ABSTRACT
Maintenance of pluripotency and specification towards a new cell fate are both dependent on precise interactions between extrinsic signals and transcriptional and epigenetic regulators. Directed methylation of cytosines by the de novo methyltransferases DNMT3A and DNMT3B plays an important role in facilitating proper differentiation, whereas DNMT1 is essential for maintaining global methylation levels in all cell types. Here, we generated single-cell mRNA expression data from wild-type, DNMT3A, DNMT3A/3B and DNMT1 knockout human embryonic stem cells and observed a widespread increase in cellular and transcriptional variability, even with limited changes in global methylation levels in the de novo knockouts. Furthermore, we found unexpected transcriptional repression upon either loss of the de novo methyltransferase DNMT3A or the double knockout of DNMT3A/3B that is further propagated upon differentiation to mesoderm and ectoderm. Taken together, our single-cell RNA-sequencing data provide a high-resolution view into the consequences of depleting the three catalytically active DNMTs in human pluripotent stem cells.
KEY WORDS: DNA methylation, DNA methyltransferases, Pluripotency, Single-cell RNA-sequencing
Summary: Combining scRNA-seq and genetic perturbations presents a powerful tool for dissecting the role of epigenetic regulators in development and disease, and has shown that disruption of DNMTs increases cell-cell variability.
INTRODUCTION
Isogenic populations of cells can exhibit substantial phenotypic variation, which can, in turn, play an important role in development and in adapting to changing external conditions (Heitzler and Simpson, 1991). Variation in gene expression, due to stochastic bursting and asymmetric division of key molecular drivers of cellular identity, accounts for a large amount of observed cell-to-cell (cell-cell) variability within a given cell type (McAdams and Arkin, 1997). Cellular heterogeneity has historically been measured using microscopy and fluorescent labeling of key markers. These techniques have high spatial and cellular resolution but rely on prior knowledge and a limited number of markers, making it difficult to assay cellular differences comprehensively. The advent of single-cell genomic methods now enables profiling of transcriptional, genetic and epigenetic variation between individual cells on a global scale that depends less on a priori hierarchies and predefined markers (Tanay and Regev, 2017).
Single-cell RNA-sequencing (scRNA-seq), in particular, has led to remarkable advances in defining and refining the myriad cell states (Shalek et al., 2013, 2014), cell types (Jaitin et al., 2014; Shekhar et al., 2016; Montoro et al., 2018) and progenitors (Treutlein et al., 2014; Olsson et al., 2016) that are present during mammalian development and differentiation (Petropoulos et al., 2016; Tang et al., 2010; Scialdone et al., 2016; Klein et al., 2015). This has been aided by computational advances in clustering and pseudotemporal ordering of single cells that have enabled accurate inference of cell states and developmental trajectories, respectively (Trapnell et al., 2014; Haghverdi et al., 2015; Street et al., 2018). From a biological perspective, scRNA-seq has allowed the role of transcriptional heterogeneity to be explored. For example, single-cell profiling of mouse embryonic stem (ES) cells has revealed sporadic expression of polycomb targeted lineage regulators and less heterogeneity among pluripotency-associated genes in 2i versus serum growth conditions (Kumar et al., 2014). These results suggest a model whereby mouse ES cells are afforded the opportunity to access lineage specification programs through stochastic expression of pluripotency factors and lineage regulators typically repressed by H3K27me3.
DNA methylation also plays an important role in maintenance of and exit from pluripotency. Variation in DNA methylation modulates metastable switching in mouse ES cells between ZFP42 low and high states (Singer et al., 2014). Three catalytically active DNA methyltransferases (DNMTs) are responsible for maintenance (DNMT1) and de novo DNA methylation (DNMT3A/3B) in mammals, and all three are essential for normal development (Smith and Meissner, 2013). DNA methylation by DNMT3A/3B plays a particularly important role during development and ES cell differentiation (Gifford et al., 2013; Ziller et al., 2018), and both catalytically active enzymes are highly expressed in undifferentiated cells. Bulk experiments have shown a limited global impact of DNMT3A/3B knockout on the global DNA methylation landscape in human ES cells (Liao et al., 2015). This limited effect may be, in part, a consequence of bulk measurements, and it remains unknown how these epigenetic regulators affect transcriptional variation at the single-cell level, including how this may bias differentiation to new cell fates. To study this, we utilized previously generated knockout cell lines (Liao et al., 2015) in the undifferentiated and differentiated states to investigate the effects of these mutations on transcription at single-cell resolution.
RESULTS
Increased cellular variation in ES cells lacking DNMT3A and DNMT3A/3B
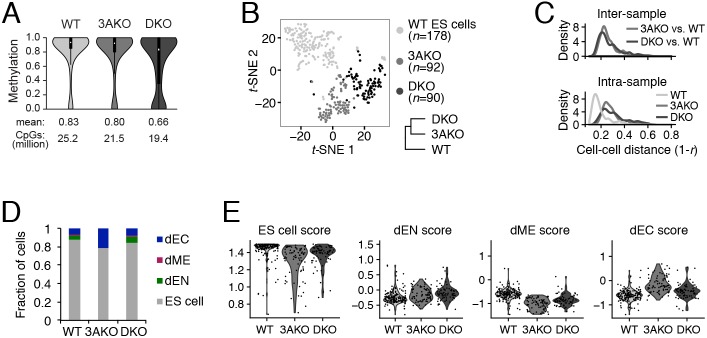
To explore the role of DNMTs in transcriptional regulation within individual cells, we used Smart-Seq2-based scRNA-seq (Picelli et al., 2014) to profile three HUES64 human ES cell lines – wild type (WT), with homozygous catalytic disruption of DNMT3A (3AKO), and with double knockout of both DNMT3A/3B (DKO) (Liao et al., 2015). Although the global decrease in methylation levels in the DKO cells is limited (Fig. 1A), they have 10-fold more differentially methylated regions than 3AKO relative to WT (Liao et al., 2015). Dimensionality reduction showed that WT, 3AKO and DKO cells mostly cluster by cell line (Fig. 1B). We found that 3AKO and DKO undifferentiated cells were equally dissimilar to WT ES cells (Fig. 1C, top), which was unexpected given the much greater similarity in the global methylation landscape between WT and 3AKO bulk samples (Liao et al., 2015). Interestingly, we noticed a significantly higher intra-sample cell-cell distance in the DKO and 3AKO populations relative to WT (P<10−15, Wilcoxon signed rank test, Fig. 1C, bottom).
Fig. 1.
Increased cellular variation in DNMT3A and DNMT3A/3B knockout ES cells. (A) Violin plot of CpG methylation for wild-type (WT), DNMT3A−/− (3AKO) and DNMT3A/3B−/− (DKO) cells averaged across two replicates. Mean methylation level and number of CpGs per sample are shown at the bottom and black boxes within the violin plots represent the interquartile range. Data were obtained from Liao et al. (2015) and are available at GEO under accession number GSE63281. (B) Dimensionality reduction of WT, 3AKO and DKO single ES cells (dots) using t-distributed stochastic neighbor embedding (t-SNE) and hierarchical clustering (bottom right) of the averaged expression profiles for sorted ES cells from each cell line. Samples 3AKO and DKO were more similar to each other than to WT ES cells. (C) Inter-sample (top) and intra-sample (bottom) density distribution of all pairwise cell-cell distances for WT, 3AKO and DKO cells. (D) Fraction of WT, 3AKO and DKO cells classified into four categories: ES cell; endoderm (dEN); mesoderm (dME); and ectoderm (dEC). (E) Violin plots of ES cell, dEN, dME and dEC scores for WT, 3AKO and DKO samples: each dot represents an in silico-sorted undifferentiated cell.
To control for the effect of background differentiation on our measure of cellular heterogeneity, we classified all cells as pluripotent, endoderm (dEN), mesoderm (dME) and ectoderm (dEC) using previously reported germ layer markers (Gifford et al., 2013; Tsankov et al., 2015a,b). We observed an increase in differentiated cells in DNMT mutant cells and a distinct bias towards ectoderm in 3AKO cells (Fig. 1D). We then in silico sorted for all undifferentiated cells and found that the intra-sample cell-cell distance using only cells classified as pluripotent was also significantly higher in the mutant cell lines relative to WT (P<10−15, Wilcoxon signed rank test, Fig. S1A). Our results were unchanged when repeating this analysis using three different cell-cell distance metrics (Euclidean, Manhattan, Spearman correlation; see Materials and Methods) and after controlling for data quality by focusing the analysis on the highest-quality ES cells (Fig. S1B,C). Among undifferentiated 3AKO and DKO cells, we also found increased variation in pluripotency, ectoderm and endoderm scores (Fig. 1E). Taken together, these results suggest increased cell-cell transcriptional variation that may affect the differentiation potential in 3AKO and DKO cells.
