generated from the VZ (Pax6+/β-gal+ cells) were BrdU-positive when BrdU was injected at E14.5 (Fig. 5I,I′, arrows and arrowheads) but not at E10.5 (Fig. 5H,H′). These findings suggest that, in Ptf1aCre/Atoh1;R26R embryos, glutamatergic DCN neurons were first generated from the Atoh1-expressing VZ and then granule-lineage cells emerged from the VZ, and further suggest that the fate-changed neurons were generated from the Atoh1-expressing VZ according to temporal schedules very similar to that in the wild-type RL. This suggests that common temporal information is shared between the VZ and RL, in terms of glutamatergic cell-type determination (see Fig. 11A).
Figure 11.
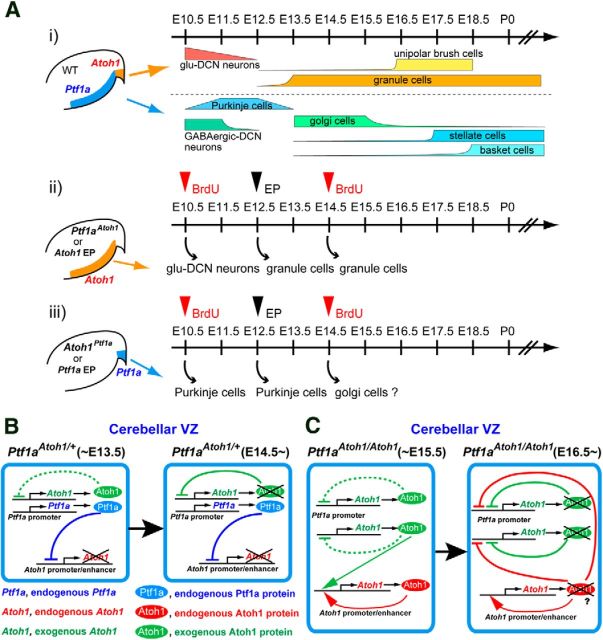
Schematic models of Atoh1 and Ptf1a function and expression in the cerebellar neuroepithelium. Ai, Temporal schedule of the neurogenesis in the cerebellar primordium of wild-type mice drawn according to previous studies. Aii, Aiii, Summary for the birthdates of ectopically produced neurons in the knock-in mice or electroporated mice that ectopically express Atoh1 (Aii) and Ptf1a (Aiii). Red arrowheads indicate BrdU administration at E10.5 or E14.5 to the knock-in mice. Black arrowheads indicate electroporation with Atoh1 or Ptf1a at E12.5 to the wild-type VZ or RL, respectively. B, C, Schematic models for Ptf1a and Atoh1 expression in the cerebellar VZ of Ptf1aAtoh1 heterozygotes and homozygotes. Left and right boxes represent cells in the VZ at indicated stages. WT, Wild-type; EP, electroporation; KI, knock-in.
Ptf1a can induce GABAergic neuron production in the RL
In wild-type mice, Ptf1a is expressed in the VZ (Fig. 1V–W′) and is necessary for the development of cerebellar GABAergic neurons, such as Purkinje, Golgi, basket, and stellate cells (Hoshino et al., 2005). However, it is unclear whether Ptf1a is sufficient to specify GABAergic lineage in the cerebellum. To test this, we used the Atoh1Ptf1a line (Fig. 6A) in which Ptf1a is designed to be ectopically expressed in the RL. Although crossing with a Cre-expressing mouse line did not delete the pgk–neo cassette, it resulted in expression of Ptf1a not only in the VZ but also ectopically in the cerebellar RL of Atoh1Ptf1a/+ embryos at E14.5 (Fig. 6C,C′, red arrowheads; data not shown), which was never observed in wild-type mice (Fig. 6B,B′). In the RL of Atoh1Ptf1a/+ embryos, ratios of Atoh1 and Ptf1a single-positive cells were 32.20 ± 2.62 and 57.66 ± 2.73%, respectively, although a small cell population was found to express both Atoh1 and Ptf1a (10.14 ± 1.63%, mean ± SEM). At later developmental stages, such as at E18.5, in the wild-type cerebella, Ptf1a expression was sparsely observed inside the cerebellum but not in the RL or EGL (Fig. 6H), which may correspond to previously reported expression of Ptf1a in the prospective white matter at late neurogenesis stages (Fleming et al., 2013). We did not observe any Ptf1a signals in the RL or EGL of the heterozygotes (Atoh1Ptf1a/+; Fig. 6I) and homozygotes (Atoh1Ptf1a/Ptf1a; data not shown), except for the sparse signals inside the cerebellum that were similarly observed in the wild-type mice. Interestingly, endogenous expression of Atoh1 was absent in the heterozygous RL and EGL (Atoh1Ptf1a/+; Fig. 6K, arrowheads) compared with wild type (Fig. 6J, white arrows). This will be discussed later.
