Summary
Despite its importance for membrane stability and pathogenicity of mammalian pathogens, functions of the O‐polysaccharide (OPS) of lipopolysaccharide (LPS) remain unclear in plant‐associated bacteria. Genetic information about OPS biosynthesis in these bacteria is largely missing. Genome analysis of various plant‐associated Pseudomonas strains revealed that one of the two known OPS biosynthesis clusters from Pseudomonas aeruginosa PAO1, the common polysaccharide antigen (CPA) gene cluster, is only conserved in some strains of the Pseudomonas fluorescens group. For the O‐specific antigen (OSA) biosynthesis cluster, the putative genomic position could be identified, but orthologues of most functional important OSA biosynthesis enzymes could not be detected. Nevertheless, orthologues of the glycosyltransferase WbpL, required for initiation of CPA and OSA synthesis in P. aeruginosa PAO1, could be identified in the analysed Pseudomonas genomes. Knockout mutations of wbpL orthologues in Pseudomonas syringae pv. tomato DC3000 (Pst) and Pseudomonas cichorii ATCC10857/DSM50259 (Pci) resulted in strains lacking the OPS. Infection experiments of Arabidopsis thaliana plants revealed a reduced entry into the leaf apoplast after spray inoculation and a reduced apoplastic amplification of Pst ∆wbpL. Stab and spray inoculation of lettuce (Lactuca sativa) leaves with Pci ∆wbpL causes reduced infection symptoms compared to the wild‐type strain. Furthermore, swarming motility was reduced in ∆wbpL mutants of Pst and Pci. This might be a possible reason for reduced bacterial titres after surface inoculation and reduced bacterial amplification in the plant. Our results imply that the presence of lipopolysaccharide OPS is required for efficient host colonization and full virulence of plant‐pathogenic Pseudomonas bacteria.
Keywords: lipopolysaccharide, motility, O‐polysaccharide, Pseudomonas syringae, virulence
Introduction
The genus Pseudomonas comprises several plant‐associated bacteria, including versatile plant pathogens that infect crops and cause substantial economic damage (Vanneste, 2017; Young, 2010). For successful infection, bacteria have to overcome various chemical and physiological barriers and induced defence responses that depend on the plant species and their habitat. Therefore, bacteria rely on various defence characteristics themselves.
In Gram‐negative bacteria, the outer membrane (OM) is crucial for bacterial survival. It confers structural stability to bacterial cells and, as a restrictive permeability barrier, protects bacteria from antimicrobial substances while facilitating material exchange with the environment (Alexander and Rietschel, 2001). Lipopolysaccharide (LPS) is an essential OM component and takes part in interactions between bacterial cells and their environment. It can mask the bacterial cell surface to avoid opsonization and possesses endotoxic activity in mammals (Lukácová et al., 2008; Needham and Trent, 2013; Trent et al., 2006).
LPS consists of three covalently linked domains with different chemical and biological properties: the lipophilic lipid A (LA) moiety, the hydrophilic oligosaccharide core region (core‐OS) and O‐polysaccharide (OPS) (Alexander and Rietschel, 2001). Commonly, the OPS is a heteropolymeric O‐specific antigen (OSA) that has a highly diverse composition among bacterial species and strains, and determines their serological and antigenic specificity. In Pseudomonas aeruginosa, the common polysaccharide antigen (CPA) can also be found instead of, or in parallel with, the OSA (Raetz and Whitfield, 2002). The length of the OPS chain ranges from one to >50 repeats and influences cell surface hydrophobicity. LPS molecules of different OPS chain length can occur in parallel to OPS‐deficient variants on a single bacterial cell (Alexander and Rietschel, 2001). OPS protects the bacterium from unfavourable environments by providing a steric as well as a diffusion barrier, e.g. to antibacterial agents that target the interior core and LA parts of LPS (Ranf, 2016). Due to its outermost localization and hydrophilic properties, OPS mediates adhesion to host surfaces and possibly host selectivity (Bogino et al., 2013). Through adherence to other bacteria, it is involved in formation of biofilms, which likely is the preferred growth mode in host tissues (Bogino et al., 2013; Dongari‐Bagtzoglou, 2008). Bacteria lacking OPS have a reduced viability and cannot persist under stress conditions, such as in a host (Raetz and Whitfield, 2002). Additionally, distribution of OPS chain length is species‐specific (King et al., 2014; Murray et al., 2006; Rahman et al., 1997; Tran et al., 2014; Van den Bosch et al., 1997). Variances in the OPS structure are commonly linked to an altered pathogenicity (Lerouge and Vanderleyden, 2002; Lukácová et al., 2008).
In P. aeruginosa, two gene clusters are known to code for enzymes that are required for the complete synthesis of the respective OPS type (Lam et al., 2011). The synthesis of the homopolymeric CPA is ABC‐transporter‐dependent and mainly takes place at the cytoplasmic face of the inner membrane (IM). The polysaccharide is built up on an undecaprenyl pyrophosphate (Und‐PP)‐carrier through addition of saccharide residues to the non‐reducing terminus by glycosyltransferases. An ABC‐transporter system transfers the fully synthesized polysaccharide to the periplasmic face of the IM (Greenfield and Whitfield, 2012). P. aeruginosa OSA is synthesized via the Wzy‐dependent pathway. Single OSA repeating units are assembled on an Und‐PP‐carrier at the cytoplasmic face of the IM and transported to the periplasmic face by the flippase Wzx. In the periplasm, the saccharide subunits are joined with the nascent OSA at its reducing terminus by the polysaccharide polymerase Wzy. The OSA chain length is regulated by the polysaccharide copolymerase Wzz (Islam and Lam, 2014). Both pathways share the initiation step, the addition of a first monosaccharide to the Und‐PP‐carrier by the glycosyltransferase WbpL (Lam et al., 2011). In the periplasm, the completed OPS is transferred from the Und‐PP‐carrier to the LA‐core‐OS unit by the OPS ligase WaaL (Whitfield et al., 1997).
LPS mutants from several members of the Rhizobiales lacking an OPS are defective in symbiosis because of premature abortion of infection threads (Carlson et al., 2010; Lerouge and Vanderleyden, 2002; Ormeño‐Orrillo et al., 2008). This suggest that OPS plays a role in root infection during symbiotic plant‐bacteria interactions. Similar to mammalian pathogens, the protective nature of OPS is considered a key factor for bacterial survival and growth in plants. Since resistance against hostile environmental conditions is a premise for a successful infection, it is assumed that OPS is essential for plant‐pathogen virulence (Kutschera and Ranf, 2019a). Several studies with OPS‐defective mutants of Erwinia spp., Ralstonia solanacearum, Xanthomonas axonopodis pv. citri and Xylella fastidiosa showed a loss or strong reduction of virulence in the host plant (Berry et al., 2009; Drigues et al., 1985; Li et al., 2014; Petrocelli et al., 2012; Rapicavoli et al., 2018; Schoonejans et al., 1987). Rudolph et al. (1989) reported a strongly reduced colonization of the leaf mesophyll and a decreased virulence of OPS‐deficient Pseudomonas syringae pv. phaseolicola. They postulated that OPS confers compatibility with the plant and determines host specificity (Rudolph, 2001). Although the molecular OPS composition of many plant‐associated bacteria has been elucidated in recent years, the genetic background of OPS biosynthesis remains unknown in many plant‐associated Pseudomonas strains. Differences in OPS chain length distribution between different pathovars/strains raise the question about the regulatory mechanisms determining the prevalence for specific OPS sizes analogous to other bacterial species (Kalynych et al., 2011; King et al., 2014; Kutschera and Ranf, 2019b; Whitfield et al., 1997).
In this work, we shed light on the genetics of OPS synthesis in P. syringae and related plant‐associated bacteria through genome and proteome sequence analysis. We identified the glycosyltransferase WbpL, which is conserved in all examined strains, as a key component of OPS synthesis. However, we could not identify a complete OPS gene cluster responsible for OPS synthesis in P. syringae and related species. Deletions of the identified wbpL orthologues in P. syringae pv. tomato DC3000 (Pst) and Pseudomonas cichorii ATCC10857/DSM50259 (Pci) resulted in OPS‐deficient mutants. Infection of Arabidopsis thaliana with Pst ∆wbpL or lettuce (Lactuca sativa) leaves with Pci ∆wbpL was delayed compared to the wild‐type strains, but neither a rapid decline of bacterial titres nor a complete avirulence of the ∆wbpL mutants was observed. Finally, our findings suggest that OPS influences motility and thereby affects colonization of plant hosts.
Results
Partial conservation of OPS‐synthesis gene clusters in phytopathogens
We conducted comparative analyses with predicted protein sequences from the genomes of various Pseudomonas species to identify LPS biosynthesis components and to reveal similarities and differences in OPS synthesis. Most P. aeruginosa strains produce two different OPS types, OSA and CPA. The corresponding genes are organized in two independent gene clusters (Hao et al., 2013; Samuel and Reeves, 2003). Putative orthologues of the proteins encoded by these OPS gene clusters were identified in the investigated predicted proteomes, followed by synteny analysis of the respective genes in the genome context.
