Abstract
Crystallography has long been the unrivaled method that can provide the atomistic structural models of macromolecules, using either X-rays or electrons as probes. The methodology has gone through several revolutionary periods, driven by the development of new sources, detectors, and other instrumentation. Novel sources of both X-ray and electrons are constantly emerging. The increase in brightness of these sources, complemented by the advanced detection techniques, has relaxed the traditionally strict need for large, high quality, crystals. Recent reports suggest high-quality diffraction datasets from crystals as small as a few hundreds of nanometers can be routinely obtained. This has resulted in the genesis of a new field of macromolecular nanocrystal crystallography. Here we will make a brief comparative review of this growing field focusing on the use of X-rays and electrons sources.
Keywords: nanocrystallography, X-ray free-electron laser, electron diffraction
1. Introduction
In the process of interrogating structure and function of molecules with probes like X-rays and electrons, it was found that putting them in a regular array amplifies the scattered signal to an extent as large as the square of the number of molecules in the array [1]. The technique for determination of the structure of protein molecules from their crystal diffraction is broadly termed as macromolecular crystallography. Conventional macromolecular crystallography needs large crystals of high quality in order to obtain diffraction maps, which can generate interpretable structures. With the emergence of new sources of both X-rays and electrons, the necessity for big crystals is in the process of being relaxed. Recently, experiments have been performed with crystals as small as a few hundred nanometers [2,3]. This opens the possibility of obtaining structures of many molecules, such as membrane proteins, for which growing high quality crystals of sufficient size has been found to be extremely challenging. The capability to obtain structures from such tiny nanocrystals has been demonstrated with both X-ray and electrons. The method of solving the structure of macromolecules with such tiny crystals has been introduced as nanocrystallography and will be the primary content of this review.
An ideal structural method for biological macromolecules would be the one that would probe a single macromolecule and provide atomistic resolution structural information. However, despite progress and significant efforts in single particle imaging methods with X-rays [4] and electrons [5], this feat has not yet been achieved and crystallography remains unrivalled for structural determination. In crystallography, the structure obtained is the average of many identical copies of the molecules, sharing the dose requirement amongst the molecules [6]. However, arranging a large number of molecules in a regular array rarely gives a defect-free crystal. The smaller the crystal, the lesser the defect is and hence, high quality data can be obtained from small crystals [7]. This goal is largely accomplished with the emergence of nanocrystallography. In this review, we strictly define nanocrystal as the crystals which are of submicron size. Keeping crystals to this size, the number of molecules over which the signal is averaged is decreased by six orders of magnitude, or more, compared to traditional standard crystallography. The field of nanocrystallography is still in its infancy. Predictions have been made that with current generation pulsed X-rays and coherent electron beams, crystals of protein molecules with as little as few tens of unit cells in each dimension will be sufficient to obtain structural insights at high resolution [8]. The field of nanocrystallography has progressed with both electrons and X-rays. Here, we will briefly review the development in both domains. A comparison between electron and X-ray crystallography applied to nanocrystals will be made in order to introduce them as complementary techniques. Challenges present in the field will be reviewed and we will offer a glance to the future of this emerging field.
2. Nanocrystal X-Ray Crystallography
The origin of X-ray crystallography dates back to 1912 when Max Von Laue detailed the physics of the interaction of crystalline material with electromagnetic radiation, specifically with X-rays [9]. Since then, the method has undergone stupendous development and has become an indispensable tool in the study of physical and chemical properties of the materials. For biological sciences, it was only after Watson and Crick showed that fiber diffraction could be useful in determining the structure of DNA, that the method started to grow exponentially [10]. This was followed by the historic results from Max Perutz and John Kendrew, solving the atomistic structure of hemoglobin and myoglobin, respectively, exploiting X-ray crystal diffraction [11,12]. In the 1970s, the first series of synchrotrons appeared and this boosted the field of protein crystallography, reducing the size of the crystals necessary to get atomistic models [13]. Another significant boost was received in the 1990s when third-generation synchrotron came into operation and started delivering microbeams in routine fashion. With this development, the necessary crystal size shrunk even further into tens of micron and the method of micro-crystallography gained wide popularity in the macromolecular crystallography community. In the last decade, the powerful femtosecond X-ray lasers have caused a revolution in the X-ray community, delivering a peak power ten orders of magnitude or more higher than previously available X-ray sources [14,15,16]. In addition to many other scientific achievements, this has also resulted in a further decrease in the necessary crystal volume. Currently, submicron crystals are routinely measured at such facilities and a new field of X-ray nanocrystallography has been further developed.
Protein nanocrystallography with the synchrotron X-rays has already been discussed in the early 2000s [17,18]. In these approaches, use of nanotechnologies have been made for the growth and characterization of nanocrystals and the micro-focused beam from the synchrotron X-rays was used for data collection. The Langmuir–Blodgett nanotemplate method appeared as a valuable method in growing stable micro/nanocrystals of proteins which had not been previously crystallized [19].
