It was previously determined that the type II secretion system (T2SS) promotes the ability of Legionella pneumophila to grow in coculture with amoebae. Here, we discerned the stage of intracellular infection that is potentiated by comparing the wild-type and T2SS mutant legionellae for their capacity to parasitize Acanthamoeba castellanii.
KEYWORDS: Acanthamoeba, LCV, Legionella pneumophila, ProA, T2SS, type II secretion, Vermamoeba, phagosome
ABSTRACT
It was previously determined that the type II secretion system (T2SS) promotes the ability of Legionella pneumophila to grow in coculture with amoebae. Here, we discerned the stage of intracellular infection that is potentiated by comparing the wild-type and T2SS mutant legionellae for their capacity to parasitize Acanthamoeba castellanii. Whereas the mutant behaved normally for entry into the host cells and subsequent evasion of degradative lysosomes, it was impaired in the ability to replicate, with that defect being first evident at approximately 9 h postentry. The replication defect was initially documented in three ways: by determining the numbers of CFU recovered from the lysates of the infected monolayers, by monitoring the levels of fluorescence associated with amoebal monolayers infected with green fluorescent protein (GFP)-expressing bacteria, and by utilizing flow cytometry to quantitate the amounts of GFP-expressing bacteria in individual amoebae. By employing confocal microscopy and newer imaging techniques, we further determined the progression in volume and shape of the bacterial vacuoles and found that the T2SS mutant grows at a decreased rate and does not attain maximally sized phagosomes. Overall, the entire infection cycle (i.e., entry to egress) was considerably slower for the T2SS mutant than it was for the wild-type strain, and the mutant’s defect was maintained over multiple rounds of infection. Thus, the T2SS is absolutely required for L. pneumophila to grow to larger numbers in its intravacuolar niche within amoebae. Combining these results with those of our recent analysis of macrophage infection, T2SS is clearly a major component of L. pneumophila intracellular infection.
INTRODUCTION
Legionella pneumophila, a ubiquitous occupant of water systems, is the main agent of Legionnaires’ disease, a potentially fatal pneumonia of increasing incidence (1, 2). The Gram-negative L. pneumophila flourishes in its aquatic environments as an intracellular parasite of amoebae, primarily species of Acanthamoeba and Vermamoeba (3–5). In the human lung, the bacterium grows in alveolar macrophages (6, 7). In both protozoan and mammalian cells, L. pneumophila avoids degradation by the host lysosomal pathway and replicates to large numbers in a membrane-bound compartment known as the Legionella-containing vacuole (LCV). This infection ultimately lyses the spent host cell, and the L. pneumophila parasite swims away in pursuit of new targets. Essential to the intracellular infection cycle is a type IV protein secretion system (T4SS), which serves to transport 300+ effectors from the LCV into the host cell’s cytoplasm (8–11).
The type II protein secretion system (T2SS) is a second major contributor to L. pneumophila pathogenesis (12, 13). T2SS facilitates infection of the lungs by promoting bacterial growth in macrophages, limiting the host’s immune response, and elaborating destructive enzymes (14–16). During infection of macrophages, the T2SS does not mediate entry or avoidance of lysosomes; rather, it promotes bacterial growth in the LCV at 4 to 12 h postentry (17). The T2SS is even more vital to L. pneumophila survival in water, promoting sliding motility, biofilm formation, and, most of all, infection of the prominent amoebal host Acanthamoeba castellanii as well as Naegleria lovaniensis, Vermamoeba vermiformis, and Willaertia magna (18–27). L. pneumophila T2SS secretes ≥25 proteins, including many enzymes, in addition to novel proteins that are unique to Legionella (12, 13, 25, 28, 29). Secreted proteins that are known to foster infection of amoebae are the acyltransferase PlaC, aminopeptidase LapA, metalloprotease ProA, RNase SrnA, and novel proteins NttA, NttC, and NttD (19, 23–25, 30, 31). At least some of the T2SS substrates, including ProA, are translocated out of the LCV into the host cytoplasm where they likely act on a wide range of targets (32).
Even though the L. pneumophila T2SS is one of the most defined T2SSs (12, 13), there remain notable gaps in our knowledge. For example, it is still not known at what stage(s) of infection of amoebae the T2SS is required. In past work, the importance of the T2SS was identified through the use of coculture systems, in which wild-type, T2SS mutant (e.g., lspF and lspDE mutants), and complemented T2SS mutant legionellae were inoculated at a multiplicity of infection (MOI) of 0.1 into wells containing amoebae, and at various times postinoculation (p.i.) aliquots taken from the tissue culture medium were plated for CFU counts (23–25, 30, 31). Since L. pneumophila does not replicate in the medium used for the coculture, increases in CFU recovery reflect bacterial entry into and growth in the amoebae. Thus, when multiple, independently derived lsp mutants displayed impaired CFU recovery relative to that of both their parental wild-type strain and complemented derivative, it was concluded that the T2SS promotes one or more stages of intracellular infection. However, it has been unclear whether the T2SS is promoting entry into the amoebal host cell, evasion of lysosomes, and/or actual replication within the intracellular compartment. Using a broader range of techniques, we now demonstrate that the L. pneumophila T2SS is not required for entry in amoebae or escape from lysosomes but promotes subsequent intracellular replication and the formation of large-volume LCVs.
RESULTS
Determining an inoculating dose for studying the interaction between the L. pneumophila T2SS and amoebae.
In order to more readily monitor the behavior of L. pneumophila within amoebae, we introduced plasmid-encoded green fluorescent protein (GFP) into wild-type strain 130b and observed a high correlation between the MOI and the levels of cell-associated GFP immediately following bacterial entry into A. castellanii (Fig. 1A, left). To discern which MOI(s) resulted in a majority of the amoebae becoming productively infected, we inoculated amoebae with the same range of MOIs (i.e., 3 to 100), waited for 10 h to allow for replication, and using flow cytometry, quantified the number of individually infected cells. With increasing MOIs, there was a corresponding rise in the proportion of infected cells; however, at MOIs of ≥50, the increase in the number of infected cells diminished, suggesting that each amoeba was infected with >1 bacterium (Fig. 1A, right). We used an MOI of 20 for the majority of later experiments as this was within the linear range of detection during the initial rounds of infection (Fig. 1A, left) and maximized the proportion of infected cells to 70 to 80% (Fig. 1A, right). Also, at the MOI of 20, an lspF mutant of strain 130b that lacks the T2SS was still impaired, as measured by a CFU-based coculture at 24, 48, and 72 h p.i., although the maximal defect observed at 72 h was not as great as that when the previously employed MOI of 0.1 was used (i.e., 100-fold versus 1,000-fold) (Fig. 1B). That the higher MOI resulted in a smaller difference between the strains is likely because at that MOI the wild-type strain had exhausted all available acanthamoebae by 48 h and as a result did not increase in CFU count over the remaining 24 h (Fig. 1C).
FIG 1.
Effect of MOI on T2SS promotion of L. pneumophila infection of amoebae. (A) A. castellanii amoebae were inoculated with wild-type strain 130b (WT) expressing plasmid-encoded GFP at increasing MOIs, and the tissue culture plates were centrifuged to promote contact of L. pneumophila with the amoebae. Following 1 h of incubation, the amoebal monolayers were washed to remove extracellular bacteria and treated with gentamicin for an additional hour to completely kill any noninternalized bacteria. The GFP intensities in the infected wells were immediately determined using a fluorescent microplate reader and plotted against the MOI (left), or the plates were incubated for an additional 10 h, at which time the infected amoebae were lifted and analyzed by flow cytometry for GFP intensity as a measure of productive infection (right). RFU, relative fluorescent units. (B) A. castellanii amoebae were infected with either the wild type or the lspF mutant strain NU275 (lspF) at the indicated MOI, as described above. At 0, 24, 48, and 72 h p.i., net bacterial growth (number of CFU at the denoted time/number of CFU at t = 0) was determined by plating aliquots of the supernatant on BCYE agar. Data in log units are presented as means with standard deviations from quadruplicate infected wells and are representative of two independent experiments. (C) A replotting of the data in panel B showing how wild-type and lspF mutant net growth is impacted by increasing the MOI from 0.1 to 20. In panels B and C, the asterisks indicate significant differences between results for the strains (Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
The T2SS is not required for L. pneumophila entry into amoebae or subsequent evasion of lysosomes.