The main CPA cluster is highly conserved among P. aeruginosa strains and consists of eight genes (rmd–wbpZ, Fig. 1). Additionally, a proximal cluster of five genes (PA5455–PA5459) is required for CPA synthesis in P. aeruginosa PAO1 and PA14 (Lam et al., 2011). BLASTP search for CPA biosynthesis enzymes, using P. aeruginosa PAO1 sequences as query, revealed that some homologues are present in the respective proteomes (Fig. 1). Sequence identities of most BLASTP hits were below 50%, whereas hits with over 98% sequence identity for all query sequences were retrieved in the P. aeruginosa PA14 proteome. For P. aeruginosa PA7, sequence identities of the respective hits ranged from 82.8% to 98.5% (main cluster) and from 41.3% to 54.7% (additional cluster). Likewise, P. fluorescens Pf0‐1, P. chlororaphis ATCC17415 and P. brassicacearum NFM421 showed sequence identities ranging from 51.0% to 88.2% for all query sequences. As previously reported (Lam et al., 2011; Silby et al., 2009), P. aeruginosa serovars, P. fluorescens Pf0‐1, P. chlororaphis ATCC17415 and P. brassicacearum NFM421 share a common organization of CPA synthesis genes (Fig. 1). No similar gene organization was observed in the remaining analysed genomes. Most of the coding sequences of the putative CPA synthesis enzyme homologues are distributed in the respective genome and a few occur in pairs. For example, genes corresponding to the homologues of the two ABC‐transporter subunits Wzt (PA5450) and Wzm (PA5451) of Pst and Pci JBC1 are located next to each other in the respective genomes. The location of these genes is similar in Pst and Pci JBC1, but the synteny in the context of the P. aeruginosa PAO1 CPA gene cluster is not conserved (Fig. 1B). According to the low sequence identities of the putative homologues, these genes likely code for ABC‐transporter subunits not involved in CPA biosynthesis. Except for the P. aeruginosa strains, P. fluorescens Pf0‐1, P. chlororaphis ATCC17415 and P. brassicacearum NFM421, the overall results of the comparative analysis indicate that the CPA cluster is not conserved in all plant‐associated Pseudomonas species. In summary, the analyses suggest an OPS synthesis mediated by enzymes not orthologous to the CPA biosynthesis in P. aeruginosa PAO1.
Figure 1.
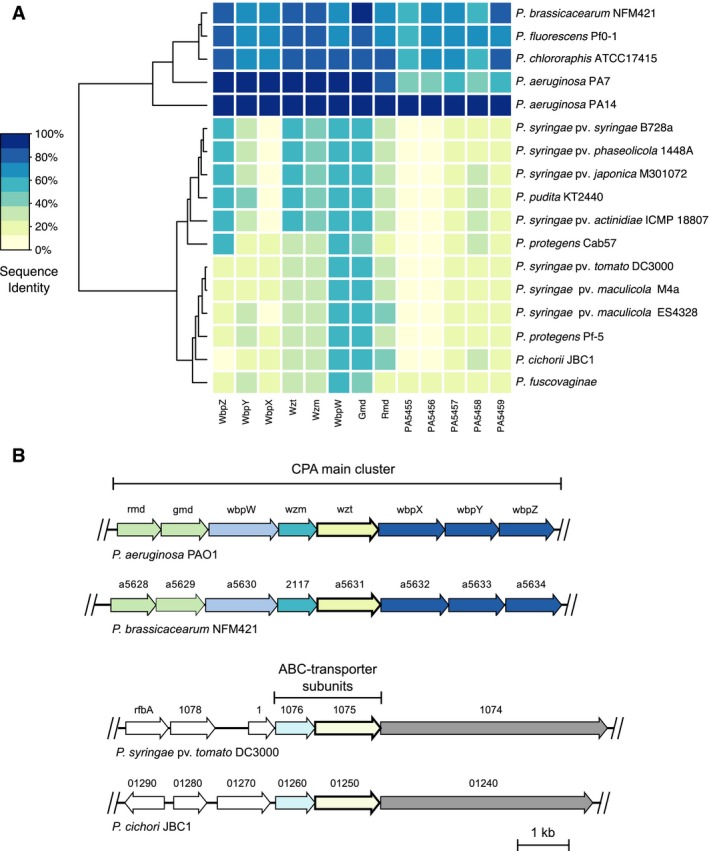
Comparative analysis of the common polysaccharide antigen (CPA) cluster. The CPA synthesis gene cluster could not be identified in most of the analysed Pseudomonas genomes. (A) Heatmap of results from a BLASTP search for homologous CPA biosynthesis enzymes in different predicted proteomes. P. aeruginosa PAO1 sequences were used as reference (e‐value cut‐off of 10−9). Phylogenetic tree shows the Euclidean distance calculated from the BLASTP identities. (B) Gene structure of the P. aeruginosa PAO1 and P. brassicacearum NFM421 CPA cluster and locus of putative ABC‐transporter subunit homologues in P. syringae pv. tomato DC3000 and P. cichorii JBC1. Colour indicates the respective gene hits in the genomes.
The OSA biosynthesis cluster is not as highly conserved as the CPA cluster due to the intrinsic variation of OSA structures of the P. aeruginosa serovars. While the cluster content differs considerably in serovar‐specific glycosyltransferases, the locus of the gene cluster and regulatory enzymes of OSA synthesis such as the chain length regulator Wzz are conserved. The OSA cluster position is defined by the proposed border genes wzz (PA3160) and wbpM (PA3141, saccharide epimerase/dehydratase). A comparative analysis of P. aeruginosa PA14 and P. aeruginosa PA7 OSA‐biosynthesis enzymes confirms these variations (Raymond et al., 2002) (Fig. 2). In other Pseudomonas genomes, orthologues of the border gene wbpM with amino acid sequence identities ranging from 68.6% to 77.0% of the corresponding predicted proteins were found in a similar gene context. In all Pseudomonas strains, orthologues of the glycosyltransferase WbpL with sequence identities ranging from 59.2% to 66.0% were identified. Sequence identities of putative WbpK orthologues were considerably lower (36.2–42.4%). In P. aeruginosa PAO1, the corresponding genes (wbpL: PA3145, wbpK: PA3146) are located upstream of wbpM. Synteny studies revealed a conserved location of the respective wbpL orthologues in all analysed Pseudomonas strains, whereas location of wbpK is only conserved in a few strains (Fig. 2B). For other proteins encoded by the P. aeruginosa PAO1 OSA cluster, the analysis of most proteomes yielded no or only low sequence identity hits that do not seem to be located in a similar OSA cluster context. Notably, genomes of P. chlororaphis ATCC17415 and P. brassicacearum NFM421 contain putative orthologues of wzz. Analysis of the P. brassicacearum NFM421 genome revealed several other putative gene orthologues of the P. aeruginosa PAO1 OSA cluster with a conserved synteny (Fig. 2B).
Figure 2.
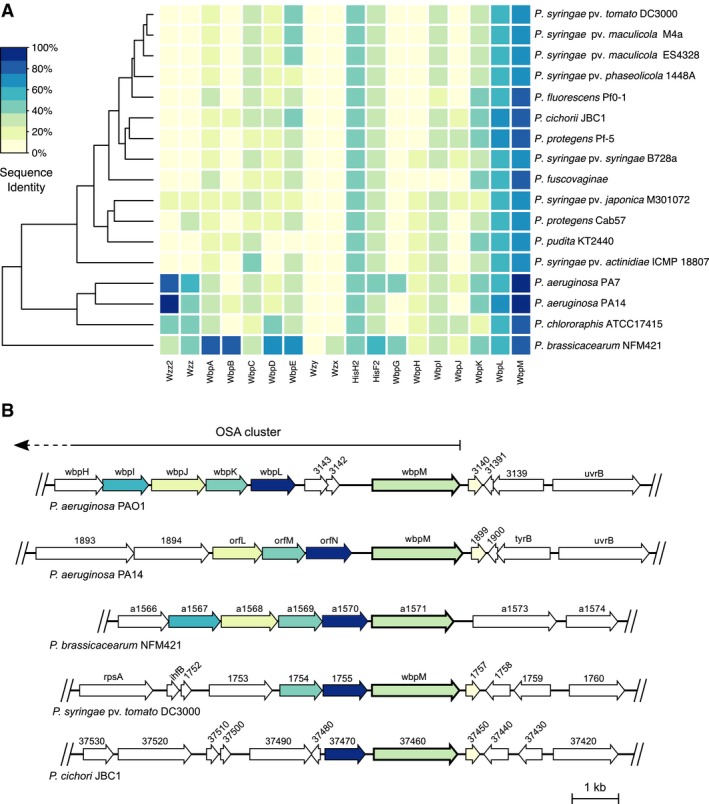
Comparative analysis of the O‐specific antigen (OSA) cluster. The putative locus and homologues of genes from the OSA synthesis cluster could be identified in Pseudomonas spp. (A) Heatmap of results from a BLASTP search for homologous OSA biosynthesis enzymes in different predicted proteomes. P. aeruginosa PAO1 sequences were used as reference (e‐value cut‐off = 10−9). Phylogenetic tree shows the Euclidean distance calculated from the BLASTP identities. (B) Gene structure of the upstream part of the P. aeruginosa PAO1 OSA cluster and hits from synteny analysis of corresponding genes in other Pseudomonas species. Colour indicates the respective gene hits in the genomes.
Overall, only two to three of 17 genes in the P. aeruginosa PAO1 OSA cluster appear to be conserved in sequence and position in the majority of the analysed Pseudomonas strains (Fig. 2B). Hence, this short cluster might not be sufficient for the synthesis of a complete OPS in these strains. Nevertheless, the high sequence identities of the putative WbpM and WbpL orthologues suggest that these proteins function in OPS synthesis. In P. aeruginosa, WbpL initiates OSA as well as CPA synthesis (Bélanger et al., 1999; Rocchetta et al., 1998, 1999). The putative WbpL orthologues of the analysed Pseudomonas strains likely also initiate OPS synthesis.
Knockout of wbpL results in disruption of OPS synthesis
Knockout of wbpL in P. aeruginosa disrupts OPS synthesis (Bélanger et al., 1999). Knockout of the putative wbpL genes in Pst and Pci should reveal if the corresponding proteins are also involved in OPS synthesis. We deleted the identified wbpL candidates in Pci (PCH70_40470 in P. cichorii JBC1) and Pst (PSPTO_1755) by replacement of the wbpL gene sequence with a gentamicin resistance cassette (GmR). Three independent transformants of each strain were analysed for alterations in OPS biosynthesis. Silver‐stained SDS‐PAGE of crude LPS preparations indicated a loss of OPS synthesis. The ∆wbpL::GmR (∆wbpL) deletion mutants showed no noticeable growth phenotypes compared to the wild‐type strains. For further analysis, LPS was isolated by hot phenol/water and phenol/chloroform/petroleum ether‐extraction from one selected transformant per strain. Bis‐Tris NuPAGE/silver staining of these LPS preparations (Fig. 3) confirmed that LPS of the Pci and Pst ∆wbpL mutants lacks OPS.
