Enteroviruses are a vast group of viruses associated with diverse human diseases, but only two of them could be controlled with vaccines, and effective antiviral therapeutics are lacking. Here, we investigated in detail the contribution of a cellular protein, GBF1, in the replication of poliovirus, a representative enterovirus. GBF1 supports the functioning of cellular membrane metabolism and is recruited to viral replication complexes upon infection. Our results demonstrate that the virus requires a limited subset of the normal GBF1 functions and reveal the elements of GBF1 essential to support viral replication under different conditions. Since diverse viruses often rely on the same cellular proteins for replication, understanding the mechanisms by which these proteins support infection is essential for the development of broad-spectrum antiviral therapeutics.
KEYWORDS: Arf activation, ArfGEF, GBF1, poliovirus, replication organelles
ABSTRACT
The replication of many positive-strand RNA viruses [(+)RNA viruses] depends on the cellular protein GBF1, but its role in the replication process is not clear. In uninfected cells, GBF1 activates small GTPases of the Arf family and coordinates multiple steps of membrane metabolism, including functioning of the cellular secretory pathway. The nonstructural protein 3A of poliovirus and related viruses has been shown to directly interact with GBF1, likely mediating its recruitment to the replication complexes. Surprisingly, viral mutants with a severely reduced level of 3A-GBF1 interaction demonstrate minimal replication defects in cell culture. Here, we systematically investigated the conserved elements of GBF1 to understand which determinants are important to support poliovirus replication. We demonstrate that multiple GBF1 mutants inactive in cellular metabolism could still be fully functional in the replication complexes. Our results show that the Arf-activating property, but not the primary structure of the Sec7 domain, is indispensable for viral replication. They also suggest a redundant mechanism of recruitment of GBF1 to the replication sites, which is dependent not only on direct interaction of the protein with the viral protein 3A but also on determinants located in the noncatalytic C-terminal domains of GBF1. Such a double-targeting mechanism explains the previous observations of the remarkable tolerance of different levels of GBF1-3A interaction by the virus and likely constitutes an important element of the resilience of viral replication.
IMPORTANCE Enteroviruses are a vast group of viruses associated with diverse human diseases, but only two of them could be controlled with vaccines, and effective antiviral therapeutics are lacking. Here, we investigated in detail the contribution of a cellular protein, GBF1, in the replication of poliovirus, a representative enterovirus. GBF1 supports the functioning of cellular membrane metabolism and is recruited to viral replication complexes upon infection. Our results demonstrate that the virus requires a limited subset of the normal GBF1 functions and reveal the elements of GBF1 essential to support viral replication under different conditions. Since diverse viruses often rely on the same cellular proteins for replication, understanding the mechanisms by which these proteins support infection is essential for the development of broad-spectrum antiviral therapeutics.
INTRODUCTION
The absolute dependence of viruses on the cellular metabolic machinery provides an attractive opportunity to develop antiviral interventions targeting cellular proteins. Such an approach could potentially yield therapeutics effective against broad classes of viruses that rely on the same host factors. Antivirals directed against stable cellular instead of easily adaptable viral proteins are also expected to have a higher barrier to the development of resistance. Yet, such a promising approach has not yet been successfully implemented. One of the major concerns in using cellular proteins as antiviral targets is potential toxicity to the host. In this regard, it would be highly desirable to be able to specifically inhibit activities/interactions of the cellular proteins that are crucial for viral infection but spare those important for normal cellular metabolism. In this paper, we systematically analyzed functional elements of the cellular protein GBF1 known to be essential for poliovirus replication in order to identify those required to support infection.
Poliovirus is a representative member of the Enterovirus genus of the Picornaviridae family of small positive-strand RNA viruses [(+)RNA viruses] infecting vertebrate hosts, including humans. Its genome RNA of ∼7,500 nucleotides (nt) in length is translated into one polyprotein which undergoes proteolytic processing by viral proteases to generate about a dozen structural and replication proteins. Replication complexes of poliovirus, as those of all (+)RNA viruses of eukaryotes, are associated with cellular membranes, which implies that at least some cellular proteins involved in membrane metabolism should be important for viral replication. Indeed, it was observed that the replication of poliovirus, as well as that of other related picornaviruses, is sensitive to a fungal metabolite brefeldin A (BFA) (1–3), a compound known to inhibit the activity of three so-called large guanine nucleotide exchange factors (GEFs) for small GTPases Arf, GBF1, BIG1, and BIG2. GEFs facilitate GDP/GTP exchange to generate GTP-bound Arfs, which then can associate with cellular membranes and regulate the recruitment of Arf-interacting effector proteins that support multiple aspects of membrane metabolism. The subsequent hydrolysis of GTP results in the dissociation of the now-inactive Arf-GDP from the membranes. Arf cycling regulates Golgi homeostasis and membrane trafficking through the secretory pathway and supports the molecular identity of membranous organelles (4, 5). BFA stabilizes a transient intermediate formed by Arf-GDP and the catalytic Sec7 domains of BIG1, BIG2, or GBF1 (but not other cellular ArfGEFs), locking the GEF molecules in a nonfunctional conformation and inhibiting Arf activation. It was established that the BFA sensitivity of poliovirus and other picornaviruses is due to the requirement for GBF1 in the RNA replication process (6, 7). Moreover, GBF1 has been shown to be an important cellular factor for the replication of such diverse (+)RNA viruses as hepatitis C virus, coronaviruses, and hepatitis E virus (8–10).
Yet, our understanding of the mechanistic role of GBF1 in the viral RNA replication process is very limited. The nonstructural protein 3A from poliovirus and a related coxsackie B3 virus was shown to strongly interact with GBF1, resulting in its recruitment to viral replication complexes (6, 11, 12). It was proposed that Arf activated by GBF1 may be either directly involved in the functioning of the replication complexes or mediate the recruitment of other effector proteins necessary to support the replication. In the case of poliovirus, it was suggested that the activated Arf may be responsible for the recruitment of a phosphatidylinositol 4 kinase III beta (PI4KIIIβ), an Arf effector and a protein whose activity is essential for the replication of many picornaviruses and other (+)RNA viruses (13–16). However, the accumulating data suggest that such straightforward interpretation of the role of GBF1 in viral replication may be misleading. The detailed investigation of the recruitment of PI4KIIIβ to the replication complexes of multiple picornaviruses demonstrated that it is independent of GBF1 and relies on interaction of the viral protein 3A with another host factor, ACBD3, which directly interacts with PI4KIIIβ (17–20).
BFA block of picornavirus replication can be relieved only by GBF1 overexpression but not by the overexpression of constantly activated Arf mutants, and small interfering RNA (siRNA) knockdown or CRISPR knockouts of different Arf isoforms individually or in combinations do not significantly affect picornavirus replication (7, 21). Moreover, it was reported that some GBF1 mutants with an inactivated Sec7 domain could support at least some level of poliovirus replication and that BFA resistance poliovirus mutants can replicate in the absence of detectable Arf activation but not in cells with GBF1 knockdown, arguing that GBF1 may be required for more than just enriching the replication membranes in Arf-GTP (22). Even the requirement for a strong 3A-GBF1 interaction is not absolute, since picornavirus mutants severely defective in such an interaction demonstrate only minor growth defects, at least in cell culture (6, 12).
In this paper, we aimed to revisit these controversial findings and identify the elements/activities of GBF1 that are essential to support poliovirus replication, as well as to understand if they can be separated from those mediating GBF1 function in cellular metabolism. To this end, we generated multiple GBF1 mutants using mutagenesis of conserved motifs, domain swapping, and alanine scanning. Our data demonstrate a much higher tolerance of GBF1 mutations by the viral replication machinery than by cellular metabolism, indicating that the virus utilizes only a small subset of GBF1 functions. However, none of the several GBF1 constructs with a mutation in the catalytic Sec7 domain rendering them defective in Arf activation could support poliovirus replication. Surprisingly, the cognate Sec7 domain of GBF1 could be substituted with that from a distantly related GEF, indicating that virus replication is dependent on the Arf activation property but not the amino acid sequence of the Sec7 domain. Our results confirm and extend the previous data that the very N-terminal sequence of GBF1 holds important determinants of the protein functioning in the replication complexes and that all the C-terminal domains of GBF1 downstream of the Sec7 domain are fully dispensable for replication of the wild-type poliovirus. However, the C-terminal HDS2 and HDS3 domains of GBF1 become important for replication of a poliovirus mutant with impaired 3A-GBF1 interaction. This discrepancy suggests a redundant mechanism of GBF1 recruitment to viral replication complexes. Such a complementary mechanism likely facilitates the access of GBF1 to the replication machinery and may underlie the resilience of poliovirus replication in diverse cell-specific environments encountered by the virus during the development of infection in a natural host.
RESULTS
Alanine substitutions within the N terminus of GBF1 do not significantly interfere with its function in poliovirus replication but affect GBF1 function in cellular secretion.