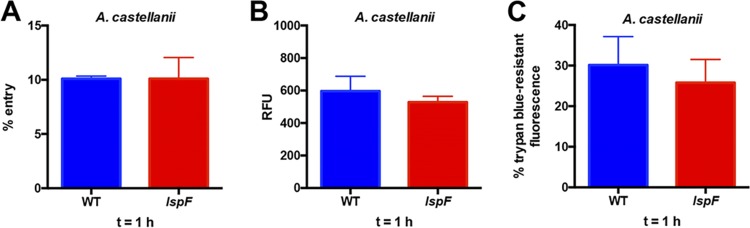
To learn if the T2SS influences the entry of L. pneumophila into amoebae, we inoculated wild-type strain 130b and its lspF mutant onto A. castellanii monolayers at an MOI of 20 and, following gentamicin treatment and washes to remove any remaining extracellular bacteria, either mechanically lysed the amoebae and plated for intracellular CFU or monitored intracellular fluorescence. No difference in levels of CFU recovery (Fig. 2A) or fluorescence (Fig. 2B) was observed, suggesting that the T2SS is not required for L. pneumophila entry into acanthamoebae. Confirming these results, there was no difference in uptake of GFP-expressing wild-type and mutant L. pneumophila bacteria when a trypan blue solution was utilized to quench extracellular fluorescence (Fig. 2C). Together, these data showed that the T2SS is not needed for L. pneumophila entry into amoebae. We next assessed whether the T2SS is important in the ensuing key step in intracellular infection, i.e., evasion of lysosomal fusion. As other investigators have reported when studying L. pneumophila infection of A. castellanii (33), LysoTracker Red, which is often used to mark lysosomes in Legionella-infected macrophages (25, 34), did not properly label lysosomes and instead accumulated throughout the amoebal cytosol. As an alternate means for labeling acanthamoebal lysosomes, we utilized 40-kDa dextran red, a fluorescent compound that when “fed” to amoebae is phagocytosed, trafficked along the endocytic pathway, and accumulated in phagolysosomes, where it is eventually digested (35–37). Thus, A. castellanii that had been pretreated with dextran red was infected with GFP-positive strains of L. pneumophila, and live-cell confocal microscopy was performed at 1 h p.i. to look for colocalization of green-fluorescent bacteria with red-fluorescent phagolysosomes (Fig. 3). Wild-type 130b was detected in a phagolysosome only ∼25% of the time; this level of lysosomal evasion is similar to what has been previously reported for that strain infecting A. castellanii (38). In contrast, >60% of dotA mutant bacteria colocalized with the dextran red label, compatible with the well-known role of the Dot/Icm T4SS in lysosomal avoidance (39, 40). Most importantly, the lspF mutant localized to the phagolysosome ∼20% of the time (Fig. 3). Given that the T2SS mutant evaded the lysosome to a degree similar to that of the parental wild type, these results indicate that the T2SS is not required for the evasion of lysosomal fusion. In sum, the T2SS is not needed for either L. pneumophila entry into or subsequent evasion of lysosomes within amoebae.
FIG 2.
Entry of wild-type and T2SS mutant L. pneumophila strains into amoebae. (A) A. castellanii amoebae were infected with either wild-type 130b (WT) or the lspF mutant NU275 (lspF) at an MOI of 20, and plates were centrifuged to promote contact of L. pneumophila with amoebae. After 1 h of incubation, A. castellanii amoebae were washed to remove extracellular bacteria and then treated with gentamicin for an additional 1 h to kill remaining extracellular bacteria. Following a final wash step, amoebae were lysed, and the numbers of released CFU were determined via dilution plating on BCYE agar. Entry is presented as the mean percentage of the inoculum that was protected from gentamicin treatment, with standard error from three independent experiments (n = 3). (B) A. castellanii amoebae were infected as described for panel A with either GFP-expressing wild-type or lspF mutant bacteria. Following the entry period and gentamicin treatment, the intracellular GFP signal corresponding to internalized L. pneumophila was determined. Data are presented as mean relative fluorescent units (RFU) from six infected wells with standard deviations and are representative of three independent experiments. (C) A. castellanii amoebae were infected with GFP-expressing wild-type or lspF mutant bacteria at an MOI of 20 as described above. Following 1 h of incubation, amoebae were washed to remove extracellular bacteria, and then a trypan blue solution was added to quench GFP signal from remaining extracellular bacteria. Entry was reported as the percentage of trypan-resistant GFP signal corresponding to internalized L. pneumophila. Data are presented as means with standard deviations from six wells and are representative of three independent experiments.
FIG 3.
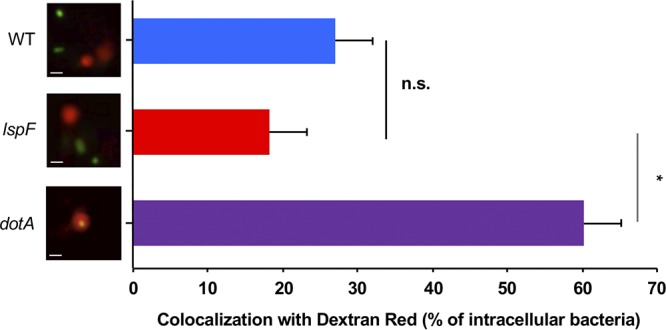
Assessment of wild-type and T2SS mutant L. pneumophila strains trafficking to lysosomes. A. castellanii amoebae were pulsed with fluorescent dextran red for 1 h and then infected with GFP-positive wild-type strain 130b (WT), lspF mutant NU275 (lspF), or dotA mutant NU428 (dotA). The infected amoebae were immediately imaged by confocal microscopy, and the percentage of intracellular bacteria colocalizing with dextran red-positive lysosomes was enumerated. Results presented show representative images on the left (green, bacteria; red, dextran red-containing lysosomes; scale bar, 1 μm) and the percentage of colocalization on the right. Quantified data are presented as means with standard errors from three independent experiments. The asterisk indicates a significant difference between results for the lspF mutant and those for the dotA mutant (Student's t test; *, P < 0.05), whereas the lspF mutant was not significantly (ns) different from the wild-type strain.
The T2SS promotes the intracellular replication of L. pneumophila within amoebal host monolayers.
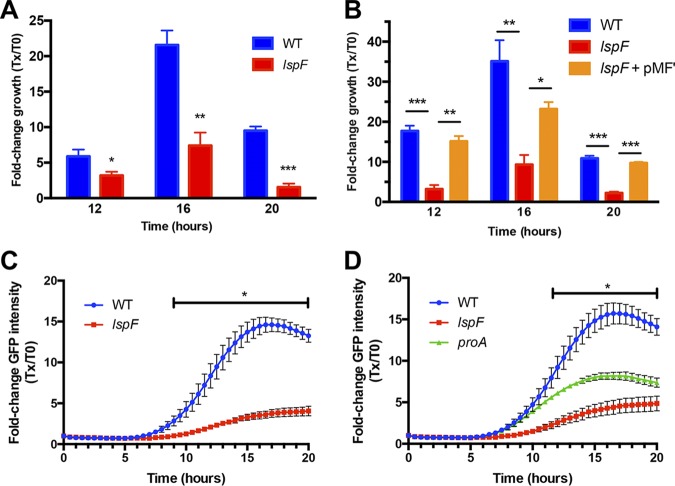
To determine if the T2SS promotes the replication phase, we infected A. castellanii with wild-type and lspF mutant L. pneumophila as described above, lysed the infected amoebal monolayers at 0, 12, 16, and 20 h p.i., and then enumerated intracellular bacteria by plating the amoebal lysates for CFU counts. The numbers of intracellular wild-type bacteria increased approximately 6-fold by 12 h and 22-fold by 16 h, before declining at 20 h due to amoebal cell lysis (Fig. 4A). In contrast, the numbers of intracellular mutant CFU increased only approximately 3-fold at 12 h, 7-fold at 16 h, and 2-fold at 20 h (Fig. 4A). Because the lspF gene is monocistronic within the L. pneumophila chromosome (13), this mutant defect was most likely due to the specific loss of lspF as opposed to a downstream gene(s) being impacted by the mutation in lspF. The importance of LspF for intracellular growth was affirmed when we repeated the assay and found that a complemented lspF mutant behaved similarly to the parental wild-type strain (Fig. 4B). These data, when coupled with prior demonstrations of complementation of T2SS mutants in the traditional coculture assay that monitored the numbers of bacteria released into supernatant of infected monolayers (23–25, 30, 31), documented that the T2SS is required for the replicative phase of L. pneumophila within amoebae.
FIG 4.
