Abstract
Introduction
Epidemic forecasting and prediction tools have the potential to provide actionable information in the midst of emerging epidemics. While numerous predictive studies were published during the 2016–2017 Zika Virus (ZIKV) pandemic, it remains unknown how timely, reproducible, and actionable the information produced by these studies was.
Methods
To improve the functional use of mathematical modeling in support of future infectious disease outbreaks, we conducted a systematic review of all ZIKV prediction studies published during the recent ZIKV pandemic using the PRISMA guidelines. Using MEDLINE, EMBASE, and grey literature review, we identified studies that forecasted, predicted, or simulated ecological or epidemiological phenomena related to the Zika pandemic that were published as of March 01, 2017. Eligible studies underwent evaluation of objectives, data sources, methods, timeliness, reproducibility, accessibility, and clarity by independent reviewers.
Results
2034 studies were identified, of which n = 73 met the eligibility criteria. Spatial spread, R0 (basic reproductive number), and epidemic dynamics were most commonly predicted, with few studies predicting Guillain-Barré Syndrome burden (4%), sexual transmission risk (4%), and intervention impact (4%). Most studies specifically examined populations in the Americas (52%), with few African-specific studies (4%). Case count (67%), vector (41%), and demographic data (37%) were the most common data sources. Real-time internet data and pathogen genomic information were used in 7% and 0% of studies, respectively, and social science and behavioral data were typically absent in modeling efforts. Deterministic models were favored over stochastic approaches. Forty percent of studies made model data entirely available, 29% provided all relevant model code, 43% presented uncertainty in all predictions, and 54% provided sufficient methodological detail to allow complete reproducibility. Fifty-one percent of predictions were published after the epidemic peak in the Americas. While the use of preprints improved the accessibility of ZIKV predictions by a median of 119 days sooner than journal publication dates, they were used in only 30% of studies.
Conclusions
Many ZIKV predictions were published during the 2016–2017 pandemic. The accessibility, reproducibility, timeliness, and incorporation of uncertainty in these published predictions varied and indicates there is substantial room for improvement. To enhance the utility of analytical tools for outbreak response it is essential to improve the sharing of model data, code, and preprints for future outbreaks, epidemics, and pandemics.
Author summary
Researchers published many studies which sought to predict and forecast important features of Zika virus (ZIKV) infections and their spread during the 2016–2017 ZIKV pandemic. We conducted a comprehensive review of such ZIKV prediction studies and evaluated their aims, the data sources they used, which methods were used, how timely they were published, and whether they provided sufficient information to be used or reproduced by others. Of the 73 studies evaluated, we found the accessibility, reproducibility, timeliness, and incorporation of uncertainty in these published predictions varied; indicating there is substantial room for improvement. We identified that the release of study findings before formal journal publication (‘pre-prints’) increased the timeliness of Zika prediction studies, but note they were infrequently used during this public health emergency. Addressing these areas can improve our understanding of Zika and other outbreaks and ensure forecasts can inform preparedness and response to future outbreaks, epidemics, and pandemics.
Introduction
Zika virus (ZIKV) is a positive-sense RNA flavivirus primarily transmitted through the Aedes aegypti mosquito [1–3]. While the majority of ZIKV infections are asymptomatic or present as a self-limiting febrile illness, strong evidence links ZIKV infection with microcephaly and a range of other birth defects, including limb deformity and retinopathy [4, 5]. ZIKV is also associated with Guillian-Barré syndrome, and a spectrum of other neurological disorders including meningoencephalitis and acute myelitis [6–9]. ZIKV was discovered in Uganda in a febrile non-human primate in 1947 [10], and the first human case was detected in Nigeria in 1953 [11]. ZIKV outbreaks were detected in Southeast Asia and the Pacific Islands in the early 21st century [12–16] followed by wide spread epidemics in the Americas from late 2014 onward, with a cumulative count of 583,144 suspected and 223,336 laboratory-confirmed Zika cases reported across 49 countries and territories by the end of 2017 [17, 18].