DNA methylation and transcript variation in DNMT3A/3B knockouts
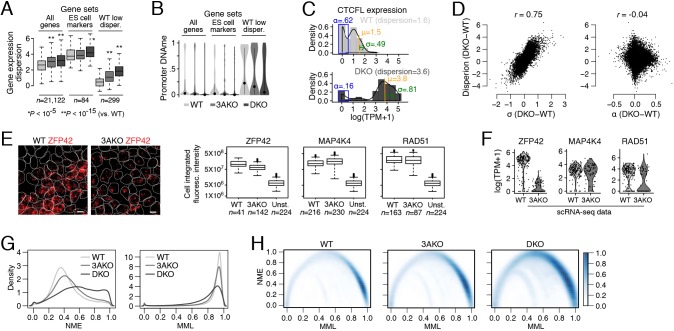
To further examine whether disruption of the de novo methyltransferases also increases global transcriptional variability, we computed the dispersion – log(variance/mean) – and standard deviation in expression for every gene within each sample population (Fig. 2A, Fig. S2A left). DKO and 3AKO showed a significant increase in transcript variation at all genes relative to WT using both metrics (P<10−15, Wilcoxon signed rank test; Fig. 2A) that associated with a corresponding decrease in mean promoter methylation level (Fig. 2B). To control for the impact of technical dropouts on our measurements of transcript variation, we explicitly modeled three parameters for the expression of each gene: the fraction of cells with no detectable expression (α), and the mean (μ) and standard deviation (σ) of expression among only detectably expressing cells (McDavid et al., 2013 and Shalek et al., 2014). As an example, the transcriptional variation of CTCFL increased in DKO versus WT cells as measured both by dispersion and by σ, whereas α decreased (Fig. 2C). As observed using dispersion, we also confirmed a global increase in transcriptional variation using σ in the DNMT mutants relative to WT cells (Fig. S2A, right). Globally, we found that difference in dispersion was highly correlated with difference in σ (r=0.75) but not with difference in α (r=−0.04, Fig. 2D), indicating that changes in the fraction of cells with detectable expression between samples does not associate with the global increase in transcriptional variation we report among DNMT mutant and WT ES cells. Consistent with the increased variation in pluripotency scores (Fig. 1E), we also observed a significant increase in standard deviation of expression across all cells and cells with detectable expression (σ) at pluripotency gene markers (P<10−6, Wilcoxon signed rank test, Fig. S2A), indicating that DNMT disruption leads to more variable expression of key pluripotency genes. We further quantified the relationship between changes in dispersion and average expression. We found 4740 and 1139 genes with a higher dispersion (difference greater than 1.5) in the DNMT mutants and WT cells, respectively; of those, 92% and 97%, respectively, also displayed a higher mean expression in the sample with higher dispersion (Fig. S2B). We confirmed the trends in transcriptional variation that we observed in the scRNA-seq data from WT and 3AKO cells for ZFP42, MAP4K4 and RAD51 using RNA fluorescence in situ hybridization (FISH; Fig. 2E,F, Fig. S2C). The standard deviation of gene expression for ZFP42 using RNA FISH was slightly higher in WT versus 3AKO, whereas the difference in transcriptional variation was more pronounced between the two conditions for MAP4K4 and RAD51. In summary, we find increased transcriptional variation in undifferentiated 3AKO and DKO cells at genes that predominantly increase in mean expression; however, this increase in transcript variation is uncorrelated with dropout rate in our scRNA-seq data.
Fig. 2.
Relationship between DNA methylation level, mean methylation entropy and transcript variation in DNMT3A and DNMT3A/3B knockouts. (A) Box plots of gene expression dispersion distribution, log(variance/mean), for all genes, ES cell markers and WT low dispersion genes for WT, 3AKO and DKO ES cells. (B) Violin plots of promoter mean CpG methylation for all genes, ES cell markers and WT low dispersion genes from WT, 3AKO and DKO ES cell bulk samples, averaged across two replicates. Dots represent the mean and lines extend at most one standard deviation from the mean. (C) Histograms of CTCFL expression in WT (top) and DKO (bottom) cells binned at intervals of 0.5 log(TPM+1) expression levels and normalized to the total cell counts. The three parameters that are estimated for CTCFL gene expression distribution (α, μ and σ) are shown in blue, orange and green, respectively. Dispersion for CTCFL increases and coincides with an increase in σ and μ as well as a decrease in α. (D) Scatter plots of the difference in dispersion, log(variance/mean) and parameters σ (left) and α (right) in DKO versus WT cells. We observe a high correlation between dispersion and σ difference (r=0.75) but not between dispersion and α difference (r=−0.04). (E) Left: representative images of RNA FISH with probes targeting ZFP42 (red) in WT (left) and 3AKO (right) ES cells. Cell segmentation is shown using white outlines. Scale bars: 10 μm. Right: box plots of ZFP42 (left), MAP4K4 (middle) and RAD51 (right) integrated probe intensity summed over the volume of the cell for WT, 3AKO and unstained (Unst.) ES cells. (F) Violin plots of log(TPM+1) gene expression for ZFP42 (left), MAP4K4 (middle) and RAD51 (right) in WT and 3AKO ES cell scRNA-seq data show similar trends in transcript variation as the RNA FISH experiment for these three genes. (G) Normalized methylation entropy (NME; left) and mean methylation level (MML; right) measured using WT, 3AKO and DKO ES cell whole genome bisulfite sequencing data across all chromosome 21 and 22 CpGs using the approach in Jenkinson et al. (2017). (H) Smoothed scatter plot with color intensity showing density of all chromosome 21 and 22 CpGs NME versus MML data. For WT, most CpGs have high MML and low NME (dark blue, bottom right). DKO CpGs with high NME spread across a lower MML (middle top of DKO plot; intensity gets darker), consistent with the global loss of methylation in the DKO sample. Box plots: boxes display the interquartile range, horizontal line within the box shows the median, whiskers extend to the most extreme data point that is no more than 1.5 times the length of the interquartile range.
Changes in DNA methylation variability have been linked to cancer risk markers, higher order chromatin organization and variability in gene expression across cancer patients (Hansen et al., 2011; Jenkinson et al., 2017; Teschendorff and Widschwendter, 2012). DNA methylation variability can be measured in phase at the individual read level using bisulfite sequencing, whereby each read can be considered to derive from a different cell. Globally, an increase in the percentage of reads displaying discordant methylation states was reported for DKO, but not for 3AKO, relative to WT ES cells (Liao et al., 2015). In addition, we measured the normalized methylation entropy (NME) (Jenkinson et al., 2017), an alternative approach based on statistical physics and information theory, and found that NME increased slightly in 3AKO and drastically in DKO relative to WT as mean methylation level decreased (Fig. 2G). Density scatter plots showed that the increase in NME was largely due to a shift of high methylation level CpGs with low entropy in WT to intermediate methylation level CpGs with high entropy in DKO (Fig. 2H). As the DKO-specific increase in NME does not appear to be proportional to the increase in transcriptional variation observed in both 3AKO and DKO, it suggests that genome-wide DNA methylation variability does not fully explain global transcript variation.
To further explore the relationship between DNA methylation and transcriptional dispersion at a single-cell level, we focused our analysis on gene promoters. We initially performed promoter epigenetic state enrichment analysis in ES cells (Gifford et al., 2013) for the most and least transcriptionally variable genes in WT, 3AKO and DKO samples, and found an association between low dispersion genes and H3K9me3-enriched or highly methylated promoters in WT cells (Fig. S2D). In contrast, whereas genes with H3K9me3-enriched promoters also correlated with genes of lowest transcriptional variance in 3AKO and DKO cells, high methylation promoters showed low to no correlation (Fig. S2D). Consistent with this result, for the least variable genes in WT ES cells, we found a significant increase in mean expression and transcriptional dispersion in 3AKO and DKO samples (P<10−15, Wilcoxon signed rank test; Fig. 2A, right) and a concomitant decrease in DNA methylation at the corresponding promoters (Fig. 2B). This implies that the expression of low dispersion genes in WT is regulated, in part, by the DNA methylation level. Although mean methylation and transcript dispersion levels appear to correlate at these genes, the same correlation is not apparent when comparing promoter mean NME and transcriptional variation globally, as measured by gene dispersion or σ (Fig. S2E,F). Together, this suggests that the increase in transcript variability observed after loss of DNMT3A/3B associates with loss of methylation at a subset of promoters but not globally.