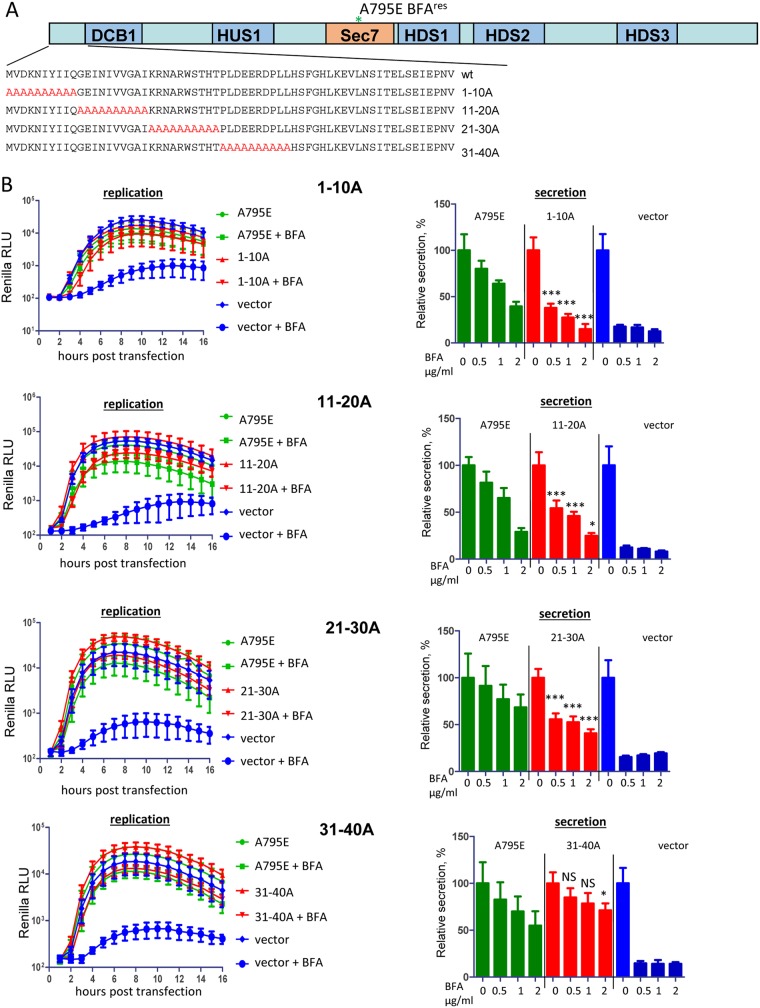
The yeast two-hybrid and coimmunoprecipitation (co-IP) data demonstrate that the N-terminal fragment of GBF1 that includes the DCB and HUS domains is likely engaged in direct protein-protein interactions with the nonstructural protein 3A of poliovirus and a related enterovirus coxsackievirus B3 (11, 12). Furthermore, the deletion of the first 37 amino acids of GBF1 (GBF1Δ37) blocks the poliovirus 3A-mediated recruitment of GBF1 to membranes and the capacity of the protein to rescue poliovirus replication from BFA inhibition, suggesting that the very N-terminal stretch of amino acids contains important determinants of GBF1 recruitment/function in replication (6). Thus, we performed an extensive mutagenesis of the N-terminal part of GBF1 to identify amino acids important for poliovirus replication. First, we substituted the authentic N-terminal amino acids with blocks of 10 Ala, generating mutants GBF1/1-10A, 11-20A, 21-30A, and 31-40A (Fig. 1A). All the mutants were created in the background of the A795E mutation in the Sec7 domain that makes GBF1 resistant to BFA (6, 7). Thus, in the presence of the drug, the endogenous GBF1 is inactivated, and the input of only the mutant protein to cellular metabolism or to viral replication can be assessed. HeLa cells were transfected with plasmids expressing GBF1 mutants and the next day transfected with a poliovirus replicon RNA with a Renilla luciferase gene substituting for the capsid protein-coding region. As a positive control, cells were transfected with a plasmid coding for GBF1 with only the BFA resistance mutation A795E, and as a negative control, cells were transfected with an empty vector. Surprisingly, in the replication assay, all the Ala scanning mutants of GBF1 were just as functional as the positive control (Fig. 1B, replication).
FIG 1.
Functional analysis of alanine scanning mutants of the N terminus of GBF1 in replication and secretion. (A) Schematic representation of the alanine substitutions in the GBF1 sequence. All GBF1 expression constructs are GFP tagged and contain the A795E BFA resistance mutation in the Sec7 domain. (B) Performance of the indicated mutants in the poliovirus replicon replication and cellular secretion assays. For the replication assay, cells were transfected with the plasmids expressing a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control). The next day, the cells were transfected with a poliovirus replicon RNA expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA. For the secretion assay, the cells were cotransfected with plasmids coding for a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control) and a plasmid coding for a secreted Gaussia luciferase. The next day, they were washed and incubated in the medium with the indicated amount of BFA, and the amount of secreted luciferase was determined after 4 h. Secretion data are normalized to the signal obtained without BFA for each construct. Statistical significance of the difference between the signal in the positive control and in the sample expressing a mutant GBF1 for corresponding concentrations of BFA is indicated. RLU, relative light units.
The capacity of the same GBF1 mutants to function in cellular metabolism was evaluated by measuring their ability to support cellular secretion. To this end, cells were cotransfected with a plasmid coding for a GBF1 mutant and a plasmid coding for Gaussia luciferase, an enzyme that has a natural sequence targeting it to the secretory pathway (23). The next day, the medium was replaced with medium containing BFA, and the amount of secreted Gaussia luciferase was determined after 4 h. The 1-10A, 11-20A, and 21-30A mutants were partially compromised in supporting secretion, while the 31-40A mutant supported an even somewhat higher level of secretion than did the wild-type GBF1 (Fig. 1B, secretion). Thus, viral replication can tolerate significant modifications of the very N-terminal fragment of GBF1, while such mutations have a more pronounced negative effect on GBF1 physiological function in secretion.
Substitutions of the N-terminal regions of GBF1 with those from BIG2 affect the function of GBF1 in poliovirus replication and cellular secretion.
To introduce more severe perturbations within the N-terminal region of GBF1, we replaced the entire N-terminal part of GBF1 up to the DCB domain with the corresponding 68-amino-acid-long fragment from BIG2, another large Arf GEF with similar domain organization, generating the 2-68/BIG2 construct (the first methionine is the same in both proteins). BIG2 localizes to the trans-Golgi network (TGN) and endosomes in cells and is incapable of rescuing poliovirus and coxsackievirus B3 replication from BFA inhibition (6, 7, 24, 25). The chimeric protein was expressed to a level similar to that of the positive control GBF1/A795E and was severely compromised in poliovirus replicon replication and nonfunctional in cellular secretion assays (data not shown).
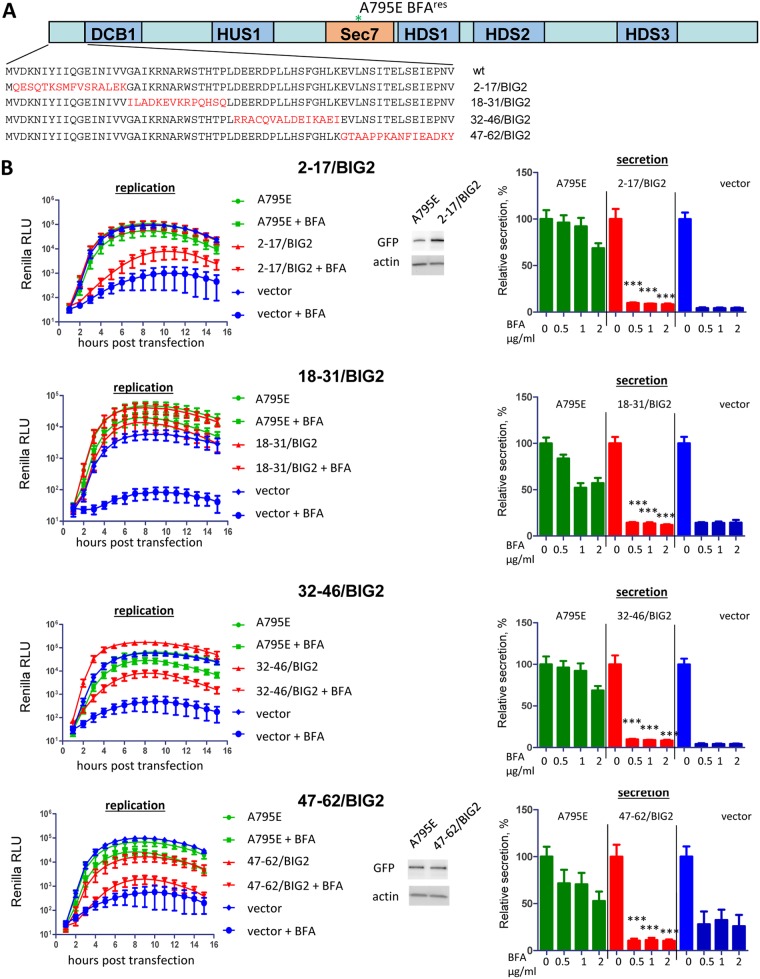
The 68-amino-acid BIG2 insert was further split into shorter stretches of 15 amino acids to generate the following four GBF1 constructs with the corresponding segments from BIG2: 2-17/BIG2, 18-31/BIG2, 32-46/BIG2, and 47-62/BIG2 (Fig. 2A). Two constructs with the BIG2-derived substitutions in the middle of the N-terminal fragment of GBF1 demonstrated either complete (18-31/BIG2) or partial (32-46/BIG2) ability to support poliovirus replication, while the ones with the most N-terminal (2-17/BIG2) and the most C-terminal (47-62/BIG2) substitutions were severely impaired in the replication assay. This suggests that either two distinct regions (positions 2 to 17 and 47 to 62) within the N terminus facilitate GBF1 function in poliovirus replication or that the two domains may interact to form a single functional interface. In contrast to their performance in viral replication, all four constructs were defective in cellular secretion (Fig. 2B). This supports the data in Fig. 1 showing that poliovirus replication is more resilient to N-terminal GBF1 mutations than its functioning in physiological context of cellular secretion.
FIG 2.
Functional analysis of GBF1/BIG2 N-terminal chimeras in replication and secretion. (A) Scheme of the BIG2-derived substitutions in the GBF1 sequence. All GBF1 expression constructs are GFP tagged and contain the A795E BFA resistance mutation in the Sec7 domain. (B) Performance of the indicated mutants in the poliovirus replicon replication and cellular secretion assays. For the replication assay, cells were transfected with the plasmids expressing a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control). The next day, the cells were transfected with a poliovirus replicon RNA expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA. Expression of the 2-17/BIG2 and 47-62/BIG2 constructs, the most compromised in the replication assay, is additionally verified by Western blotting in the samples from the corresponding experiments. For the secretion assay, the cells were cotransfected with plasmids coding for a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control) and a plasmid coding for a secreted Gaussia luciferase. The next day, they were washed and incubated in the medium with the indicated amount of BFA, and the amount of secreted luciferase was determined after 4 h. Secretion data are normalized to the signal obtained without BFA for each construct. The statistical significance of the difference between the signal in the positive control and that in the sample expressing a mutant GBF1 for corresponding concentrations of BFA is indicated.