Figure 3.
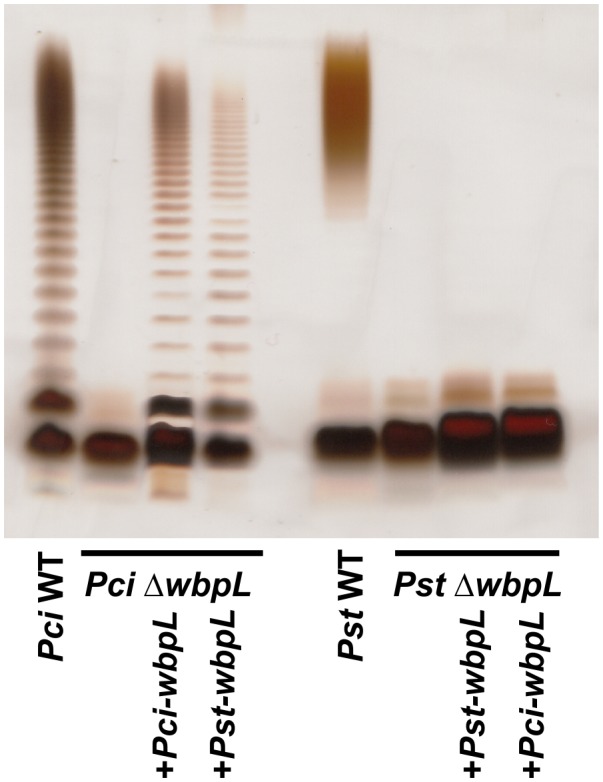
12% Bis‐Tris NuPAGE gel and silver staining of lipopolysaccharide (LPS) preparations from Pseudomonas cichorii ATCC10857/DSM50259 (Pci) and Pseudomonas syringae pv. tomato DC3000 (Pst) wild‐type (WT), ∆wbpL::GmR (∆wbpL) deletion mutant strains and mutant strains complemented with plasmid‐expressed wbpL orthologues from Pci or Pst. LPS was isolated by hot phenol‐water extraction followed by a phenol‐chloroform‐petroleum ether extraction. 2 μg of ∆wbpL LPS, for all other LPS 4 μg were applied per lane.
Complementation of the gene knockout with Pci‐wbpL or Pst‐wbpL partially restores OPS synthesis in Pci ∆wbpL
Complementation strains of Pst ∆wbpL and Pci ∆wbpL were generated to exclude polar effects of the GmR insertion. Complementation plasmids containing the wbpL orthologue from Pci (PCH70_40470 in P. cichorii JBC1) or Pst (PSPTO_1755) under control of the GmR promoter were transferred into the respective ∆wbpL mutant strain. SDS‐PAGE analysis of crude LPS preparations from two transformants per mutant strain indicated successful reconstitution of OPS biosynthesis in the complemented Pci ∆wbpL but not in Pst ∆wbpL strains. LPS isolation by hot phenol/water and phenol/chloroform/petroleum ether‐extraction and Bis‐Tris NuPAGE/silver staining confirmed the recovery of OPS biosynthesis in Pci ∆wbpL strains containing the Pci‐wbpL expression plasmid (Fig. 3). However, LPS preparation of Pst ∆wbpL strain complemented with the Pst‐wbpL gene did not show reconstitution of OPS synthesis. To assess whether the Pst‐wbpL complementation plasmid is functional and transient expression of the putative WbpL orthologues in Pst is sufficient to complement the LPS phenotype, Pci ∆wbpL was transformed with the Pst‐wbpL complementation plasmid and Pst ∆wbpL with the Pci‐wbpL complementation plasmid (cross‐complementation). SDS‐PAGE and silver staining of crude LPS preparations from two transformants each suggested that the cross‐complemented Pci ∆wbpL but not Pst ∆wbpL strains showed a recovery of OPS biosynthesis. Analysis of LPS isolated by hot phenol/water and phenol/chloroform/petroleum ether‐extraction and Bis‐Tris NuPAGE/silver‐staining confirmed this result (Fig. 3). The presence of LPS substituted with OPS in Pci ∆wbpL cross‐complemented with Pst‐wbpL demonstrates that the Pst‐wbpL complementation plasmid is functional, and transient expression of the Pst WbpL orthologue principally complements the Pci ∆wbpL LPS phenotype, although apparently not to the full extent of the endogenous WbpL. LA and core‐OS‐OPS were isolated from Pci ∆wbpL + Pci‐wbpL by mild acid hydrolysis (Ranf et al., 2015) and respective molecular structures were analysed and compared with wild‐type Pci LPS. No substantial structural alterations could be observed (as judged by mass spectrometry of core‐OS and 1H NMR of core‐OS with OPS; data not shown). Notably, hydrolysis of the LPS from the wild‐type strain resulted in a weight‐to‐weight ratio of 2:1 for the core‐OS versus core‐OS‐OPS, whereas for LPS from the complemented strain a ratio of 10:1 was observed. This was further corroborated by NuPAGE analysis using different LPS concentrations. A five‐times higher LPS concentration of the LPS isolated from the Pci ∆wbpL + Pci‐wbpL complementation strain compared to Pci wild‐type LPS shows a similar band pattern of OPS‐substituted LPS molecules with a simultaneous increase of staining intensity for OPS‐deficient LPS. A similar effect was observed for the LPS isolated from the Pci ∆wbpL + Pst‐wbpL complementation strain (Fig. S1). In summary, this indicates that the complementation was only partially successful as the complementation strains contain less LPS substituted with OPS.
To confirm the OPS‐loss in the Pst ∆wbpL complementation strain (+Pst‐wbpL) and to exclude inefficient staining as cause for the visual absence of signals in the high molecular weight region (as observed with crude extract of Pst (Kutschera and Ranf, 2019b), the LPS preparations of the Pst wild‐type, the ∆wbpL and the complemented ∆wbpL strain were fractionated by gel permeation chromatography (GPC) with a deoxycholate (DOC)‐containing buffer (Fig. 4A–C). NuPAGE analysis of LPS pools 1 and 2 from Pst showed signals in the high molecular weight region, while pool 3 contained OPS‐deficient LPS (Fig. 4D). GPC chromatograms and subsequent NuPAGE analysis of the ∆wbpL (Fig. 4B,D) and of the Pst ∆wbpL complementation strain (+Pst‐wbpL; Fig. 4C,D) prove the absence of OPS‐substituted LPS species. The sole presence of OPS‐deficient LPS in the complemented strain (pool 4 in Fig. 4C) shows that the OPS synthesis in Pst ∆wbpL was not reconstituted by transient wbpL expression.
Figure 4.
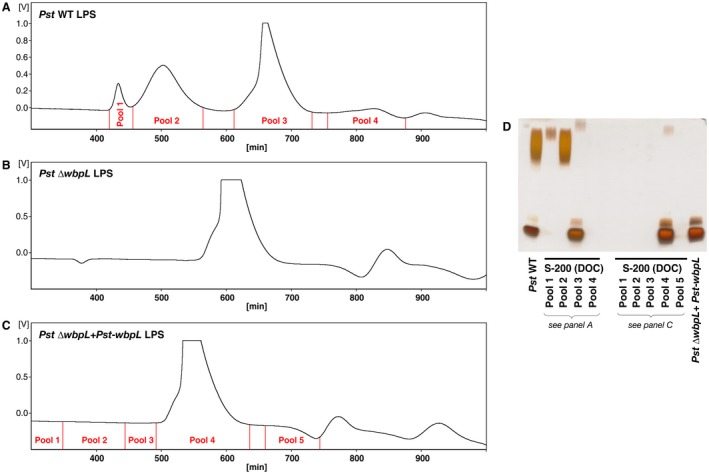
Fractionation of lipopolysaccharide (LPS) from Pseudomonas syringae pv. tomato DC3000 (Pst) (A) wild‐type (WT), (B) ∆wbpL and (C) complemented (∆wbpL + Pst‐wbpL) strain on Sephacryl S‐200 HR using a desoxycholate (DOC)‐containing buffer. The relevant regions of representative GPC chromatograms are depicted, including the chosen fractionation. In case of ∆wbpL + Pst‐wbpL strain, material of two comparable GPC runs were combined for further assays. (D) Silver‐stained 12% Bis‐Tris NuPAGE gel of the indicated combined column pools and the respective starting material of A and C. 4 μg of LPS were applied per lane.
Pst ∆wbpL and Pci ∆wbpL are less virulent than the wild‐type strains
Arabidopsis thaliana plants were infected with Pst wild‐type and Pst ∆wbpL strains to evaluate effects of the WbpL knockout and the resulting OPS‐deficiency on virulence. Spray inoculation revealed a reduced bacterial titre in plants treated with Pst ∆wbpL compared to plants treated with wild‐type Pst (Fig. 5A). This difference had already been observed 4 h post‐inoculation (hpi). Bacterial titres were strongly increased at 72 hpi in both wild‐type and ∆wbpL mutant‐infected plants. The OPS‐deficient Pst ∆wbpL mutant, however, amplified to lower titres than the wild‐type strain. Spray inoculation mimics natural infection, since bacteria need to overcome stomatal immunity and actively enter the leaf interior. Thus, the initial differences in bacterial titres suggest an impaired capability of Pst ∆wbpL cells to enter the host tissue. To determine bacterial amplification in the leaf apoplast, A. thaliana leaves were pressure‐infiltrated with bacterial suspensions. Accordingly, wild‐type and mutant Pst bacterial titres were comparable at 4 hpi. At 72 hpi, Pst ∆wbpL titres were significantly lower than Pst wild‐type titres (Fig. 5B), suggesting that the apoplastic amplification rate of the mutant bacteria is reduced. Thus, the ability to enter the host as well as colonization of the apoplast is affected by the wbpL knockout and the concomitant OPS loss. Long‐term monitoring of A. thaliana inoculated with both Pst strains by pressure‐infiltration of leaves revealed that Pst ∆wbpL causes typical infection symptoms, but delayed compared to the wild‐type strain (Video S1). Stab inoculation of lettuce midrib with Pci ∆wbpL resulted in reduced lesion size compared to the wild‐type strain. Similarly, Pci ∆wbpL caused less severe disease symptoms when sprayed on lettuce leaves (Fig. 5C,D).