The Director-General of the World Health Organization declared the ZIKV pandemic a public health emergency of international concern (PHEIC) on February 1, 2016 [19]. The urgency for immediate coordinated global response was further accelerated by the Olympic and Paralympic games set to take place in Rio De Janeiro, Brazil during August 2016 [20]. As public health and medical research efforts for Zika increased across the Americas, scientists developed mathematical models to anticipate further outbreak spread, evaluate possible control measures, and gain insight into outbreak dynamics. These models used a range of data sources including case counts, relative vector abundance and distribution, population age structure, human mobility, climate information, viral sequence and serological data, and internet ‘big data’ streams. A range of statistical and mathematical models predicted the spread and other epidemic dynamics of ZIKV, as well as the burden of its complications [21–26].
While the WHO PHEIC status was lifted in November 2016 and the neotropical Zika pandemic has waned, the forecasting activities during the pandemic have not been systematically examined, particularly whether the studies were published in a manner and time-frame that was actionable during the Zika pandemic [27]. Such an exercise is critical, not only due to the ongoing risk of Zika globally [28], but also to inform modeling efforts for future major epidemics. We therefore undertook a systematic review to identify all published ZIKV prediction and forecasting studies during a time period which encompassed the PHEIC period and the peak and waning phase of the epidemic in the Americas. The first aim of this systematic review was to identify all published models that predicted, forecasted, or simulated any ecological or epidemiological phenomenon about the Zika pandemic and describe the predicted phenomena, the range of data sources used, and the modeling methods employed. This first aim sought to characterize the methods and data employed to answer key questions during the epidemic and to identify potentially underutilized data or methods. The second aim was to evaluate key scientific characteristics of these studies, including (i) accessibility and timeliness of the publication, (ii) reproducibility of the methods and access to the statistical code and data, and (iii) clarity of the presentation of the prediction results, including uncertainty in prediction estimates. The third aim was to describe the funding structure and major contributing sectors, such as government, industry, non-governmental organizations, or academia, behind these publications.
Methods
The PRISMA and Cochrane systematic review guidelines were adopted [29]. A panel of 12 investigators developed the systematic review protocol including the eligibility criteria and the data abstraction tool. No formal protocol was published for this systematic review.
Literature search strategy
We conducted a literature review using EMBASE and MEDLINE (PubMed) to identify all potentially eligible studies, which predicted or forecasted phenomena of the ZIKV pandemic. In MEDLINE we performed a highly sensitive search solely using the term “Zika.” A complementary search in EMBASE used a more specific ontology: “Zika AND (forecasting OR prediction OR model OR modeling OR modelling OR risk OR estimating OR dynamics) NOT mouse.” Both database searches were limited to articles published as of March 1, 2017, and the MEDLINE searches were restricted to publications released between February 1, 2016 and March 1, 2017. We used this end date to capture those models published during a time-frame of major operational relevance (that is, during the early, peak, and waning phase of the Americas epidemic). We complemented these database search results with ‘grey literature,’ including hand-searched bibliographies of major Zika epidemiological review articles [17, 30, 31] and contacting experts in the field of Zika modeling to identify any studies which may have been missed by the above search strategies.
Screening and eligibility determination
Using a two-reviewer system (with consensus for disagreements and conferral with a 3rd party adjudicator if a consensus was unable to be reached), all articles identified through the above literature search were screened by reviewing the title and abstract to remove all articles that clearly did not meet the eligibility criteria (below). The full text of the remaining articles was reviewed by two reviewers, with a third reviewer if a consensus was not reached by the first two reviewers. Eligibility was based on the following inclusion and exclusion criteria:
Inclusion criteria
We included studies that forecasted, predicted, or simulated any epidemiological or ecological phenomena about the Zika pandemic (including studies regarding previous outbreaks and epidemics, and regions outside the Americas), including but not limited to spatial spread risk, host and ecological range, disease and complication burden, economic impact transmission, and other epidemic dynamics. We didn’t require studies to explicitly present a future phenomenon risk, and we included time agnostic estimations of key epidemic parameters (for instance R0) and other phenomena.