Intracellular replication of wild-type and T2SS mutant L. pneumophila strains in amoebae. (A and B) Monolayers of A. castellanii amoebae were infected with GFP-expressing wild-type strain 130b (WT), lspF mutant NU275 (lspF), or the complemented lspF mutant NU275(pMF1) (lspF/lspF+) at an MOI of 20. After centrifugation and a 1-h incubation to facilitate uptake, gentamicin was added to kill remaining extracellular bacteria. The infected amoebae were then lysed at 12, 16, or 20 h postinoculation, and intracellular CFU were enumerated via plating to obtain the fold change in growth relative to the level at time zero (Tx/T0). The data are presented as means with standard deviations from triplicate infected wells and are representative of four (A) or two (B) independent experiments, and significant differences between results for the strains were obtained at each time point (Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (C and D) Monolayers of A. castellanii amoebae were infected as described above with GFP-expressing wild-type strain 130b (WT), lspF mutant NU275 (lspF), or proA mutant AA200 expressing plasmid-encoded GFP (proA). The GFP fluorescence from L. pneumophila was monitored kinetically every 30 min for the next 20 h, and the fluorescence values obtained were normalized to the GFP signal at t = 0 following gentamicin treatment. The data are presented as means with standard errors from three (n = 3) independent experiments, and the lines with asterisks indicate the time intervals in which significant differences were observed between wild-type- and mutant-infected amoebae (Student's t test; P < 0.05).
Having confirmed the role of the T2SS in intracellular replication through the use of a traditional CFU-based assay, we investigated the applicability of a GFP-based assay for monitoring the growth of wild-type and T2SS mutant legionellae. Thus, we infected A. castellanii with GFP-expressing wild-type and lspF mutant L. pneumophila bacteria as described above and then monitored increases in the GFP signal over time. The signal from the wild-type-infected monolayers doubled by ∼8 h p.i. and continued to increase by 8-fold at 12 h and 15-fold at 16 to 20 h, at which time it plateaued, indicating that a single round of infection is completed by ∼20 h (Fig. 4C). In contrast, the GFP signal from lspF mutant-infected A. castellanii did not double until ∼12 h p.i. and then increased only 4-fold by 20 h (Fig. 4C). Significant differences in the fluorescence intensities obtained between wild-type and mutant infections were observed at 9 h p.i. and beyond; for example, by 12 h, there was an ∼4-fold difference in GFP intensities between wild-type- and mutant-infected amoebae, and this difference was sustained throughout the remainder of infection (Fig. 4C). In agreement with the confirmed role that T2SS has in the intra-amoebal growth of L. pneumophila, a second, independently derived T2SS mutant which lacks the two-gene operon lspDE (13) displayed a similar defect in the GFP-based assay (see Fig. S1 in the supplemental material). Because increases in GFP fluorescence paralleled those of CFU counts for both the wild-type and mutant strains (e.g., Fig. 4A compared to Fig. 4C), the GFP-based assay is a good assessment of intracellular bacterial replication and not simply of induction of amoebal autofluorescence. In order to further document the utility of the GFP-based assay, we investigated the behavior of a mutant lacking a specific T2SS substrate. One of the best-characterized substrates of the L. pneumophila T2SS is the metalloprotease ProA, which degrades a wide range of exogenous proteins as well as cleaving and activating other T2SS-dependent exoenzymes (25). It had been previously determined that when a proA mutant is inoculated into cultures of A. castellanii, fewer CFU are recovered from the supernatants of the infected monolayers (19). Thus, we tested whether a proA mutant would also exhibit an intracellular growth defect in the GFP-based assay. The fluorescence of both wild-type strain 130b and its proA mutant doubled by ∼8 h, suggesting that ProA is not required for the initiation of replication, unlike what was observed for the T2SS overall (Fig. 4D). However, the proA mutant did subsequently display a lower growth rate and was unable to reach the high levels of intracellular GFP seen for the wild-type infection (Fig. 4D). The GFP signal from wild-type-infected wells significantly increased from 12 to 20 h, unlike that of the proA mutant-infected wells. Thus, ProA likely acts after the onset of replication to promote intracellular replication to high numbers between 12 and 20 h.
The T2SS promotes the formation of large phagosomes within L. pneumophila-infected amoebae.
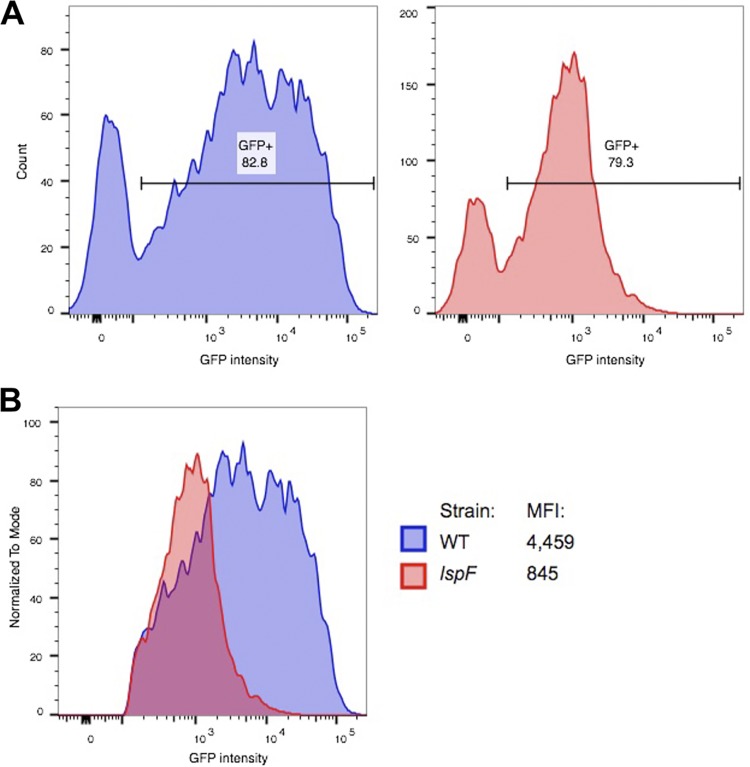
Although the previous experiments demonstrated an overall role for the T2SS in intracellular replication of amoebae, it did not provide us with single-cell resolution. On the one hand, the defect observed for the T2SS mutant could be due to the strain replicating more slowly in all of the infected amoebae and thereby failing to reach numbers that are comparable to those of the wild-type strain. On the other hand, the mutant might be unable to establish productive infection in many of the amoebae but is fully capable of replicating to high numbers within a certain subset of amoebae. To begin to distinguish between these alternative scenarios, we collected A. castellanii amoebae after 12 h of infection and assessed the bacterial load within the amoebae via flow cytometry. To first determine the establishment of replication niches, we quantitated the proportion of productively infected amoebae, based on single-cell GFP intensity. Overall, the proportions of GFP-positive cells were equivalent between the strains, with ∼80% of A. castellanii amoebae containing replicating bacteria (Fig. 5A). However, a much higher GFP signal per cell was observed for the wild-type-infected than for the mutant-infected A. castellanii; for example, there was a 5-fold difference in the median fluorescence intensities (Fig. 5B). These data indicated that the T2SS is required for the ability of L. pneumophila to grow to large numbers within (all of) its amoebal host cells.
FIG 5.
Flow cytometric analysis of A. castellanii amoebae infected with wild-type and T2SS mutant L. pneumophila. (A) A. castellanii amoebae were inoculated with either wild-type strain 130b (WT) expressing plasmid-encoded GFP or lspF mutant NU275 expressing GFP (lspF), as described in the legend of Fig. 4. Infected amoebae were incubated for an additional 12 h and collected for flow cytometry, with the results from wild-type infection shown on the left and those from mutant infection on the right. The GFP-positive interval gate was drawn such that it excluded signal from uninfected amoebae. The percentage of GFP-positive amoebae is indicated above each interval gate. (B) Histogram overlay of the GFP-positive populations from panel A to compare the GFP intensities of individual infected amoebae. The median fluorescence intensity (MFI) for each of the infections is indicated. The data presented are representative of three independent experiments.