To examine what types of cells are generated from ectopic Ptf1a-expressing RL, we used mice carrying a transgene designed to express Cre recombinase under the control of an Atoh1 enhancer (Helms et al., 2000; Machold and Fishell, 2005; Fujiyama et al., 2009). In the cerebellum of Tg–Atoh1–Cre;R26R and Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R mice, Cre was expressed in the RL and EGL, resembling the expression pattern of endogenous Atoh1 (Fig. 6D–G). Accordingly, β-gal-positive cells were observed in the EGL and DCN (Fig. 6L,M), mimicking the distribution pattern reported previously for cells in the Atoh1 lineage (Machold and Fishell, 2005; Wang et al., 2005). In addition, in the RL of Atoh1Ptf1a/+;Tg–Atoh1–Cre, Cre was colocalized with Neph3, a direct target gene of the transcription factor Ptf1a, which perfectly mimics the expression of Ptf1a in the cerebellum (Nishida et al., 2010, Mizuhara et al., 2010; Fig. 6F,G, arrowheads).
We immunostained the cerebella of Tg–Atoh1–Cre;R26R and Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R mice with β-gal and cell type-specific markers at E18.5 (Fig. 6L–S′). In the Tg–Atoh1–Cre;R26R mice, β-gal-positive cells were negative for GABA (GABAergic neuron marker), calbindin (Purkinje cell marker), and Pax2 (marker for GABAergic interneurons, such as Golgi, basket, and stellate cells; Fig. 6L′,N′,P′) but positive for glutaminase (glutamatergic neurons; Fig. 6L′, yellow arrowheads), consistent with the fact that only glutamatergic neurons but not GABAergic neurons are produced from the wild-type RL. However, in the Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R mice, some β-gal-positive cells were immunoreactive to GABA, calbindin, and Pax2 (Fig. 6M′,O′,Q′, white arrowheads), suggesting that the ectopic Ptf1a-expressing RL produced some GABAergic neurons, such as Purkinje cells and Pax2-positive GABAergic interneurons. Furthermore, in the Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R mice, the EGL was much thinner than that of wild type (Fig. 6R–S′, white bars), although some β-gal-positive cells were immunoreactive to Pax6 (Fig. 6S′, arrowheads) as found in the control cerebellum (Fig. 6R′, arrowheads). These findings indicate that Ptf1a is sufficient to induce GABAergic neurons, such as Purkinje cells and Pax2-positive interneurons, when ectopically expressed in the cerebellar RL and therefore suggest that Ptf1a has the ability to switch neuroepithelial cells from glutamatergic neuron progenitors to GABAergic neuron progenitors. In wild-type mice, this ability of Ptf1a may confer the capability to generate GABAergic neurons on neuroepithelial cells of the VZ during normal development. Interestingly, few to no β-gal-positive cells in Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R mice were stained with both glutaminase and GABA (Fig. 6M′). Although we observed a small population of cells expressing both Atoh1 and Ptf1a in the RL of Atoh1Ptf1a/+mice (Fig. 6C,C′), this result suggests that they could not differentiate into cells with glutamatergic and GABAergic characteristics.
Birthdate analyses of ectopically produced GABAergic neurons
Birthdating studies using [3H]thymidine and BrdU (Chan-Palay et al., 1977; Batini et al., 1992; De Zeeuw and Berrebi, 1995; Sultan et al., 2003; Leto et al., 2006; Sudarov et al., 2011), as well as adenovirus (Hashimoto and Mikoshiba, 2003) revealed that each type of neuron is generated at distinct developmental stages. For GABAergic neurons, in mice, Purkinje cells are produced at early stages (E10.5–E13.5), GABAergic DCN neurons at early stages (E10.5–E11.5), Golgi cells at middle stages (E13.5 to perinatal, peak at approximately E13.5–E15.5), and stellate/basket cells at later stages (approximately E13.5 to perinatal, peak at approximately E17.5 to perinatal). To investigate when the fate-changed neurons were generated from the ectopic Ptf1a-expressing RL, we examined Atoh1Ptf1a/+ pregnant mice crossed with Tg–Atoh1–Cre;R26R mice that were given two intraperitoneal injections of BrdU with a 30 min interval at E10.5 or E14.5. At E18.5, the embryos were fixed, sectioned, and triple stained with BrdU, β-gal, and cell type-specific markers, such as calbindin and Pax2 (Fig. 7A).
Figure 7.