Widespread transcriptional repression and super-enhancer misregulation
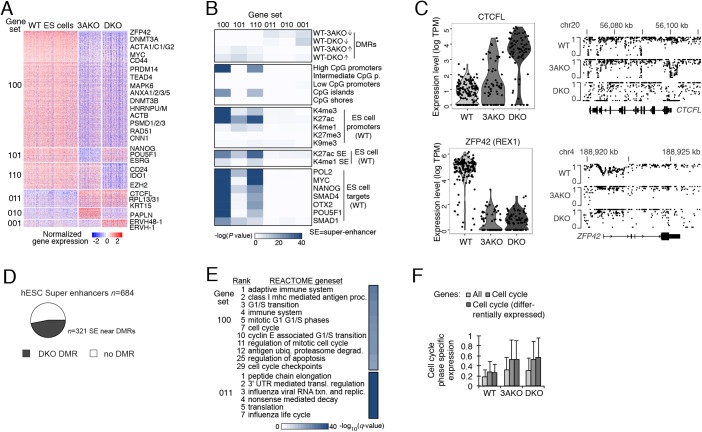
To better understand the regulatory changes that underlie the observed transcriptional dynamics in the DNMT3A/3B mutants, we identified all three-way differentially expressed genes between WT, 3AKO and DKO sorted samples (Fig. 3A). We found that the vast majority of genes were repressed (1964) rather than activated (470) relative to WT, which was somewhat unexpected given that loss of methylation is typically more associated with gene activation. Among the most downregulated genes in human ES cells, we observed a number of zinc fingers and important pluripotency transcription factors (TFs), including ZFP42, PRDM14, NANOG, POU5F1 and MYC (Table S1). Interestingly, the latter three TFs showed lower expression in 3AKO than DKO despite the DKO being generated through a DNMT3B deletion in the DNMT3A knockouts (Liao et al., 2015). We also found a number of housekeeping genes with reduced expression, including those encoding actin, heterogeneous nuclear ribonucleoproteins (HNRNPs) and proteasome genes.
Fig. 3.
Global transcriptional repression and altered regulation in DNMT3A and DNMT3A/3B knockout ES cells. (A) Differentially expressed genes (right; rows) for sorted populations of WT, 3AKO and DKO ES cells (columns). Genes are separated into six gene sets [left: 100 (n=1443), 101 (n=191), 110 (n=330), 011 (n=229), 010 (n=143) and 001 (n=98)], where 1 or 0 indicates high or low expression for the respective condition (order: WT, 3AKO, DKO). (B) Genomic enrichment analysis for gene sets (columns) defined in panel A against CpG density features, epigenetic and TF binding data collected in matching WT ES cells (Gifford et al., 2013; Tsankov et al., 2015b). (C) Top: distribution (dots indicate individual cells) of CTCFL expression (left) and the corresponding CpG methylation levels at the CTCFL locus for WT, 3AKO and DKO ES cells. Bottom: ZFP42 cellular expression (left) and promoter methylation (right) as described above. (D) Of all 684 H1 ES cell super-enhancers (Hnisz et al., 2013), 321 (47%) are located within 1 kb of a DKO DMR (displayed in black). In total, 734 DKO DMRs (of 44,244 total) were associated with super-enhancers, and are defined as regions with difference in methylation>0.6 relative to WT, with P<0.01 (F-test). (E) Functional enrichment analysis for the gene sets defined in panel A against the REACTOME database. (F) Distribution of cell cycle phase-specific expression for sorted WT, 3AKO and DKO ES cells considering all genes, cell cycle annotated genes and differentially expressed cell cycle annotated genes. Error bars indicate one standard deviation. DMR, differentially methylated region; K, lysine on histone 3; me3, tri-methylation; ac, acetylation; me1, mono-methylation.
We next performed a comprehensive search for promoter enrichment against published DNA methylation, histone modification and TF binding data from matched samples (Fig. 3B) to explore the potential underlying mechanism (Gifford et al., 2013; Tsankov et al., 2015b; Liao et al., 2015). We found a significant association between loss of promoter methylation and expression increase, as illustrated at the CTCFL locus (Fig. 3C, top). Surprisingly, we also identified 152 and 82 promoters that increased in DNA methylation (e.g. ZFP42; Fig. 3C, bottom) in 3AKO and DKO, respectively, which overlapped significantly with decreased expression in the mutants (Fig. 3B). Genes repressed in the knockouts frequently had high CpG-dense promoters that were enriched for active histone modification in WT cells (H3K27ac and, to a lesser degree, H3K4me3 and H3K4me1; Fig. 3B). DNMT3A and DNMT3B have previously been shown to occupy active enhancers, and knockdowns of the de novo methyltransferases reduced super-enhancer activity and disrupted homeostasis in epidermal stem cells (Rinaldi et al., 2016). In our dataset, repressed genes (e.g. NANOG, POU5F1) associated significantly with upstream H3K27ac and H3K4me1 super-enhancers, suggesting a similar role for the de novo methyltransferases at super-enhancers in human ES cells. We also found that nearly half of human ES cell super-enhancers (Hnisz et al., 2013) showed drastic changes in methylation levels in DKO (Fig. 3D, Fig. S3A), suggesting that DNMT3A/3B shape the methylation landscape near super-enhancers. We further identified high enrichment for in vivo binding of a number of key pluripotency associated TFs upstream of 3AKO and DKO repressed genes, including MYC, NANOG, and POU5F1. As these factors occupy 76% of downregulated gene promoters in WT ES cells, a large fraction of the repressed phenotype may be mediated by their decreased expression and/or activity in the mutants. Taken together, loss of DNMT3A and DNMT3A/3B appears to interfere with normal super-enhancer activity upstream of pluripotency associated regulators, leading to downregulation of these core ES cell TFs and their downstream targets.
Loss of DNMT3A/3B alters cell cycle gene expression
We next performed gene set enrichment analysis and observed that genes upregulated in the 3AKO and DKO mutants included those encoding a number of ribosomal proteins (e.g. RPL13/31) that are associated with the influenza life cycle and viral RNA transcription and replication (Fig. 3E, bottom). Combined with the observation that ERVH48-1 and ERVH-1 are also upregulated in the DKO, these changes in expression point to increased activity of endogenous retroviral elements. Interestingly, we also found that downregulated genes associated with a number of cell cycle categories, including gene sets related to G1/S transition and the establishment of checkpoints (Fig. 3E, top).
To investigate possible cell cycle alterations in the DNMT3A/3B mutants, we identified all differentially expressed cell cycle annotated genes in the WT, 3AKO and DKO ES cell samples (Fig. S3B). We found decreased expression relative to WT ES cells in a number of key cell cycle genes (e.g. TP53, MCM2/3/4/5/6 and ORC1/2/5, Fig. S3B) that were also downregulated during normal differentiation (Gifford et al., 2013). We also observed downregulation of CDK4/6 and upregulation of CCND1 in the 3AKO, which has previously been observed during normal ectoderm differentiation (Gifford et al., 2013). Although the proportion of cells in different phases of the cell cycle is similar for all three samples (Fig. S3C), we found a global shift from constant to phase-specific cell cycle expression in the DNMT3A/3B mutants at all genes and, especially, at ones annotated to have cell cycle function (Fig. 3F). Taken together, our data show a global change in expression of cell cycle-associated genes upon DNMT3A/3B loss with increases in phase-specific cell cycle expression that suggest the establishment of a regulated G1/S transition and cell cycle checkpoints relative to WT human ES cells.