Although discussed theoretically already in 2000 [20], the field of X-ray nanocrystallography with ultrashort (shorter than the damage formation in the sample) pulses was experimentally verified with the seminal paper from Henry Chapman and colleagues where they shot streams of submicron-sized crystals of photosystem II with the ultrashort ultra-bright pulses of the LCLS X-ray free-electron laser [2]. In these experiments, crystals as small as ~200 nm were shot to collect diffraction data. This method of shooting streams of tiny crystals with pulses of X-ray laser is widely known as serial femtosecond crystallography (SFX) [21,22]. The initial trials resulting in the low-resolution diffraction datasets using the SFX method were soon extended to reach atomic resolution [23,24,25,26]. In vivo protein crystallization found its way to nanocrystallography with the seminal works of Koopmann et al., [27]. Further, the structure of membrane protein from nanocrystals grown in lipidic cubic phase was solved with SFX [28,29]. Johansson et al., demonstrated the usefulness of the technique to solve the structure of the photosynthetic reaction center [30] and the same year Barends et al. demonstrated the solution of a de novo protein crystal structure [31]. Demicri et al. demonstrated the extension of the method to solve structures from ribosome microcrystals [32]. Young et al. reported the structure of photosystem II and the substrate binding at room temperature [33]. Room temperature studies of bacterial phytochrome were reported by Edlund et al. [34] and the crystal structure of CO-Bound Cytochrome C oxidase was reported at room temperature [35]. The crystal structure of rhodopsin bound to Arrestin was reported by Kang et al. [36]. Utilizing the ultrashort pulse duration of the X-ray FEL, the method soon found its application in several exciting results in time-resolved studies [37,38]. Time-resolved studies were also made on Photosystem II by Kupitz et al. [39]. Structures of riboswitch intermediate states performed by mix and inject XFEL nanocrystallography were demonstrated [40]. In cellulo crystallography of alcohol oxidase in yeast cells has been performed [41] and nanocrystallography measurement of early stage synthetic malaria pigments has been reported by Dilanian et al. [42]. Interesting applications of the nanocrystallography of in vivo crystallization of Cathepsin B were performed [43]. Recent experiments on mosquito larvicide BinAB nanocrystals have also been reported [44]. Gati et al. have reported 2 Å resolution structure from native nanocrystalline granulovirus occlusion bodies [45]. Though most of these data have been collected in on-flight mode (from samples in a liquid jet), a few instances of goniometer-based data collection have also been reported for X-ray nanocrystallography [46]. Furthermore, the use of two-dimensional nanosized crystals has been reported in a few instances [47]. The X-ray nanocrystallography, so far, has been realized predominantly with the X-ray free-electron lasers and significant developments in sample delivery and data analysis have been made to support this development [48,49]. A schematic of the protein nanocrystallography with XFEL is shown in Figure 1. With the emergence of the new diffracted limited storage rings [50], supplemented by supremely efficient nano-focusing optics, we may soon be witnessing the realization of nanocrystallography at next-generation synchrotrons as well. Such a development would make X-ray nanocrystallography more accessible to a large number of users and will make an even more pronounced impact on the structural biology community.
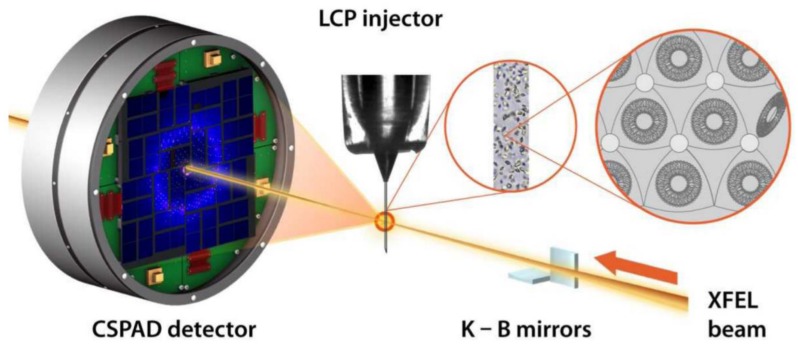
Figure 1.
Experimental setup for serial femtosecond crystallography (SFX) data collection using an LCP injector. Microcrystals dispersed in LCP are injected as a continuous column of 20–50 μm diameter and intersected with 1.5 μm diameter pulsed XFEL beam focused by Kirkpatrick–Baez (KB) mirrors. Single pulse diffraction patterns are collected at 120 Hz using a CSPAD detector. (Adapted from Liu et al. [28]).
Data treatment for X-ray serial nanocrystallography differs from the one used for traditional single crystal diffraction methods. The diffraction data are collected from many randomly oriented crystals and each diffraction pattern is considered as “stills”. Existing auto-indexing algorithms are used for finding the orientation of each crystal. The coherent nature of the source can result in the presence of the intensities from the shape transforms of the crystal around the Bragg peak. Optimized peak searching algorithms are used for such diffraction data. Due to the inhomogeneity in the crystal shape, size and orientation Monte–Carlo integration of the intensities around the Bragg peak is performed to obtain the structure factor, which can be used to obtain the electron density of the molecule of interest. A detailed description of the data treatment for XFEL nanocrystallography can be found in Kirian et al. [51].
3. Nanocrystal Electron Crystallography
The history of electron crystallography is relatively new. The endeavor of electron crystallography began with the realization by Aaron Klug that the phases can be extracted from the Fourier transform of the electron micrographs [52]. His efforts on the development of electron crystallography and the study of nucleic acid complexes were recognized with the Nobel Prize for chemistry in 1982. For protein crystals, it began only in the 1970s when Richard Henderson used it in solving the structure of bacteriorhodopsin from their naturally occurring two-dimensional crystals [53]. In 2005, the structure of two-dimensional Aquaporin was solved to a resolution of 1.9 Å [54]. The field of electron crystallography remains largely limited to two-dimensional crystals, although some instances of electron crystallography of microcrystals were reported in the mid-1970s by Dorset et al. [55]. Recent developments in electron source, electron optics, and detection techniques, as well as in software, have enabled electron crystallography of three-dimensional micro-crystals. This field is widely known as MicroED [56]. Owing to the fact that electrons scatter strongly, a crystal of submicron size is preferred to avoid the loss of information by multiple scattering effects. Structures from crystals, as small as a few hundred of nanometers, have been demonstrated with electron diffraction recently. This marks the beginning of the highly potential field of electron nanocrystallography.
The MicroEd technique in the last few years has largely focused on methodology development. After the demonstration of electron crystallography for three dimensional crystals in 2013 by Shi et al. [57], significant progress has been made in data acquisition and treatment. Large efforts have also been made in crystal preparation, such as producing the lamella of the crystals in order to reduce the thickness of the crystals in the beam propagation direction [58,59]. The recent progress in electron crystallography has not only been facilitated by the developments in source and detection techniques. Revolutionary developments have also been made in the techniques for sample delivery to the electron beam. The flash freezing technique, commonly employed in the cryo electron microscopy system, has been recently recognized with the Nobel Prize for Chemistry in 2017 [60]. By this technique it is possible to keep the sample in the near-native condition and minimize radiation damage issues. These developments have been translated to the electron crystallography community. Many reviews on MicroEd have been written in recent times [61,62,63], where additional information can be found. Some of the recent works on MicroED include solving the structure of the toxic core of alpha-synuclein [64], building the atomic model with charges [65], solving the structures of HIV-1 Gag CTD-SP1 [66], atomic resolution structures from fragmented protein crystals [67], and solving the previously unobserved polymorph of hen egg-white lysozyme [68]. Xu et al. recently reported the solution of the structure of new protein R2lox using MicroED [69]. Recently, structures from microcrystals embedded in the lipidic cubic phase have been solved using MicroED [70].