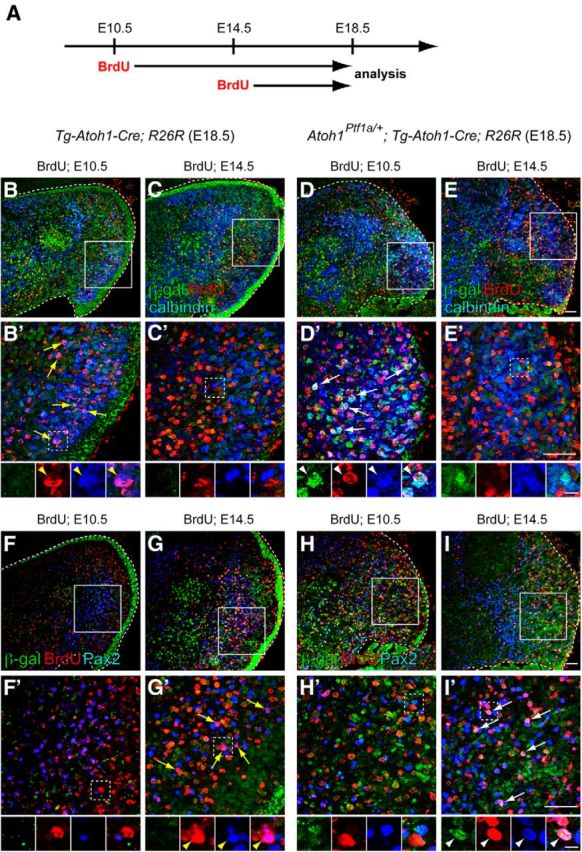
Birthdate analyses of GABAergic neurons ectopically produced from the RL. A, Scheme for the birthdate analyses by BrdU incorporation. Pregnant mice (E10.5 or E14.5) were given intraperitoneal injections of BrdU, and the embryos (Tg–Atoh1–Cre;R26R and Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R) were fixed at E18.5, followed by immunostaining. B–I, Triple immunolabeling with β-gal (green), BrdU (red), and cell type-specific markers (blue), such as calbindin (B–E) and Pax2 (F–I). B′–I′, High-magnification views of rectangular regions in B–I, respectively. Small panels below B′–I′ are single colored or merged pictures corresponding to the rectangular regions in B′–I′. Yellow arrows and arrowheads indicate colocalization of BrdU and the marker. White arrows and arrowheads indicate triple colocalization of β-gal, BrdU, and calbindin or Pax2. All are sagittal sections. Top is dorsal, and left is rostral. Scale bars: B–I′, 50 μm; small panels below B′–I′, 10 μm.
Consistent with the previous birthdating studies, in the E18.5 control embryos (Tg–Atoh1–Cre;R26R) administered BrdU at E10.5, many BrdU-positive cells were labeled with calbindin (Purkinje cells; Fig. 7B,B′, yellow arrows and arrowheads), whereas only a few BrdU-positive cells were stained with Pax2 (Fig. 7F,F′). We believe that this small number of Pax2-positive cells are GABAergic interneurons in the DCN, because they are reported to emerge at early neurodevelopmental stages (Sudarov et al., 2011). In the E18.5 control embryos administered BrdU at E14.5, many BrdU-positive cells expressed Pax2 (Fig. 7G,G′, yellow arrows and arrowheads) but not calbindin (Fig. 7C,C′), also consistent with previous knowledge that Purkinje cells are generated only at early neurogenesis stages (E10.5–E13.5), but Pax2-positive interneurons can emerge at later stages (Maricich and Herrup, 1999; Hashimoto and Mikoshiba, 2003).
In E18.5 embryos of Atoh1Ptf1a/+;Tg–Atoh1–Cre;R26R, we found that some Purkinje cells generated from the Ptf1a-expressing RL (calbindin+/β-gal+ cells) incorporated BrdU when BrdU was injected at E10.5 (Fig. 7D,D′, white arrows and arrowheads) but not at E14.5 (Fig. 7E,E′). Conversely, some of the Pax2-positive interneurons derived from the ectopically Ptf1a-expressing RL (Pax2+/β-gal+ cells) were immunoreactive to BrdU when BrdU was administered at E14.5 (Fig. 7I,I′, white arrows and arrowheads) but not at E10.5 (Fig. 7H,H′). Thus, the temporal generation of ectopically produced Purkinje cells and Pax2-positive interneurons resemble that of those normally generated from the wild-type VZ (Fig. 7B–C′,F–G′), suggesting an existence of a common temporal nature of the neuroepithelium between the RL and VZ (see Fig. 11A).
Transient expression of Atoh1 and Ptf1a induce ectopic production of glutamatergic and GABAergic neurons in the cerebellum
In mice that carry the knock-in alleles (Ptf1aAtoh1/+and Atoh1Ptf1a/+), ectopic expression of Ptf1a and Atoh1 continued for several days. To test whether a transient and ectopic expression of Atoh1 (in the VZ) and Ptf1a (in the RL) can induce ectopic production of glutamatergic and GABAergic neurons, respectively, we introduced Atoh1- and Ptf1a-expressing vectors into the wild-type cerebellar neuroepithelium by means of in utero electroporation (Inoue and Krumlauf, 2001; Saito and Nakatsuji, 2001, Kawauchi et al., 2003; Fig. 8). Two vectors, Atoh1–IRES–EGFP and Ptf1a–IRES–EGFP, which were designed to express Atoh1 and Ptf1a with EGFP, were electroporated into E12.5 cerebellar neuroepithelium, and embryos were then fixed at E14.5, followed by immunostaining. The transfected cells were distinguishable by EGFP signals. This “2 d interval” between the date of electroporation and fixation enabled us to deduce the site of electroporation; with longer intervals, it is often difficult to determine the site of electroporation.
Figure 8.