Figure 5.
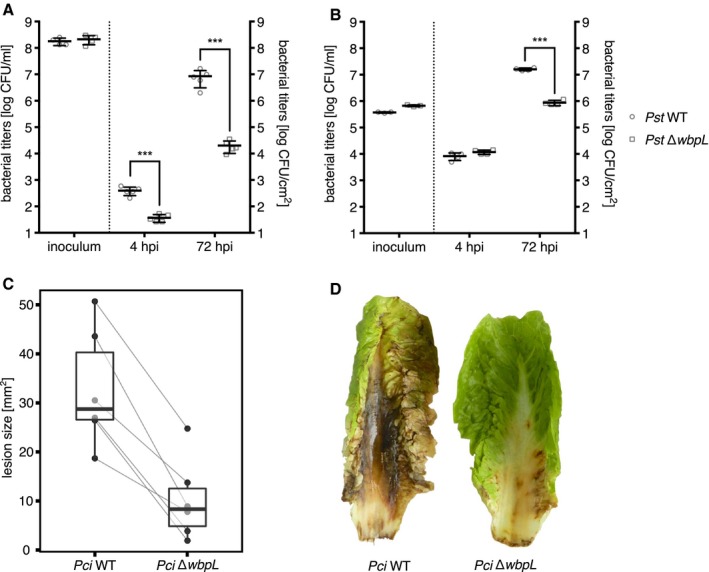
Infection experiments with Pseudomonas syringae pv. tomato DC3000 (Pst) and P. cichorii ATCC10857/DSM50259 (Pci) wild‐type (WT) and ∆wbpL deletion mutants. Arabidopsis thaliana plants were inoculated by spraying (A, n = 5, mean ± SEM) or syringe‐infiltration of leaves (B, n = 4, mean ± SEM) with Pst WT or Pst ∆wbpL suspension. Samples were taken 4 and 72 h post‐inoculation (hpi). (P < 0.001, multiple t‐test, experiments were repeated at least three times with similar results). (C) Lettuce midrib was stab infected with Pci WT and Pci ∆wbpL and the size of the emerging lesions was measured 2 days post‐inoculation (dpi) (Fig. S2A, n = 6, P < 0.05, Wilcoxon signed‐rank test, experiment was repeated at least three times with similar results). (D) Lettuce leaves were sprayed with Pci WT or Pci ∆wbpL suspensions and disease symptoms were monitored over time. The picture was taken 9 dpi. The original image is provided in the supporting information (Fig. S2B).
WbpL knockout diminishes swarming motility
The ability to move actively is crucial for the lifestyle of plant‐associated bacteria. In particular, swarming motility, collective and directional movements of bacteria on a surface, are important for plant colonization (Haefele and Lindow, 1987; Harshey, 2003; Zeng et al., 2010). The swarming motility of Pst and Pci wild‐type and ∆wbpL strains was therefore determined in vitro. Swarming motility of ∆wbpL mutants was significantly reduced compared to the wild‐type in both strains (Fig. 6). For Pci ∆wbpL strains complemented with the respective native Pci‐wbpL and Pst‐wbpL, a significant increase of migration area could be observed compared to the mutant strain. In accordance with the lack of complementation of the LPS phenotype, transient expression of the Pst or Pci wbpL gene in Pst ∆wbpL did not reconstitute the motility phenotype. These results show that a loss of the OPS diminishes swarming motility in both bacterial strains.
Figure 6.
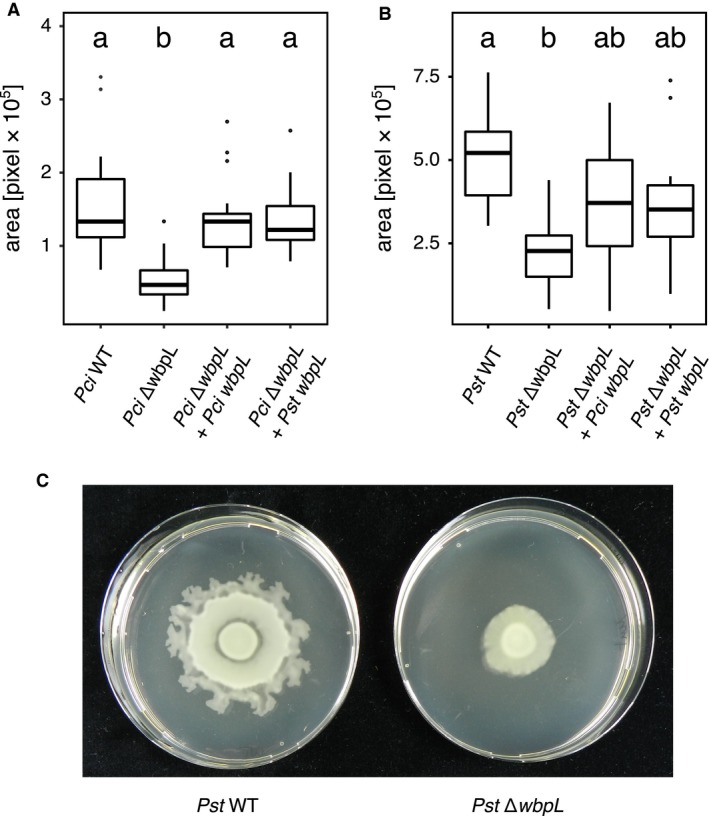
Comparison of swarming motility of Pseudomonas syringae pv. tomato DC3000 (Pst WT, A, n = 11, exemplary images in C) and Pseudomonas cichorii ATCC10857/DSM50259 (Pci WT, B, n = 16) with the respective ∆wbpL::GmR (∆wbpL) and Pci WbpL (Pci‐wbpL) or Pst WbpL (Pst‐wbpL) expressing complementation strains. Data from three independent experiments were pooled. Letters indicate statistical difference (P < 0.05) calculated by a Kruskal–Wallis rank sum test and post hoc pairwise comparisons using the Nemenyi‐test with chi‐squared approximation for independent samples.
Discussion
LPS is a major virulence factor for many pathogenic Gram‐negative bacteria. OPS, a molecular substructure of LPS, has various well‐investigated virulence functions in mammalian hosts, such as complement resistance (Murray et al., 2006) and influence on biofilm formation (Murphy et al., 2014). However, little is known about OPS biosynthesis in plant‐associated bacteria and the role of OPS in plant infection. Here, we report that most of the analysed plant‐associated Pseudomonas strains lack orthologues of the P. aeruginosa CPA biosynthesis gene cluster. We identified a putative but short orthologue of the P. aeruginosa OSA biosynthesis gene cluster, which includes the conserved glycosyltransferase WbpL. By genetic knockout and complementation, we show that WbpL is essential for OPS synthesis in Pst and Pci. Infection assays of A. thaliana and lettuce with OPS‐deficient Pst or Pci strains, respectively, revealed a reduction but not a loss of virulence. Finally, our results indicate a possible link between loss of the OPS, reduced swarming motility and reduced virulence in planta.
The genetic background of OPS synthesis in phytopathogenic Pseudomonas differs from P. aeruginosa
OPS structures of plant‐associated Pseudomonas species show a low monosaccharide variability compared to mammalian pathogens (Molinaro et al., 2009). The structural similarity of rhamnose‐rich P. aeruginosa CPA and the rhamnose‐rich chemical composition of OPS from other Pseudomonas strains suggests a genetic analogy. In our genetic analysis, however, we could not identify a CPA cluster orthologue in most of the investigated Pseudomonas genomes. Besides P. aeruginosa PA14 and PA7, only P. fluorescens Pf0‐1, P. chlororaphis ATCC17415 and P. brassicacearum NFM421 apparently contain a complete orthologue of the P. aeruginosa PAO1 CPA cluster (Lam et al., 2011). Notably, in contrast to P. aeruginosa CPA, many of the known OPS structures from plant‐associated Pseudomonas strains contain l‐rhamnose, not d‐rhamnose (Molinaro et al., 2009; Zdorovenko and Veremeichenko, 2001; Zdorovenko et al., 2001). Nevertheless, P. aeruginosa core‐OS contains an l‐rhamnose residue that is transferred by the rhamnosyltransferases MigA (PA0705) or WapR (PA5000). Possibly, one specific rhamnosyltransferase, potentially orthologous to MigA or WapR, could be responsible for the synthesis of l‐rhamnose‐rich OPS. Alternatively, stereochemical differences in OPS structure could be enabled by OPS synthesis by a different, yet unknown, set of genes.