The BIG2-derived N-terminal inserts in the constructs 2-17/BIG2 and 47-62/BIG2, the most severely compromised in the poliovirus replication assay, were further split into 4 amino acid blocks. All these constructs except for one with the substitution of the GBF1 amino acids in positions 9 to 12 (QGEI) for those derived from BIG2 (FVSR) could rescue poliovirus replication as well as did the positive control (data not shown). Since we are interested in finding GBF1 mutants that might be compromised in supporting viral replication but still functional in the cellular metabolism, the construct 9-12/BIG2 was also analyzed in a cellular secretion assay. However, 9-12/BIG2 proved to be completely nonfunctional in supporting secretion (data not shown). Collectively, our data demonstrate that the very N-terminal segment of GBF1 holds important determinants mediating protein functioning in poliovirus replication, and that poliovirus replication requirements for the N terminus of GBF1 are much less stringent than those of the secretory pathway.
The ability of the N-terminal GBF1 mutants to support poliovirus replication does not strictly correlate with interaction with the viral protein 3A.
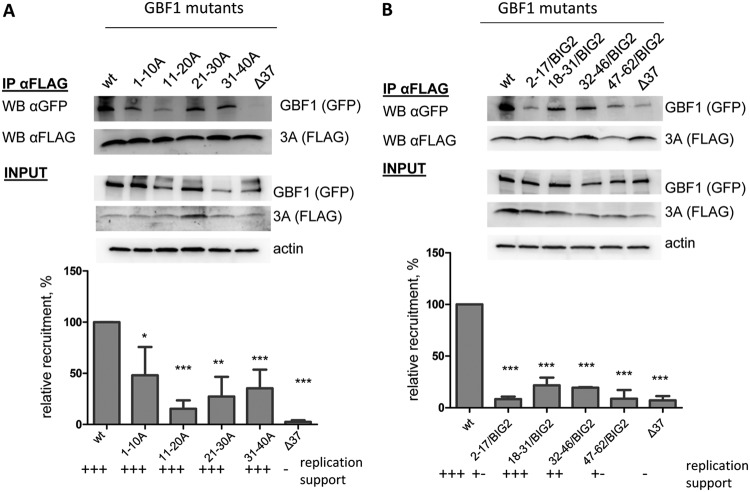
It is believed that direct interaction of the viral protein 3A with GBF1 accounts for the relocalization of GBF1 to the replication organelles where it supports the functioning of the replication complexes (11, 12). Thus, we used a co-IP assay to evaluate whether the mutations in the N terminus of GBF1 that did not support replication could have interfered with 3A interaction. HeLa cells were cotransfected with a plasmid expressing a GBF1 mutant and a plasmid expressing poliovirus protein 3A with a FLAG-Y insert after the sixth amino acid in the N terminus of the protein (3A-FLAG-Y). This insert was previously found to be fully compatible with poliovirus replication (26, 27). As a positive control for 3A-GBF1 interaction, cells were cotransfected with plasmids expressing 3A-FLAG-Y and wild-type (wt) GBF1, and for a negative control, cells were cotransfected with plasmids expressing 3A-FLAG-Y and the GBF1Δ37 mutant lacking 37 amino acids at the N terminus. Since all GBF1 constructs were expressed as N-terminal green fluorescent protein (GFP) fusions, anti-GFP antibodies were used to detect mutant GBF1 molecules. Surprisingly, all of the GBF1 mutants containing 10-amino-acid-long Ala substitutions in the N terminus, which were positive in the replication assay, had a noticeably decreased interaction with 3A compared to the positive control (Fig. 3A).
FIG 3.
Effects of mutations in the N terminus of GBF1 on interaction with the viral protein 3A. (A) Co-IP of 3A-FLAG with GFP fusions of alanine scanning mutants of GBF1. (B) Co-IP of 3A-FLAG with GFP fusions of GBF1/BIG2 chimeras. Cells were cotransfected with plasmids coding for corresponding GFP-tagged GBF1 mutants and a plasmid coding for 3A-FLAG-Y. IP was performed with anti-FLAG resin. GBF1s were detected with anti-GFP antibodies, and 3A was detected with anti-FLAG antibodies. Actin in the lysates is shown as a loading control. Relative recruitment is calculated by normalizing the GBF1-to-3A signal ratio in the pulldown material of the mutants to that of the positive-control (wt) sample. Each bar is an average of the results from at least three independent experiments. The performance of the corresponding GBF1 constructs in the replication assay is indicated.
The interaction of 3A with 2-68/BIG2 GBF1 mutant with a long BIG2-derived substitution, which could not support poliovirus replication, was reduced to an undetectable level (data not shown). All of the N-terminal GBF1 mutants containing 15-amino-acid-long inserts derived from BIG2 were also severely compromised in the interaction with 3A, irrespective of whether they supported replication or not (Fig. 3B). However, the lowest level of GBF1-3A interaction was repeatedly observed with constructs 2-17/BIG2 and 47-62/BIG2, which were almost negative in the replication assay (Fig. 3B). Collectively, these data are consistent with the previous observations that the 3A-GBF1 interaction generally correlates with the ability of GBF1 to support replication, although it can vary significantly without negative effect on the viral replication, at least in cell culture (6, 12, 27).
A functional Sec7 domain is critically important for poliovirus replication.
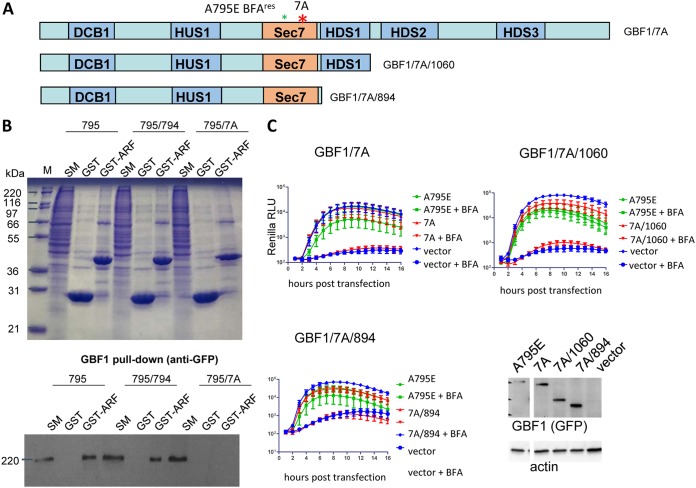
It was previously reported that GBF1 with the mutation E794K that inactivates the nucleotide exchange function of the Sec7 domain cannot support the replication of poliovirus and coxsackievirus B3 (6, 7). However, these data could not be unambiguously interpreted as a strict dependence of the viral replication machinery on activated Arf. The E794K mutation is believed to lock the GBF1-Arf-GDP complex in an inactive conformation, similar to the action of BFA (28, 29), and the GBF1/E794K mutant remains associated with membranes significantly longer than the functional wild-type GBF1 (30–32). Moreover, it was previously observed that GBF1/E794K mutants truncated just after the Sec7 domain, or even a GBF1 N-terminal fragment truncated before the Sec7 domain, could support at least some level of poliovirus replication in the presence of BFA, suggesting an Arf-independent contribution of GBF1 to the replication complexes (22). These findings are consistent with an alternative explanation that the virus requires the GBF1 protein irrespective of its Arf activation property, and that the E794K mutant could not be recruited to the replication complexes, perhaps by being too strongly stabilized on membranes. We recently generated and characterized a GBF1 mutant with a substitution of amino acids 883 to 889 to alanines (GBF1/7A) in the loop after the J helix of the Sec7 domain. The GBF1/7A mutant is predicted not to bind Arf-GDP and, therefore, should lack Arf activation capacity. Importantly, in contrast to the GBF1/E794K mutant, GBF1/7A retains the membrane binding dynamics of the wt protein and is not stabilized on membranes (33).
We generated a full-length GBF1 and two truncated constructs containing the 7A mutation within the context of the E795A mutation conferring BFA resistance (Fig. 4A). To confirm that the 7A mutation prevents the interaction of GBF1 with Arf, we performed a GBF1 pulldown assay with purified Arf1-glutathione S-transferase (Arf1-GST) that demonstrated that Arf1-GST interacts with wt GBF1 and the GBF1/A794K mutant but does not interact with the GBF1/7A mutant, as predicted (Fig. 4B). We then tested in a replication assay the full-length GBF1/7A construct, as well as 1060 and 894 truncated 7A constructs, similar to those previously shown to be partially functional in poliovirus replication in the contexts of the A794K Sec7-inactivating mutation (22). None of the GBF1 constructs with the 7A mutation could support poliovirus replication, even though they were expressed at a somewhat higher level than the positive control GBF1/A795E (Fig. 4C). Thus, we conclude that the Arf-activating function of GBF1 is essential for viral replication.
FIG 4.
GBF1 mutants unable to activate Arf are defective in replication. (A) Schematic of GBF1 constructs containing the inactivating 7A mutation. All GBF1 expression constructs are GFP tagged. (B) GST-Arf1-GBF1 pulldown assay. GST or GST-Δ17 ARF1 was immobilized on glutathione beads and incubated with lysate from cells (designated SM [starting material]) expressing GFP-tagged GBF1/A795E, GBF1/A795E/A794K, or GBF1/A795E/7A. The bound material was analyzed by SDS-PAGE and the gel either stained with Coomassie blue (top) or transferred to nitrocellulose (NC) and immunoblotted with anti-GFP antibodies (bottom). GBF1/795/7A does not bind the ARF substrate, whereas GBF1/795 and GBF1/795/794 bind ARF. (C) Performance of the corresponding mutants in a poliovirus replication assay. Cells were transfected with the plasmids expressing a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control). The next day, the cells were transfected with a poliovirus replicon RNA expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA. For the secretion assay, the cells were cotransfected with plasmids coding for a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control) and a plasmid coding for a secreted Gaussia luciferase. The next day, they were washed and incubated in the medium with the indicated amount of BFA, and the amount of secreted luciferase was determined after 4 h. Expression of the 7A constructs in the samples from the corresponding replication experiments is additionally verified by Western blotting. Representative positive-control (A795E) and negative-control (vector) samples are shown.