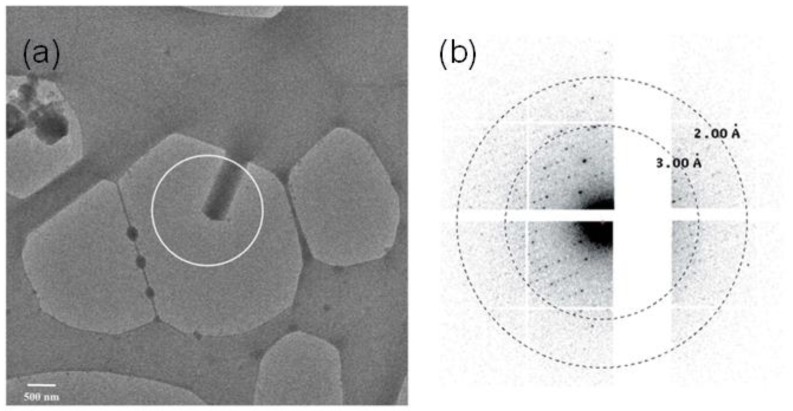
An ideal case for electron crystallography would be with nanocrystals, as multiple scattering events are expected to be minimized with such samples. With the use of nanocrystals, no intense sample preparation, such as ion beam milling, will be required and nanocrystallography represents a very promising niche for electron crystallography. A few instances of electron crystallography with such intrinsic nanocrystals have been reported. In 2013, Igor et al. reported the first such case [71]. Lysozyme nanocrystals with a thickness of the order of 100 nm were reported to collect diffraction maps at a resolution higher than 2 Å. In 2016, Sawaya et al. reported the first ab initio structure determined from prion nanocrystals at atomic resolution [72]. In 2017, Clabbers et al., reported electron crystallography with crystals as small as 140 nm3 in volume. It was reported that such crystals diffracted to 2.1 Å resolution [3]. An image of the nanocrystal and the diffraction pattern reported in the article is shown in Figure 2a,b respectively. Electron nanocrystallography has also been performed with automated diffraction tomography [73,74] and rotation electron diffraction [75]. However, these techniques, so far, have been limited to the nanocrystals of inorganic and organic materials which are relatively radiation-hard compared to the biological samples.
Figure 2.
(a) Electron micrograph of a nanocrystal and (b) the diffraction from the selected area in the micrograph. (Adapted from Clabbers et al. [3]).
Data analysis in electron nanocrystallography is not straightforward. Relatively flatter Ewald’s sphere, imprecise tilt measurement, and distortion in measured intensities by the electron lens makes data analysis challenging with the existing software used for single-crystal X-ray crystallography. For the first experiments in MicroED, where the still diffraction patterns were recorded, the authors used self-developed software to perform the data reduction [76]. Lately, collection of electron diffraction in continuous rotation mode has been performed [77,78]. With this mode of operation, and making specific changes to the electron crystallography, standard software such as MOSFLM [79], DIALS [80], and XDS [81] have been reported to be used in the treatment of electron crystallography data. Following the data integration, merging, and scaling, standard crystallographic suites can be used for phasing and structure refinement.
Crystallography with electrons is not limited to the continuous sources. Ultrafast electron sources are also under constant development [82]. Several recent works have been reported on ultrafast electron nanocrystallography of inorganic and small molecule crystals [83,84,85]. For macromolecules, a notable improvement in the source parameters, such as flux and coherence, is needed in order to realize ultrafast electron nanocrystallography.
4. Advantage of Nanocrystallography
Although the field of nanocrystallography has emerged in parallel with nanotechnology, some of its advantages are already obvious. The primary benefit of nanocrystallography is that it can be applied to the broad category of molecules for which high-quality crystals do not grow into sufficiently big sizes suitable for standard synchrotron-based X-ray crystallography. Such molecules could be membrane proteins, intrinsically disordered proteins, and molecular complexes. The possibility of getting atomic structures from nanocrystals also opens the application of crystallography to in vivo studies of naturally occurring nanocrystals, which is preferable for the study of the structure and function of proteins in their naturally occurring state. Nanocrystallography also opens the possibility of solving the structure of proteins, naturally occurring in trace amounts.
The field of nanocrystallography has also opened up the possibility of exploiting novel phasing options. Coherent phasing of crystals, which was envisioned by David Sayre in the early 1950s, can now be realized with the introduction of nanocrystallography, as highly brilliant coherent sources of X-rays and electrons become available [86,87]. Such phasing would relax the necessity of adding heavy atoms to the molecules during crystallization or making use of multiple wavelength measurements. Such a direct phasing method would also not need any homologous structures to solve the phase.
Smaller volume crystals are also desired for pump–probe crystallography. In a small sample, a larger fraction of the molecules can also be reached when the pump beam has limited penetration depth. This is, for example, the case when terahertz radiation is used to activate the sample. In such demanding cases of pump–probe experiments, a small sample volume ensures that the molecules in the crystals are homogeneously excited by the pump field. This allows a more accurate interpretation of the structure and function of the protein in time-resolved studies.
Protein crystals are never perfect, they usually occur in poorly aligned blocks of ordered arrangements. In crystallographic terms, they are always mosaic. In standard continuous rotation crystallography, mosaicity is supposed to present additional challenges in the structural solution and the larger the crystal size, the higher the mosaicity. This is unarguably true when seeding is done in growing crystals of suitable size to be used for single crystal X-ray diffraction. With the introduction of nanocrystallography, the mosaicity of the crystal has less of an effect on the structure solution [88]. In addition to these obvious benefits, many presently unseen benefits of nanocrystallography can be expected to appear as the field develops towards maturity.