Aberrant expression following ES cell differentiation of DNMT3A/3B knockouts
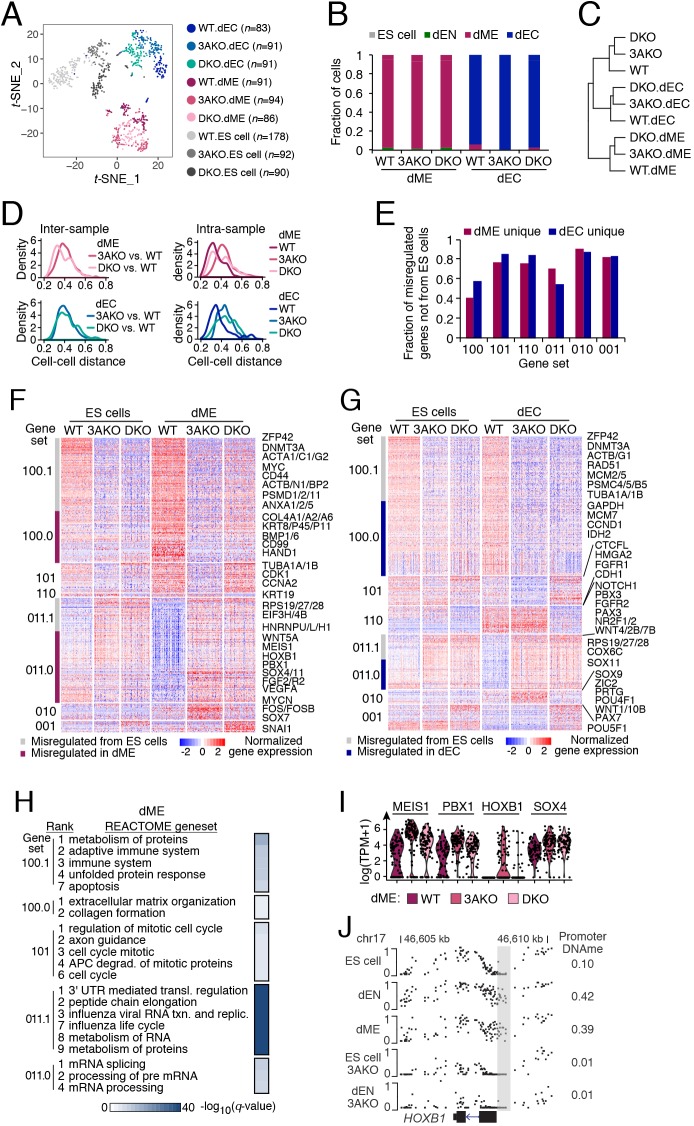
To investigate whether and how the observed transcriptional changes in the knockouts affect cellular specification, we differentiated all three cell lines for 5 days towards dME and dEC followed by scRNA-seq. Dimensionality reduction showed that cells clustered primarily by cell type and sample identity (Fig. 4A). We observed a similar proportion of dME- and dEC-positive cells between knockout and WT samples following differentiation (Fig. 4B). The spread of dEC scores was similar across WT, 3AKO and DKO dEC samples, whereas variation in dME scores was slightly greater in the 3AKO dME sample relative to WT and DKO (Fig. S4A). Population averaged transcriptomes for all samples clustered by cell type (Fig. 4C) and showed that dEC samples were more similar to ES cells than dME samples, which is consistent with the inherent dEC bias we noted in 3AKO and DKO ES cells (Fig. 1D,E). In line with our ES cell results, we found that the DKO dME/dEC cells were slightly more similar to WT dME/dEC than 3AKO dME/dEC cells (Fig. 4D, left). We also observed an increase in intra-sample cell distance in the knockouts versus WT for both dME and dEC (Fig. 4D, right), although the difference was not as pronounced as in ES cells (Fig. 1C).
Fig. 4.
Transcriptional changes and misregulation in DNMT3A/3B knockout cells during ES cell differentiation. (A) Dimensionality reduction of wild-type (WT), DNMT3A knockout (3AKO) and DNMT3A/3B knockout (DKO) single ES, mesoderm (dME) and ectoderm (dEC) cells (dots) using t-distributed stochastic neighbor embedding (t-SNE). Number of cells is shown in parentheses. (B) Fraction of WT, 3AKO and DKO mesoderm (left) and ectoderm (right) cells classified into four cell types (ES cell, dEN, dME, dEC). (C) Hierarchical clustering of the averaged expression profiles for all sorted samples. (D) Inter-sample (left) and intra-sample (right) density distribution of all pairwise cell-cell distances (1–Pearson correlation coefficient) for WT, 3AKO and DKO dME (top) and dEC (bottom) cells. (E) Fraction of differentially expressed genes in dME (red) and dEC (blue) that were not already present in ES cells, or are dME/dEC unique. Gene sets are defined in the legend for F. (F) Differentially expressed genes (right; rows) for sorted population of WT, 3AKO and DKO ES and mesoderm cells (columns). Genes are separated into eight gene sets (left: 100.1, 100.0, 101, 110, 011.1, 011.0, 010 and 001), for which 1 or 0 indicates high or low expression, respectively, for each condition (order: dME WT, 3AKO, DKO). Suffix .1 indicates inherited from ES cells whereas .0 indicates dME unique. (G) Differentially expressed genes (right; rows) for sorted population of WT, 3AKO and DKO ES and ectoderm cells (columns). Genes are separated into eight gene sets as described above. (H) Functional enrichment analysis for the dME gene sets defined in F against the REACTOME database. (I) Distribution of gene expression, log(TPM+1), for selected TFs aberrantly expressed in 3AKO and DKO dME cells, relative to WT. Dots represent cells. (J) CpG methylation levels at the HOXB1 locus for ES, dEN, dME, 3AKO ES and 3AKO dEN cells. The HOXB1 promoter is highlighted with a gray bar and the mean promoter methylation level is listed on the right.
We then identified all differentially expressed genes in WT, 3AKO and DKO dME and dEC samples and compared them with the ES cell populations. We found that in dME 36% and in dEC 34% of differentially expressed genes had a similar change in expression in the ES cell mutants, including 59% in dME and 42% in dEC of the genes repressed in both ES cell knockouts (Fig. 4E). These genes included ZFP42 (Fig. S4B), actin family genes and proteasome genes (Fig. 4F,G, gene set 100.1), and associated with some of the same functional categories as repressed genes in the ES cell knockouts (e.g. protein metabolism, immune system, apoptosis; Fig. 4H). In dME, a number of genes associated with extracellular matrix organization and collagen formation (BMP1, COL4A1/2/6) were downregulated uniquely in dME DNMT mutants and not in ES cells (Fig. 4H, gene set 100.0). We saw a similar enrichment for translation and viral response for upregulated genes in both ES cell and dME knockouts versus WT (Fig. 4H, gene set 011.1, e.g. RPS19/27/28), and enrichment for mRNA processing and splicing pathways for dME-specific knockout activated genes (Fig. 4H, gene set 011.0).
To investigate the underlying mechanisms that may explain the transcriptional changes in the dME and dEC knockouts, we overlapped the promoter epigenetic state with differentially expressed gene categories. We found that genes that gain methylation in the three germ layers are also upregulated in DKO but not in 3AKO dME, suggesting that DNMT3B may compensate for DNMT3A loss at these lineage-specific targets (Fig. S4C). This trend was also notable in dEC but to a lesser degree (Fig. S4D). Further, we observed enrichment of high-CpG promoters (HCP) and CpG islands for inherited repressed genes (100.1) but not for dME- or dEC-specific repressed genes (100.0). Finally, we found an enrichment for genes downstream of super-enhancers being misregulated in dEC knockouts relative to WT, including FGFR1 (gene set 101), SOX11 (011) and NR6A1 (010). In dME, we found an association between dME upstream super-enhancers and dME-specific gene repression (gene set 100.0), including COL4A1/2, KRT8, CD99, and the TF HAND1, which may point to a cell type-specific role of DNMT3A/3B at dME super-enhancers. Moreover, downregulation of HAND1 may mediate further downstream repression at its dME targets, as we observed for core TFs POU5F1 and NANOG in undifferentiated DNMT3A/3B knockouts.