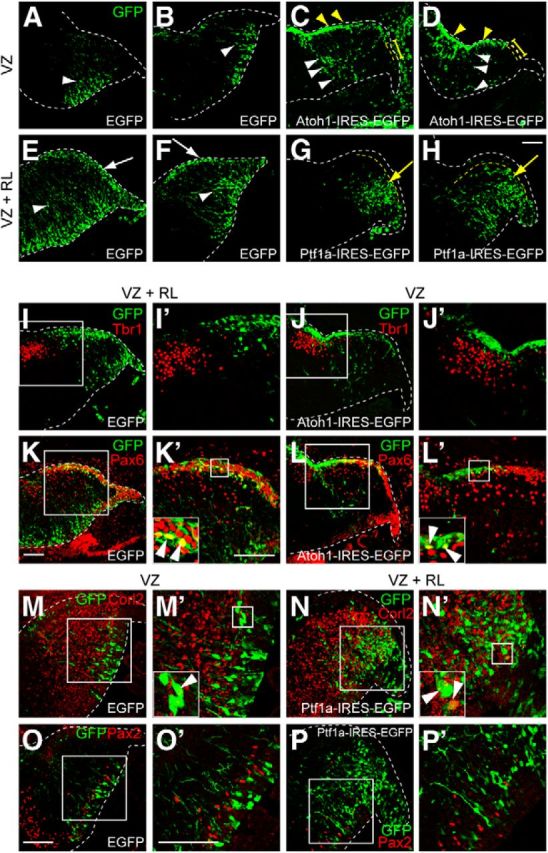
Transient introduction of Atoh1 and Ptf1a into the developing cerebellum. Cerebellar primordium electroporated with the indicated vector at E12.5 and killed at E14.5. A–H, Electroporated cells were labeled with GFP (green). White arrowheads indicate radially migrating cells from the VZ. White arrows show tangentially migrating cells from the RL. Yellow arrowheads and bars indicate the rostral and caudal EGL, respectively. Yellow arrows show radially migrating cells that avoided the EGL. I–P, Double immunostaining with GFP (green) and indicated markers (red). I′–P′, High magnification of the boxed region in I–P, respectively. Insets in K′–N′, High-magnification pictures in the boxed regions. Arrowheads indicate colocalization of GFP and the markers. All are sagittal sections. Top is dorsal, and left is rostral. Scale bars, 100 μm.
Because of technical limitations, it is quite difficult to specifically target the RL by in utero electroporation as a result of its small size. However, it is possible to specifically target the VZ without introducing vectors into the RL. Therefore, we performed two types of electroporations: (1) electroporation only into the VZ (Fig. 8A–D,J,J′,L–M′,O,O′); and (2) electroporation into both the VZ and the RL (Fig. 8E–H,I,I′,K,K′,N,N′,P,P′). When EGFP was introduced only into the VZ of E12.5 cerebella, the EGFP-positive cells migrate radially (Fig. 8A,B, white arrowheads). Their position and shape identify them as likely GABAergic neurons. When EGFP was electroporated into both the VZ and RL, in addition to GABAergic neuron-like cells (Fig. 8E,F, white arrowheads), some EGFP-positive cells were observed that were located at the superficial-most region and seemed to be derived from the RL (Fig. 8E,F, white arrows). From their position and shape, those cells in the superficial-most region are most likely glutamatergic neurons.
When Atoh1 was electroporated only into the VZ, we often found many EGFP-labeled cells in the superficial region of the cerebellar primordium corresponding to the rostral EGL (Fig. 8C,D, yellow arrowheads), whereas electroporated cells were rarely observed in the caudal EGL (Fig. 8C,D, yellow bars) or in the RL, suggesting that these Atoh1 electroporated cells in the rostral EGL were derived from the VZ. That is also supported by the fact that electroporated cells were continuously observed from the VZ to the rostral EGL (Fig. 8C,D, white arrowheads). This type of distribution pattern was never observed with the control EGFP vector (Fig. 8A,B). Immunohistochemistry revealed that some of the Atoh1-electroporated cells were positive for Pax6 (Fig. 8L,L′, white arrowheads) but not for Tbr1 (Fig. 8J,J′). However, when control EGFP was introduced only into the VZ, electroporated cells were never labeled with Pax6 (data not shown), consistent with our knowledge that glutamatergic neurons were never produced from the VZ. These findings suggest that transient expression of Atoh1 in the VZ at E12.5 can induce ectopic production of glutamatergic neurons, such as granule cells, but not glutamatergic DCN neurons (see Fig. 11A). A similar discrepancy as to glutamatergic DCN neurons and granule cells was observed in the cerebella in which the VZ and RL were electroporated with EGFP at E12.5. Some EGFP-positive cells were labeled with Pax6 (Fig. 8K,K′, arrowheads) but not with Tbr1 (Fig. 8I,I′), consistent with previous findings that glutamatergic DCN neurons leave the RL earlier than E12.5 and granule cells later than E12.5 (Machold and Fishell, 2005; Wang et al., 2005). Together with our finding that some ectopically produced glutamatergic DCN neurons in Ptf1aCre/Atoh1;R26R were labeled with E10.5-introduced BrdU (Fig. 5D,D′), we believe that transient introduction of Atoh1 to the VZ at E10.5 may result in ectopic production of glutamatergic DCN neurons. However, technical difficulties prevent in utero electroporation studies at such an early developmental stage.