5. Challenges in Nanocrystallography
There are challenging issues in nanocrystallography as well. A common issue in micro and macro crystallography is to find suitable conditions to grow protein crystals of sufficiently large size. Similar challenges persist with nanocrystallography. Apart from the actual growth of the crystal, a critical challenge arises in the screening of the crystal, as no noninvasive technique exists to observe such tiny crystals. Alignment of the nanocrystals with the micro-focused or nano-focused X-ray or electron beam presents additional challenges. The use of a large number of crystals for SFX experiments has also raised concerns about sample consumption. This concern persists, although the increase in XFEL rep rates to the MHz range [89] and the development of sample delivery systems with flow rates optimized for the different repetition rates of different X-ray sources [90], as well as concepts for online hit finding [91], are addressing these topics. Sample clogging in the jet and the possibility of mechanical damage to the crystal are some other challenges in femtosecond X-ray nanocrystallography. Additionally, growing such a large number of crystals with homogeneous sizes is a daunting task. X-ray nanocrystallography with solid support presents challenges such as reduced signal to noise ratio and modulation of the diffraction signal by the refracted signal from the support.
With the smaller number of molecules in the beam and the trade-offs necessary to minimize the radiation damage (unless ultrashort pulses are used), the number of electrons and photons reaching the detector after interacting with the sample is often relatively small. This brings challenges to the detection techniques as all signals reaching the detector should be recorded and differentiated from the various sources of noise. With the emergence of the new field of nanocrystallography, new challenges also materialize on the side of data analysis and interpretation. In response to this, a completely new set of data analysis tools have been introduced for the X-ray free-electron laser-based nanocrystallography [92,93,94]. For electron diffraction experiments, methods to circumvent the multiple scattering have to be developed. Furthermore, the progress in the development of dynamic refinement tools has to be accelerated. Several groups have responded to these challenges and are making progress at an exciting pace [95,96,97]. A summary of the current challenges in nanocrystallography is presented in Table 1. With all these challenges addressed, the techniques will be more ready for the study of the structure and function of new proteins.
Table 1.
Challenges in nanocrystallography.
| Nanocrystallography | |
|---|---|
| Screening, crystal identification | Need for development of less invasive tools for screening and crystal identification. |
| Detector Requirements | The photons reaching detector are sparse with small crystals. Necessity of sample background reduction. |
| Radiation damage | Methods to minimize radiation damage in nanocrystals with continuous and pulsed X-ray and electron sources have to be found. One approach is to use ultrashort pulses |
| Data analysis | Challenging due to datasets from multiple crystals in unknown orientation. |
6. Electron Vs. X-Rays
For a long time, electron and X-ray diffraction have been seen as rivaling techniques. Both techniques are well-described with their individual merits and demerits in various earlier literature. Here we present both tools as complementary techniques for nanocrystallography.
Compared to X-rays, electrons scatter strongly [98]. This presents both advantages and challenges with electrons. They can scatter efficiently from a small volume of molecules. However, for the size of the sample from which high resolution structural information can be obtained, the data may still be affected by multiple scattering effects. Radiation damage is another issue that can be especially serious with electrons and it can lead to low resolution or false interpretation of the structures. Several key technologies have been introduced in the past decades to solve this issue. However, the employment of such techniques will prevent room temperature measurements, which are important to obtain the physiologically relevant structures. Although the development of ultrafast electron sources is in a state of rapid progress, the field of electron nanocrystallography is still largely limited to static studies or studies of structures relevant to slow phenomena.
Due to the fact that the electron interacts with the Coulomb potential in the atom, it has proven to be useful in locating the position of hydrogen atoms and other ions in the structure more precisely. This is otherwise extremely challenging with the X-rays. Furthermore, the availability of instruments for electron nanocrystallography in university scale laboratories makes it a more readily accessible resource than the X-ray free-electron lasers used for X-ray nanocrystallography.
X-ray nanocrystallography, in present days is mainly realized with X-ray free-electron laser. These sources use extremely short pulses with very high intensity. In the SFX experiment the nanocrystals are ionized and destroyed after the scattered field leaves the crystal. In this method, generally termed “diffraction before destruction” the effects of radiation damage on the sample structure are limited as long as the X-ray pulse duration is sufficiently short. [16,99]. The use of ultrashort pulses makes it ideal for time-resolved studies, but the large number of shots needed for structural determination comes with the additional challenges in data management and treatment. Statistical scaling of intensities from the partial reflections in the data may lead to misinterpretations of the data. On the good side, the measurements can be done at room temperature. This makes it more likely to obtain physiologically relevant structures. All in all, electron and X-ray nanocrystallography have to be taken as complementary tools to take full advantage of the growing field of nanocrystallography. A summary of the comparison of the state of the art X-ray and electron nanocrystallography is presented in Table 2.
Table 2.
Comparison of X-ray and electron nanocrystallography.
| X-Ray Nanocrystallography | Electron Nanocrystallography | |
|---|---|---|
| Source/accessibility | Large Facilities/Less accessible | Laboratory Sources/Frequent accessibility |
| Sample consumption | Large to moderate with serial crystallography | Less |
| Radiation damage | Minimal with X-ray FEL | Large and limits the highest attainable resolution. However, can be overcome by merging a large number of low dose datasets |
| Room temperature studies | Possible with current technologies | Not possible with current technologies |
| Ultrafast Time resolved studies | Possible | Not possible |
| Sensitivity to ions and H-atom | Less sensitive | Highly sensitive |
7. Future Prospects in Nanocrystallography
Nanocrystallography, both with electrons and X-rays, is a field in its infancy and further developments in crystal growth, sample screening and delivery, data collection, and data analysis have to be made before it becomes a routine method for solving the structure of proteins. Nanocrystals are suitable to study the static and dynamic structure of the molecules that are difficult to grow into larger crystals. Progress in screening techniques has to be made in order to ensure that nanocrystallography becomes a standard tool in structural biology. Work on data analysis in nanocrystallography can result in novel phasing methods that could solve issues with several protein crystals that cannot otherwise be phased. Pulsed X-ray and electron sources offer new possibilities for time-resolved studies when combined with nanocrystallography. This development is presently well underway at X-ray FELs. New laser-driven X-ray and electron sources operating in the femtosecond pulsed regime, like those developed for user operation at the ELI Beamlines facility, will offer additional capacities for pump–probe experiments at a high level of synchronization and control. Overall, the field of nanocrystallography is expanding to complement other biophysical techniques in the quest to increase our knowledge of the macromolecules.