Mutations in the conserved elements of C-terminal domains of GBF1 do not affect viral replication.
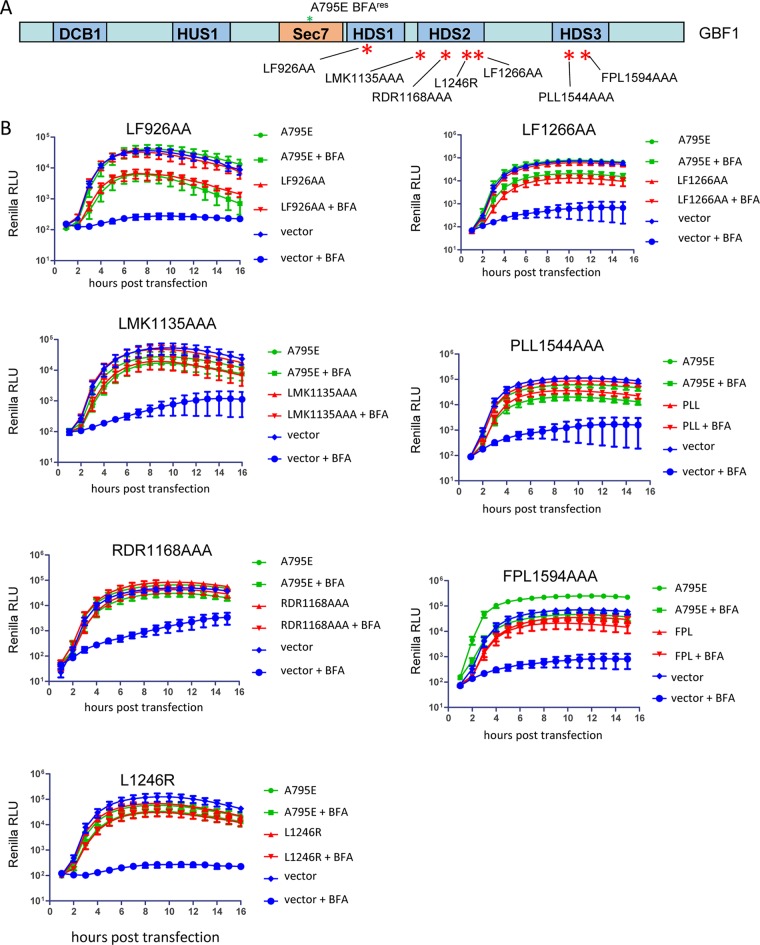
GBF1 orthologs regulate membrane trafficking in all eukaryotes, and they all contain several domains showing a high level of sequence and structure conservation among diverse taxa, separated by less-conserved linkers (34). Previously, we generated a series of mutations in the highly conserved regions of the HDS1, HDS2, and HDS3 domains of GBF1 targeting the most conserved residues. The HDS1 domain was recently found to target GBF1 to phosphoinositide phosphate (PIP)-enriched membranes, and the LF926AA mutation (number corresponds to the position of the first amino acid changed in the GBF1 sequence) disrupted this property and inhibited GBF1 function in secretion (35). Similarly, the HDS2 mutants L1246R, LF1266AA, and RDR1168AAA were severely compromised in supporting the cellular secretory pathway and targeting to the Golgi, while the HDS2 mutant LMK1135AAA behaved like a positive control in a secretion assay (23). HDS3 mutants FPL1594AAA and PLL1544AAA also inhibit GBF1 association with Golgi membranes and do not support secretion (our unpublished data). Here, we tested these GBF1 mutants (Fig. 5A) for their ability to support poliovirus replication. As shown in Fig. 5B, all the GBF1 mutants tested, including those previously shown to be severely compromised in cellular metabolism, supported poliovirus replication as well as the positive control.
FIG 5.
Functional analysis of GBF1 mutants targeting conserved elements in the C-terminal noncatalytic domains in replication and secretion. (A) Scheme of the mutations in the GBF1 sequence. All GBF1 expression constructs are GFP tagged and contain the A795E BFA resistance mutation in the Sec7 domain. (B) Performance of the corresponding mutants in a poliovirus replicon replication assay. Cells were transfected with the plasmids expressing a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control). The next day, the cells were transfected with a poliovirus replicon RNA expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA.
These data are consistent with the previously reported results that truncation of the whole C-terminal part of GBF1 downstream of the HDS1 domain does not interfere with GBF1 function in poliovirus replication complexes (22) and confirm that virus-specific function(s) of GBF1 require only a subset of the interactions/activities of this protein essential for cellular metabolism.
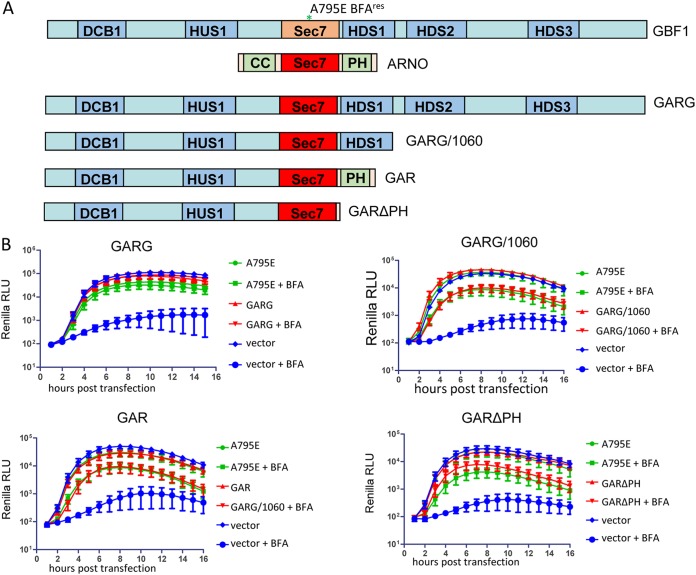
GBF1 constructs containing the Sec7 domain from the ArfGEF ARNO are fully functional in poliovirus replication.
To determine whether viral replication requires the cognate Sec7 domain of GBF1 or whether any active Sec7 domain would work, we generated chimeric constructs where the Sec7 domain of GBF1 was replaced with the Sec7 domain of another ArfGEF, ARNO. ARNO is responsible for Arf activation on the plasma membrane, where it regulates actin cytoskeleton-dependent cellular migration and endosomal trafficking (36–38). Thus, ARNO engages in a completely different set of interactions than does GBF1, which functions at the endoplasmic reticulum (ER)-Golgi interface. Apart from the Sec7 domain, ARNO and GBF1 do not share any significant homology, and the C-terminal part of ARNO contains only a pleckstrin homology (PH) domain responsible for targeting to the plasma membrane (4). Importantly, the amino acid sequence conservation between ARNO and GBF1 Sec7 domains is only 44%, indicating that substituting the Sec7 domains can be used to test the requirement for the GBF1 sequences in poliovirus replication. Due to the different amino acid sequence, ARNO is intrinsically resistant to BFA. We generated a construct (GARG, from GBF1-ARNO-GBF1), where the GBF1 Sec7 domain was substituted with that from ARNO (Fig. 6A). We have shown previously that GARG targets to the Golgi and supports secretion as well as wild-type GBF1 (39). Importantly, the GARG chimeric protein also was fully functional in supporting poliovirus replication (Fig. 6B). Thus, poliovirus replication has no specific requirement for GBF1 Sec7 domain but rather can be supported by any Sec7 domain capable of activating Arfs.
FIG 6.
GBF1 constructs containing a Sec7d from another GEF support poliovirus replication. (A) Scheme of the GBF1/ARNO chimeras. All GBF1 expression constructs are GFP tagged. (B) Analysis of the corresponding constructs in a poliovirus replicon replication assay. Cells were transfected with the plasmids expressing a corresponding GBF1 mutant, a full-length GBF1 A795E (positive control), or an empty vector (negative control). The next day, the cells were transfected with a poliovirus replicon RNA expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA.
Next, we tested if the GBF1-ARNO Sec7 substitution can function in the context of the 1060 truncation, i.e., when GARG is terminated after the HDS1 domain (GARG/1060) (Fig. 6A). The truncated protein retained the full capacity to support poliovirus replication (Fig. 6B).
It was previously observed that a GBF1 construct truncated just after the Sec7 domain (GBF1/894) is much less efficient in supporting poliovirus replicon replication than the GBF1/1060 construct truncated after the HDS1 domain (22). This phenomenon suggests two non-mutually exclusive possibilities, that either the HDS1 domain contains important elements necessary for proper functioning of GBF1 in replication, or HDS1 presence at the C terminus of the protein stabilizes the functionally important Sec7 domain. To see if the HDS1 domain of GBF1 provides any specific contribution to poliovirus replication, we generated a chimeric construct where the Sec7-HDS1 fragment of GBF1 was substituted with Sec7-PH domains of ARNO (GAR, from GBF1-ARNO) (Fig. 6A). GAR supported poliovirus replication similar to the GBF1 positive control (Fig. 6B), thus ruling out the need for HDS1-specific interactions for the functioning of the replication complexes. To assess if the PH domain in this construct is important, we generated a truncated mutant, GARΔPH, with just 10 amino acids of PH after the end of the ARNO Sec7 domain (Fig. 6A). This protein was also fully functional in the replication assay, conforming the HDS1-independent role of GBF1 in poliovirus replication (Fig. 6B), at least under the conditions of HeLa cells and a wt poliovirus RNA (see below).