As described above, when we introduced control EGFP into both the RL and the VZ at E12.5, we observed tangential migration of the GFP-positive cells from the RL to the EGL (Fig. 8E,F, white arrows). However, such cells were never observed when the Ptf1a–IRES–EGFP expression vector was electroporated into both the RL and VZ (Fig. 8G,H). In Ptf1a-electroporated cells, their migration terminated in the region beneath the EGL (Fig. 8G,H, yellow arrows), resembling the migratory behavior of Purkinje cells. Immunostaining revealed that many of the Ptf1a-electroporated cells in the vicinity of the RL expressed Corl2 (an early Purkinje cell marker; Minaki et al., 2008; Fig. 8N,N′, white arrowheads) but not Pax2 (Fig. 8P,P′), whereas control EGFP-electroporated cells derived from the RL did not express these markers (data not shown). A similar discrepancy as to Purkinje cells and Pax2-positive interneurons was observed in the cerebella in which the VZ was electroporated with EGFP at E12.5. Some EGFP-positive cells were labeled with Corl2 (Fig. 8M,M′, arrowheads) but in very rare cases with Pax2 (Fig. 8O,O′), consistent with the previous finding that the major population of neurons generated at E12.5 is Purkinje cells and only a very small fraction of Pax2-positive interneurons (probably GABAergic DCN interneurons) can be generated at this stage (Maricich and Herrup, 1999; Hashimoto and Mikoshiba, 2003). Moreover, in Ptf1a-electroporated brains, we rarely observed electroporated cells expressing Pax6 or Tbr1 (data not shown). These findings suggest that transient expression of Ptf1a in the RL at E12.5 can induce ectopic production of GABAergic neurons, such as Purkinje cells (see Fig. 11A).
Ptf1a suppresses endogenous Atoh1 expression in the cerebellar VZ
As described above, ectopic Atoh1 expression in the VZ was weakly observed in the Ptf1aAtoh1/+ and strongly in the Ptf1aAtoh1/Atoh1 cerebella (Fig. 1C,D, arrows). To determine whether the endogenous Atoh1 transcripts from the wild-type Atoh1 allele (Atoh1+) were expressed in the VZ of mice containing the Ptf1aAtoh1 allele, we performed in situ hybridization to embryonic cerebella with a probe corresponding to the 3′ UTR of Atoh1 that detects only the endogenous Atoh1 transcripts (Fig. 9A–D,I–L). Interestingly, endogenous Atoh1 transcripts were ectopically detected in the VZ of E13.5 cerebellar primordia of Ptf1aAtoh1/Atoh1 (Fig. 9C, arrow) but not in the Ptf1aAtoh1/+ mice (Fig. 9B, white arrowhead). Although Ptf1a protein was detected in the VZ of both wild-type and Ptf1aAtoh1/+ mice (Fig. 9E,F, arrows), its expression was lost in the VZ of Ptf1aAtoh1/Atoh1 (Fig. 9G). These facts lead us to the possibility that Ptf1a directly or indirectly suppresses the expression of the endogenous Atoh1 gene. This is supported by the observation that endogenous Atoh1 transcripts were ectopically expressed in the VZ of Ptf1a null (Ptf1aCre/Cre) and Ptf1aCre/Atoh1 cerebella (Fig. 9D,L, arrows), in which Ptf1a expression was lost (Fig. 9H; data not shown). At various developmental stages, we performed in situ hybridization with the Atoh1 3′ UTR probe to cerebella of different genotypes (Fig. 9I–L; data not shown).
To test whether Ptf1a suppresses the expression of endogenous Atoh1, the Ptf1a–IRES–EGFP expression vector was introduced into wild-type cerebellum at E12.5 by means of in utero electroporation. At E14.5, the ratio of Atoh1-expressing cells in the Ptf1a-electroporated cells in the RL was significantly reduced (Fig. 10E–H,I) compared with the control-transfected cells (Fig. 10A–D,I), confirming that Ptf1a can suppress the expression of Atoh1 in the developing cerebellum (35.08 ± 5.98% in control, 7.27 ± 2.41% in Ptf1a-electroporated samples, p < 0.01). Consistently, in the RL of Atoh1Ptf1a/+, only a small number of cells expressed both Atoh1 and Ptf1a (Fig. 6C,C′, 10.14 ± 1.63%). This suppression machinery may explain our observation that Atoh1 expression in Atoh1Ptf1a/+ disappeared at earlier stages (at E18.5; Fig. 6K, arrowheads) than that in the control mice (Fig. 6J, arrows).
We also introduced Atoh1–IRES–EGFP (or the control) vector into the wild-type VZ at E12.5 and examined embryos at E13.5. Immunostaining revealed that the ratio of Ptf1a-expressing cells in the Atoh1-electroporated cells was significantly reduced in the VZ (Fig. 10N–Q,R), whereas control transfected cells prominently expressed Ptf1a (Fig. 10J–M,R; 33.92 ± 6.46% in control, 5.95 ± 1.64% in Atoh1-electroporated animals, p < 0.01). This result suggests that Atoh1 suppresses the expression of Ptf1a in the developing cerebellum.
It has been reported that Atoh1 expression is positively controlled by autoregulation via its enhancer (Helms et al., 2000). This implies that, in the VZ of Ptf1aAtoh1/Atoh1 cerebellum, ectopically expressed Atoh1 protein from the Ptf1aAtoh1 allele may positively regulate the enhancer activity of endogenous Atoh1, leading to ectopic expression of endogenous Atoh1 in the VZ in which Ptf1a is not expressed. In contrast, in the Ptf1aAtoh1/+ embryos, endogenous Atoh1 expression in the VZ may be suppressed by endogenous expression of Ptf1a from the intact Ptf1a allele, although Atoh1 is exogenously expressed from the Ptf1aAtoh1 allele. Furthermore, ectopically expressed Atoh1 in the Ptf1aAtoh1/+ and Ptf1aAtoh1/Atoh1 cerebellum may negatively regulate the enhancer of the Ptf1aAtoh1 allele, resulting in earlier disappearance of exogenous expression of Atoh1 at later stages (Figs. 1, 11B,C). Figure 9M represents the schematic pictures for the expression of Ptf1a and Atoh1 from endogenous and exogenous alleles in the VZ of cerebellar primordium (Fig. 9M).