Acknowledgments
We would like to thank Dr. Vitaly Polovinkin for several fruitful discussions. J.A. acknowledges support from the Chalmers Area of Advance in Material Science.
Funding
This research was funded by Czech Academy of Sciences (Grant number: MSM100101901),Czech Science Foundation project No.17-00973S, Structural dynamics of biomolecular systems (CZ.02.1.01/0.0/0.0/15_003/0000447) and advanced research using high-intensity laser produced photons and particles (CZ.02.1.01/0.0/0.0/16_019/0000789) from the European Regional Development Fund. The results of the Project LQ1606 were obtained with the financial support of the Ministry of Education and Youth and Sports of Czech Republic, as part of targeted support from the National Program of Sustainability II.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Drenth J. Principles of Protein X-Ray Crystallography. 3rd ed. Springer; New York, NY, USA: 2006. [Google Scholar]
- 2.Chapman H.N., Fromme P., Barty A., White T.A., Kirian R.A., Aquila A., Hunter M.S., Schulz J., DePonte D.P., Weierstall U., et al. Femtosecond x-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clabbers M.T.B., Genderen E.V., Wan W., Wiegers E.L., Gruene T., Abrahams J.P. Protein structure determination by electron diffraction using a single three dimensional nanocrystal. Acta Cryst. Sect. D. 2017;73:738–748. doi: 10.1107/S2059798317010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aquila A., Barty A., Bostedt C., Boutet S., Carini G., dePonte D., Drell P., Doniach S., Downing K.H., Earnest T., et al. The linac coherent light source single particle imaging road map. Struct. Dyn. 2015;2:041701. doi: 10.1063/1.4918726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai X.C., McMullan G., Scheres S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Duke E. Macromolecular crystallography at synchrotron radiation sources: Current status and future developments. Proc. R. Soc. A. 2010;466:3421–3452. doi: 10.1098/rspa.2010.0448. [DOI] [Google Scholar]
- 7.Cusack S., Belrhali H., Bram A., Burghammer M., Perrakis A., Riekel C. Small Is Beautiful: Protein Micro-Cryst. Nat. Struct. Biol. 1998;5:634–637. doi: 10.1038/1325. [DOI] [PubMed] [Google Scholar]
- 8.Hunter M.S., DePonte D.P., Shapiro D.A., Kirian R.A., Wang X., Starodub D., Marchesini S., Weierstall U., Doak R.B., Spence J.C., et al. X-ray diffraction from membrane protein nanocrystals. Biophys. J. 2011;100:198–206. doi: 10.1016/j.bpj.2010.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laue M.V. Eine quantitative prüfung der theorie für die interferenz-erscheinungen bei Röntgenstrahlen. Ann. Phys. 1913;346:989–1002. doi: 10.1002/andp.19133461005. [DOI] [Google Scholar]
- 10.Watson J.D., Crick F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 11.Perutz M.F., Rossman M.G., Cullis A.F., Muirhead H., Will G., North A.C. Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-Å resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 12.Kendrew J.C., Dickerson R.E., Strandberg B.E., Hart R.G., Davies D.R., Philips D.C., Shore V.C. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 13.Dauter Z., Jaskolski M., Wlodawer A. Impact of synchrotron radiation on macromolecular crystallography: A personal view. J. Synchrotron Radiat. 2010;17:433–444. doi: 10.1107/S0909049510011611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emma P., Akre R., Arthur J., Bionta R., Bostedt C., Bozek J., Brachmann A., Bucksbaum P., Coffee R., Decker F.J., et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nat. Photon. 2010;4:641–647. doi: 10.1038/nphoton.2010.176. [DOI] [Google Scholar]
- 15.Yabashi M., Tanaka H., Ishikawa T. Overview of the SACLA facility. J. Synchrotron Radiat. 2015;22:477–484. doi: 10.1107/S1600577515004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altarelli M., Brinkmann R., Chergui M., Decking W., Dobson B., Düsterer S., Grübel G., Graeff W., Graafsma H., Hajdu J., et al. The European X-Ray Free-Electron Laser Technical Design Report. DESY; Hamburg, Germany: 2007. [DOI] [Google Scholar]
- 17.Pechkova E., Nicolini C. Protein nanocrystallography: A new approach to structural proteomics. Trends Biotechnol. 2004;22:117–122. doi: 10.1016/j.tibtech.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Nicolini C., Pechkova E. Nanocrystallography: An emerging technology for structural proteomics. Expert Rev. Proteom. 2004;1:253–256. doi: 10.1586/14789450.1.3.253. [DOI] [PubMed] [Google Scholar]
- 19.Pechkova E., Nicolini C. Langmuir-Blodgett nanotemplates for protein crystallography. Nat Protoc. 2017;12:2570–2589. doi: 10.1038/nprot.2017.108. [DOI] [PubMed] [Google Scholar]
- 20.Neutze R., Wout R., van der Spoel D., Weckert E., Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 21.Schlichting I. Serial femtosecond crystallography: The first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson L.C., Stauch B., Ishchenko A., Cherezov V. A Bright Future for Serial Femtosecond Crystallography with XFELs. Trends Biochem. Sci. 2017;42:749–762. doi: 10.1016/j.tibs.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nango E., Royant A., Kubo M., Nakane T., Wickstrand C., Kimura T., Tanaka T., Tono K., Song C.Y., Tanaka R., et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science. 2016;354:1552–1557. doi: 10.1126/science.aah3497. [DOI] [PubMed] [Google Scholar]