The C-terminal HDS2 and HDS3 domains of GBF1 are important for viral replication under conditions of limited 3A-GBF1 interaction.
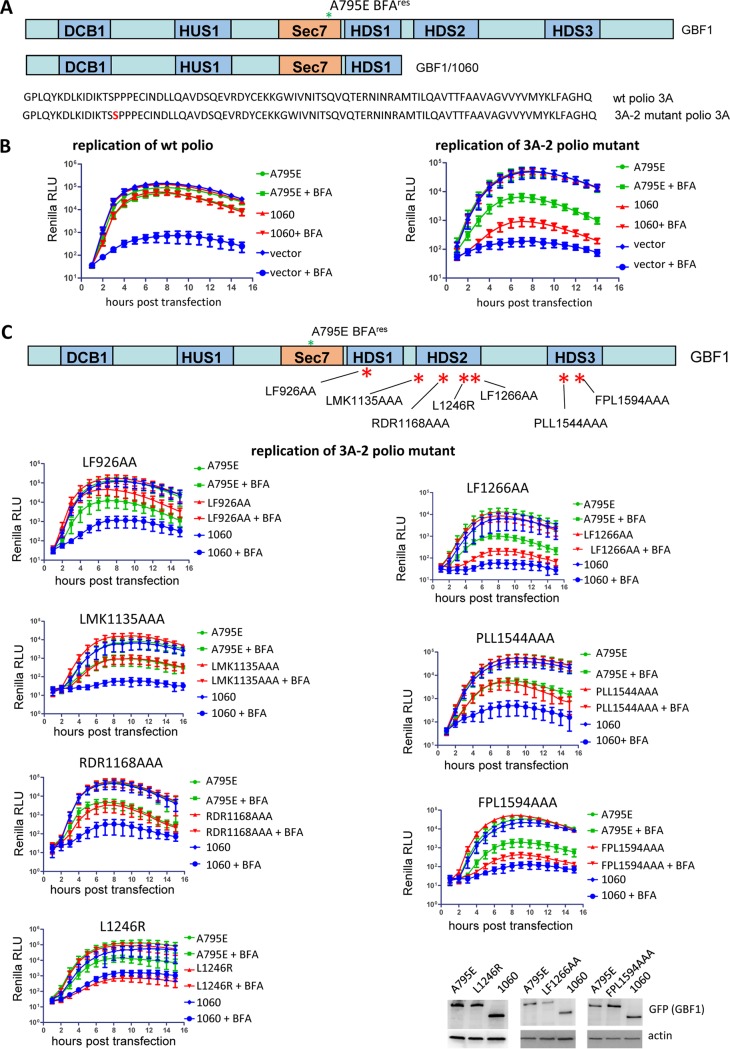
The surprising tolerance of poliovirus replication to varied levels of 3A-GBF1 interaction suggests that there may be an additional mechanism facilitating GBF1 recruitment to the replication complexes. The intrinsic propensity of GBF1 to bind membranes with specific lipid and protein compositions could be an important component of such a mechanism, positioning the protein where it can be easily accessed by the viral replication machinery. To test this possibility and to identify the GBF1 domains responsible, we used a poliovirus replicon containing the 3A-2 mutation, in which an additional serine is inserted at position 15 within the 3A sequence (Fig. 7A). The 3A-2 virus demonstrates a cold-sensitive phenotype, much higher sensitivity to BFA inhibition, a slightly reduced replication level, and a delay in the development of CPE compared to the wt virus (6, 40, 41). Importantly, the 3A-2 mutant and similar coxsackievirus B3 constructs are severely compromised in interactions with GBF1 (6, 7, 42).
FIG 7.
C-terminal GBF1 mutants show differential capacity to support replication of a wild-type versus the 3A-2 mutant replicon. (A) Top, scheme of the full-length and 1060 truncated GBF1 constructs; bottom, amino acid sequence of 3As from the wt poliovirus and 3A-2 mutant. All GBF1 expression constructs are GFP tagged and contain the A795E BFA resistance mutation in the Sec7 domain. (B) Performance of the full-length and 1060 truncated GBF1 constructs in a replication assay with a wt replicon and a replicon bearing the 3A-2 mutation, severely inhibiting 3A-GBF1 interaction. Cells were transfected with the plasmids expressing a full-length GBF1 A795E, a 1060 GBF1 A795E truncated construct, or an empty vector (negative control). The next day, the cells were transfected with a wt or 3A-2 poliovirus replicon RNAs expressing Renilla luciferase and incubated in the presence or absence of 1 μg/ml BFA. (C) Performance of the GBF1 mutants in the HDS1, HDS2, and HDS3 domains in a replication assay with a replicon containing 3A-2 mutation. Scheme of the mutation positions in the GBF1 domains is shown. All GBF1 expression constructs are GFP tagged and contain the A795E BFA resistance mutation in the Sec7 domain. Cells transfected with the full-length GBF1 A795E serve as a positive control, and those transfected with the 1060 truncated GBF1 A795 serve as a negative control. Expression of constructs L1246R, LF1266AA, and FPL1594AAA, severely impaired in rescuing the replication of the 3A-2 poliovirus replicon, is additionally verified by Western blotting in the samples from the corresponding experiments.
We compared the abilities of different GBF1 mutants to support the replication of the wild-type and 3A-2 replicon. As shown in Fig. 7B, the replication of a wild-type replicon was efficiently supported by both the full-length GBF1 and a construct truncated after the HDS1 domain (A795E/1060), consistent with data in this study (see Fig. 6) and reported elsewhere (22). In contrast, the replication of the 3A-2 poliovirus mutant could be efficiently supported by the full-length GBF1, although the level of rescue was lower than with the wt poliovirus, in accordance with previously reported data (6, 42), but not by the A795E/1060 truncated GBF1 construct (Fig. 7B). Thus, the C-terminal domains of GBF1 are dispensable for the replication of wild-type virus but become important when GBF1-3A interaction is diminished.
To identify the domains within the C terminus of GBF1 important for the replication of 3A-2, we examined the ability of C-terminal GBF1 mutants, which were fully functional in replication of the wild-type replicon (see Fig. 5), to support 3A-2 replication. GBF1 with the mutation LF926AA in the HDS1 domain consistently rescued replication of the 3A-2 replicon somewhat better than the positive control (Fig. 7C). GBF1 with the mutation LMK1135AAA or RDR1168AAA in the HDS2 domain and mutation PLL1544AAA in the HDS3 domain rescued replication of the 3A-2 replicon just like the positive control (Fig. 7C). However, the mutation FPL1594AAA in HDS3 and the mutations LF1266AA and especially L1246R in HDS2, severely impaired the capacity of GBF1 to rescue replication of the 3A-2 replicon (Fig. 7B).
Thus, the C-terminal HDS2 and HDS3 domains of GBF1, while dispensable for the replication of the wild-type replicon, hold important determinants facilitating the recruitment/function of the protein in replication complexes under conditions of reduced interaction between GBF1 and the viral protein 3A.
The C-terminal domains of GBF1 functionally alleviate the defects of GBF1-3A interaction in poliovirus replication.
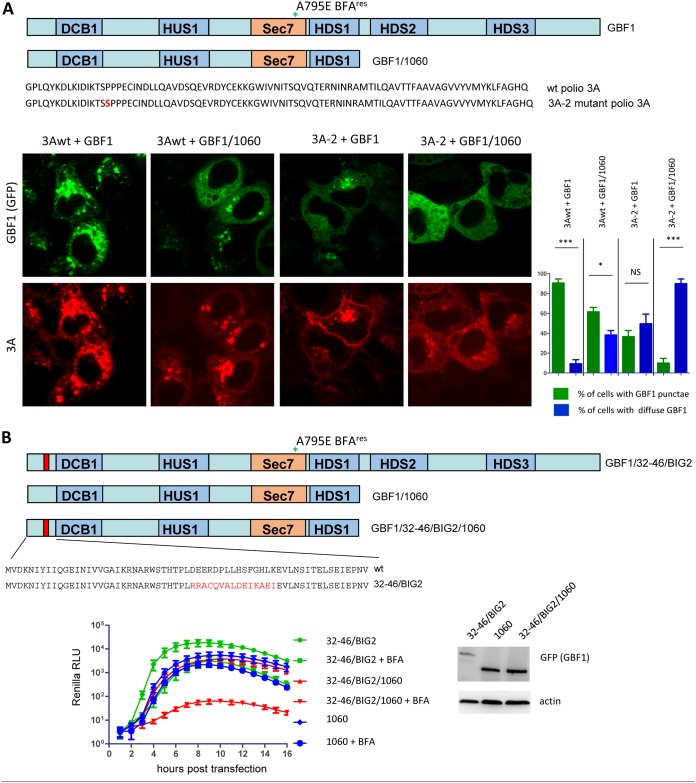
To further investigate the role of C-terminal domains of GBF1 in its recruitment by 3A, we cotransfected HeLa cells with plasmids expressing either wt or 3A-2 mutant poliovirus 3A proteins and plasmids expressing either a full-length GBF1 or a truncated GBF1/1060 construct lacking HDS2 and HDS3 (Fig. 8A). In cells coexpressing wt 3A and a full-length GBF1 construct, GBF1 signal was concentrated in bright agglomerates in virtually every cell, and such a distribution almost perfectly mirrored the 3A signal. In cells coexpressing wt 3A and the truncated version of GBF1, the GBF1 punctae became smaller, and they were present in only about half of the cells. A significant amount of 3A and GBF1 signals were distributed in an ER-like reticular pattern. A similar distribution of 3A and GBF1 signals was observed also in cells coexpressing 3A-2 mutant and a full-length GBF1, although both the GBF1 and the 3A agglomerates were noticeably smaller than those observed in cells expressing wt 3A. However, removing the GBF1 C-terminal domains resulted in an almost complete loss of the punctate GBF1 distribution in cells coexpressing the 3A-2 mutant protein (Fig. 8A). Thus, combinations of 3A and GBF1 corresponding to those found in functional replication complexes (wt 3A plus full-length or truncated GBF1, or 3A-2 plus full-length GBF1) demonstrated a significant amount of colocalizing GBF1 and 3A signals, while in cells coexpressing a nonfunctional combination (3A-2 plus truncated GBF1), such punctate colocalization was completely lost.