We also found a number of TFs with important roles in developmental processes and oncogenesis to be aberrantly expressed in the dEC and dME DNMT mutants relative to WT. In dEC, genes encoding key TFs associated with ectoderm lineage development were specifically downregulated in DKO (e.g. PAX3, NR2F1/2), upregulated in both mutants (e.g. SOX11) or upregulated in 3AKO relative to WT (e.g. SOX9, ZIC2, POU4F1, PAX7; Fig. S4E). Moreover, key pluripotency TFs, such as POU5F1, with a promoter that is focally methylated during differentiation, and PRDM1, along with POU domain TFs POU5F2 and POU2F2, were specifically upregulated in the DKO dEC sample relative to WT (Fig. S4E, bottom). This was accompanied by an increase in the median and standard deviation of ES cell scores observed in dEC DKO cells versus WT (Fig. S4F). In dME, TFs MEIS1, PBX1, HOXB1 and SOX4 were upregulated in the 3AKO cells relative to WT (Fig. 4I). As the promoter methylation of HOXB1 increases drastically during ES cell differentiation towards dEN and dME, and this gain in dEN depends on the catalytic activity of DNMT3A (Fig. 4J), it is likely that the HOXB1 promoter methylation is misregulated in a similar manner in dME cells lacking DNMT3A. Although we do not observe a change in promoter methylation for the genes encoding TFs MEIS1 and SOX4, their expression is correlated with HOXB1 (r=0.2, P value<0.05; Pearson) implying that these TFs are co-regulated as part of the same gene expression program.
Finally, we observed aberrant expression for a number of cell cycle annotated genes and key transcriptional regulators in the knockouts after dME and dEC differentiation. In dME mutants, and especially in 3AKO, we observed downregulation of mitosis-associated genes CDK1 and CCNA2 (Fig. S4G) as well as cell cycle-associated genes linked to lineage choice (CCND1, CCND3, CDKN1A). In dEC mutants, a number of S-phase genes were downregulated (MCM2/5/7) as well as CCND1 (Fig. 4G), which acts to block endoderm formation in late G1 phase (Pauklin and Vallier, 2013) and promotes neuroectoderm cell fate (Pauklin et al., 2016). In both dME and dEC, we observed a similar proportion of cells in different phases of the cell cycle (Fig. S4H) and levels of phase-specific expression (Fig. S4I).
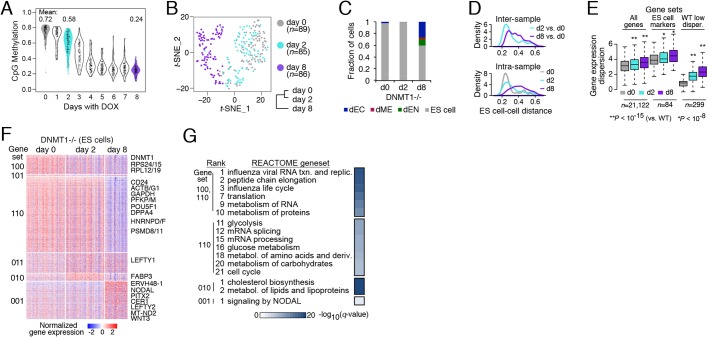
Loss of DNMT1 triggers increased transcript variation and differentiation
To complement our results from the de novo DNA methyltransferases, we explored the effects of loss of the maintenance enzyme DNMT1. Loss of DNMT1 results in a global loss of DNA methylation rather than the limited dynamics we find in the DNMT3 knockouts (Liao et al., 2015). Specifically, we utilized our previously established doxycycline-inducible downregulation of DNMT1 system and collected live cells every day for 8 days for single cell methylation profiling and at day 0, 2 and 8 following doxycycline treatment for scRNA-seq. We observed global loss of methylation in all the profiled cells beginning at day 2, which plateaued at a minimum at around day 6 to 8 (Fig. 5A). Dimensionality reduction of the scRNA-seq revealed a high similarity between most day 0 and day 2 cells, with a gradual departure from WT for some day 2 cells and all day 8 cells (Fig. 5B). Quantifying cell type identity (Fig. 5C) showed an increase in cells exiting pluripotency at day 8, with a preference of escape towards ectoderm, as observed in 3AKO and DKO. We found that both the population average similarity between in silico-sorted undifferentiated samples (Fig. 5B, bottom right) and the inter-sample ES cell distance versus day 0 (Fig. 5D, top) increased with time after doxycycline induction. We also observed an increase in intra-sample cell-cell distance at day 2 and day 8 compared with day 0 (Fig. 5D, bottom), and note that the heterogeneity at day 8 exceeds that found in the DNMT3A/3B knockout ES cells (Fig. 1C). Variation in gene expression also increased at day 2 and day 8 for all genes, pluripotent markers and the least variable WT genes (P<10−8, Wilcoxon signed rank test; Fig. 5E). Our results were consistent after controlling for differences in data quality and sequencing depth between samples. We again found an association between genes with the lowest expression dispersion and high methylation promoter occupancy at day 0, and this enrichment gradually decreased at day 2 and day 8, with downregulation of DNMT1 and concurrent global loss of methylation (Fig. S5A), as we observed for 3AKO and DKO versus WT ES cells.
Fig. 5.
Increased transcript variation and differentiation upon loss of DNMT1. (A) Violin plot of single cell methylation data, where each dot represents the average CpG methylation level per cell. Cells were collected for scRNA-seq after 0, 2 and 8 days of doxycycline treatment. (B) Dimensionality reduction of day 0, 2 and 8 single cells (dots) using t-distributed stochastic neighbor embedding (t-SNE) and hierarchical clustering (bottom right) of the averaged expression profiles for in silico-sorted ES cell populations. (C) Fraction of cells classified into four categories (ES cell, dEN, dME, dEC) following 0, 2 and 8 days of doxycycline treatment. (D) Inter-sample (top) and intra-sample (bottom) density distribution of all pairwise cell-cell distances for in silico-sorted ES cells at day 0, 2 and 8. (E) Box plots of gene expression dispersion distribution at all genes, ES cell markers, and WT low dispersion genes for sorted ES cell populations at day 0, 2 and 8. (F) Differentially expressed genes (right; rows) for sorted population of ES cells at day 0, 2 and 8 (columns). Genes are separated into six gene sets [left: 100 (n=337), 101 (n=36), 110 (n=1301), 011 (n=349), 010 (n=139) and 001 (n=644)], where 1 or 0 indicates high or low expression for the respective condition (day 0, 2 and 8). (G) Functional enrichment analysis for the gene sets defined in F against the REACTOME database.
To gain insight into the functional changes induced by downregulation of DNMT1, we identified all differentially expressed genes in ES cells collected at day 0, 2 and 8 (Fig. 5F). The majority (1638 of 2631; 62%) of day 8 differentially expressed genes were repressed relative to day 0, as observed in 3AKO and DKO ES cells. We found downregulation as early as day 2 of a number of ribosomal protein genes (e.g. RPS24/15, RPL12/19) associated with influenza life cycle (Fig. 5G, gene set 100). At day 8, we observed a small downregulation of POU5F1 and other pluripotency-associated genes (e.g. CD24, DPPA4) and concomitant activation of NODAL signaling genes, including NODAL, CER1, LEFTY1/2 and downstream TF PITX2 (Yoshioka et al., 1998). We also note a shift in expression from glycolysis genes (e.g. GAPDH, PFKP/M) at day 0/2 to lipid metabolism at day 2 (e.g. FABP3, FADS2), to oxidative phosphorylation genes (MT-ND2, MT-ND4L) at day 8 (Fig. 5F,G). Finally, we observed changes in cell cycle regulation for ES cells that survived loss of methylation, including an increase in fraction of G2/M cells (Fig. S5B) and increase in cell cycle phase-specific expression (Fig. S5C). These changes might reflect a longer G2/M phase needed for methylation maintenance fidelity and compacting of chromosomes. Taken together, we observe repression at most differentially expressed genes and an increase in differentiation, as well as cellular and gene expression variation in ES cells upon loss of DNMT1.
DISCUSSION
Pluripotent stem cells are a powerful model to explore the targets and role of epigenetic regulators. We have previously generated knockout human ES cell lines for the three catalytically active DNA methyltransferases (Liao et al., 2015). With the advance of single-cell technologies, we wanted to explore the effects of these knockouts within individual cells to better understand how the subtle changes in the undifferentiated state translate to substantial disruptions upon exit from pluripotency (Ziller et al., 2018). Using our scRNA-seq approach, we observed a global increase in cellular and gene expression variation for all DNMT mutants. As variability has been linked to the ability of a cell to evolve and adapt to a changing environment (Heitzler and Simpson, 1991), our results suggest that disruption of DNMTs may increase cellular plasticity. It would therefore be interesting in the future to explore the effects of this by tracking individual cells using molecular barcoding (Chan et al., 2019).