The search for orthologues of P. aeruginosa OSA synthesis proteins yielded hits in all analysed predicted proteomes and uncovered a putative syntenic region of the OSA cluster in the Pseudomonas genomes. Compared to the complete cluster in P. aeruginosa PAO1 (17 genes), most of these cluster‐orthologues contained only two to three genes. The two genes conserved in all Pseudomonas genomes code for an epimerase (WbpM) and a glycosyltransferase (WbpL) that possess a generic function for OPS synthesis in P. aeruginosa. However, these very short clusters are unlikely to facilitate synthesis of a complete OPS. The lack of Wzx, Wzy and Wzz/Wzz2, which define the Wzy‐dependent OSA biosynthesis pathway in P. aeruginosa PAO1, indicates a Wzy‐independent OPS synthesis in most of the analysed strains (Islam and Lam, 2014; Kalynych et al., 2011). Two investigated P. syringae pv. tomato OPS‐chains consist of an l‐rhamnose backbone with lateral N‐acetyl‐d‐fucosamine (d‐FucNAc) residues (Knirel et al., 1993, 1998), whereas P. cichorii OPS is composed of alternating N‐acetyl‐l‐fucosamine (l‐FucNAc) and N‐acetyl‐d‐quinovosamine (d‐QuiNAc) (Jimenez‐Barbero et al., 2002) saccharide units. Their generally heteropolymeric structure contradicts a synthesis through an ABC transporter‐dependent pathway or S. enterica serovar Borreze‐specific synthase‐dependent pathway. The partially conserved cluster raises questions about the evolution of OPS biosynthesis in Pseudomonas. Our results may suggest that in the analysed genomes the majority of the OPS cluster genes were lost or are differently organized. Taken together, additional, as yet unknown, genes, possibly organized in another cluster, could be required for OPS synthesis in these plant‐associated Pseudomonas strains.
WbpL has a conserved function in Pseudomonas
In P. aeruginosa, the glycosyltransferase WbpL (PA3145) catalyses the initiating step of CPA and OSA biosynthesis. Knockout of wbpL leads to synthesis of LPS lacking OPS in P. aeruginosa (Bélanger et al., 1999; Rocchetta et al., 1998, 1999) and P. putida (Junker et al., 2001). Our analyses indicate that WbpL function is generally conserved in Pseudomonas and its knockout results in OPS‐deficient LPS in both Pst and Pci. While transient complementation of Pci ∆wbpL confirms WbpL function in OPS synthesis, no plasmid‐driven complementation of OPS synthesis could be achieved in Pst ∆wbpL. However, transient expression of Pst‐wbpL in Pci ∆wbpL reconstitutes OPS synthesis and demonstrates its conserved function in OPS synthesis. The unsuccessful Pst ∆wbpL complementation might further indicate an influence of wbpL expression level on OPS initiation. Accordingly, we observed an increase of OPS‐deficient LPS species in Pci ∆wbpL + Pci‐wbpL compared to the wild‐type. Possibly, the initiation frequency decreases due to a changed wbpL expression level and leads to the altered substitution of LPS with OPS. Bronner et al. (1994) observed a similar gene dosage effect when transiently expressing OPS‐synthesis components in Escherichia coli and assumed that increasing the copy number of the ABC‐transporter components relative to the polysaccharide synthesis enzymes leads to a reduction of the average OPS length. Since the precise coordination of the ratio of OPS‐synthesis components appears to be crucial, alterations in wbpL expression might disturb OPS synthesis initiation. Thus, ectopic wbpL expression in Pst ∆wbpL mutants might not result in reconstitution of OPS synthesis. It cannot be excluded that polar effects of the wbpL knockout on expression of the wbpK gene, which is located directly upstream of wbpL, lead to the loss of OPS but with regard to WbpK function it seems rather unlikely. It is assumed that WbpK catalyses the conversion of UDP‐4‐keto‐d‐QuiNAc to UDP‐d‐FucNAc in P. aeruginosa, which is an essential step for the synthesis of d‐FucNAc‐containing OPS (King et al., 2009). The OPS of Pst, however, contains only non‐stoichiometric d‐FucNAc side chains (Knirel et al., 1993, 1998).
O‐polysaccharide‐deficient Pst and Pci strains are viable in planta and cause disease symptoms
To date, only a few reports on the function of OPS during plant infection exist and most mechanistic insights derive from bacteria–host interaction studies in mammals. For most mammalian pathogens, loss of OPS has a negative impact on virulence (Raetz and Whitfield, 2002). Several studies report a diminished symbiosis capability of plant root symbionts (Carlson et al., 2010; Lerouge and Vanderleyden, 2002; Ormeño‐Orrillo et al., 2008) and reduced virulence of plant‐pathogenic bacteria (Berry et al., 2009; Drigues et al., 1985; Li et al., 2014; Petrocelli et al., 2012; Rapicavoli et al., 2018; Schoonejans et al., 1987) that are defective in OPS synthesis. These findings endorse the hypothesis that OPS is also essential for plant‐pathogen virulence. An OPS‐deficient P. syringae pv. syringae 61 ∆galU mutant strain shows reduced viability in planta (Deng et al., 2010). GalU is required for synthesis of the monosaccharide precursor UDP‐glucose. Knockout of galU leads to OPS‐loss due to truncation of core‐OS in Pseudomonas but also affects other pathways, such as exopolysaccharide synthesis and protein glycosylation, which might additionally impair virulence (DeLucia et al., 2011; Kocincova and Lam, 2011; Liao et al., 2014; Molinaro et al., 2009). By contrast, a targeted disruption of OPS‐synthesis by knockout of the OPS‐specific glycosyltransferase wbpL allows to specifically address the role of OPS in plant–microbe interactions.
Here, we used the well‐established Pst‐A. thaliana pathosystem and infected lettuce leaves with Pci to study the role of OPS during infection processes. Infection experiments with the OPS‐deficient Pst ∆wbpL mutant revealed a reduced entry into, and slower amplification within, the leaf apoplast, but not a loss of virulence. Indeed, compared to Pst‐infected leaves, disease symptoms in Pst ∆wbpL‐infected leaves develop more slowly but eventually reach similar severity at later time points. Pci ∆wbpL causes lesions with reduced sizes in wounded lettuce midrib and less severe infection symptoms in lettuce leaves after surface inoculation. Apparently, OPS‐deficient ∆wbpL strains can still amplify in planta and cause disease symptoms (Fig. 5 and Video S1). Recent reports describe an earlier recognition of OPS‐defective X. fastidiosa by plant innate immunity (Rapicavoli et al., 2018). Possibly, the Pst ∆wbpL might trigger A. thaliana immunity earlier than wild‐type bacteria. Therefore, initial amplification of bacteria within the plant might be delayed until the pathogen can overcome defence responses, e.g. with type III‐secreted effectors like AvrPtoB and AvrPto, and eventually cause disease (Macho and Zipfel, 2015; Wei and Collmer, 2018). Additionally, OPS‐deficient mutants could be more sensitive to plant antimicrobial metabolites. OPS may provide a general protection against plant immune recognition and defence responses (Ranf et al., 2015). Spray inoculation shows that the ability to enter host tissues is reduced for Pst ∆wbpL. Similarly, disease symptoms on lettuce leaves spray‐inoculated with Pci ∆wbpL are strongly reduced compared to leaves sprayed with wild‐type Pci. Hence, reduced motility of the ∆wbpL strain might additionally affect its amplification rate and dissemination in planta. Taken together, this suggests that the OPS‐defective mutants are not impaired in their ability to cause disease but that the reduced virulence correlates with a reduced capacity to colonize host tissue.
Loss of the O‐polysaccharide influences bacterial motility
Entering of host tissue through stomata requires active movement by the Pst cells (Zeng et al., 2010). Here, we show a possible link between reduced bacterial titre in spray‐inoculated plants and impaired motility of Pst ∆wbpL, as observed in in vitro swarming motility assays. Several studies report reduced motility for Gram‐negative bacteria lacking OPS (Huang et al., 2006; Kim and Surette, 2005; Petrocelli et al., 2012). However, it is not clear whether this is due to a motility‐enabling effect of the OPS or a general change of the bacterial cell wall composition and polarity. For example, in P. aeruginosa PAO1, a truncated LPS structure leads to a decrease of flagellar motility due to modulation of cell‐surface attachment (Lindhout et al., 2009). Although flagella and LPS synthesis share some precursor synthesis steps, flagellar assembly was not linked with genetic alterations leading to OPS‐deficient LPS. However, attachment to abiotic surfaces was increased and motility was impaired (Lindhout et al., 2009). Furthermore, reduced motility and increased surface attachment seem to be linked to an increased biofilm formation in OPS‐defective bacterial strains (Lee et al., 2010). Bacterial motility is essential for entering the host tissue and for epiphytic fitness (Haefele and Lindow, 1987; Tans‐Kersten et al., 2001; Zeng et al., 2010). We observed that a loss of OPS impairs motility, which in turn may contribute to reduced virulence in planta. Future studies of possible alterations in surfactant release or surface composition and polarity will clarify how OPS influences motility during plant–bacteria interactions. The OPS chain length distribution varies significantly between different Pseudomonas species, pathovars and strains (Kutschera and Ranf, 2019b). The modulation of OPS chain length might therefore constitute a mechanism for fine‐tuning of cell‐surface polarity to facilitate motility as well as adhesion and biofilm formation in the context of host adaption.
Experimental Procedures
Plant growth conditions
Arabidopsis thaliana ecotype Col‐0 plants were grown on soil in climate chambers under short day conditions with 8 h of light, 22/18 °C (day/night) and 55% relative humidity.
Bacterial strains and growth conditions
Pst, Pci and respective mutants were grown at 26 °C in King’s B (KB) medium liquid culture with shaking (230 rpm) or on KB and low‐salt lysogeny broth (LSLB) agar plates with 2% (w/v) agar. E. coli DH5α and S17‐1 were grown at 37 °C in lysogeny broth (LB) liquid culture with shaking (230 rpm) or on LB agar plates with 2% (w/v) agarose.