- 24.Boutet S., Lomb L., Williams G.J., Barends T.R., Aquila A., Doak R.B., Weierstall U., DePonte D.P., Steinbrener J., Shoeman R.L., et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., Shen J.-R. Native structure of photosystem II at 1.95A° resolution viewed by femtosecond X-ray pulses. Nature. 2015;517:99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- 26.Masuda T., Suzuki M., Inoue S., Song C., Nakane T., Nango E., Tanaka R., Tono K., Joti Y., Kameshima T., et al. Atomic resolution structure of serine protease proteinase K at ambient temperature. Sci. Rep. 2017;7:45604. doi: 10.1038/srep45604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopmann R., Cupelli K., Reduce L., Nass K., Deponte D.P., White T.A., Stellato F., Rehders D., Liang M., Andreasson J., et al. In vivo protein crystallization opens new routes in structural biology. Nat. Methods. 2012;9:259–262. doi: 10.1038/nmeth.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W., Wacker D., Gati C., Han G.W., James D., Wang D., Nelson G., Weierstall U., Katritch V., Barty A., et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weierstall U., James D., Wang C., White T.A., Wang D., Liu W., Spence J.C., Doak R.B., Nelson G., Fromme P., et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson L.C., Arnlund D., Katona G., White T.A., Barty A., DePonte D.P., Shoeman R.L., Wickstrand C., Sharma A., Williams G.J., et al. Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nat. Commun. 2013;4:2911. doi: 10.1038/ncomms3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barends T.R.M., Foucar L., Botha S., Doak R.B., Shoeman R.L., Nass K., Koglin J.E., Williams G.J., Boutet S., Messerschmidt M., et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505:244–247. doi: 10.1038/nature12773. [DOI] [PubMed] [Google Scholar]
- 32.Demirci H., Sierra R.G., Laksmono H., Shoeman R.L., Botha S., Barends T.R., Nass K., Schlichting I., Doak R.B., Gati C., et al. Serial femtosecond X-ray diffraction of 30S ribosomal subunit microcrystals in liquid suspension at ambient temperature using an X-ray free-electron laser. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013;69:1066–1069. doi: 10.1107/S174430911302099X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young I.D., Ibrahim M., Chatterjee R., Gul S., Fuller F., Koroidov S., Brewster A.S., Tran R., Alonso-Mori R., Kroll T., et al. Structure of photosystem II and substrate binding at room temperature. Nature. 2016;540:453–457. doi: 10.1038/nature20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edlund P., Takala H., Claesson E., Henry L., Dods R., Lehtivuori H., Panman M., Pande K., White T., Nakane T., et al. The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography. Sci. Rep. 2016;6:35279. doi: 10.1038/srep35279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishigami I., Zatsepin N.A., Hikita M., Conrad C.E., Nelson G., Coe J.D., Basu S., Grant T.D., Seaberg M.H., Sierra R.G., et al. Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature. Proc. Natl. Acad. Sci. USA. 2017;114:8011–8016. doi: 10.1073/pnas.1705628114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y., Zhou X.E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T.A., Yefanov O., Han G.W., et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aquila A., Hunter M.S., Doak R.B., Kirian R.A., Fromme P., White T.A., Andreasson J., Arnlund D., Bajt S., Barends T.R., et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Opt. Express. 2012;20:2706–2716. doi: 10.1364/OE.20.002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenboer J., Basu S., Zatsepin N., Pande K., Milathianaki D., Frank M., Hunter M., Boutet S., Williams G.J., Koglin J.E., et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kupitz C., Basu S., Grotjohann I., Fromme R., Zatsepin N.A., Rendek K.N., Hunter M.S., Shoeman R.L., White T.A., Wang. D., et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stagno J.R., Liu Y., Bhandari Y.R., Conrad C.E., Panja S., Swain M., Fan L., Nelson G., Li C., Wendel D.R., et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017;541:242–246. doi: 10.1038/nature20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobi A.J., Passon D.M., Knoops K., Stellato F., Liang M., White T.A., Seine T., Messerschmidt M., Chapman H.N., Wilmanns M. In cellulo serial crystallography of alcohol oxidase crystals inside yeast cells. IUCrJ. 2016;3:88–95. doi: 10.1107/S2052252515022927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dilanian R.A., Streltsov V., Coughlan H.D., Quiney H.M., Martin A.V., Klonis N., Dogovski C., Boutet S., Messerschmidt M., Williams G.J., et al. Nanocrystallography measurements of early stage synthetic malaria pigment. J. Appl. Cryst. 2017;50:1533–1540. doi: 10.1107/S1600576717012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redecke L., Nass K., DePonte D.P., White T.A., Rehders D., Barty A., Stellato F., Liang M., Barends T.R.M., Boutet S., et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339:227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colletier J.P., Sawaya M.R., Gingery M., Rodriguez J.A., Cascio D., Brewster A.S., Michels-Clark T., Hice R.H., Coquelle N., Boutet S., et al. De novo phasing with X-ray laser reveals mosquito larvicide BinAB structure. Nature. 2016;539:43–47. doi: 10.1038/nature19825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gati C., Oberthuer D., Yefanov O., Bunker R.D., Stellato F., Chiu E., Yeh S.M., Aquila A., Basu S., Bean R., et al. Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser. Proc. Natl. Acad. Sci. USA. 2017;114:2247–2252. doi: 10.1073/pnas.1609243114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen A.E., Soltis S.M., González A., Aguila L., Alonso-Mori R., Barnes C.O., Baxter E.L., Brehmer W., Brewster A.S., Brunger A.T., et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc. Natl. Acad. Sci. USA. 2014;111:17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedrini B., Tsai C.J., Capitani G., Padeste C., Hunter M.S., Zatsepin N.A., Barty A., Benner W.H., Boutet S., Feld G.K., et al. 7 Å resolution in protein two-dimensional-crystal X-ray diffraction at Linac Coherent Light Source. Philos. Trans. R Soc. Lond. B Biol. Sci. 2014;369:20130500. doi: 10.1098/rstb.2013.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grünbein M.L., Nass Kovacs G. Sample delivery for serial crystallography at free-electron lasers and synchrotrons. Acta Cryst. D Struct. Biol. 2019;75:178–191. doi: 10.1107/S205979831801567X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H., Spence J.C.H. XFEL data analysis for structural biology. Quant Biol. 2016;4:159. doi: 10.1007/s40484-016-0076-z. [DOI] [Google Scholar]