We also found two somewhat unexpected effects in the double knockout ES cells. First, we found widespread repression in gene expression upon loss of DNMT3A and DNMT3A/3B in the undifferentiated cells, particularly at genes associated with CpG islands and with H3K27ac super-enhancers. In epidermal stem cells, knockdown of the de novo methyltransferases triggers a reduction of super-enhancer activity (Rinaldi et al., 2016) and this may occur through a similar mechanism in human ES cells, albeit at different loci. In support of our findings, in epidermal stem cells we also observe 7765 genes that are downregulated versus 2136 upregulated (1.4 fold difference) in the DNMT3A knockdown versus control (Rinaldi et al., 2016). Secondly, we do observe a gain of DNA methylation at selected sites in the 3AKO and DKO ES cells. As the latter are derived from the 3AKO this may be a consequence of DNMT3B activity. Known DNMT3B targets include germline genes and it will be interesting to explore how and why these additional loci are targeted in the mutant ES cells.
Differentiation of 3AKO and DKO towards mesoderm and ectoderm showed that the knockout repressed genes were largely inherited from ES cells. We also observed that dME DNMT mutant repressed genes associated with super-enhancers in a mesoderm-specific manner. As core TFs (NANOG, POU5F1 in ES cells; HAND1 in mesoderm) are associated with super-enhancers, we provide evidence that DNMT3A/3B disruption may lead to decreased expression of key cell identity TFs and their downstream targets. Furthermore, we find upregulation of a number of key developmental and oncogenic TFs in 3AKO mesoderm (e.g. MEIS1/2, PBX1, HOXB1, SOX4) and 3AKO ectoderm (SOX11, SOX9, ZIC2, POU4F1, PAX7). DNMT3A is often mutated in human tumors (Kim et al., 2013), has been shown to act as a first hit mutation (Shlush et al., 2014), and its loss in hematopoietic stem cells and the epidermis promotes leukemia and squamous cell carcinoma formation, respectively (Rinaldi et al., 2017; Yang et al., 2016). It will be interesting in the future to further explore the possible role of increased transcriptional variability in tumor initiation and progression. Taken together, we show that combining scRNA-seq and genetic perturbations presents a powerful tool for dissecting the role of epigenetic regulators in development and disease.
MATERIALS AND METHODS
Human ES cell culture
Cell culture was carried out as reported previously (Tsankov et al., 2015b). Briefly, we chose the National Institutes of Health-approved, male human ES cell line HUES64 because it has maintained a stable karyotype over many passages and is able to differentiate well into mesoderm and ectoderm. The cells are frequently tested for mycoplasma and identity for the knockout cell lines was confirmed through genotyping PCR. ES cells were maintained on ∼15,000 cells/cm2 irradiated murine embryonic fibroblasts (MEFs, MTI-GlobalStem) and cultured in 20% KnockOut Serum Replacement (KSR, Life Technologies), 200 mM Glutamax (Life Technologies), 1× Minimal Essential Medium (MEM) Non-essential Amino Acids Solution (Life Technologies), 10 μg/ml basic fibroblast growth factor (bFGF, Millipore), 55 µM β-mercaptoethanol in Knockout Dulbecco's Modified Eagle Medium (KO DMEM, Life Technologies). ES cells were passaged every 4-5 days using 1 mg/ml Collagenase IV (Life Technologies). All human ES cell work has been approved by the Harvard University ESCRO (#E00021).
Directed differentiation of human ES cells towards mesoderm and ectoderm
When human ES cells reached 60-70% confluency on MEFs, the cells were plated as clumps on 6-well plates coated with Matrigel (Life Technologies) in mTeSR1 basal medium (Stemcell Technologies). We maintained the cells for 3 days in feeder-free culture and then induced directed differentiation towards mesoderm and ectoderm. For the first 24 h of mesoderm differentiation, cells were cultured in DMEM/F12 medium supplemented with 100 ng/ml Activin A (R&D Systems), 10 ng/ml bFGF (Millipore), 100 ng/ml BMP4 (R&D Systems), 100 ng/ml VEGF (R&D Systems), 0.5% fetal bovine serum (Hyclone), 200 mM GlutaMax (Life Technologies), 0.2× MEM Non-essential Amino Acids Solution (Life Technologies) and 55 µM β-mercaptoethanol. From 24 to 120 h of mesoderm differentiation, Activin A was removed from the culture. To induce ectoderm differentiation, cells were cultured for 5 days in DMEM/F12 differentiation media supplemented with 2 µM TGFβ inhibitor (Tocris, A83-01), 2 µM WNT3A inhibitor (Tocris, PNU-74654), 2 µM Dorsomorphin BMP inhibitor (Tocris), 55 µM β-mercaptoethanol, 1× MEM Non-essential Amino Acids Solution (Life Technologies) and 15% KOSR (Life Technologies). Media were changed daily. Before inducing differentiation, we manually removed the differentiated cell clumps.
Cell collection and fluorescence-activated cell sorting
Cells were treated with StemPro Accutase (Life Technologies, #A1110501) for 5 min, quenched in MEF medium and pelleted using centrifugation [5 min, 1000 rpm (94 g)]. Media was aspirated and cell pellets were washed once in PBS. RNA was immediately stabilized by resuspending the cells in RNAprotect Cell Reagent (∼100 μl per 100,000 cells, Qiagen, #76526) and 1 μl of RNaseOUT Recombinant Ribonuclease Inhibitor (Life Technologies, #10777-019). Before sorting, cells in RNAprotect Cell Reagent were diluted in ∼1.5 ml PBS (pH 7.4; no calcium, no magnesium, no phenol red; Life Technologies, #10010-049). Also, 5 μl of lysis buffer, composed of a 1/500 dilution of Phusion HF buffer (New England Biolabs, #B0518S) was aliquoted in Eppendorf 96-well skirted plates (VWR, #95041-430). Cells were sorted individually in each well of 96-well plates using the FACSAria II flow cytometer (BD Biosciences), avoiding doublets and cell debris. After sorting, plates were immediately sealed, spun down, frozen on dry ice and stored at −80°C.
Cell culture, fixation and FISH
Human ES cells were dissociated to single cells using Accutase (Life Technologies, A11105-01), and 30,000 cells were plated per well of 96-well imaging plate coated with Geltrex (Gibco) in mTeSR1 media. The culture media were changed daily and fixed on the third day when cells were ∼90% confluent. Before fixing they were stained with 2uM CFSE for 20 min in the incubator. The cells were fixed in 4% formaldehyde solution while covered with aluminum foil for 30 min at room temperature and then dehydrated in 50%, 70% and 100% ethanol for 2 min each concentration. The plates were stored in 100% ethanol in a −20°C chest freezer.
Following fixation, expression levels of three different mRNA transcripts were measured in situ using RNA-FISH probes (Thermo Fisher Scientific) as previously described (Shalek et al., 2013). Briefly, the ViewRNA ISH Cell Assay Kit (Invitrogen) was performed to stain cells according to the manufacturer's recommendations. Following staining, cells were imaged on an Olympus IX83 inverted microscope using 405 nm excitation for the DAPI stain and 647 nm excitation for the RNA-FISH probes. To quantify RNA expression, single cells were segmented using CellProfiler, and their total probe content was summed over the volume of the cell. Integrated probe intensity box plots were generated to confirm qualitative agreement between RNA-FISH and scRNA-seq.
scRNA-seq
Following sorting, 96-well plates of single cells were whole-transcriptome amplified using a Smart-Seq2-based approach, as previously described (Trombetta et al., 2014). Cell lysates were first cleaned with 2.2× volume AMPure XP SPRI beads (Beckman Coulter). Reverse transcription and PCR were then performed on the samples. Following whole-transcriptome amplification, PCR products were cleaned with 0.9× volume SPRI beads and eluted into 20 μl of water. Concentration of cDNA in the resulting solution was determined using a Qubit 3.0 Fluorimeter (Thermo Fisher Scientific) and analyzed using a high sensitivity DNA chip for BioAnalyzer (Agilent Technologies). Whole-transcriptome amplification products were diluted to a concentration of 0.1 to 0.4 ng/μl and tagmented and amplified using Nextera XT DNA Sample preparation reagents (Illumina). Tagmentation was performed according to the manufacturer's instructions, modified to use one quarter of the recommended volume of reagents, extended tagmentation time to 10 min and extended PCR time to 60 s. PCR primers were ordered from Integrated DNA Technologies. Primer sequences: 3′ SMART CDS Primer IIA: 5′ AAGCAGTGGTATCAACGCAGAGTACT(30)VN; SMARTer II A oligonucleotide: 5′ AAGCAGTGGTATCAACGCAGAGTACATrGrGrG; IS PCR primer: 5′ AAGCAGTGGTATCAACGCAGAGT. Nextera products were then cleaned with 0.9× volume of SPRI beads and eluted in water. The library was quantified using Qubit and analyzed using a high-sensitivity DNA chip. The library was diluted to 2.2 pM and sequenced on a NextSeq 500 (Illumina).