In summary, Atoh1 and Ptf1a seem to mutually suppress their expression, which may contribute to the generation of their non-overlapping neuroepithelial domains in the cerebellar neuroepithelium.
Discussion
Previously, our studies demonstrated that Ptf1a and Atoh1 were expressed in discrete neuroepithelial regions in a non-overlapping manner throughout r1–r8 and are required for generation of distinct subtypes of neurons (Hori and Hoshino, 2012; Hoshino, 2012, Hoshino et al., 2012). This raised the possibility that these transcription factors confer specific spatial identities along the dorsoventral axis on the neuroepithelium, which enables each neuroepithelial domain to produce specific subtypes of neurons. However, this hypothesis remained unproven. Although we and others had clarified the requirement of these transcription factors in the generation of specific subtypes of neurons, it had not been shown whether they were sufficient to produce them.
In this study, we generated two lines of knock-in alleles (Ptf1aAtoh1 and Atoh1Ptf1a) in which Atoh1 and Ptf1a were designed to be ectopically expressed in the cerebellar VZ and RL, respectively. As expected, ectopic expression of Atoh1 and Ptf1a was observed in the VZ and RL; however, this ectopic expression did not persist but ceased a little earlier than that of endogenous Ptf1a and Atoh1 in the wild-type mice during development.
In mice carrying the Ptf1aAtoh1 allele, we observed that glutamatergic neurons, such as glutamatergic DCN neurons and granule cells, were generated from the cerebellar VZ in which Atoh1 was ectopically expressed. Furthermore, transient introduction of Atoh1 into the VZ succeeded in generating granule cells from the VZ. These findings suggest that Atoh1 is sufficient to change the fate of neurons produced from the VZ, from GABAergic to glutamatergic ones. Another explanation may be that Atoh1 can give the VZ cells an ability to enter the EGL in which those cells are instructed to differentiate into granule cells. However, including this notion, our results suggest that Atoh1 is sufficient to confer the ability to produce glutamatergic neurons on the neuroepithelium.
In mice carrying the Atoh1Ptf1a allele, we observed that GABAergic neurons, such as Purkinje cells and Pax2-positive interneurons, were produced from the RL in which Ptf1a was ectopically expressed. Consistently, transient introduction of Ptf1a into the RL by in utero electroporation led to production of Purkinje cells. These results suggest that Ptf1a is sufficient to change the fate of neurons produced from the RL, from glutamatergic to GABAergic, and further suggest that Ptf1a is sufficient to confer the ability to produce GABAergic neurons on the neuroepithelium. Altogether, these facts imply that Atoh1 and Ptf1a can confer specific spatial identities on the cerebellar neuroepithelium.
Although some clarification of the machinery governing GABAergic and glutamatergic neuronal subtype specification by transcription factors has been provided previously, the molecular mechanisms that specify each GABAergic (e.g., Purkinje, Golgi, basket, stellate cells, etc.) or glutamatergic (e.g., granule, UBCs, and glutamatergic DCN neurons) subtype remained unclear. Birthdating studies using [3H]thymidine, BrdU (Chan-Palay et al., 1977; Batini et al., 1992; De Zeeuw and Berrebi, 1995; Sultan et al., 2003; Leto et al., 2006), adenovirus (Hashimoto and Mikoshiba, 2003), and genetic fate-mapping studies (Machold and Fishell, 2005; Wang et al., 2005; Englund et al., 2006; Sudarov et al., 2011) revealed that each type of neuron is generated at a distinct developmental stage. With regard to GABAergic neurons, Purkinje cells are produced early (E10.5–E13.5 in mice), Golgi cells a little later (E13.5 to postnatal day 0), and stellate/basket cells mainly perinatally. As to glutamatergic neurons, glutamatergic DCN neurons leave the cerebellar RL at early stages (E10.5–E12.5) and granule cells and UBCs at middle to late stages (granule cell, E13.5–postnatal stages; UBC, E13.5–E18.5). In addition, somatic recombination-based clonal analyses suggested that Purkinje, Golgi, and basket/stellate cells, as well as some DCN neurons (probably GABAergic), belong to the same lineage (Mathis et al., 1997; Mathis and Nicolas, 2003). These data indicate that, other than spatial information, some temporal information in the neuroepithelium may be involved in specification of neuronal types in the RL and VZ. Actually, we showed recently that the transcription factors Olig2 and Gsx1 regulate the temporal information of neuroepithelial cells of the cerebellar VZ, controlling the production of Purkinje cells and GABAergic interneurons (Seto et al., 2014).