FIG 8.
C-terminal part of GBF1 can compensate for the defects in 3A-GBF1 interaction. (A) The C-terminal part of GBF1 facilitates its recruitment by 3A. Top, scheme of the full-length and 1060 truncated GBF1 constructs and amino acid sequence of 3As from the wt poliovirus and 3A-2 mutant. HeLa cells were cotransfected with plasmids expressing wt 3A or 3A-2 mutant proteins together with plasmids expressing either full-length or 1060 truncated GBF1 constructs. The next day, the cells were fixed and stained for 3A upon mild permeabilization, and the distribution of GBF1 signal in 3A-expressing cells was evaluated. Graph shows quantification of three independent experiments, with at least 200 cells counted for each sample. Statistical significance is indicated. (B) The C-terminal part of GBF1 can functionally compensate for the defect of GBF1-3A interaction in poliovirus replication. Top, scheme of a full-length GBF1 construct with substitution of amino acids 32 to 46 for those derived from a corresponding segment of BIG2; 1060 truncated GBF1 construct with wt N-terminal sequence; and 1060 truncated GBF1 construct with BIG2-derived substitution of amino acids 32 to 46. HeLa cells were transfected with the plasmids expressing indicated GBF1 constructs. The next day, the cells were transfected with a wt poliovirus replicon RNA coding for the Renilla luciferase gene, and incubated in the presence or absence of 1 μg/ml of BFA. Expression of the GBF1 constructs was verified by Western blot.
Our data support the double targeting model of GBF1 recruitment to the replication complexes where the C-terminal domains of GBF1 can compensate for deficiencies in 3A-GBF1 interactions. To further test this model, we made a truncated GBF1 construct lacking the C-terminal domains and incorporating the substitution of amino acids 32 to 46 for those derived from BIG2 (GBF1/32-46/BIG2/1060), and we tested its ability to support replication of the wt poliovirus replicon (Fig. 8B). The full-length GBF1 construct with 32-46/BIG2 substitution was capable of supporting poliovirus replication but was strongly compromised in interaction with 3A (see Fig. 2 and 3). Consistent with the previous data, both the 1060 construct with the wt N terminus and the full-length construct with the 32-46/BIG2 substitution could efficiently support poliovirus replication. However, when the 32-46/BIG2 substitution was introduced into the 1060 truncated context, such a construct was completely nonfunctional in the replication assay (Fig. 8B).
Thus, recruitment of GBF1 to the replication complexes relies on both 3A-GBF1 interaction and membrane targeting mediated by the C-terminal domains. Functional replication complexes may be formed if either of these mechanisms is compromised but not when both are inactivated at the same time.
DISCUSSION
The cellular protein GBF1 that supports functioning of the secretory pathway has long been known to be recruited to the replication complexes of poliovirus and related viruses and to mediate the sensitivity of infection to the fungal metabolite BFA (1–3, 6, 12). The replication-independent expression of poliovirus polyprotein in HeLa cells in the presence of BFA as well as the experiments in an in vitro system that allows separation of individual steps in the viral replication cycle revealed that GBF1 is required for proper assembly and/or functioning of the membrane-associated viral replication complexes (6, 43). However, the exact mechanism by which GBF1 supports the viral replication machinery remains unknown. GBF1 is a large protein of ∼200 kDa, containing six conserved domains, and in this study, we aimed to identify GBF1 elements necessary to sustain viral replication.
Previous co-IP and yeast two-hybrid assays established that the N-terminal ∼60-kDa fragment of GBF is sufficient for direct interaction with the 3A protein of coxsackievirus B3, a close relative of poliovirus (7, 12), and that the deletion of the first 37 amino acids of GBF1 blocks such 3A-GBF1 interaction (6). Here, we report that the N terminus of GBF1 can tolerate significant modifications, such as replacements of long stretches of the protein sequence with alanines, without a noticeable effect on its function in poliovirus replication. Interestingly, such extensive modifications of GBF1 were also compatible with its function in the cellular secretory pathway. Substitutions of N-terminal fragments of GBF1 with segments derived from a related protein, BIG2, which has a similar domain architecture, proved to be more detrimental. None of the four tested 16-amino-acid-long substitutions were tolerated in the secretion assay, but only two of them, the one substituting amino acids 2 to 17 and the one replacing amino acids 47 to 62, severely inhibited the replication-supporting function of GBF1. Further dissecting the BIG2-derived segments revealed that the substitution of only four GBF1 amino acids in positions 9 to 12 significantly inhibits viral replication.
The most studied function of GBF1 is its guanidine nucleotide exchange activity required for activation of the small GTPase Arf. This activity is mediated by the Sec7 domain of GBF1 and is the target of BFA and similar molecules that inhibit the cellular secretory pathway (reviewed in references 34, 44, and 45). GBF1 with the A794K mutation in the Sec7 domain locking GBF1 and Arf-GDP in a nonfunctional conformation does not support the replication of either poliovirus or coxsackie B3 virus (6, 7, 22). Here, we used a novel mutation in the Sec7 domain that prevents binding of Arf-GDP to GBF1 and found that similarly to the inactive A794K mutant, GBF1 constructs containing the 7A mutation could not support poliovirus replication. Collectively, these data strongly suggest that activated Arf is required for viral replication, although its mechanistic contribution in the replication process is unknown. The importance of Arf activation for poliovirus replication is also supported by the fact that a chimeric protein consisting of only the N-terminal half of GBF1 and a Sec7 domain from the Arf GEF ARNO was fully functional in the replication assay. One can envision that activated Arf molecules on the replication membranes may serve to recruit other cellular proteins, or that the presence of activated Arf may be necessary to keep cellular or even viral proteins in a functional conformation. These data seem to contradict previous reports that demonstrated at least partial functionality of certain truncated GBF1 species, either containing the A794K mutation or completely lacking the Sec7 domain, in poliovirus replication (22). Such a discrepancy may reflect the existence of some Arf activation-independent role of GBF1 in poliovirus replication, which may to a certain extent be sustained in the context of A794K (or in the constructs lacking the entire Sec7 domain) but not in the context of a 7A mutation. It is also possible that different modes of inactivating Sec7 domains, i.e., trapping of ArfGDP in an inactive conformation like in the case of the A794K mutation, or preventing ArfGDP binding like in the case of 7A mutation, result in different conformation and/or cellular localization of GBF1 molecules.
It has been shown that the specificity of GEFs, including GBF1, toward activation of certain Arfs is mediated by cellular localization of GEFs rather than by specificity of their Sec7 domains (39). It is likely that in infected cells such targeting signals are no longer functional, and the spectrum of Arfs activated by GBF1 may be different than that in noninfected cells, thus probably explaining the resilience of enterovirus replication to knockout or knockdown of expression of Arfs normally activated by GBF1 (7, 21).
Protein 3A of picornaviruses has increasingly been recognized as an important player in organizing the membranous environment suitable for viral replication. In poliovirus and related viruses, the interaction of 3A with GBF1 appears to be required for the recruitment of this cellular factor to the replication complexes (6, 7, 11, 12). In addition, 3A proteins of diverse picornaviruses directly interact with another cellular protein, ACBD3, and this interaction is important for recruitment to the replication complexes of a phosphatidylinositol kinase, PI4KIIIβ (19, 46–49). The data available so far, such as the accumulation of distinct resistance mutations upon propagation of picornaviruses in the presence of inhibitors of GBF1 or PI4KIIIβ, as well as PI4KIIIβ-independent recruitment of GBF1 and vice versa (50–52), indicate that GBF1- and PI4KIIIβ-controlled processes in the replication complexes are mutually independent, in spite of being coordinated by the same viral protein. This poses a question of whether the same molecule of protein 3A, which in poliovirus is only 87 amino acids long, simultaneously interacts with both ACBD3 and GBF1, or if there are different populations of 3As in the replication complexes that participate in different sets of interactions with cellular and/or viral proteins.
While the 3A-GBF1 interaction appears to be important for poliovirus replication, numerous findings indicate that the strength of such an interaction can vary significantly without a dramatic effect on virus replication. This suggests that there may be a significant excess of GBF1 available in cells, at least in the conventional cell culture models used for the study of picornaviruses, or that there is an axillary mechanism targeting GBF1 to the replication membranes that may compensate for the weakened 3A-GBF1 interaction. We previously found that mutations in the conserved regions of GBF1 located in the C-terminal noncatalytic domains of the protein prevent its targeting to Golgi membranes and inhibit GBF1 function in the secretory pathway (35, 53). When we tested such mutants in poliovirus replication, all fully supported replication of the wild-type replicon, but several C-terminal GBF1 mutants were severely compromised in supporting the replication of a 3A-2 replicon carrying a mutation that strongly diminishes the 3A-GBF1 interaction (6, 7, 42). The mechanism of how the C-terminal domains of GBF1 facilitate recruitment of GBF1 to the replication complexes is, however, far from obvious and requires further investigation.