Genomic analysis
The annotated P. aeruginosa PAO1 genome [GenBank assembly accession: GCA_000006765.1, Stover et al. (2000)] was used as reference to identify orthologous LPS biosynthesis genes in other genomes. Reciprocal BLAST experiments of protein‐coding regions were conducted against the proteomes listed in Table S1. A python script was established using BLASTP to identify orthologues of the query sequences in each proteome set up as individual local databases. The sequence of the first hit was retrieved by the algorithm and sequence identity was calculated from the quotient of hit sequence length and corresponding identities. Heatmaps were created from sequence identity values of the BLAST results and dendrograms were calculated from the corresponding Euclidean distances. Scripts are available online (https://gitlab.com/alexander.kutschera/quickblast). Gene synteny was analysed using SyntTax with standard settings (Oberto, 2013).
Gene knockout in Pst and Pci
Knockout plasmids were constructed using the pGGKO‐blue plasmid derived from pK18mobsacB (oligonucleotides: Table S2). Flanking sequences (800–1000 bp) up‐ and downstream of the target gene were PCR‐amplified from genomic DNA (oligonucleotides; Table S2) and inserted into the pGGKO‐blue backbone by Golden Gate cloning using BpiI. Precursor knockout plasmids were transformed into E. coli DH5α cells and isolated after amplification using standard protocols. The GmR gene was amplified from plasmid pPS856 (Hoang et al., 1998) and inserted between the flanking sequences (oligonucleotides; Table S2). Knockout plasmids were amplified in E. coli DH5α, verified by restriction enzyme digestion and sequencing. Mutants of Pst and Pci were generated as described (Kvitko and Collmer, 2011). Mutants were verified by PCR and amplicon sequencing (oligonucleotides; Table S2).
Knockout complementation in Pst and Pci
Complementation plasmids are based on the backbone of the pGGKO‐blue plasmid. They contain the respective complementing gene under control of the promoter from the GmR cassette of pPS856 (oligonucleotides; Table S2). Gene sequences were PCR‐amplified from genomic DNA. Together with the promoter, they were inserted into the pGGKO‐blue backbone by Golden Gate cloning using BpiI. Complementation plasmids were amplified in E. coli DH5α, verified by restriction analysis and sequencing, and transformed into E. coli S17‐1 cells. They were used to conjugate the plasmids to Pst or Pci via biparental mating as described above.
LPS extraction and analysis
Extraction of crude LPS
Crude LPS was extracted from bacterial cells as described by Hitchcock and Brown (1983). Two millilitres of a KB liquid overnight culture (A 600 of 1.0) was harvested and washed with 0.15 M NaCl solution. The cells were resuspended in 1 mL lysis buffer (2 M Tris‐HCl pH 6.8, 4% (w/v) SDS, 8% (v/v) 2‐mercaptoethanol, 10% (w/v) glycerol, 0.02% (w/v) bromophenol blue in Millipore‐grade water (MP‐water)) and denatured at 100 °C for 5 min. After cooling to RT, 20 μL proteinase K (10 mg/mL, Sigma‐Aldrich, St Louis, MO, USA) was added to the suspension and incubated at 50 °C overnight.
Hot phenol‐water and phenol‐chloroform‐petroleum ether extraction of LPS
Preparation, purification and yields of LPS from bacterial cells are specified in the supporting information. Briefly, bacteria were grown in KB medium at 26 °C under shaking. Harvested cells were washed with solvents and enzyme digested. LPS was isolated via hot phenol‐water extraction and in most cases subsequent phenol‐chloroform‐petroleum ether extraction as described by Westphal and Jann (1965) and Galanos et al. (1969), respectively.
Gel permeation chromatography purification of LPS preparations
LPS of Pst was further purified by gel permeation chromatography (GPC) on Sephacryl S‐400 HR (GE Healthcare) on a column (2.5 × 120 cm) as described (Jimenez‐Barbero et al., 2002; Kutschera et al., 2019). Purification of 24.3 mg in one run yielded 11.3 mg of purified LPS. Selected LPS preparations were further fractionated on Sephacryl S‐200 HR (GE Healthcare, Chicago, IL, USA) on a column (1.5 × 120 cm) using a desoxycholate (DOC)‐containing buffer as described (Kutschera et al., 2019; Peterson and McGroarty, 1985). The exact procedure and the respective yields are specified in the supporting information.
Urea SDS‐PAGE, Bis‐Tris NuPAGE and silver staining
Urea SDS‐PAGE or Bis‐Tris NuPAGE and silver staining were performed as described (Kittelberger and Hilbink, 1993) with minor modifications. The detailed procedure is described in the supporting information.
Mild acidic hydrolysis of LPS
Mild acidic hydrolysis of LPS was performed as described (Ranf et al., 2015) with the following modification: re‐extraction of the water phase was performed four times with CHCl3.
Bacterial infection assay
Bacterial infection experiments with Pst and Pci strains were performed as described by Zipfel et al. (2004) and Starkey and Rahme (2009), respectively. Briefly, for spray inoculation, leaves of 6‐week‐old A. thaliana plants or leaves from store‐bought romaine lettuce were inoculated by spraying with a 108 CFU/mL suspension of Pst or Pci wild‐type and ∆wbpL bacteria containing 0.04% Silwet‐L77 (Lehle Seeds) and 10 mM MgCl2. For pressure infiltration, suspensions of Pst or the Pst ∆wbpL mutant strain with 10 mM MgCl2 were infiltrated into A. thaliana leaves with a needleless syringe at 106 CFU/mL if not stated otherwise. Stab inoculation of lettuce midrib with Pci strains (104 CFU/mL) was performed as described by Starkey and Rahme (2009). Infected A. thaliana leaves were harvested 4 and 72 h after inoculation. Bacterial CFU in the inoculum and leaf samples were determined by counting of colonies’ serial dilution on LSLB plates supplemented with 75 μg/mL rifampicin. Disease symptoms on the lettuce leaves and midrib were monitored over time. The size of the emerging lesions after stab inoculation was measured from images using Fiji (Schindelin et al., 2012).
Bacteria swarming motility assay
Protocols described by Burch et al. (2012) and Tremblay and Déziel (2008) were adapted and modified for the Pst and Pci swarming motility assessment. Fifty millilitres of freshly prepared KB medium containing 0.4% (w/v) agarose was poured into Petri dishes and dried for 45 min. For complementation strains, 50 μg/mL kanamycin was added to the medium. Bacterial cells were harvested from 3 mL KB overnight cultures by centrifugation (2000 g, RT, 5 min) and resuspended in phosphate‐buffered saline to an OD600 of 3.0. 5 μL of the suspension was spotted on the middle of a dried agar plate and dried for 5 min. After sealing, the plates were incubated for 16 h at 26 °C in light, photographed and bacterial motility was digitally assessed using Fiji (Schindelin et al., 2012).
Supporting information
Fig. S1 12% Bis‐Tris NuPAGE gel and silver staining of lipopolysaccharide (LPS) preparations from Pseudomonas cichorii ATCC10857/DSM50259 (Pci) wild‐type (WT), ∆wbpL::Gm R (∆wbpL) deletion mutant strains complemented with plasmid‐expressed wbpL orthologue from either Pci (+Pci‐wbpL) or Pst (+Pst‐wbpL). LPS was isolated by hot phenol‐water extraction followed by a phenol‐chloroform‐petroleum ether extraction. The amount of applied LPS is indicated below the respective lane.
Fig. S2 Exemplary images of (A) lettuce midrib stab infected and (B) lettuce leaves spray inoculated with Pseudomonas cichorii ATCC10857/DSM50259 wild‐type (WT) and ∆wbpL deletion mutants.
Table S1 Proteomes used for BLAST experiments.
Table S2 Oligonucleotides.
Text S1 Experimental procedure details. Hot phenol‐water and phenol‐chloroform‐petroleum ether extraction of LPS; GPC of LPS preparations using a desoxycholate‐containing buffer (DOC‐GPC), urea SDS‐PAGE, Bis‐Tris NuPAGE and silver staining.
Video_S1 Time lapse video of Arabidopsis thaliana plants infected with Pseudomonas syringae pv. tomato DC3000 wild‐type (WT) and ∆wbpL deletion mutants (∆). Leaves were inoculated by syringe‐infiltration with 107 CFU/mL Pst WT or Pst ∆wbpL suspension. Experiment was repeated two times with 106 CFU/mL bacterial suspension and one time with spray inoculation (108 CFU/mL) with similar results.
Acknowledgements
This work is funded by the German Research Foundation through Emmy Noether programme RA 2541/1‐1 (S.R.). We thank B. Kunz and H. Käßner (RC Borstel) for technical assistance and Uwe Mamat (RC Borstel) for his valuable comments on the manuscript. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Contributor Information
Alexander Kutschera, Email: alexander.kutschera@tum.de.