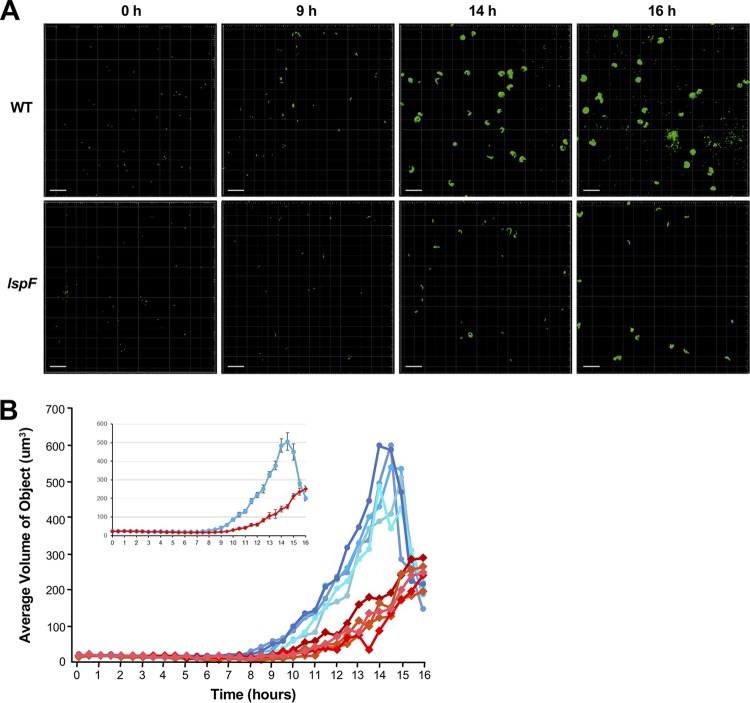
To study this further, we analyzed the size of individual LCVs, utilizing a combination of confocal microscopy and imaging techniques that, to our knowledge, has not been previously applied to the study of L. pneumophila or intracellular infection of amoebae. Acanthamoebae were synchronously infected with GFP-positive wild-type and lspF mutant bacteria, and over a 16-h infection cycle, z-stack images of the bacteria and phagosomes were taken every 30 min and then reconstructed into three-dimensional (3D) objects using image analysis software (Fig. S2A). The volume of the 3D objects associated with wild-type infection began to increase at 9 h p.i. (Fig. 6A and B and Fig. S2B), compatible with the timing of increased bacterial numbers as measured by the overall fluorescence of the infected monolayer (Fig. 4C). The size of the wild-type vacuoles increased over the next 5 h and reached a peak at 14 h p.i. (Fig. 6A and B and Fig. S2B). The timing for peak phagosome size was a few hours earlier than the timing for maximal bacterial load in the monolayer (Fig. 4C), suggesting that the LCV is spacious at times but eventually becomes filled with bacteria. Between 14 and 16 h, the average volume of the wild-type objects decreased, indicative of lysis of many of the LCVs/amoebal hosts and the release of free bacteria (Fig. 6A and B and Fig. S2B). The volume of the 3D objects associated with infection by the lspF mutant began to increase ∼2 h later than had been the case for wild-type infection (Fig. 6B and Fig. S2B). Moreover, over the next ∼5 h, the object volume for the mutant increased more slowly than that of the wild type. Thus, the volume of the mutant objects was significantly less than that of the wild type beginning at 9 h p.i. and persisting over the subsequent 6 h (Fig. 6A and B and Fig. S2B). For example, at 14.5 h, the average volume for the mutant was approximately 3 times less than that of the wild type, indicating that the mutant phagosome was significantly smaller. By the end of the 16-h time period, the average volume of the mutant objects was comparable to that of the wild-type strain. This was due to lysis events liberating free bacteria from the wild-type phagosomes, bringing the average volume of objects down dramatically, and was not due to the mutant achieving a similarly sized phagosome (Fig. 6A and B and Fig. S2B). From these data, we estimated the growth rate constant of the wild-type and T2SS mutant object/phagosomes, using the final time point of growth as 14 h, which is when the wild type peaked, and the initial time point of growth at 8 h, when the wild type first started to grow. We calculated a growth rate constant of 0.42 ± 0.03/h for wild-type phagosomes, which was significantly different from the rate constant of 0.29 ± 0.03/h for the lspF mutant phagosomes (Student's t test; P = 0.02). From these data, we also calculated the doubling time for the volume of wild-type phagosomes as 1.5 ± 0.1 h, while the mutant phagosomes doubled approximately 1.5 times more slowly, in 2.2 ± 0.2 h (Student's t test; P = 0.04). The growth rates were consistent across three independent experiments (Fig. 6A and Fig. S2B). In sum, the comparison between wild-type and mutant bacteria by the method of volume analysis agreed well with the results obtained from the more traditional method of CFU count determination and the assay for overall GFP fluorescence.
FIG 6.
Volume analysis of bacteria and phagosomes within L. pneumophila-infected A. castellanii. Amoebae were infected with GFP-positive wild-type strain 130b (WT) or lspF mutant NU275 (lspF), and z-stack images from confocal microscopy were generated every 30 min for 16 h. The images were then converted into 3D objects for volume analysis (see Fig. S1A in the supplemental material). (A) Fields of view at 0, 9, 14, and 16 h postinoculation, displaying the green fluorescent objects, whether individual bacteria or bacteria-containing phagosomes. The data presented are representative of three independent experiments. Scale bar, 30 μm. (B) The volumes of the GFP-positive objects from five fields of view are graphed over time, following infection with wild-type (blue lines) or T2SS mutant (red lines) bacteria. The inset shows the average values of the five fields of view graphed with the standard error of the mean. Data presented are from a representative experiment, with the data from two additional independent experiments appearing in Fig. S1B. The difference between the values for the average volume of objects over time for the wild type and the lspF mutant was statistically significant (ANOVA; P = 0.02).
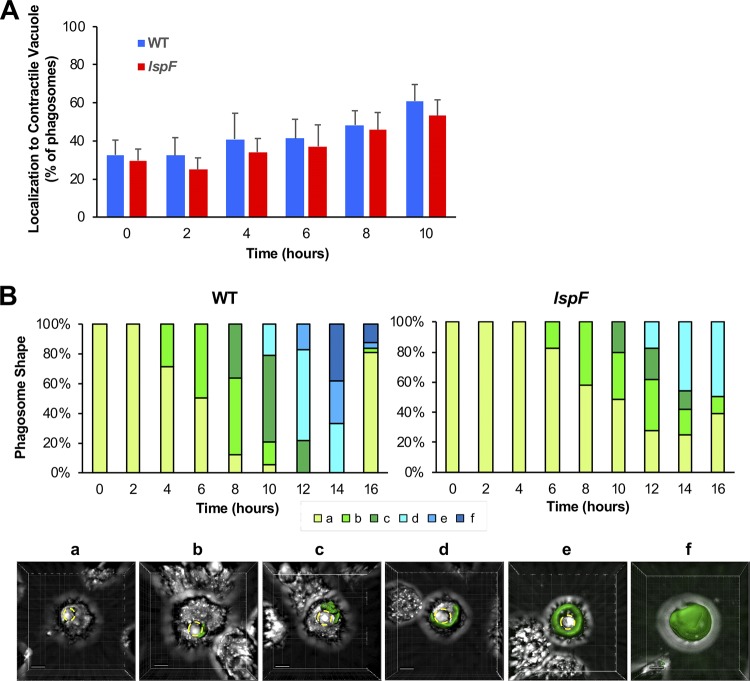
In the course of the experiment designed to measure the volume of the wild-type versus mutant LCV, we also had the opportunity to observe the localization and shape of the bacterial phagosomes. Interestingly, we noticed that over time the wild-type phagosomes and the mutant phagosomes localized in close proximity to the A. castellanii contractile vacuole (Fig. 7A). Over the 16-h time period, there was a progression in the shape of the phagosomes localizing near the contractile vacuole, with the phagosomes ultimately wrapping around the amoebal organelle before filling the spent host cell (Fig. 7B). The progression in the shape of the lspF mutant phagosome was slowed, presumably a reflection of the T2SS mutant’s decreased growth rate (Fig. 7B, top panel). The phagosomes that did not localize near the contractile vacuole remained mostly spherical for both the wild type and mutant (Fig. S3).
FIG 7.
Localization and shape of the wild-type and T2SS mutant L. pneumophila phagosomes. A. castellanii amoebae were infected with GFP-positive wild-type strain 130b (WT) or lspF mutant NU275 (lspF), and z-stack images were generated every 30 min and then converted into 3D objects. (A) Quantification of the localization of GFP-positive phagosomes to the amoebal contractile vacuole following infection with wild-type (WT) or lspF mutant bacteria. (B) Distinct phagosome shapes (defined as a to f) were observed and quantitated as a percentage of the total phagosomes over the course of the infection, as displayed in the top panel for the wild type and the mutant. Representative images of these shapes are presented in the lower panel (scale bar, 5 μm), showing GFP-expressing wild-type phagosomes localizing to and then enveloping the contractile vacuole (demarcated by yellow dashed lines) before filling the entire amoebal cell. The data presented in panels A and B are pooled from three replicate experiments.
The T2SS promotes L. pneumophila growth through multiple rounds of intracellular infection of amoebae.