- 50.Eriksson M., Friso J.V., Quitmann C. Diffraction-limited storage rings—A window to the science of tomorrow. J. Synchrotron Rad. 2014;21:837–842. doi: 10.1107/S1600577514019286. [DOI] [PubMed] [Google Scholar]
- 51.Kirian R.A., Wang X., Weierstall U., Schmidt K.E., Spence J.C.H., Hunter M., Fromme P., White T., Chapman H.N., Holton J. Femtosecond Protein Nanocrystallography-Data Anal. Methods. Opt. Express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Rossier D.J., Klug A. Reconstruction of three dimensional structures from Electron Micrographs. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 53.Henderson R., Unwin P.N.T. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 54.Gonen T., Cheng Y., Sliz P., Hiroaki Y., Fujiyoshi Y., Harrison S.C., Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorset D.L., Parsons D.F. Electron diffraction from single, fully-hydrated, ox-liver catalase microcrystals. Acta Cryst. A. 1975;31:210–215. doi: 10.1107/S0567739475000423. [DOI] [Google Scholar]
- 56.Nannenga B.L., Gonen T. MicroED: A versatile cryoEM method for structure determination. Emerg. Top. Life Sci. 2018;2:1–8. doi: 10.1042/ETLS20170082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi D., Nannenga B.L., Iadanza M.G., Gonen T. Three-dimensional electron crystallography of protein microcrystals. Elife. 2013;2:e01345. doi: 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duyvesteyn H.M.E., Kotecha A., Ginn H.M., Hecksel C.W., Beale E.V., de Haas F., Evans G., Zhang P., Chiu W., Stuart D.I. Machining protein microcrystals for structure determination by electron diffraction. Proc. Natl. Acad. Sci. USA. 2018;115:9569–9573. doi: 10.1073/pnas.1809978115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X., Zhang S., Zhang J., Sun F. In situ protein micro-crystal fabrication by cryo-FIB for electron diffraction. Biophys. Rep. 2018;4:339–347. doi: 10.1007/s41048-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson R.F., Walker M., Siebert C.A., Muench S.P., Ranson N.A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods. 2016;100:3–15. doi: 10.1016/j.ymeth.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nannenga B.L., Gonen T. MicroED opens a new era for biological structure determination. Curr. Opin. Struct. Biol. 2016;40:128–135. doi: 10.1016/j.sbi.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martynowycz M.W., Gonen T. From electron crystallography of 2D crystals to MicroED of 3D crystals. Curr. Opin. Colloid Interface Sci. 2018;34:9–16. doi: 10.1016/j.cocis.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nannenga B.L., Gonen T. The cryo-EM method microcrystal electron diffraction (MicroED) Nat. Methods. 2019;16:369–379. doi: 10.1038/s41592-019-0395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez J.A., Ivanova M.I., Sawaya M.R., Cascio D., Reyes F.E., Shi D., Sangwan S., Guenther E.L., Johnson L.M., Zhang M., et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yonekura K., Kato K., Ogasawara M., Tomita M., Toyoshima C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proc. Natl. Acad. Sci. USA. 2015;112:3368–3373. doi: 10.1073/pnas.1500724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Purdy M.D., Shi D., Chrustowicz J., Hattne J., Gonen T., Yeager M. MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc. Natl. Acad. Sci. USA. 2018;115:13258–13263. doi: 10.1073/pnas.1806806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De la Cruz M.J., Hattne J., Shi D., Seidler P., Rodriguez J., Reyes F.E., Sawaya M.R., Cascio D., Weiss S.C., Kim S.K., et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat. Methods. 2017;14:399–402. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lanza A., Margheritis E., Mugnaioli E., Cappello V., Garau G., Gemmi M. Nanobeam precession-assisted 3D electron diffraction reveals a new polymorph of hen egg-white lysozyme. IUCrJ. 2019;6:178–188. doi: 10.1107/S2052252518017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu H., Lebrette H., Clabbers M., Zhao J., Griese J.J., Zou X., Högbom M. Solving a new R2lox protein structure by microcrystal electron diffraction. Sci. Adv. 2019;5:eaax4621. doi: 10.1126/sciadv.aax4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu L., Bu G., Jing L., Shi D., Gonen T., Liu W., Nannenga B.L. Structure determination from lipidic cubic phase embedded microcrystals by MicroED. bioRxiv. 2019:724575. doi: 10.1101/724575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nederlof I., Li Y.W., Marin V.H., Abrahams J.P. Imaging protein three-dimensional nanocrystals with cryo-EM. Acta Cryst. 2013;D69:852–859. doi: 10.1107/S0907444913002734. [DOI] [PubMed] [Google Scholar]
- 72.Sawaya M.R., Rodriguez J., Cascio D., Collazo M.J., Shi D., Reyes F.E., Hattne J., Gonen T., Eisenberg D.S. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc. Natl. Acad. Sci. USA. 2016;113:11232–11236. doi: 10.1073/pnas.1606287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kolb U., Mugnaioli E., Gorelik T.E. Automated electron diffraction tomography—A new tool for nano crystal structure analysis. Cryst. Res. Technol. 2011;46:542–554. doi: 10.1002/crat.201100036. [DOI] [Google Scholar]