Processing of scRNA-seq data
RNA-seq reads were first trimmed using Trimmomatic (Bolger et al., 2014). Trimmed reads were aligned to the RefSeq hg38 genome and transcriptome (GRCh38.2) using Bowtie2 (Langmead and Salzberg, 2012) and TopHat (Trapnell et al., 2009), respectively. The resulting transcriptome alignments were processed using RSEM to estimate the abundance of RefSeq transcripts (Li and Dewey, 2011), in transcripts per million reads mapped (TPM). All cells with fewer than 2000 detectable transcripts (TPM>1) were removed from further analysis. Expression levels for gene i in sample j were quantified as Ei,j=log(TPMi,j+1). Relative expression level for gene i was computed within each subpopulation S as Eri,Sj = Ei,Sj − Êi,S, where Êi,S = average[Ei,S1…Sn] or the mean expression of that gene across all cells within subpopulation S.
Unsupervised dimensionality reduction
To visualize cells in 2-dimensional space, we first performed principal component analysis (PCA) using the Seurat R package version 2.0 as previously described (Satija et al., 2015) using highly variable genes of mean expression ≥1. We then determined the statistically significant principal components by calculating 1000 random permutations of 1% of genes in the data. We used all significant principal components (P<10−10) as input to non-linear dimensionality reduction via t-distributed stochastic neighbor embedding (t-SNE).
Classification of cells into ES cells, endoderm, mesoderm and ectoderm
We calculated ES cell, endoderm (dEN), mesoderm (dME) and ectoderm (dEC) scores for all cells by using the AddModuleScore function in Seurat with default parameters for the top 50 most uniquely expressed markers for the ES cell, dEN, dME and dEC purified populations (Gifford et al., 2013) that were also present in the scRNA-seq data. Uniqueness was defined as in previous studies (Tsankov et al., 2015a). Cells were then classified into one of four cell types, based on the maximal ES cell, dEN, dME or dEC score. We obtained in silico-sorted populations of ES cells by filtering out all cells collected at day 0 that had an ES cell score ≥max[dEN score, dME score, dEC score]. We defined dEN, dME and dEC in silico-sorted populations similarly.
Hierarchical clustering of sorted samples
Relative expression values for all genes was averaged across all ES or dME cells for the defined subpopulations (WT ES cells, 3AKO, DKO, DNMT1−/− day 0, 2, 8). The mean relative expression values were then clustered using hierarchical clustering, average linkage, and 1−Pearson correlation coefficient (r) of all non-zero values as a distance metric.
Inter- and intra-sample cell-cell distance
Inter-sample cell-cell distance was computed by comparing all pairs of cells between two samples, using 1−Pearson correlation coefficient (r) of all non-zero values as a distance metric. Intra-sample cell-cell distance was computed by comparing all pairs of cells within a sample using the same distance metric. Cells in each comparison were in silico sorted to contain only ES (Figs 1, 2, 3 and 5), dME or dEC cells (Fig. 4). Before computing the distance, all cells were quantile normalized to control for the total number of transcripts detected per cell. The same approach was applied for other distance metrics (Euclidean, Manhattan and 1-Spearman correlation).
Gene expression dispersion analysis
To assay the level of transcriptional variation per gene, we first quantile normalized TPMs for all cells within each sample population and then computed the dispersion or log(variance/mean) for all genes. We also performed quantile normalization before computing other measures of transcript variation.
Normalized methylation entropy analysis
To compare the variability of methylation levels at the individual CpG and read level, we used ‘informMe’, an information-theoretic approach that uses the Ising model of statistical physics to generate mean methylation levels and normalized methylation entropy per CpG. We ran informME for WT, 3AKO and DKO data, for all CpGs located on chromosomes 21 and 22, and used R to plot the respective levels of mean methylation level (MML) and normalized methylation entropy (NME).
Three-way differential expression analysis
Differential expression was tested across all possible pairwise comparisons (100, 101, 110, 011, 010 and 001) of three samples, where 1 or 0 indicates high or low expression for the respective sample (e.g. WT, 3AKO and DKO ES cells). To measure differential expression, we used the likelihood-ratio test for single-cell gene expression (McDavid et al., 2013) as implemented in the Seurat R package, requiring a P value≤10−8 and 1.22-fold change. For ease of visualization, differentially expressed genes were then combined into gene sets representing all possible three-way comparisons (100, 101, 110, 011, 010 and 001) and gene expression was row normalized across cells. Genes were only included in one gene set that had the highest P value in differential expression. The same analysis was performed to compare WT, 3AKO and DKO dME/dEC cells and day 0, 2 and 8 DNMT1-depleted ES cells.
Genomic region enrichment analysis
We assessed the significance of overlap of any gene set against a number of predefined genomic regions that can be mapped to their nearest downstream gene. Significance was calculated using the hypergeometric distribution with Bonferroni correction for multiple hypotheses testing. The resulting P value was −log() transformed and displayed for a number of genomic regions (rows), including CpG density features, epigenetic, and TF binding data collected in matching WT, ES, or dME cells (Gifford et al., 2013; Tsankov et al., 2015b). This analysis was performed for gene sets predefined using the three-way differential expression analysis as well as for high and low dispersion set of genes for all samples.
Gene set enrichment analysis
Gene sets enrichment analysis (http://software.broadinstitute.org/gsea/) was performed on defined gene sets above selecting only for common pathways from the REACTOME database (http://www.reactome.org/).
Cell cycle differential expressed genes and phase classification
To show differentially expressed cell cycle annotated genes, we performed the three-way differential expression analysis as described above solely for genes related to the cell cycle (Whitfield et al., 2002; Kanehisa and Goto, 2000). We used a less stringent threshold for displaying cell cycle annotated differentially expressed genes of P value≤10−4. For visualization, cells (columns) within each sample were ordered according to progress in the cell cycle, as previously described (Kowalczyk et al., 2015), starting with M/G1 cells on the left and ending with G2/M cells on the right. Expression values were averaged using a 20-cell window.
To assign cells according to cell cycle phase, we used a similar approach to that previously described (Tirosh et al., 2016). Briefly, we defined cell cycle phase-specific markers for G1/S, S, G2/M for ES, dME and dEC cells separately, keeping only genes in each predefined cell cycle phase gene set (Whitfield et al., 2002) if they had a correlation r≥0.3 with the average gene set expression. The most predictive markers for M/G1 phase cells were key markers with a low expression in the other phases (G1/S, S, G2/M). Cells were quantile normalized in expression, which preserves the order in expression levels between genes within a cell. We then measured the cell cycle phase score of each cell as the average relative expression Eri,Sj of the selected cell cycle phase markers, where the M/G1 score was multiplied by −1, as it consisted of lowly expressed cell cycle markers for other phases. We used these scores to assign single cells to phases of the cell cycle, according to their maximal score for the four cell cycle phases.
Cell cycle phase-specific expression
Phase-specific expression for each gene i that peaked in expression in phase j (for example j=M/G1) was defined as Ei,j = Ei,M/G1 = Êi,M/G1−average[Êi,G1/S, Êi,S, Êi,G2/M], where Êi,j represents the average expression of gene in all cells classified as phase j of the cell cycle for a given sorted population of cells. This analysis was repeated for all genes that peaked in expression in one of four possible phases of the cell cycle (M/G1, G1/S, S and G2/M). Bar plots for cell cycle phase-specific expression in different cell types display the mean phase specific expression for a given gene set (all genes, cell cycle genes or differentially expressed cell cycle genes); error bars represent one standard deviation.