We next sought to determine the intracellular growth characteristics of the T2SS mutant over a longer time period. The goal of this endeavor was 2-fold. First, it would allow us to determine whether the lspF mutant is capable of eventually reaching the same level of intracellular growth as the wild-type strain, given that the mutant phagosome size appeared to be still on the rise at the end of the 16-h time course used in our previous experiment (Fig. 6B and Fig. S2B). Second, it would discern whether the T2SS mutant defect changes, either increasing or decreasing in magnitude, over the course of multiple rounds of intracellular infection. To perform this long-term experiment, we imaged infected cells every hour for 48 h using wide-field microscopy and calculated the area of the phagosomes over time. During the first 16 h, the bacterial strains behaved similarly to how they acted in the volume experiment. That is, the area of the wild-type phagosomes began to increase at ∼8 h p.i., peaked at ∼14 h, and then dramatically declined within the subsequent 2-h period, whereas the expansion of the lspF mutant phagosomes was delayed and occurred at a reduced rate (Fig. 8 and Fig. S4). Beyond 16 h, the lspF mutant phagosomes continued to increase in size for another 4 to 6 h, but, notably, they never reached the magnitude of the wild-type phagosomes (Fig. 8 and Fig. S4). Nonetheless, after reaching initial peak size, the average area of the mutant phagosomes declined, indicative of egress from the amoebal hosts. This decline occurred at a lower rate than that of the wild-type phagosomes, taking ∼8 h rather than ∼2 h (Fig. 8 and Fig. S4). Thus, the entire infection cycle (i.e., entry to egress) was considerably slower for the T2SS mutant than it was for the wild-type strain. A similar result was obtained when we examined a second, independently derived T2SS (lspDE) mutant (Fig. S5). Thus, this analysis indicated that the T2SS is required for the formation of maximally sized phagosomes. In this same experiment, the kinetics of reinfection by wild-type L. pneumophila were readily apparent, with the reemergence of large phagosomes occurring ∼10 h after the end of the first round of intracellular infection (Fig. 8 and Fig. S4). That this occurred ∼4 h faster in the second round of infection suggested that the bacteria emerging from the amoebae are more infectious than are the bacteria obtained from cultures grown on bacteriological medium, i.e., buffered charcoal-yeast extract (BCYE) agar plates. There was also a trend toward the size of the phagosome being larger in the second round of infection than in the first round of infection, further suggesting an adaptation on the part of the legionellae, the effects of a higher MOI, and/or an increased permissiveness of the amoebal host cells (due to prolonged culturing). The lspF mutant did not appear to be as capable of rapid reinfection and subsequent growth, as evidenced by the longer time period between its first and second peaks in phagosome size (Fig. 8 and Fig. S4). However, in the second round of intracellular infection, the area of the mutant phagosomes did reach that of the wild-type phagosomes, suggesting, once again, a possible adaptation by the bacteria and/or an increased permissiveness of the amoebae. Overall, the importance of T2SS for L. pneumophila infection of A. castellanii was evident over multiple rounds of intracellular infection.
FIG 8.
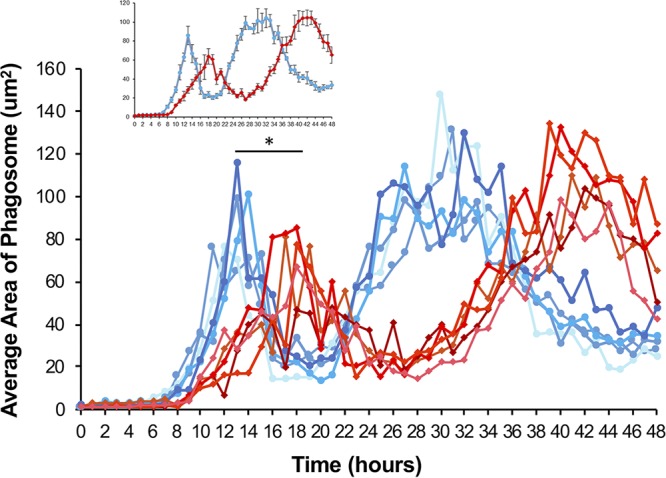
Area analysis of phagosomes within L. pneumophila-infected A. castellanii over a 48-h time period. Amoebae were infected with GFP-positive wild-type strain 130b (WT) or lspF mutant NU275 (lspF), and wide-field images were generated every hour for 48 h. Analysis of areas of fluorescence was then performed using Nikon Elements software. The areas of intracellular GFP-positive phagosomes from five fields of view are graphed over time, following infection with wild-type (blue lines) or T2SS mutant (red lines) bacteria. The inset shows the average values of the five fields of view graphed with the standard error of the mean. Data presented are from a representative experiment, with the data from two additional independent experiments appearing in Fig. S3 in the supplemental material. The asterisk indicates a significant difference between wild-type and lspF mutant average phagosome areas (Student's t test; *, P < 0.05).
DISCUSSION
Without question, aquatic amoebae, including Acanthamoeba species, represent a key replication niche for L. pneumophila and other Legionella species in the environment (3, 4, 41). Moreover, amoebae harboring intracellular legionellae may contribute to the inoculum that initiates human lung infection (42, 43). Therefore, deciphering how L. pneumophila infects and grows within amoebae is critical to understanding the transmission and possibly prevention of Legionnaires’ disease. Although the L. pneumophila T2SS has long been known to promote infection of aquatic amoebae, based upon the results of traditional coculture methods (18, 19), the data presented here clarify, for the first time, the stage of intracellular infection at which the secretion system primarily operates. While not being required for bacterial entry into A. castellanii or for the evasion of the amoebal lysosome, the T2SS is needed for both the efficient initiation of intracellular replication at ∼8 h p.i. and for the ability to grow to large numbers within the LCV over the ensuing 8 to 12 h. This type of role for the T2SS in amoebal hosts is reminiscent of what we previously documented for the T2SS during L. pneumophila infection of macrophages (17), reinforcing the long-held belief that the ability of L. pneumophila to infect macrophages is a result of its prior, natural adaptation to niches within amoebae (44–47). By utilizing newer microscopic techniques and software analyses, we were able to closely link the numbers of intracellular bacteria (as measured by the recovery of CFU from infected amoebae and fluorescence from GFP-expressing bacteria) to the physical size of the LCV. This, in turn, allowed us to identify a significant difference in the intracellular growth rate between wild-type and T2SS mutant bacteria. Finally, the importance of the secretion system in infection of acanthamoebae was documented over multiple rounds of intracellular infection.
We surmise that the T2SS promotes intracellular growth within amoebae through the combined action of multiple secreted effectors, as partly evidenced here by the demonstration of a required role for the ProA protease in the ability of L. pneumophila to grow intracellularly to large numbers between 12 and 20 h p.i. Presumably, other secreted proteins account for the importance of the T2SS in the early stages of replication at 8 to 12 h; candidate effectors include the aminopeptidase LapA, acyltransferase PlaC, and novel proteins NttA, NttC, NttD, NttE, and NttF (13, 23–25). Because of the enzymatic activities of ProA, LapA, PlaC, and possibly other T2SS substrates, it is quite likely that the T2SS promotes intracellular replication by mediating the degradation of foodstuffs (e.g., proteins and peptides) that come to reside within the lumen of the LCV (25, 48). However, since ProA and potentially other T2SS effectors can translocate out of the LCV and localize to the cytosolic side of the bacterial vacuole in infected A. castellanii (32), it is also conceivable that T2SS-dependent proteins alter amoebal function in broader ways. Recently, we determined that, in infected macrophages, the T2SS promotes the association of Rab1B with the LCV, a process that is linked to the recruitment of ER-derived vesicles to the LCV (17). Thus, among many scenarios, the T2SS may be manipulating an A. castellanii equivalent of Rab1B and organelle trafficking to facilitate L. pneumophila growth within its amoebal host. Although our study was based on the analysis of A. castellanii, we posit that the role of the T2SS in promoting the expansion of bacterial numbers within the LCV extends to other amoebal hosts such as Naegleria, Vermamoeba, and Willaertia species (23, 24).
In the course of analyzing the T2SS mutant by confocal microscopy, we observed that many of the LCVs, both wild-type and mutant, localized to and then enveloped the amoebal contractile vacuole. To our knowledge, this colocalization and progression in phagosome shape have not been reported before. The contractile vacuole is known for its ability to expel water from the amoebal cell for the purpose of osmotic regulation (49, 50). However, recent studies done on the soil amoeba Dictyostelium discoideum revealed the presence of the metal (iron) exporter Nramp2 on the contractile vacuole (51), suggesting that the colocalization of the LCV with this amoebal vacuole might help to promote bacterial growth though iron acquisition. When we extended our microscopic analysis to include multiple rounds of intracellular infection, we saw evidence of the legionellae, both wild type and T2SS mutant, becoming more infectious (i.e., faster increases in phagosome size and larger phagosomes) over time. This finding harkens back to an earlier study which had found that L. pneumophila bacteria emerging from A. castellanii are more infectious for human monocytes than plate-grown legionellae (52).