Stefanie Ranf, Email: ranf@wzw.tum.de.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Alexander, C. and Rietschel, E.T. (2001) Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7, 167–202. [PubMed] [Google Scholar]
- Bélanger, M. , Burrows, L.L. and Lam, J.S. (1999) Functional analysis of genes responsible for the synthesis of the B‐band o antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology, 145, 3505–3521. [DOI] [PubMed] [Google Scholar]
- Berry, M.C. , McGhee, G.C. , Zhao, Y. and Sundin, G.W. (2009) Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora . FEMS Microbiol. Lett. 291, 80–87. [DOI] [PubMed] [Google Scholar]
- Bogino, P.C. , Oliva, M.D.l.M. , Sorroche, F.G. , and Giordano, W. (2013) The role of bacterial biofilms and surface components in plant‐bacterial associations. Int. J. Mol. Sci. 14, 15838–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner, D. , Clarke, B.R. and Whitfield, C. (1994) Identification of an ATP‐binding cassette transport system required for translocation of lipopolysaccharide O‐antigen side‐chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol. Microbiol. 14, 505–519. [DOI] [PubMed] [Google Scholar]
- Burch, A.Y. , Shimada, B.K. , Mullin, S.W.A. , Dunlap, C.A. , Bowman, M.J. and Lindow, S.E. (2012) Pseudomonas syringae coordinates production of a motility‐enabling surfactant with flagellar assembly. J. Bacteriol. 194, 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, R.W. , Forsberg, L.S. and Kannenberg, E.L. (2010) Lipopolysaccharides in Rhizobium–legume symbioses. Subcell. Biochem. 53, 339–386. [DOI] [PubMed] [Google Scholar]
- Delucia, A.M. , Six, D.A. , Caughlan, R.E. , Gee, P. , Hunt, I. , Lam, J.S. and Dean, C.R. (2011). Lipopolysaccharide (LPS) inner‐core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. MBio, 2, e00142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W.‐L. , Lin, Y.‐C. , Lin, R.‐H. , Wei, C.‐F. , Huang, Y.‐C. , Peng, H.‐L. and Huang, H.‐C. (2010) Effects of galU mutation on Pseudomonas syringae–plant interactions. Mol. Plant‐Microbe Interact. 23, 1184–1196. [DOI] [PubMed] [Google Scholar]
- Dongari‐Bagtzoglou, A. (2008) Pathogenesis of mucosal biofilm infections: challenges and progress. Expert Rev. Anti. Infect. Ther. 6, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigues, P. , Demery‐Lafforgue, D. , Trigalet, A. , Dupin, P. , Samain, D. and Asselineau, J. (1985) Comparative studies of lipopolysaccharide and exopolysaccharide from a virulent strain of Pseudomonas solanacearum and from three avirulent mutants. J. Bacteriol. 162, 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanos, C. , Lüderitz, O. and Westphal, O. (1969) A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9, 245–249. [DOI] [PubMed] [Google Scholar]
- Greenfield, L.K. and Whitfield, C. (2012) Synthesis of lipopolysaccharide O‐antigens by ABC transporter‐dependent pathways. Carbohydr. Res. 356, 12–24. [DOI] [PubMed] [Google Scholar]
- Haefele, D.M. and Lindow, S.E. (1987) Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae . Appl. Environ. Microbiol. 53, 2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y. , King, J.D. , Huszczynski, S. , Kocíncová, D. and Lam, J.S. (2013) Five new genes are important for common polysaccharide antigen biosynthesis in Pseudomonas aeruginosa . MBio, 4, e00631–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey, R.M. (2003) Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. [DOI] [PubMed] [Google Scholar]
- Hitchcock, P.J. and Brown, T.M. (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver‐stained polyacrylamide gels. J. Bacteriol. 154, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, T.T. , Karkhoff‐Schweizer, R.R. , Kutchma, A.J. and Schweizer, H.P. (1998) A broad‐host‐range flp‐FRT recombination system for site‐specific excision of chromosomally‐located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene, 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Huang, T.‐P. , Somers, E.B. and Wong, A.C.L. (2006) Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide‐coupled biosynthetic genes in Stenotrophomonas maltophilia . J. Bacteriol. 188, 3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, S.T. and Lam, J.S. (2014) Synthesis of bacterial polysaccharides via the Wzx/Wzy‐dependent pathway. Can. J. Microbiol. 60, 697–716. [DOI] [PubMed] [Google Scholar]
- Jimenez‐Barbero, J. , Castro, C.D. , Evidente, A. , Molinaro, A. , Parrilli, M. , and Surico, G. (2002) Structural determination of the O‐specific chain of the lipopolysaccharide from Pseudomonas cichorii . Eur. J. Org. Chem. 2002, 1770–1775. [Google Scholar]
- Junker, F. , Rodríguez‐Herva, J.J. , Duque, E. , Ramos‐González, M.I. , Llamas, M. and Ramos, J.L. (2001) A WbpL mutant of Pseudomonas putida DOT‐T1E strain, which lacks the O‐antigenic side chain of lipopolysaccharides, is tolerant to organic solvent shocks. Extremophiles, 5, 93–99. [DOI] [PubMed] [Google Scholar]
- Kalynych, S. , Ruan, X. , Valvano, M.A. and Cygler, M. (2011) Structure‐guided investigation of lipopolysaccharide O‐antigen chain length regulators reveals regions critical for modal length control. J. Bacteriol. 193, 3710–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W. and Surette, M.G. (2005) Prevalence of surface swarming behavior in Salmonella . J. Bacteriol. 187, 6580–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.D. , Kocíncová, D. , Westman, E.L. and Lam, J.S. (2009) Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa . Innate Immun. 15, 261–312. [DOI] [PubMed] [Google Scholar]
- King, J.D. , Berry, S. , Clarke, B.R. , Morris, R.J. and Whitfield, C. (2014) Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. USA. 111, 6407–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelberger, R. and Hilbink, F. (1993) Sensitive silver‐staining detection of bacterial lipopolysaccharides in polyacrylamide gels. J. Biochem. Biophys. Methods. 26, 81–86. [DOI] [PubMed] [Google Scholar]
- Knirel, Y.A. , Shashkov, A.S. , Paramonov, N.A. , Zdorovenko, G.M. , Solyanic, L.P. and Yakovleva, L.M. (1993) Somatic antigens of pseudomonads: structure of the O‐specific polysaccharide of Pseudomonas syringae pv. tomato 140(R). Carbohydr. Res. 243, 199–204. [DOI] [PubMed] [Google Scholar]
- Knirel, Y.A. , Ovod, V.V. , Zdorovenko, G.M. , Gvozdyak, R.I. and Krohn, K.J. (1998) Structure of the O polysaccharide and immunochemical relationships between the lipopolysaccharides of Pseudomonas syringae pathovar tomato and pathovar maculicola . Eur. J. Biochem. 258, 657–661. [DOI] [PubMed] [Google Scholar]
- Kocincova, D. and Lam, J.S. (2011) Structural diversity of the core oligosaccharide domain of Pseudomonas aeruginosa lipopolysaccharide. Biochemistry, 76, 755–760. [DOI] [PubMed] [Google Scholar]
- Kutschera, A. and Ranf, S. (2019a) The multifaceted functions of lipopolysaccharide in plant–bacteria interactions. Biochimie, 159, 93–98. [DOI] [PubMed] [Google Scholar]
- Kutschera, A. and Ranf, S. (2019b). Variation of the O‐polysaccharide length distribution in plant‐associated Pseudomonas strains. figshare. 10.6084/m9.figshare.8208932.v2. [DOI] [Google Scholar]
- Kutschera, A. , Dawid, C. , Gisch, N. , Schmid, C. , Raasch, L. , Gerster, T. , Schäffer, M. , Smakowska‐Luzan, E. , Belkhadir, Y. , Vlot, A.C. , Chandler, C.E. , Schellenberger, R. , Schwudke, D. , Ernst, R.K. , Dorey, S. , Hückelhoven, R. , Hofmann, T. and Ranf, S. (2019) Bacterial medium‐chain 3‐hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science, 364, 178–181. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. and Collmer, A. (2011) Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Methods Mol. Biol. 712, 109–128. [DOI] [PubMed] [Google Scholar]
- Lam, J.S. , Taylor, V.L. , Islam, S.T. , Hao, Y. and Kocíncová, D. (2011) Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.‐W. , Jeong, S.‐Y. , In, Y.‐H. , Kim, K.‐Y. , So, J.‐S. and Chang, W.‐S. (2010) Lack of O‐polysaccharide enhances biofilm formation by Bradyrhizobium japonicum . Lett. Appl. Microbiol. 50, 452–456. [DOI] [PubMed] [Google Scholar]
- Lerouge, I. and Vanderleyden, J. (2002) O‐antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol. Rev. 26, 17–47. [DOI] [PubMed] [Google Scholar]
- Li, C.‐H. , Wang, K.‐C. , Hong, Y.‐H. , Chu, T.‐H. , Chu, Y.‐J. , Chou, I.‐C. , Lu, D.‐K. , Chen, C.‐Y. , Yang, W.‐C. , Lin, Y.‐M. and Cheng, C.‐P. (2014) Roles of different forms of lipopolysaccharides in Ralstonia solanacearum pathogenesis. Mol. Plant–Microbe Interact. 27, 471–478. [DOI] [PubMed] [Google Scholar]
- Liao, C.‐T. , Du, S.‐C. , Lo, H.‐H. and Hsiao, Y.‐M. (2014) The galU gene of Xanthomonas campestris pv. campestris is involved in bacterial attachment, cell motility, polysaccharide synthesis, virulence, and tolerance to various stresses. Arch. Microbiol. 196, 729–738. [DOI] [PubMed] [Google Scholar]
- Lindhout, T. , Lau, P.C.Y. , Brewer, D. and Lam, J.S. (2009) Truncation in the core oligosaccharide of lipopolysaccharide affects flagella‐mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology, 155, 3449–3460. [DOI] [PubMed] [Google Scholar]
- Lukácová, M. , Barák, I. and Kazár, J. (2008) Role of structural variations of polysaccharide antigens in the pathogenicity of Gram‐negative bacteria. Clin. Microbiol. Infect. 14, 200–206. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2015) Targeting of plant pattern recognition receptor‐triggered immunity by bacterial type‐III secretion system effectors. Curr. Opin. Microbiol. 23, 14–22. [DOI] [PubMed] [Google Scholar]
- Molinaro, A. , Newman, M.‐A. , Lanzetta, R. and Parrilli, M. (2009) The structures of lipopolysaccharides from plant‐associated Gram‐negative bacteria. Eur. J. Org. Chem. 2009, 5887–5896. [Google Scholar]