- 74.Kolb U., Krysiak Y., Plana-Ruiz S. Automated electron diffraction tomography – development and applications. Acta Cryst. Sect. Bstruct. Sci. Cryst. Eng. Mater. 2019;75:463–474. doi: 10.1107/S2052520619006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wan W., Sun J.L., Su J., Hovmoller S., Zou X.D. Three-dimensional rotation electron diffraction: Software RED for automated data collection and data processing. J. Appl. Cryst. 2013;46:1863–1873. doi: 10.1107/S0021889813027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iadanza M.G., Gonen T. A suite of software for processing MicroED data of extremely small protein crystals. J. Appl. Cryst. 2014;47:1140–1145. doi: 10.1107/S1600576714008073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nannenga B.L., Shi D., Leslie A., Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods. 2014;11:927–930. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hattne J., Reyes F.E., Nannenga B.L., Shi D., de la Cruz M.J., Leslie A.G., Gonen T. MicroED data collection and processing. Acta Cryst. Sect. Afound. Adv. 2015;71:353–360. doi: 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leslie A.G.W., Powell H.R. Evolving Methods for Macromolecular Crystallography. Springer; Dordrecht, The Netherlands: 2007. Processing diffraction data with mosflm. [Google Scholar]
- 80.Clabbers M., Gruene T., Parkhurst J.M., Abrahams J.P., Waterman D.G. Electron diffraction data processing with DIALS. Acta Cryst. Sect. Dstruct. Biol. 2018;74:506–518. doi: 10.1107/S2059798318007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kabsch W. XDS. Acta Cryst. Sect. Dbiol. Cryst. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dwyer J.R., Hebeisen C.T., Ernstorfer R., Harb M., Deyirmenjian B.V., Jordan R.E., Miller R.J.D. Femtosecond electron diffraction: Making the molecular movie. Philos. Trans. A Math Phys. Eng. Sci. 2006;364:741–778. doi: 10.1098/rsta.2005.1735. [DOI] [PubMed] [Google Scholar]
- 83.Ruan C.Y., Vigliotti F., Lobastov V.A., Chen S., Zewail A.H. Ultrafast electron crystallography: Transient structures of molecules, surfaces, and phase transitions. Proc. Natl. Acad. Sci. USA. 2004;101:1123–1128. doi: 10.1073/pnas.0307302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang D.S., Baum P., Zewail A.H. Ultrafast electron crystallography of the cooperative reaction path in vanadium dioxide. Struct. Dyn. 2016;3:034304. doi: 10.1063/1.4953370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen S., Seidel M.T., Zewail A.H. Ultrafast Electron Crystallography of Phospholipids. Angew. Chem. Int. Ed. 2006;45:5154–5158. doi: 10.1002/anie.200601778. [DOI] [PubMed] [Google Scholar]
- 86.Spence J.C.H., Kirian R.R., Wang X., Weierstall U., Schmidt K.E., White T., Barty A., Chapman H.N., Marchesini S., Holton J. Phasing of coherent femtosecond X-ray diffraction from size-varying nanocrystals. Opt. Express. 2011;19:2866–2873. doi: 10.1364/OE.19.002866. [DOI] [PubMed] [Google Scholar]
- 87.Kirian R.A., Bean R.J., Beyerlein K.R., Yefanov O.M., White T.A., Barty A., Chapman H.N. Phasing coherently illuminated nanocrystals bounded by partial unit cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130331. doi: 10.1098/rstb.2013.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gallagher-Jons M., Ophus C., Bustillo K.C., Boyer D.R., Panova O., Glynn C., Zee C.T., Ciston J., Mancia K.C., Minor A.M., et al. Nanoscale mosaicity revealed in peptide microcrystals by scanning electron nanodiffraction. Commun. Biol. 2019;18:2–26. doi: 10.1038/s42003-018-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiedorn M.O., Oberthür D., Bean R., Schubert R., Werner N., Abbey B., Aepfelbacher M., Adriano L., Allahgholi A., Al-Qudami N., et al. Megahertz serial crystallography. Nat. Commun. 2018;9:4025. doi: 10.1038/s41467-018-06156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weierstall U. Liquid sample delivery techniques for serial femtosecond crystallography. Phil. Trans. R. Soc. B. 2014;369 doi: 10.1098/rstb.2013.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jonsson H.O., Caleman C., Andreasson J., Tımneanu N. Hit detection in serial femtosecond crystallography using X-ray spectroscopy of plasma emission. IUCrJ. 2017;4:778–784. doi: 10.1107/S2052252517014154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.White T.A., Mariani V., Brehm W., Yefanov O., Barty A., Beyerlein K.R., Chervinskii F., Galli L., Gati C., Nakane T., et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 2016;49:680–689. doi: 10.1107/S1600576716004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kabsch W. Processing of X-ray snapshots from crystals in random orientations. Acta Cryst. Sect. Dbiol. Cryst. 2014;70:2204–2216. doi: 10.1107/S1399004714013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ginn H.M., Brewster A.S., Hattne J., Evans G., Wagner A., Grimes J.M., Sauter N.K., Sutton G., Stuart D.I. A revised partiality model and post-refinement algorithm for X-ray free-electron laser data. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1400–1410. doi: 10.1107/S1399004715006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Palatinus L., Brázda P., Boullay P., Perez O., Klementová M., Petit S., Eigner V., Zaarour M., Mintova S. Hydrogen positions in single nanocrystals revealed by electron diffraction. Science. 2017;355:6321. doi: 10.1126/science.aak9652. [DOI] [PubMed] [Google Scholar]
- 96.Palatinus L., Corrêa C.A., Steciuk G., Jacob D., Roussel P., Boullay P., Klementová M., Gemmi M., Kopeček J., Domeneghetti M.C. Structure refinement using precession electron diffraction tomography and dynamical diffraction: Tests on experimental data. Acta Cryst. A. 2013;69:171–188. doi: 10.1107/S010876731204946X. [DOI] [PubMed] [Google Scholar]
- 97.Jansen J., Tang D., Zandbergen H.W., Schenk H. MSLS, a Least-Squares Procedure for Accurate Crystal Structure Refinement from Dynamical Electron Diffraction Patterns. Actacrystallogr. A. 1998;54:91–101. doi: 10.1107/S0108767397010489. [DOI] [Google Scholar]
- 98.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/S003358350000305X. [DOI] [PubMed] [Google Scholar]
- 99.Barty A., Caleman C., Aquila A., Timneanu N., Lomb L., White T.A., Andreasson J., Arnlund D., Bajt S., Barends T.R., et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]