Supplementary Material
Acknowledgements
We thank all members of the Meissner and Shalek laboratories for their support and feedback.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.M.T., A.K.S., A.M.; Methodology: M.H.W., V.A., A.M.T., A.A., J.C., S.J.A., B.E.M., R.S.D., A.K.S., A.M.; Validation: M.H.W., V.A., A.M.T., S.J.A., B.E.M., R.S.D., A.K.S., A.M.; Formal analysis: A.M.T., J.C., M.H.W., S.J.A.; Investigation: A.M.T., A.K.S., A.M.; Resources: M.H.W., V.A., A.A., J.C., Z.D.S., T.S.M.; Data curation: A.M.T., A.A., J.C.; Writing - original draft: A.M.T., M.H.W., A.K.S., A.M.; Writing - review & editing: A.M.T., J.C., A.K.S, A.M.; Visualization: A.M.T.; Supervision: A.K.S., A.M.; Project administration: A.K.S., A.M.; Funding acquisition: A.K.S., A.M.
Funding
A.M.T. is supported, in part, by the Icahn School of Medicine at Mount Sinai internal seed funding and the Chan Zuckerberg Initiative (CZI). A.M. was supported by the National Institutes of Health (NIH) (P01GM099117 and 1DP3K111898) and the Max Planck Society. A.K.S. was supported, in part, by the Searle Scholars Program, the Beckman Young Investigator Program, the Pew-Stewart Scholars Program for Cancer Research, a Sloan Fellowship in Chemistry and the NIH (2RM1HG006193). Deposited in PMC for release after 12 months.
Data availability
All data have been deposited in GEO under accession number GSE134483.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.174722.supplemental
References
- Bolger A. M., Lohse M. and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114 -2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. M., Smith Z. D., Grosswendt S., Kretzmer H., Norman T. M., Adamson B., Jost M., Quinn J. J., Yang D. and Jones M. G. (2019). Molecular recording of mammalian embryogenesis. Nature 570, 77-82. 10.1038/s41586-019-1184-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford C. A., Ziller M. J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A. K., Kelley D. R., Shishkin A. A., Issner R. et al. (2013). Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149-1163. 10.1016/j.cell.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Buettner F. and Theis F. J. (2015). Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989-2998. 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- Hansen K. D., Timp W., Bravo H. C., Sabunciyan S., Langmead B., Mcdonald O. G., Wen B., Wu H., Liu Y., Diep D. et al. (2011). Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768 10.1038/ng.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P. and Simpson P. (1991). The choice of cell fate in the epidermis of Drosophila. Cell 64, 1083-1092. 10.1016/0092-8674(91)90263-X [DOI] [PubMed] [Google Scholar]
- Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A. and Young R. A. (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934-947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin D. A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A. et al. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776-779. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson G., Pujadas E., Goutsias J. and Feinberg A. P. (2017). Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat. Genet. 49, 719 10.1038/ng.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27-30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Kim Y. R., Yoo N. J. and Lee S. H. (2013). Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS 121, 85-94. 10.1111/j.1600-0463.2012.02940.x [DOI] [PubMed] [Google Scholar]
- Klein A. M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D. A. and Kirschner M. W. (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187-1201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M. S., Tirosh I., Heckl D., Rao T. N., Dixit A., Haas B. J., Schneider R. K., Wagers A. J., Ebert B. L. and Regev A. (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860-1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. M., Cahan P., Shalek A. K., Satija R., Daleykeyser A. J., Li H., Zhang J., Pardee K., Gennert D., Trombetta J. J. et al. (2014). Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516, 56-61. 10.1038/nature13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. and Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Karnik R., Gu H., Ziller M. J., Clement K., Tsankov A. M., Akopian V., Gifford C. A., Donaghey J., Galonska C. et al. (2015). Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47, 469-478. 10.1038/ng.3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcadams H. H. and Arkin A. (1997). Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA 94, 814-819. 10.1073/pnas.94.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdavid A., Finak G., Chattopadyay P. K., Dominguez M., Lamoreaux L., Ma S. S., Roederer M. and Gottardo R. (2013). Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461-467. 10.1093/bioinformatics/bts714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro D. T., Haber A. L., Biton M., Vinarsky V., Lin B., Birket S. E., Yuan F., Chen S., Leung H. M., Villoria J. et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319 10.1038/s41586-018-0393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Venkatasubramanian M., Chaudhri V. K., Aronow B. J., Salomonis N., Singh H. and Grimes H. L. (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698-702. 10.1038/nature19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S. and Vallier L. (2013). The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135-147. 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Madrigal P., Bertero A. and Vallier L. (2016). Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev. 30, 421-433. 10.1101/gad.271452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsgard D., Reinius B., Deng Q., Panula S. P., Codeluppi S., Reyes A. P., Linnarsson S., Sandberg R. and Lanner F. (2016). Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 167, 285 10.1016/j.cell.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O. R., Björklund A. K., Winberg G., Sagasser S. and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171-181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J. I., Matallanas D., von Kriegsheim A. et al. (2016). Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell 19, 491-501. 10.1016/j.stem.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Avgustinova A., Martín M., Datta D., Solanas G., Prats N. and Benitah S. A. (2017). Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-γ. eLife 6, e21697 10.7554/eLife.21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N. K., Macaulay I. C., Marioni J. C. and Göttgens B. (2016). Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 289-293. 10.1038/nature18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A. K., Satija R., Adiconis X., Gertner R. S., Gaublomme J. T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D. et al. (2013). Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236-240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek A. K., Satija R., Shuga J., Trombetta J. J., Gennert D., Lu D., Chen P., Gertner R. S., Gaublomme J. T., Yosef N. et al. (2014). Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363-369. 10.1038/nature13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K., Lapan S. W., Whitney I. E., Tran N. M., Macosko E. Z., Kowalczyk M., Adiconis X., Levin J. Z., Nemesh J., Goldman M. et al. (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308-1323.e30. 10.1016/j.cell.2016.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush L. I., Zandi S., Mitchell A., Chen W. C., Brandwein J. M., Gupta V., Kennedy J. A., Schimmer A. D., Schuh A. C., Yee K. W. et al. (2014). Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer Z. S., Yong J., Tischler J., Hackett J. A., Altinok A., Surani M. A., Cai L. and Elowitz M. B. (2014). Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319-331. 10.1016/j.molcel.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. D. and Meissner A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204-220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E. and Dudoit S. (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A. and Regev A. (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331-338. 10.1038/nature21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K. and Surani M. A. (2010). Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6, 468-478. 10.1016/j.stem.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E. and Widschwendter M. (2012). Differential variability improves the identification of cancer risk markers in DNA methylation studies profiling precursor cancer lesions. Bioinformatics 28, 1487-1494. 10.1093/bioinformatics/bts170 [DOI] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S. M., Wadsworth M. H. II, Treacy D., Trombetta J. J., Rotem A., Rodman C., Lian C., Murphy G. et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189-196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S. and Rinn J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381-386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D. G., Wu A. R., Neff N. F., Mantalas G. L., Espinoza F. H., Desai T. J., Krasnow M. A. and Quake S. R. (2014). Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371-375. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta J. J., Gennert D., Lu D., Satija R., Shalek A. K. and Regev A. (2014). Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1-4.22.17. 10.1002/0471142727.mb0422s107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A. M., Akopian V., Pop R., Chetty S., Gifford C. A., Daheron L., Tsankova N. M. and Meissner A. (2015a). A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 33, 1182-1192. 10.1038/nbt.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A. M., Gu H., Akopian V., Ziller M. J., Donaghey J., Amit I., Gnirke A. and Meissner A. (2015b). Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344-349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., Alexander K. E., Matese J. C., Perou C. M., Hurt M. M., Brown P. O. et al. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977-2000. 10.1091/mbc.02-02-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Rodriguez B., Mayle A., Park H. J., Lin X., Luo M., Jeong M., Curry C. V., Kim S.-B., Ruau D. et al. (2016). DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell 29, 922-934. 10.1016/j.ccell.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., Inoue T., Ohuchi H., Semina E. V., Murray J. C. et al. (1998). Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94, 299-305. 10.1016/S0092-8674(00)81473-7 [DOI] [PubMed] [Google Scholar]
- Ziller M. J., Ortega J. A., Quinlan K. A., Santos D. P., Gu H., Martin E. J., Galonska C., Pop R., Maidl S., Di Pardo A. et al. (2018). Dissecting the functional consequences of de novo DNA methylation dynamics in human motor neuron differentiation and physiology. Cell Stem Cell 22, 559-574.e9. 10.1016/j.stem.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.