- Murphy, K. , Park, A.J. , Hao, Y. , Brewer, D. , Lam, J.S. and Khursigara, C.M. (2014) Influence of O‐polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 196, 1306–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, G.L. , Attridge, S.R. and Morona, R. (2006) Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham, B.D. and Trent, M.S. (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11, 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberto, J. (2013) SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinform. 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormeño‐Orrillo, E. , Rosenblueth, M. , Luyten, E. , Vanderleyden, J. and Martínez‐Romero, E. (2008) Mutations in lipopolysaccharide biosynthetic genes impair maize rhizosphere and root colonization of Rhizobium tropici CIAT899. Environ. Microbiol. 10, 1271–1284. [DOI] [PubMed] [Google Scholar]
- Peterson, A.A. and McGroarty, E.J. (1985) High‐molecular‐weight components in lipopolysaccharides of Salmonella typhimurium, Salmonella minnesota, and Escherichia coli . J. Bacteriol. 162, 738–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocelli, S. , Tondo, M.L. , Daurelio, L.D. and Orellano, E.G. (2012) Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS One, 7, e40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz, C.R.H. and Whitfield, C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M.M. , Guard‐Petter, J. and Carlson, R.W. (1997) A virulent isolate of Salmonella enteritidis produces a Salmonella typhi‐like lipopolysaccharide. J. Bacteriol. 179, 2126–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf, S. (2016) Immune sensing of lipopolysaccharide in plants and animals: Same but different. PLoS Pathog. 12, e1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf, S. , Gisch, N. , Schäffer, M. , Illig, T. , Westphal, L. , Knirel, Y.A. , Sánchez‐Carballo, P.M. , Zähringer, U. , Hückelhoven, R. , Lee, J. and Scheel, D. (2015) A lectin S‐domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana . Nat. Immunol. 16, 426–433. [DOI] [PubMed] [Google Scholar]
- Rapicavoli, J.N. , Blanco‐Ulate, B. , Muszyński, A. , Figueroa‐Balderas, R. , Morales‐Cruz, A. , Azadi, P. , Dobruchowska, J.M. , Castro, C. , Cantu, D. and Roper, M.C. (2018) Lipopolysaccharide O‐antigen delays plant innate immune recognition of Xylella fastidiosa . Nat. Commun. 9, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C.K. , Sims, E.H. , Kas, A. , Spencer, D.H. , Kutyavin, T.V. , Ivey, R.G. , Zhou, Y. , Kaul, R. , Clendenning, J.B. and Olson, M.V. (2002) Genetic variation at the O‐antigen biosynthetic locus in Pseudomonas aeruginosa . J. Bacteriol. 184, 3614–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta, H.L. , Burrows, L.L. , Pacan, J.C. and Lam, J.S. (1998) Three rhamnosyltransferases responsible for assembly of the A‐band D‐rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, Wbpl, is required for the initiation of both A‐band and B‐band lipopolysaccharide synthesis. Mol. Microbiol. 28, 1103–1119. [DOI] [PubMed] [Google Scholar]
- Rocchetta, H.L. , Burrows, L.L. and Lam, J.S. (1999) Genetics of O‐antigen biosynthesis in Pseudomonas aeruginosa . Microbiol. Mol. Biol. Rev. 63, 523–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, K. (2001). Thirty‐five years of phytobacteriology research with special emphasis on pathogenicity of Pseudomonas syringae In: Plant Pathogenic Bacteria: Proceedings of the 10th International Conference on Plant Pathogenic Bacteria, Charlottetown, Prince Edward Island, Canada, July 23–27, 2000, (De Boer S.H., ed.), pp. 109–117. Dordrecht, Netherlands: Springer. [Google Scholar]
- Rudolph, K.W.E. , Gross, M. , Neugebauer, M. , Hokawat, S. , Zachowski, A. , Wydra, K. and Klement, Z. (1989) Extracellular polysaccharides as determinants of leaf spot diseases caused by Pseudomonads and Xanthomonads In: Graniti A., Durbin R.D., Ballio A. (eds) Phytotoxins and Plant Pathogenesis, pp. 177–218. Berlin Heidelberg: Springer. [Google Scholar]
- Samuel, G. and Reeves, P. (2003) Biosynthesis of O‐antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O‐antigen assembly. Carbohydr. Res. 338, 2503–2519. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J.‐Y. , White, D.J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. and Cardona, A. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonejans, E. , Expert, D. and Toussaint, A. (1987) Characterization and virulence properties of Erwinia chrysanthemi lipopolysaccharide‐defective, ϕEC2‐resistant mutants. J. Bacteriol. 169, 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby, M.W. , Cerdeño‐Tárraga, A.M. , Vernikos, G.S. , Giddens, S.R. , Jackson, R.W. , Preston, G.M. , Zhang, X.‐X. , Moon, C.D. , Gehrig, S.M. , Godfrey, S.A.C. , Knight, C.G. , Malone, J.G. , Robinson, Z. , Spiers, A.J. , Harris, S. , Challis, G.L. , Yaxley, A.M. , Harris, D. , Seeger, K. , Murphy, L. , Rutter, S. , Squares, R. , Quail, M.A. , Saunders, E. , Mavromatis, K. , Brettin, T.S. , Bentley, S.D. , Hothersall, J. , Stephens, E. , Thomas, C.M. , Parkhill, J. , Levy, S.B. , Rainey, P.B. and Thomson, N.R. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens . Genome Biol. 10, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey, M. and Rahme, L.G. (2009) Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat. Protoc. 4, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C.K. , Pham, X.Q. , Erwin, A.L. , Mizoguchi, S.D. , Warrener, P. , Hickey, M.J. , Brinkman, F.S. , Hufnagle, W.O. , Kowalik, D.J. , Lagrou, M. , Garber, R.L. , Goltry, L. , Tolentino, E. , Westbrock‐Wadman, S. , Yuan, Y. , Brody, L.L. , Coulter, S.N. , Folger, K.R. , Kas, A. , Larbig, K. , Lim, R. , Smith, K. , Spencer, D. , Wong, G.K. , Wu, Z. , Paulsen, I.T. , Reizer, J. , Saier, M.H. , Hancock, R.E. , Lory, S. and Olson, M.V. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- Tans‐Kersten, J. , Huang, H. and Allen, C. (2001) Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183, 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E.N.H. , Papadopoulos, M. and Morona, R. (2014) Relationship between O‐antigen chain length and resistance to colicin E2 in Shigella flexneri . Microbiology, 160, 589–601. [DOI] [PubMed] [Google Scholar]
- Tremblay, J. and Déziel, E. (2008) Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J. Basic Microbiol. 48, 509–515. [DOI] [PubMed] [Google Scholar]
- Trent, M.S. , Stead, C.M. , Tran, A.X. and Hankins, J.V. (2006) Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12, 205–223. [DOI] [PubMed] [Google Scholar]
- Van den Bosch, L. , Manning, P.A. and Morona, R. (1997) Regulation of O‐antigen chain length is required for Shigella flexneri virulence. Mol. Microbiol. 23, 765–775. [DOI] [PubMed] [Google Scholar]
- Vanneste, J.L. (2017) The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 55, 377–399. [DOI] [PubMed] [Google Scholar]
- Wei, H.‐L. and Collmer, A. (2018) Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Mol. Plant Pathol. 19, 1779–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal, O. and Jann, K. (1965) Bacterial lipopolysaccharide‐extraction with phenol water and further application of procedure In: Methods in Carbohydrate Chemistry, Vol. 1 (Whistler R.L., BeMiller J.N. and Wolfrom M.L., eds), pp. 83–91. New York: Academic Press. [Google Scholar]
- Whitfield, C. , Amor, P.A. and Köplin, R. (1997) Modulation of the surface architecture of Gram‐negative bacteria by the action of surface polymer:lipid A–core ligase and by determinants of polymer chain length. Mol. Microbiol. 23, 629–638. [DOI] [PubMed] [Google Scholar]
- Young, J.M. (2010) Taxonomy of Pseudomonas syringae . J. Plant Pathol. 92, S5–S14. [Google Scholar]
- Zdorovenko, G.M. and Veremeichenko, S.N. (2001) Comparative characterization of the lipopolysaccharides of different Pseudomonas fluorescens biovar I strains. Microbiology, 70, 441–450. [Google Scholar]
- Zdorovenko, E.L. , Zatonsky, G.V. , Zdorovenko, G.M. , Pasichnyk, L.A. , Shashkov, A.S. and Knirel, Y.A. (2001) Structural heterogeneity in the lipopolysaccharides of Pseudomonas syringae with O‐polysaccharide chains having different repeating units. Carbohydr. Res. 336, 329–336. [DOI] [PubMed] [Google Scholar]
- Zeng, W. , Melotto, M. and He, S.Y. (2010) Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 21, 599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 12% Bis‐Tris NuPAGE gel and silver staining of lipopolysaccharide (LPS) preparations from Pseudomonas cichorii ATCC10857/DSM50259 (Pci) wild‐type (WT), ∆wbpL::Gm R (∆wbpL) deletion mutant strains complemented with plasmid‐expressed wbpL orthologue from either Pci (+Pci‐wbpL) or Pst (+Pst‐wbpL). LPS was isolated by hot phenol‐water extraction followed by a phenol‐chloroform‐petroleum ether extraction. The amount of applied LPS is indicated below the respective lane.
Fig. S2 Exemplary images of (A) lettuce midrib stab infected and (B) lettuce leaves spray inoculated with Pseudomonas cichorii ATCC10857/DSM50259 wild‐type (WT) and ∆wbpL deletion mutants.
Table S1 Proteomes used for BLAST experiments.
Table S2 Oligonucleotides.
Text S1 Experimental procedure details. Hot phenol‐water and phenol‐chloroform‐petroleum ether extraction of LPS; GPC of LPS preparations using a desoxycholate‐containing buffer (DOC‐GPC), urea SDS‐PAGE, Bis‐Tris NuPAGE and silver staining.
Video_S1 Time lapse video of Arabidopsis thaliana plants infected with Pseudomonas syringae pv. tomato DC3000 wild‐type (WT) and ∆wbpL deletion mutants (∆). Leaves were inoculated by syringe‐infiltration with 107 CFU/mL Pst WT or Pst ∆wbpL suspension. Experiment was repeated two times with 106 CFU/mL bacterial suspension and one time with spray inoculation (108 CFU/mL) with similar results.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