In this study, we obtained suggestive data that imply that the RL and VZ share common temporal information or common temporal identities during development. For example, glutamatergic DCN neurons and granule cells derived from Atoh1-expressing VZ of Ptf1aAtoh1 animals were labeled with BrdU when BrdU was administered at E10.5 and E14.5, respectively. Moreover, in utero electroporation of Atoh1 into the VZ at E12.5 induced production of granule cells but not glutamatergic DCN neurons. Conversely, ectopically produced Purkinje cells and Pax2 interneurons (probably Golgi cells) were labeled with BrdU administration at E10.5 and E14.5, respectively, in animals carrying the Atoh1Ptf1a allele. In utero electroporation of Ptf1a into the RL at E12.5 resulted in production of Purkinje cells but not Pax2-positive interneurons. Thus, the birthdays of ectopically produced neurons are similar to those of corresponding neurons that are normally generated in the wild-type mice (Fig. 11A). These findings suggest that there are common temporal identities between the RL and the VZ.
In the electroporation experiments, we observed that Ptf1a and Atoh1 mutually suppressed each other's expression in the cerebellar neuroepithelium. Accordingly, we observed that Atoh1 was ectopically expressed in the cerebellar VZ of Ptf1a-null mutants. This is also consistent with the previous observation that cells produced from the VZ in the Ptf1a-null mutants exhibited granule cell-like features, although ectopic expression of Atoh1 in the VZ was not reported in that study (Pascual et al., 2007). However, we did not observe ectopic expression of Ptf1a in the RL of Atoh1-null mutants (our unpublished data). This may indicate that there are additional molecules that regulate the expression of Ptf1a in the RL. Because both Atoh1 and Ptf1a were reported to act as transcriptional activators (Gazit et al., 2004; Wiebe et al., 2007), the mutual suppression machinery may be indirect. We believe that the mutual suppression machinery of Ptf1a and Atoh1 may contribute to formation of distinct non-overlapping domains in the cerebellar neuroepithelium.
It is believed that spatiotemporal regulation of features of neuroepithelium is important to distinctly produce a variety of neuronal types. We showed that Ptf1a and Atoh1 play pivotal roles to confer spatial information in the cerebellum, but further studies are required to understand the molecular machinery that specifies neuroepithelial identities, especially from a temporal point of view.
Footnotes
This work is supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, the Naito Foundation, Intramural Research Grants 24-12 and 25-3 for Neurological and Psychiatric Disorders of the National Center of Neurology and Psychiatry (NCNP), and Health Science Research Grant for Research on Psychiatric and Neurological Diseases and Mental Health (H23-001) from the Japanese Ministry of Health, Labor, and Welfare. We are grateful to Dr. Kageyama (Kyoto University, Kyoto, Japan) for in situ probe, Dr. Ono (KAN Research Institute, Kobe, Japan) for anti-Corl2 and anti-Neph3 antibodies, Dr. Watanabe (Hokkaido University, Hokkaido, Japan) for anti-glutaminase antibody, Dr. Imura (Foundation for Biomedical Research and Innovation, Kobe, Japan) for anti-GFP antibody, Dr. Sasai (RIKEN CDB, Kobe, Japan) for anti-Atoh1 antibody, and Dr. Wright (Vanderbilt University School of Medicine, Nashville, TN) for Ptf1aCre mouse. We thank Dr. Ruth Yu, Dr. Kawauchi (St. Jude Children's Research Hospital, Memphis, TN), and Dr. Fujiyama (National Institute of Neuroscience, NCNP, Tokyo, Japan) for comments. We also thank Dr. Masuyama (NCNP) for plasmids.
The authors declare no competing financial interests.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Embryonic development of the rat cerebellum. II. Translocation and regional distribution of the deep neurons. J Comp Neurol. 1985;231:27–41. doi: 10.1002/cne.902310104. [DOI] [PubMed] [Google Scholar]
- Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batini C, Compoint C, Buisseret-Delmas C, Daniel H, Guegan M. Cerebellar nuclei and the nucleocortical projections in the rat: retrograde tracing coupled to GABA and glutamate immunohistochemistry. J Comp Neurol. 1992;315:74–84. doi: 10.1002/cne.903150106. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Chan-Palay V, Palay SL, Brown JT, Van Itallie C. Sagittal organization of olivocerebellar and reticulocerebellar projections: autoradiographic studies with 35S-methionine. Exp Brain Res. 1977;30:561–576. doi: 10.1007/BF00237645. [DOI] [PubMed] [Google Scholar]
- De Zeeuw CI, Berrebi AS. Postsynaptic targets of Purkinje cell terminals in the cerebellar and vestibular nuclei of the rat. Eur J Neurosci. 1995;7:2322–2333. doi: 10.1111/j.1460-9568.1995.tb00653.x. [DOI] [PubMed] [Google Scholar]