It is known that replication organelles of picornaviruses, including those of poliovirus, are highly enriched in PI4P (13). Yet, the mutation LF926AA in the HDS1 domain of GBF1, previously found to impair targeting of GBF1 to PIP-containing membranes (35), did not decrease the replication of the 3A-2 mutant and even somewhat outperformed the positive control. The most severe negative effect on the replication of the 3A-2 replicon was observed with the L1246R mutation of a conserved amino acid in the HDS2 domain of GBF1. This mutation was originally identified in zebrafish embryos with severe vasculature development abnormality (54). It was later established that such a mutation in the human GBF1 makes the protein incapable of supporting the secretory pathway and results in dramatic differences in the localization of the mutant protein compared to the normal GBF1 (53). Thus, a mislocalized GBF1 with the L1246R mutation was fully functional in replication of a wt replicon but completely incapable of supporting replication under conditions when GBF1 interaction with the viral protein 3A was impaired. This suggests that the membrane-targeting determinants of GBF1 C terminus provide an additional mechanism facilitating GBF1 recruitment/function in the replication complexes. Supporting the existence of a redundant targeting mechanism of GBF1 to poliovirus replication complexes is the incompatibility of a substitution of 16 amino acids in the N-terminal region of GBF1 reducing its interaction with 3A and truncation of the C-terminal domains, neither of which by itself is detrimental to replication. Thus, poliovirus can replicate when either the 3A-GBF1 interaction or the proper cellular targeting of GBF1 is inhibited but not when both are compromised at the same time. This likely explains the surprising tolerance of at least some picornaviruses, including poliovirus, to the variability of the level of direct 3A-GBF1 interaction (6, 11). Whether specific membrane targeting can compensate for a complete loss of GBF1-3A interaction, or if at least some level of such direct interaction is absolutely essential for replication remains to be established.
In general, these and previous findings demonstrate that poliovirus replication can be supported by GBF1 species completely nonfunctional in cellular metabolism, including extensively truncated mutants, indicating that the virus needs only a subset of GBF1 activities/interactions essential for GBF1 normal function in the cell. Given that GBF1 is an essential factor for the replication of diverse (+)RNA viruses (8–10), it will be interesting to determine if these diverse viruses rely on the same mechanistic contribution(s) of GBF1 to the replication process. The identification of the molecular processes by which GBF1 supports viral replication may allow interventions to target GBF1 performance specifically in infected cells. This may open novel directions in the development of antiviral therapeutics.
MATERIALS AND METHODS
Cells and reagents.
HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) high-glucose modification supplemented with 1 mM sodium pyruvate and nonessential amino acids. Brefeldin A (BFA) was from Sigma-Aldrich, and a 4 mg/ml stock solution was prepared in dimethyl sulfoxide (DMSO) and stored at −80°C.
Plasmids.
All GBF1 expression plasmids were made based on the pYFP-GBF1 plasmid described previously (30). Numbers in the names of truncated GBF1 constructs indicate the last amino acid. An ARNO Sec7 domain fragment and ARNO Sec7-PH fragment were amplified from a plasmid coding for a FLAG-tagged human ARNO protein kindly provided by Julie Donaldson (NIH). Fragments with Ala scanning mutations and N-terminal GB1-BIG2 chimeras were synthesized by the GeneArt service (Invitrogen) and cloned into the pVenus-GBF1 plasmid. The pXpa-RenR plasmid coding for a poliovirus replicon with Renilla luciferase substituting for the capsid proteins was described previously (55). pCMV-GLUC, coding for Gaussia luciferase, was from New England BioLabs. Plasmid pCI-3A-FLAG, coding for poliovirus 3A with FLAG-Y tag under the control of cytomegalovirus (CMV) promoter, is described in reference 26, and plasmids pCI-3A and pCI-3A-2 coding for the 3A wt and 3A-2 mutant, respectively, are constructed similarly. Cloning details are available upon request. Plasmid transfections were performed with Mirus 2020 DNA transfection reagent (Mirus Bio), according to the manufacturer’s protocol. The expression of GBF1 constructs was confirmed by GFP fluorescence, and expression of the constructs that were significantly compromised in supporting poliovirus replication was additionally verified by Western blotting.
Antibodies.
Mouse monoclonal anti-poliovirus 3A antibodies were described in reference 56. Rabbit polyclonal anti-GFP antibodies were from Abcam; rabbit polyclonal anti-FLAG antibodies were from Thermo Fisher. Mouse monoclonal anti-β-actin antibodies conjugated with horseradish peroxidase (HRP) and agarose beads conjugated with anti-FLAG mouse monoclonal M13 antibody were from Sigma-Aldrich.
Immunofluorescence assay.
HeLa cells grown on coverslips were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min and washed three times with PBS. Staining was performed under mild permeabilization conditions by incubating cells with primary and secondary antibodies in 0.02% saponin (Sigma-Aldrich) in PBS containing 5% fetal bovine serum (FBS) as a blocking agent. Confocal images were taken with a Zeiss LSM 510 microscope.
Replication assay.
A poliovirus replicon replication assay was performed essentially as described by Viktorova et al. (57). Briefly, HeLa cells were transfected in suspension with a GBF1-expressing plasmid and then plated on a 96-well plate so that the cells in all corresponding wells are transfected similarly. The next day, the cells were transfected with a purified poliovirus replicon RNA using Mirus mRNA transfection reagent (Mirus Bio). The cells were incubated after replicon transfection in the medium containing EnduRen cell-permeable Renilla luciferase substrate (Promega) in the presence or absence of 1 μg/ml BFA. Renilla luciferase signal was monitored in live cells every hour with Tecan M1000 or Molecular Devices iD5 multifunctional plate readers equipped with a thermostatic incubation chamber. Each data point on the replication graph is an average signal from at least 12 wells.
Secretion assay.
A secretion assay was performed as described in reference 53. Briefly, HeLa cells were cotransfected in a suspension with a 9:1 mass ratio of a GBF1-expressing plasmid and pCMV-GLUC and then plated on a 96-well plate, so that the cells in all corresponding wells are transfected similarly. The next day, the cells were washed three times with a serum-free medium and then incubated in a fresh medium supplemented with the indicated amount of BFA for 4 h. At this time point, 20 μl of the medium was used to monitor the amount of secreted Gaussia luciferase using a Gaussia luciferase assay kit (New England BioLabs). Secretion data were normalized to the signal obtained from the cells incubated without BFA for each sample. Each data point is an average of the signal from 8 wells.
Arf1-GST-GBF1 pulldown assay.
For the preparation of recombinant Δ17 ARF1, GSTΔ17 ARF1 was expressed in BL21(DE3)/pLysS Escherichia coli (Promega, Madison, WI) and purified on Pierce glutathione agarose (Thermo Scientific, IL, USA), according to the manufacturer’s directions.
For ARF binding, HeLa cells transfected with GFP-tagged GBF1/795, GBF1/795/794, or GBF1/795/7A were lysed in 50 mM Bis-Tris (pH 7.2), 6 N HCl, 50 mM NaCl, 10% (wt/vol) glycerol, 0.001% Ponceau S, and 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) containing a Complete protease inhibitor mixture tablet, EDTA-free (Santa Cruz Biotechnology, Santa Cruz, CA). Lysates were precleared by centrifugation at 15,000 rpm for 30 min at 4°C. The supernatants (100 μl) were incubated with GST-17-residue NH2-terminal deletion mutant ([delta 17]ARF1) prebound to glutathione-Sepharose 4B beads for 1 h at room temperature and processed for SDS-PAGE. Starting material (SM) was loaded as 10 μl of the original supernatant.
Coimmunoprecipitation.
HeLa cells grown on 35-mm petri dishes were cotransfected with pCI-3A-FLAG and a GBF1 expression plasmid at the mass ratio of 1:1, and the next day, co-IP was performed using anti-FLAG agarose beads (Sigma-Aldrich), according to the manufacturer’s protocol. After a final wash, the proteins were released by heating the beads in the Laemmli SDS protein sample buffer for 5 min at 95°C. The proteins were resolved on a 4 to 15% Tris-glycine gel gradient (Bio-Rad), and 3A and GBF1 constructs were detected in Western blots with anti-FLAG and anti-GFP antibodies, respectively. Digital images of Western blots developed with ECL Select luminescent substrate (GE Healthcare) were obtained with a C500 imager (Azure Biosystems) and analyzed using the ImageStudio software (Li-Cor). Relative recruitment is calculated by normalizing the GBF1-to-3A signal ratio in the pulldown material of the mutants to that of the positive-control (wt) sample. Each bar is an average of at least three independent experiments.
Statistical analysis.
Data presentation and unpaired two-tailed t test statistical analysis were performed using the GraphPad Prism software package. Plots show mean values and standard deviation. Statistical significance is indicated as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, nonsignificant.
ACKNOWLEDGMENTS
This work was supported by the NIH (grant R01GM122802 to E.S. and grant R01AI125561 to G.A.B.) and the NSF (grant MCB-1615607 to E.S.).