In summary, the results presented significantly advance our understanding of the role of the T2SS in L. pneumophila intracellular infection and the natural history of Legionnaires’ disease. Moreover, the insights gained here have implications for many other Gram-negative bacteria, including human, animal, and plant pathogens, that encode T2SSs and reside within host cells (12, 13, 53). Finally, the microscopic methodologies that we used to obtain 3D images of the size and shape of the LCV may prove useful for other Legionella studies as well as the investigation of other intracellular parasites.
MATERIALS AND METHODS
Bacterial strains, amoebae, and media.
L. pneumophila 130b (ATCC BAA-74; also known as strain AA100 or Wadsworth) served as the wild type and parent for all mutants (26). The lspF mutant NU275, complemented lspF mutant NU275(pMF), lspDE mutant NU258, proA mutant AA200, and dotA mutant NU428 were described previously (14, 17, 30, 54). GFP-expressing variants of strain 130b and the lspF and lspDE mutants were also previously obtained, whereas a GFP-expressing version of the proA mutant was newly made by introducing pGFP into AA200 by electroporation (17). Legionellae were routinely grown at 37°C on BCYE agar (26). For infections utilizing GFP-expressing bacteria, L. pneumophila strains were grown on BCYE agar containing isopropyl-β-D-thiogalactopyranoside (IPTG) in order to induce production of the fluorescent reporter (17). A. castellanii amoebae (ATCC 30234) were maintained as described previously (23). As recommended by the ATCC, acanthamoebae were routinely maintained in 712 protease peptone-yeast extract-glucose (PYG) medium at 35°C and then infected in PYG medium that lacked the glucose component (PY medium). Unless otherwise noted, chemicals were from Sigma.
Assessment of bacterial entry into amoebae.
To assess the extent of L. pneumophila entry into amoebae, a variation on the gentamicin protection assay was done (17, 55, 56). Briefly, the bacterial strains were added to confluent monolayers of A. castellanii (1 × 105 per well) in 24-well plates at an MOI equal to 20, and then the infection was synchronized by centrifuging the plate at 400 × g for 10 min at room temperature. After the legionellae were allowed to enter amoebae for 1 h at 35°C, the inoculated wells were washed three times with 1 ml of PY medium to remove the majority of extracellular bacteria. Gentamicin was next added at 200 μg/ml in PY medium for 1 h to kill any remaining extracellular bacteria. The wells were washed twice with 1 ml of PY medium to remove the antibiotic, and the infected monolayers (n = 4) were lysed in 1 ml of phosphate-buffered saline (PBS) by being passaged seven times through a blunt-end 27-gauge needle (catalog no. 901-27-050; CML Supply) attached to a 1-ml Luer lock syringe (catalog no. 309628; Becton, Dickinson). Importantly, the needle was pressed flush against the bottom of the plastic culture well such that amoebae were mechanically sheared upon expulsion from the syringe. That all of the amoebae were lysed was confirmed by microscopic examination. Finally, the numbers of internalized bacteria were determined by plating the lysates for CFU counts on BCYE agar. Entry was measured as the percentage of intracellular CFU recovered from the monolayers compared to the number of CFU in the inoculum. As a second means for monitoring bacterial entry, we utilized a trypan blue quenching-based entry assay, which we along with others have previously described for L. pneumophila (17, 57–59). To that end, monolayers of A. castellanii (5 × 104) were seeded into black, clear-bottom, 96-well microtiter plates (Falcon) in 100 μl of Lo-Flo medium (Formedium) for 30 min to facilitate attachment. L. pneumophila strains were added at an MOI of 20 in 100 μl of Lo-Flo medium, and the infection was synchronized by centrifuging the plates at 450 × g for 10 min. Entry was allowed to occur for 1 h at 35°C. Fluorescence was read at an excitation of 485 nm and emission of 530 nm to determine the total GFP prior to quenching. The medium was removed by pipetting, and 50 μl of trypan blue solution was added to each well for 1 min. Finally, the trypan blue solution was removed with a multichannel pipette, and a final fluorescence reading was taken at an excitation of 485 nm and emission of 530 nm. Entry was defined as the percentage of trypan blue-resistant GFP fluorescence over total GFP fluorescence, after subtracting out background fluorescence from uninfected wells.
Assays for bacterial replication within amoebae.
To examine the ability of L. pneumophila strains to grow in coculture with A. castellanii, as before (23, 29, 60), amoebae were seeded in 24-well tissue culture wells (1 × 105 per well) for 30 min to allow attachment and then inoculated with the bacteria at the MOI indicated in Fig. 1B. At various times p.i., the numbers of legionellae were enumerated by plating serial dilutions of aliquots from the well supernatants on BCYE agar. Since L. pneumophila does not replicate in the medium (23), increases in extracellular CFU counts result from intracellular replication. In order to specifically examine intracellular growth, we utilized GFP-expressing bacteria in two different assays (61, 62). First, to monitor intracellular replication over time, we seeded 5 × 104 A. castellanii amoebae into black, 96-well microtiter plates for 30 min in 100 μl of Lo-Flo medium. GFP-expressing L. pneumophila strains were then added at an MOI of 20 in 100 μl of Lo-Flo medium, and the infection was synchronized by low-speed centrifugation at 400 × g for 10 min at room temperature. The plate was incubated at 35°C for 1 h to facilitate bacterial entry, and then the wells were washed gently three times with 100 μl of Lo-Flo medium. The infected wells were next treated with 200 μg/ml gentamicin for 1 h to kill any remaining extracellular bacteria. Following two additional washes with 100 μl of Lo-Flo medium to remove the gentamicin, Lo-Flo medium supplemented with IPTG (0.5 mM) and chloramphenicol (5 μg/μl) was added into the wells to induce GFP expression within the intracellular bacteria and promote the maintenance of the GFP-expressing plasmid throughout the infection period. The plate was incubated at 35°C in a fluorescent microplate reader (BioTek). GFP fluorescence (excitation at 485 nm and emission at 530 nm) was monitored kinetically every 30 min for 20 h, thereby documenting the level of bacterial growth across the entire infected monolayer. Second, in order to monitor the bacterial burden within individual amoebae, 5 × 105 A. castellanii amoebae were seeded in 24-well plates for 30 min in 1 ml of PY medium and then inoculated with GFP-expressing L. pneumophila according to the same protocol as described for the 96-well assay, except that the inoculating volume was 1 ml per well of a 24-well plate. Following 1 h of entry and 1 h of gentamicin treatment, amoebae were washed twice with 1 ml of PY medium to remove remaining antibiotic, and then fresh PY medium supplemented with chloramphenicol and IPTG was added back. Following incubation at 35°C, the infected amoebae were lifted from the well by gently pipetting up and down, centrifuged at low speed (500 × g) for 3 min at room temperature, and resuspended in 1 ml of PBS. Amoebae were immediately analyzed by flow cytometry using an LSR II flow cytometer (Becton, Dickinson), with ∼10,000 events acquired while monitoring forward-scattered light (FSC) and side-scattered light (SSC), as well as fluorescence following excitation with a 488-nm laser line and a fluorescein isothiocyanate (FITC) detection filter set. L. pneumophila-specific GFP signal was determined by gating exclusive of background GFP signal from uninfected amoebae. Finally, in order to quantitate the numbers of viable intracellular legionellae at various times p.i., infected monolayers of A. castellanii were subjected to mechanical lysis, as was done for the entry assays (above), and CFU counts were determined by plating serial dilutions of the lysates on BCYE agar. Thus, the fold change in CFU count relative to that at time zero (t = 0) was determined.
Assay for phagosome-lysosome fusion.
A total of 6 × 105 A. castellanii amoebae in 1 ml of Lo-Flo medium were seeded in wells of a four-well Lab-Tek chambered no. 1 borosilicate cover glass system (ThermoFisher) and allowed to adhere for 1 h. The amoebae were pulsed with 40-kDa dextran red (D1829; ThermoFisher) at a final concentration of 25 μg per ml for 1 h to allow the dextran to be phagocytosed and trafficked to the amoebal lysosomes. The wells were gently washed three times with 1 ml of Lo-Flo medium in order to remove extracellular fluorescent dextran. L. pneumophila bacteria that had been grown for 3 days on BCYE agar were resuspended to 5 × 107 CFU per ml in Lo-Flo medium, and 600 μl of the cell suspension was added to each well, giving an MOI of 50. The infected amoebae were imaged approximately 15 min later using a Nikon W1 spinning-disk confocal microscope with a 60× oil immersion objective (numerical aperture, 1.4). At least 100 amoebae from three replicate experiments were visualized, and colocalization of bacteria with dextran-loaded lysosomes was quantified as previously described (35–37). For experiments using LysoTracker, the amoebae were initially pulsed with 50 nM LysoTracker Red (ThermoFisher) for 1 h (63) before subsequent infection (as described above). Although various concentrations of LysoTracker Red up to 1,000 nM were attempted, lysosome punctae were not clearly visualized under any conditions.