- Dun XP. Origin of climbing fiber neurons and the definition of rhombic lip. Int J Dev Neurosci. 2012;30:391–395. doi: 10.1016/j.ijdevneu.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JT, He W, Hao C, Ketova T, Pan FC, Wright CC, Litingtung Y, Chiang C. The Purkinje neuron acts as a central regulator of spatially and functionally distinct cerebellar precursors. Dev Cell. 2013;27:278–292. doi: 10.1016/j.devcel.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- Gazit R, Krizhanovsky V, Ben-Arie N. Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development. 2004;131:903–913. doi: 10.1242/dev.00982. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Mikoshiba K. Mediolateral compartmentalization of the cerebellum is determined on the “birth date” of Purkinje cells. J Neurosci. 2003;23:11342–11351. doi: 10.1523/JNEUROSCI.23-36-11342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol. 1997;7:40–47. doi: 10.1016/S0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Helms AW, Gowan K, Abney A, Savage T, Johnson JE. Overexpression of MATH1 disrupts the coordination of neural differentiation in cerebellum development. Mol Cell Neurosci. 2001;17:671–682. doi: 10.1006/mcne.2000.0969. [DOI] [PubMed] [Google Scholar]
- Hori K, Hoshino M. GABAergic neuron specification in the spinal cord, the cerebellum, and the cochlear nucleus. Neural Plast. 2012;2012:921732. doi: 10.1155/2012/921732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M. Molecular machinery governing GABAergic neuron specification in the cerebellum. Cerebellum. 2006;5:193–198. doi: 10.1080/14734220600589202. [DOI] [PubMed] [Google Scholar]
- Hoshino M. Neuronal subtype specification in the cerebellum and dorsal hindbrain. Dev Growth Differ. 2012;54:317–326. doi: 10.1111/j.1440-169X.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Seto Y, Mayumi Y. Specification of cerebellar and precerebellar neurons. In: Manto M, Gruol D, Schmahmann J, Koibuchi N, Rossi F, editors. Handbook of the cerebellum and cerebellar disorders. Vol 1. New York: Springer; 2013. pp. 75–87. [Google Scholar]
- Inoue T, Krumlauf R. An impulse to the brain–using in vivo electroporation. Nat Neurosci. 2001;4:1156–1158. doi: 10.1038/nn1101-1156. [DOI] [PubMed] [Google Scholar]
- Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat Cell Biol. 2006;8:17–26. doi: 10.1038/ncb1338. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knöfler M, Ledermann B, Bürki K, Berney C, Zoerkler N, Hagenbüchle O, Wallauer PK. The bHLH protein PTF1–p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Leto K, Carletti B, Williams IM, Magrassi L, Rossi F. Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J Neurosci. 2006;26:11682–11694. doi: 10.1523/JNEUROSCI.3656-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol. 1999;41:281–294. doi: 10.1002/(SICI)1097-4695(19991105)41:2<281::AID-NEU10>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Mathis L, Nicolas JF. Progressive restriction of cell fates in relation to neuroepithelial cell mingling in the mouse cerebellum. Dev Biol. 2003;258:20–31. doi: 10.1016/S0012-1606(03)00098-8. [DOI] [PubMed] [Google Scholar]
- Mathis L, Bonnerot C, Puelles L, Nicolas JF. Retrospective clonal analysis of the cerebellum using genetic laacZ/lacZ mouse mosaics. Development. 1997;124:4089–4104. doi: 10.1242/dev.124.20.4089. [DOI] [PubMed] [Google Scholar]
- Minaki Y, Nakatani T, Mizuhara E, Inoue T, Ono Y. Identification of a novel transcriptional corepressor, Corl2, as a cerebellar Purkinje cell-selective marker. Gene Expr Patterns. 2008;8:418–423. doi: 10.1016/j.gep.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Mizuhara E, Minaki Y, Nakatani T, Kumai M, Inoue T, Muguruma K, Sasai Y, Ono Y. Purkinje cells originate from cerebellar ventricular zone progenitors positive for Neph3 and E-cadherin. Dev Biol. 2010;338:202–214. doi: 10.1016/j.ydbio.2009.11.032. [DOI] [PubMed] [Google Scholar]
- Nishida K, Hoshino M, Kawaguchi Y, Murakami F. Ptf1a directly controls expression of immunoglobulin superfamily molecules Nephrin and Neph3 in the developing central nervous system. J Biol Chem. 2010;285:373–380. doi: 10.1074/jbc.M109.060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martínez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Seto Y, Nakatani T, Masuyama N, Taya S, Kumai M, Minaki Y, Hamaguchi A, Inoue YU, Inoue T, Miyashita S, Fujiyama T, Yamada M, Chapman H, Campbell K, Magnuson MA, Wright CV, Kawaguchi Y, Ikenaka K, Takebayashi H, Ishiwata S, Ono Y, Hoshino M. Temporal identity transition from Purkinje cell progenitors to GABAergic interneuron progenitors in the cerebellum. Nat Commun. 2014;5:3337. doi: 10.1038/ncomms4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sudarov A, Turnbull RK, Kim EJ, Lebel-Potter M, Guillemot F, Joyner AL. Ascl1 genetics reveals insights into cerebellum local circuit assembly. J Neurosci. 2011;31:11055–11069. doi: 10.1523/JNEUROSCI.0479-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan F, Czubayko U, Thier P. Morphological classification of the rat lateral cerebellar nuclear neurons by principal component analysis. J Comp Neurol. 2003;455:139–155. doi: 10.1002/cne.10443. [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wiebe PO, Kormish JD, Roper VT, Fujitani Y, Alston NI, Zaret KS, Wright CV, Stein RW, Gannon M. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol Cell Biol. 2007;27:4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Terao M, Terashima T, Fujiyama T, Kawaguchi Y, Nabeshima Y, Hoshino M. Origin of climbing fiber neurons and their developmental dependence on Ptf1a. J Neurosci. 2007;27:10924–10934. doi: 10.1523/JNEUROSCI.1423-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]