REFERENCES
- 1.Maynell LA, Kirkegaard K, Klymkowsky MW. 1992. Inhibition of poliovirus RNA-synthesis by brefeldin-A. J Virol 66:1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irurzun A, Perez L, Carrasco L. 1992. Involvement of membrane traffic in the replication of poliovirus genomes–effects of brefeldin-A. Virology 191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 3.Cuconati A, Molla A, Wimmer E. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J Virol 72:6456–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova JE. 2007. Regulation of arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 5.Nie ZZ, Hirsch DS, Randazzo PA. 2003. Arf and its many interactors. Curr Opin Cell Biol 15:396–404. doi: 10.1016/S0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 6.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanke KHW, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld F. 2009. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol 83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farhat R, Ankavay M, Lebsir N, Gouttenoire J, Jackson CL, Wychowski C, Moradpour D, Dubuisson J, Rouille Y, Cocquerel L. 2018. Identification of GBF1 as a cellular factor required for hepatitis E virus RNA replication. Cell Microbiol 20:e12804. doi: 10.1111/cmi.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verheije MH, Raaben M, Mari M, Lintelo EGT, Reggiori F, van Kuppeveld FJM, Rottier PJM, de Haan C. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog 4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wessels E, Duijsings D, Lanke KHW, Melchers WJG, Jackson CL, van Kuppeveld F. 2007. Molecular determinants of the interaction between coxsackievirus protein 3A and guanine nucleotide exchange factor GBF1. J Virol 81:5238–5245. doi: 10.1128/JVI.02680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KHW, Klumperman J, Henke A, Jackson CL, Melchers WJG, van Kuppeveld F. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev Cell 11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Schaar HM, Leyssen P, Thibaut HJ, de Palma A, van der Linden L, Lanke KHW, Lacroix C, Verbeken E, Conrath K, MacLeod AM, Mitchell DR, Palmer NJ, van de Poël H, Andrews M, Neyts J, van Kuppeveld FJM. 2013. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase III beta. Antimicrob Agents Chemother 57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spickler C, Lippens J, Laberge MK, Desmeules S, Bellavance E, Garneau M, Guo T, Hucke O, Leyssen P, Neyts J, Vaillancourt FH, Decor A, O’Meara J, Franti M, Gauthier A. 2013. Phosphatidylinositol 4-kinase III beta is essential for replication of human rhinovirus and its inhibition causes a lethal phenotype in vivo. Antimicrob Agents Chemother 57:3358–3368. doi: 10.1128/AAC.00303-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P-2 on the Golgi complex. Nat Cell Biol 1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 17.Xiao X, Lei XB, Zhang ZZ, Ma YJ, Qi JL, Wu C, Xiao Y, Li L, He B, Wang JW. 2017. Enterovirus 3A facilitates viral replication by promoting phosphatidylinositol 4-kinase III beta-ACBD3 interaction. J Virol 91:e00791-17. doi: 10.1128/JVI.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J 31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa-Sasaki K, Nagashima S, Taniguchi K, Sasaki J. 2018. Model of OSBP-mediated cholesterol supply to Aichi virus RNA replication sites involving protein-protein interactions among viral proteins, ACBD3, OSBP, VAP-A/B, and SAC1. J Virol 92:e01952-17. doi: 10.1128/JVI.01952-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klima M, Toth DJ, Hexnerova R, Baumlova A, Chalupska D, Tykvart J, Rezabkova L, Sengupta N, Man P, Dubankova A, Humpolickova J, Nencka R, Veverka V, Balla T, Boura E. 2016. Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein. Sci Rep 6:23641. doi: 10.1038/srep23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferlin J, Farhat R, Belouzard S, Cocquerel L, Bertin A, Hober D, Dubuisson J, Rouille Y. 2018. Investigation of the role of GBF1 in the replication of positive-sense single-stranded RNA viruses. J Gen Virol 99:1086–1096. doi: 10.1099/jgv.0.001099. [DOI] [PubMed] [Google Scholar]
- 22.Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. 2010. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell Microbiol 12:1463–1479. doi: 10.1111/j.1462-5822.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Yamaji R, Adamik R, Takeda K, Togawa A, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. 2000. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci U S A 97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin HW, Morinaga N, Noda M, Nakayama K. 2004. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell 15:5283–5294. doi: 10.1091/mbc.e04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teterina NL, Lauber C, Jensen KS, Levenson EA, Gorbalenya AE, Ehrenfeld E. 2011. Identification of tolerated insertion sites in poliovirus non-structural proteins. Virology 409:1–11. doi: 10.1016/j.virol.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E. 2011. Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J Virol 85:4284–4296. doi: 10.1128/JVI.02398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mossessova E, Corpina RA, Goldberg J. 2003. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell 12:1403–1411. doi: 10.1016/S1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- 29.Renault L, Guibert B, Cherfils J. 2003. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 30.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. 2005. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell 16:1213–1222. doi: 10.1091/mbc.e04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szul T, Garcia-Mata R, Brandon E, Shestopal S, Alvarez C, Sztul E. 2005. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6:374–385. doi: 10.1111/j.1600-0854.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhao XH, Claude A, Chun J, Shields DJ, Presley JF, Melancon P. 2006. GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J Cell Sci 119:3743–3753. doi: 10.1242/jcs.03173. [DOI] [PubMed] [Google Scholar]
- 33.Lowery J, Szul T, Seetharaman J, Jian XY, Su M, Forouhar F, Xiao R, Acton TB, Montelione GT, Lin HL, Wright JW, Lee E, Holloway ZG, Randazzo PA, Tong L, Sztul E. 2011. Novel C-terminal motif within Sec7 domain of guanine nucleotide exchange factors regulates ADP-ribosylation factor (ARF) binding and activation. J Biol Chem 286:36898–36906. doi: 10.1074/jbc.M111.230631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders N, Jurgens G. 2008. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol Life Sci 65:3433–3445. doi: 10.1007/s00018-008-8227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner JM, Bhatt JM, Lee E, Styers ML, Ivanova AA, Kahn RA, Sztul E. 2018. The ARF guanine nucleotide exchange factor GBF1 is targeted to Golgi membranes through a PIP-binding domain. J Cell Sci 131:jcs210245. doi: 10.1242/jcs.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank S, Upender S, Hansen SH, Casanova JE. 1998. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem 273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 37.Santy LC, Casanova JE. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol 154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shmuel M, Santy LC, Frank S, Avrahami D, Casanova JE, Altschuler Y. 2006. ARNO through its coiled-coil domain regulates endocytosis at the apical surface of polarized epithelial cells. J Biol Chem 281:13300–13308. doi: 10.1074/jbc.M513723200. [DOI] [PubMed] [Google Scholar]
- 39.Bhatt JM, Hancock W, Meissner JM, Kaczmarczyk A, Lee E, Viktorova E, Ramanadham S, Belov GA, Sztul E. 2019. Promiscuity of the catalytic Sec7 domain within the guanine nucleotide exchange factor GBF1 in ARF activation, Golgi homeostasis, and effector recruitment. Mol Biol Cell 30:1523–1535. doi: 10.1091/mbc.E18-11-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berstein HD, Baltimore D. 1988. Poliovirus mutant that contains a cold-sensitive defect in viral-RNA synthesis. J Virol 62:2922–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodd DA, Giddings TH, Kirkegaard K. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J Virol 75:8158–8165. doi: 10.1128/jvi.75.17.8158-8165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viktorova EG, Nchoutmboube J, Ford-Siltz LA, Belov GA. 2015. Cell-specific establishment of poliovirus resistance to an inhibitor targeting a cellular protein. J Virol 89:4372–4386. doi: 10.1128/JVI.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fogg MH, Teterina NL, Ehrenfeld E. 2003. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J Virol 77:11408–11416. doi: 10.1128/jvi.77.21.11408-11416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaczmarek B, Verbavatz JM, Jackson CL. 2017. GBF1 and Arf1 function in vesicular trafficking, lipid homoeostasis and organelle dynamics. Biol Cell 109:391–399. doi: 10.1111/boc.201700042. [DOI] [PubMed] [Google Scholar]
- 45.Wright J, Kahn RA, Sztul E. 2014. Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci 71:3419–3438. doi: 10.1007/s00018-014-1602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyoo H, van der Schaar HM, Dorobantu CM, Rabouw HH, Strating J, van Kuppeveld FJM. 2019. ACBD3 is an essential pan-enterovirus host factor that mediates the interaction between viral 3A protein and cellular protein PI4KB. mBio 10:e02742-18. doi: 10.1128/mBio.02742-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lei XB, Xiao X, Zhang ZZ, Ma YJ, Qi JL, Wu C, Xiao Y, Zhou Z, He B, Wang JW. 2017. The Golgi protein ACBD3 facilitates enterovirus 71 replication by interacting with 3A. Sci Rep 7:44592. doi: 10.1038/srep44592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klima M, Chalupska D, Różycki B, Humpolickova J, Rezabkova L, Silhan J, Baumlova A, Dubankova A, Boura E. 2017. Kobuviral non-structural 3A proteins act as molecular harnesses to hijack the host ACBD3 protein. Structure 25:219–230. doi: 10.1016/j.str.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 49.McPhail JA, Ottosen EH, Jenkins ML, Burke JE. 2017. The molecular basis of Aichi virus 3A protein activation of phosphatidylinositol 4 kinase III beta, PI4KB, through ACBD3. Structure 25:121–131. doi: 10.1016/j.str.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 50.Dorobantu CM, Ford-Siltz LA, Sittig SP, Lanke KHW, Belov GA, van Kuppeveld FJM, van der Schaar HM. 2015. GBF1-and ACBD3-independent recruitment of PI4KIII beta to replication sites by rhinovirus 3A proteins. J Virol 89:1913–1918. doi: 10.1128/JVI.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorobantu CM, van der Schaar HM, Ford LA, Strating JR, Ulferts R, Fang Y, Belov G, van Kuppeveld FJ. 2014. Recruitment of PI4KIIIbeta to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1. J Virol 88:2725–2736. doi: 10.1128/JVI.03650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ford Siltz LA, Viktorova EG, Zhang B, Kouiavskaia D, Dragunsky E, Chumakov K, Isaacs L, Belov GA. 2014. New small-molecule inhibitors effectively blocking picornavirus replication. J Virol 88:11091–11107. doi: 10.1128/JVI.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pocognoni CA, Viktorova EG, Wright J, Meissner JM, Sager G, Lee E, Belov GA, Sztul E. 2018. Highly conserved motifs within the large Sec7 ARF guanine nucleotide exchange factor GBF1 target it to the Golgi and are critical for GBF1 activity. Am J Physiol Cell Physiol 314:C675–C689. doi: 10.1152/ajpcell.00221.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Wu XT, Yao LK, Yan L, Zhang L, Qiu JH, Liu XF, Jia SJ, Meng AM. 2017. Impairment of cargo transportation caused by gbf1 mutation disrupts vascular integrity and causes hemorrhage in zebrafish embryos. J Biol Chem 292:2315–2327. doi: 10.1074/jbc.M116.767608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol 81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasamontes L, Egger D, Bienz K. 1986. Production of monoclonal and monospecific antibodies against non-capsid proteins of poliovirus. J Gen Virol 67:2415–2422. doi: 10.1099/0022-1317-67-11-2415. [DOI] [PubMed] [Google Scholar]
- 57.Viktorova EG, Khattar S, Samal S, Belov GA. 2018. Poliovirus replicon RNA generation, transfection, packaging, and quantitation of replication. Curr Protoc Microbiol 48:15H.4.1–15H.4.15. doi: 10.1002/cpmc.47. [DOI] [PMC free article] [PubMed] [Google Scholar]