Microscopic analysis of the size, shape, and localization of Legionella-containing vacuoles.
A total of 6 × 105 A. castellanii amoebae were seeded in a four-well Lab-Tek chambered system and allowed to adhere for 1 h. GFP-expressing wild-type and mutant L. pneumophila bacteria that had been grown on BCYE agar were diluted to 1 × 107 CFU per ml in Lo-Flo medium. Six hundred microliters of the bacterial suspension was added to each well of A. castellanii (MOI of 10), and the chamber was incubated at 35°C for 2 h. The wells were gently washed with 1 ml of Lo-Flo medium three times to remove extracellular bacteria, and 1 ml of fresh Lo-Flo medium with 500 μM IPTG was added back to the wells. The slide was incubated in a humidified chamber at 35°C for 16 h, and five fields of view (area for one field of view, 110,513 μm2) were imaged every 30 min using a Nikon W1 spinning-disk confocal microscope with a 40× air objective (numerical aperture, 0.95). Ten z-planes of GFP and bright field were imaged to capture the full phagosome over time. The z-stacks of GFP bacteria were reconstructed into 3D objects for volume analysis using Imaris imaging software (Bitplane, South Windsor, CT) (64). The average volume per object, which includes phagosomes and bacteria, was calculated for every time point. The images from the volume analysis experiment were also used to assess localization of the Legionella-containing vacuoles to the amoebal contractile vacuole (37). The average percentage of bacteria localizing to the contractile vacuole was enumerated from three replicate experiments for three fields of view at 0, 2, 4, 6, 8, and 10 h p.i. The images were further used to gauge the shapes of phagosomes localizing near the contractile vacuole. Six distinct shapes were observed and categorized as a to f. Shapes from three replicate experiments and three fields of view at 0, 2, 4, 6, 8, 10, 12, 14, and 16 h p.i. were enumerated. In order to assess the area of the LCV over an extended period of time, A. castellanii amoebae were infected as described above for the volume analysis. The slide was incubated in a humidified chamber (within a Nikon Biostation IMQ) at 35°C for 48 h, and five fields of view (area for one field of view, 393,216 μm2) were imaged every hour using a Nikon Biostation with a 20× objective (numerical aperture, 0.8). The average area of phagosomes and intracellular bacteria was calculated for every time point using NIS-Elements software (Nikon Imaging, Inc.) (65).
Calculations for the growth rate constant and doubling time of LCVs.
The growth rate constant, r, of wild-type and lspF mutant objects/phagosomes was calculated using the equation A = Pert, where A equals the volume at the final time point of growth (14 h), P equals the volume at the initial time point of growth (8 h), t equals the total time (6 h), and r is the growth rate constant (66, 67).
The doubling time (G) of wild-type and lspF mutant objects/phagosomes was calculated using the equation G = t/(3.3 log A/P), where t equals the total time (6 h), A equals the volume at the final time point of growth (14 h), and P equals the volume at the initial time point of growth (8 h) (66, 67). The growth rate constant and doubling time were calculated for wild-type and lspF mutant phagosomes for all three independent experiments and averaged.
Statistical procedures.
In all experiments, each sample or condition was assessed using at least three technical replicates. The resultant values obtained were presented as the means and standard deviations or the standard errors of the means. Statistical analysis was applied using Student's t test or one-way analysis of variance (ANOVA), as appropriate. P values are presented in the figure legends or main text. Repeat experiments were routinely performed for confirmation, as noted in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Cianciotto lab, past and present, for helpful advice. We also thank Constadina Arvanitis and David Kirchenbuechler for assistance with imaging and data analysis.
R.C.W. was supported in part by NIH-NIAID T32 AI0007476. Confocal microscopy at the Northwestern University Center for Advanced Microscopy is supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. This work was supported by NIH grant R01 AI043987 awarded to N.P.C.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00374-19.
REFERENCES
- 1.Mercante JW, Winchell JM. 2015. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28:95–133. doi: 10.1128/CMR.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkelman RL, Pruden A. 2017. Prevention of Legionnaires’ disease in the 21st century by advancing science and public health practice. Emerg Infect Dis 23:1905–1907. doi: 10.3201/eid2311.171429. [DOI] [Google Scholar]
- 3.Cianciotto NP, Hilbi H, Buchrieser C. 2013. Legionnaires’ disease, p 147–217. In Rosenberg E, DeLong EF, Stackebrandt E, Thompson F, Lory S (ed), The prokaryotes. Human microbiology, 4th ed Springer, New York, NY. [Google Scholar]
- 4.Boamah DK, Zhou G, Ensminger AW, O'Connor TJ. 2017. From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front Cell Infect Microbiol 7:477. doi: 10.3389/fcimb.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swart AL, Harrison CF, Eichinger L, Steinert M, Hilbi H. 2018. Acanthamoeba and Dictyostelium as cellular models for Legionella infection. Front Cell Infect Microbiol 8:61. doi: 10.3389/fcimb.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherwood RK, Roy CR. 2016. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol 70:413–433. doi: 10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- 7.Steiner B, Weber S, Hilbi H. 2017. Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int J Med Microbiol 308:49–57. doi: 10.1016/j.ijmm.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA, Pupko T, Shuman HA, Segal G. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong KC, Ghosal D, Chang YW, Jensen GJ, Vogel JP. 2017. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc Natl Acad Sci U S A 114:8077–8082. doi: 10.1073/pnas.1621438114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escoll P, Song OR, Viana F, Steiner B, Lagache T, Olivo-Marin JC, Impens F, Brodin P, Hilbi H, Buchrieser C. 2017. Legionella pneumophila modulates mitochondrial dynamics to trigger metabolic repurposing of infected macrophages. Cell Host Microbe 22:302–316. doi: 10.1016/j.chom.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Valero L, Rusniok C, Carson D, Mondino S, Perez-Cobas AE, Rolando M, Pasricha S, Reuter S, Demirtas J, Crumbach J, Descorps-Declere S, Hartland EL, Jarraud S, Dougan G, Schroeder GN, Frankel G, Buchrieser C. 2019. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci U S A 116:2265–2273. doi: 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianciotto NP, White RC. 2017. The expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun 85:e00014-17. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White RC, Cianciotto NP. 5 June 2019. Assessing the impact, genomics, and evolution of type II secretion across a large, medically-important genus: the Legionella type II secretion paradigm. Microb Genom doi: 10.1099/mgen.0.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossier O, Starkenburg S, Cianciotto NP. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect Immun 72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy-Simandle K, Stewart CR, Dao J, Debroy S, Rossier O, Bryce PJ, Cianciotto NP. 2011. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect Immun 79:1984–1997. doi: 10.1128/IAI.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallama CA, McCoy-Simandle K, Cianciotto NP. 2017. The type II secretion system of Legionella pneumophila dampens the MyD88 and Toll-like receptor 2 signaling pathway in infected human macrophages. Infect Immun 85:e00897-16. doi: 10.1128/IAI.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RC, Cianciotto NP. 2016. Type II secretion is necessary for optimal association of the Legionella-containing vacuole with macrophage Rab1B but enhances intracellular replication mainly by Rab1B-independent mechanisms. Infect Immun 84:3313–3327. doi: 10.1128/IAI.00750-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liles MR, Edelstein PH, Cianciotto NP. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol 31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 19.Hales LM, Shuman HA. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect Immun 67:3662–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossier O, Cianciotto NP. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect Immun 69:2092–2098. doi: 10.1128/IAI.69.4.2092-2098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Söderberg MA, Rossier O, Cianciotto NP. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J Bacteriol 186:3712–3720. doi: 10.1128/JB.186.12.3712-3720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söderberg MA, Dao J, Starkenburg S, Cianciotto NP. 2008. Importance of type II secretion for Legionella pneumophila survival in tap water and amoebae at low temperature. Appl Environ Microbiol 74:5583–5588. doi: 10.1128/AEM.00067-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyson JY, Pearce MM, Vargas P, Bagchi S, Mulhern BJ, Cianciotto NP. 2013. Multiple Legionella pneumophila type II secretion substrates, including a novel protein, contribute to differential infection of amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect Immun 81:1399–1410. doi: 10.1128/IAI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson JY, Vargas P, Cianciotto NP. 2014. The novel Legionella pneumophila type II secretion substrate NttC contriubtes to infection of amoebae Hartmannella vermiformis and Willaertia magna. Microbiology 160:2732–2744. doi: 10.1099/mic.0.082750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White RC, Gunderson FF, Tyson JY, Richardson KH, Portlock TJ, Garnett JA, Cianciotto NP. 2018. Type ii secretion-dependent aminopeptidase LapA and acyltransferase PlaC are redundant for nutrient acquisition during Legionella pneumophila intracellular infection of amoebas. mBio 9:e00528-18. doi: 10.1128/mBio.00528-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart CR, Burnside DM, Cianciotto NP. 2011. The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol 193:5971–5984. doi: 10.1128/JB.05405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan C, Prashar A, So J, Tang P, Low DE, Terebiznik M, Guyard C. 2011. Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun 79:2168–2181. doi: 10.1128/IAI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce MM, Cianciotto NP. 2009. Legionella pneumophila secretes an endoglucanase that belongs to the family-5 of glycosyl hydrolases and is dependent upon type II secretion. FEMS Microbiol Lett 300:256–264. doi: 10.1111/j.1574-6968.2009.01801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossier O, Dao J, Cianciotto NP. 2008. The type II secretion system of Legionella pneumophila elaborates two aminopeptidases as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl Environ Microbiol 74:753–761. doi: 10.1128/AEM.01944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossier O, Dao J, Cianciotto NP. 2009. A type II-secreted ribonuclease of Legionella pneumophila facilitates optimal intracellular infection of Hartmannella vermiformis. Microbiology 155:882–890. doi: 10.1099/mic.0.023218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truchan HK, Christman HD, White RC, Rutledge NS, Cianciotto NP. 2017. Type II secretion substrates of Legionella pneumophila translocate out of the pathogen-occupied vacuole via a semi-permeable membrane. mBio 8:e00870-17. doi: 10.1128/mBio.00870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandyopadhyay P, Sumer EU, Jayakumar D, Liu S, Xiao H, Steinman HM. 2012. Implication of proteins containing tetratricopeptide repeats in conditional virulence phenotypes of Legionella pneumophila. J Bacteriol 194:3579–3588. doi: 10.1128/JB.00399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect Immun 75:723–735. doi: 10.1128/IAI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeger EM, Thuma M, Fernandez-Moreira E, Jacobs E, Schmitz M, Helbig JH. 2010. Lipopolysaccharide of Legionella pneumophila shed in a liquid culture as a nonvesicular fraction arrests phagosome maturation in amoeba and monocytic host cells. FEMS Microbiol Lett 307:113–119. doi: 10.1111/j.1574-6968.2010.01976.x. [DOI] [PubMed] [Google Scholar]
- 36.Olofsson J, Axelsson-Olsson D, Brudin L, Olsen B, Ellström P. 2013. Campylobacter jejuni actively invades the amoeba Acanthamoeba polyphaga and survives within non digestive vacuoles. PLoS One 8:e78873. doi: 10.1371/journal.pone.0078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Henst C, Scrignari T, Maclachlan C, Blokesch M. 2016. An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. ISME J 10:897–910. doi: 10.1038/ismej.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirillo SL, Yan L, Littman M, Samrakandi MM, Cirillo JD. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667–1677. doi: 10.1099/00221287-148-6-1667. [DOI] [PubMed] [Google Scholar]
- 39.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 41.Thomas V, McDonnell G, Denyer SP, Maillard JY. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev 34:231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 42.Brieland JK, Fantone JC, Remick DG, LeGendre M, McClain M, Engleberg NC. 1997. The role of Legionella pneumophila-infected Hartmannella vermiformis as an infectious particle in a murine model of Legionnaire’s disease. Infect Immun 65:5330–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berk SG, Ting RS, Turner GW, Ashburn RJ. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol 64:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cianciotto NP, Fields BS. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci U S A 89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abu Kwaik Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol 62:2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal G, Shuman HA. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67:2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson MS, Hammer BK. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol 54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 48.Best A, Jones S, Abu Kwaik Y. 2018. Mammalian solute carrier (SLC)-like transporters of Legionella pneumophila. Sci Rep 8:8352. doi: 10.1038/s41598-018-26782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siddiqui R, Khan NA. 2012. Biology and pathogenesis of Acanthamoeba. Parasit Vectors 5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bozzaro S, Buracco S, Peracino B. 2013. Iron metabolism and resistance to infection by invasive bacteria in the social amoeba Dictyostelium discoideum. Front Cell Infect Microbiol 3:50. doi: 10.3389/fcimb.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peracino B, Buracco S, Bozzaro S. 2013. The Nramp (Slc11) proteins regulate development, resistance to pathogenic bacteria and iron homeostasis in Dictyostelium discoideum. J Cell Sci 126:301–311. doi: 10.1242/jcs.116210. [DOI] [PubMed] [Google Scholar]
- 52.Cirillo JD, Cirillo SLG, Yan L, Bermudez LE, Falkow S, Tompkins LS. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanism and enhances virulence of Legionella pneumophila. Infect Immun 67:4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korotkov KV, Sandkvist M. 15 February 2019. Architecture, function, and substrates of the type II secretion system. EcoSal Plus doi: 10.1128/ecosalplus.ESP-0034-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moffat JF, Edelstein PH, Regula DP Jr, Cirillo JD, Tompkins LS. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol 12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 55.Cianciotto NP, Eisenstein BI, Mody CH, Toews GB, Engleberg NC. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun 57:1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M, Conover GM, Isberg RR. 2008. Legionella pneumophila EnhC is required for efficient replication in tumor necrosis factor a-stimulated macrophages. Cell Microbiol 10:1906–1923. doi: 10.1111/j.1462-5822.2008.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charpentier X, Gabay JE, Reyes M, Zhu JW, Weiss A, Shuman HA. 2009. Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog 5:e1000501. doi: 10.1371/journal.ppat.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. 2011. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun 79:1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allombert J, Lazzaroni JC, Bailo N, Gilbert C, Charpentier X, Doublet P, Vianney A. 2014. Three antagonistic cyclic di-GMP-catabolizing enzymes promote differential Dot/Icm effector delivery and intracellular survival at the early steps of Legionella pneumophila infection. Infect Immun 82:1222–1233. doi: 10.1128/IAI.01077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Declerck P, Behets J, Delaedt Y, Margineanu A, Lammertyn E, Ollevier F. 2005. Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb Ecol 50:536–549. doi: 10.1007/s00248-005-0258-0. [DOI] [PubMed] [Google Scholar]
- 61.Tiaden AN, Kessler A, Hilbi H. 2013. Analysis of Legionella infection by flow cytometry. Methods Mol Biol 954:233–249. doi: 10.1007/978-1-62703-161-5_14. [DOI] [PubMed] [Google Scholar]
- 62.Buckley CM, Heath VL, Gueho A, Bosmani C, Knobloch P, Sikakana P, Personnic N, Dove SK, Michell RH, Meier R, Hilbi H, Soldati T, Insall RH, King JS. 2019. PIKfyve/Fab1 is required for efficient V-ATPase and hydrolase delivery to phagosomes, phagosomal killing, and restriction of Legionella infection. PLoS Pathog 15:e1007551. doi: 10.1371/journal.ppat.1007551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chazotte B. 2011. Labeling lysosomes in live cells with LysoTracker. Cold Spring Harb Protoc 2011:pdb.prot5571. doi: 10.1101/pdb.prot5571. [DOI] [PubMed] [Google Scholar]
- 64.Patel NB, Hinojosa JA, Zhu M, Robertson DM. 2018. Acceleration of the formation of biofilms on contact lens surfaces in the presence of neutrophil-derived cellular debris is conserved across multiple genera. Mol Vis 24:94–104. [PMC free article] [PubMed] [Google Scholar]
- 65.Walton S, Hofmeyr MD, van der Horst G. 2013. Accurate automated quantitative imaging of tortoise erythrocytes using the NIS image analysis system. Biotech Histochem 88:242–249. doi: 10.3109/10520295.2013.765594. [DOI] [PubMed] [Google Scholar]
- 66.Neidhardt FC. 1999. Bacterial growth: constant obsession with dN/dt. J Bacteriol 181:7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Powell EO. 1956. Growth rate and generation time of bacteria, with special reference to continuous culture. J Gen Microbiol 15:492–511. doi: 10.1099/00221287-15-3-492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.