Exclusion criteria
Did not include original analyses (e.g. review articles, perspective pieces, editorials, recommendations, and guidelines)
Duplicated studies
Animal and mosquito in-vivo pre-clinical models (e.g mouse, non-human primates)
In vitro studies
Descriptive epidemiological publications (e.g. describing case positive proportions, total case numbers, descriptive mapping of incidence by geographic information systems)
positive proportions, total case numbers, descriptive mapping of
incidence by geographic information systems)
Models which only examined causality of ZIKV in Guillain-Barré Syndrome (GBS) or microcephaly (rather than estimating risk or burden, for example)
Studies which only modeled non-ZIKV arboviruses, unless the central aim of the study was to explicitly forecast or predict ZIKV phenomena based on the known dynamics of other arboviruses
Data abstraction, collation, and analysis
Data were abstracted from the full texts by 12 reviewers (single-reviewer abstraction) across the domains of (i) objectives and study population, (ii) methodology and reproducibility, (iii) accessibility, timeliness, and other bibliometrics of eligible studies, and (iv) author affiliation and funding sources (S1 Table). In addition, the availability of preprint manuscripts was assessed using the pre-print search webtool search.bioPreprint [32], a server which identifies preprints from arXiv, bioRxiv, F1000Research, PeerJ Preprints, and Wellcome Open Research. Additionally, we manually searched arXiv and bioRxiv archives to confirm pre-print availability. These pre-print repositories are distinct from the advanced electronic publications made available by most journals after acceptance and peer review. Such ‘grey literature’ review extended beyond the cut-off date for the main literature database searches. A two-reviewer approach was used to ascertain whether eligible studies were made available as pre-print. From the abstracted data, descriptive analyses (medians, IQR, ranges, and proportions), and limited hypothesis testing were performed using Stata version 13.0 (StataCorp, College Station, TX, USA).
Results
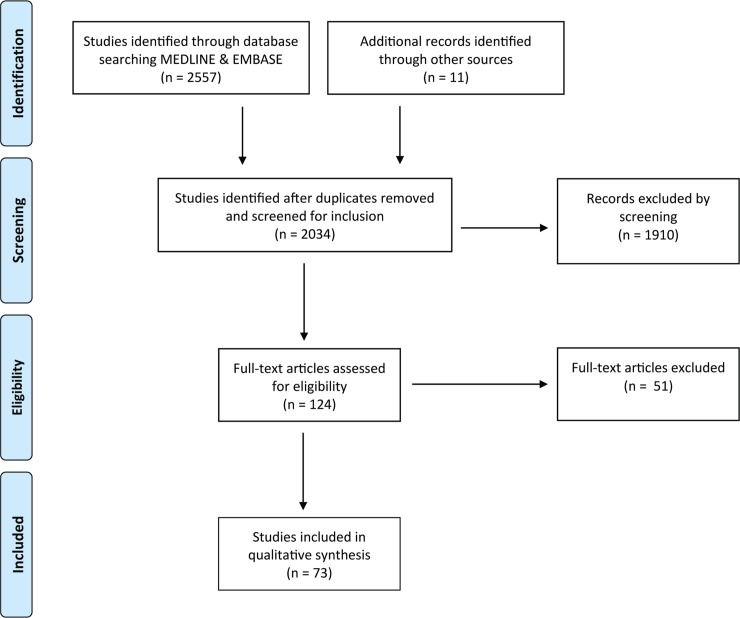
Of 2034 studies identified, 73 articles published predominantly from 2016 to 2017 met the inclusion criteria (Fig 1) [20–26, 28, 33–97]. The most commonly predicted phenomena were spatial spread (34%), followed by R0 (basic reproductive number) or RE (effective reproductive number) (29%), epidemic dynamics (peak size/timing, final size and trajectory) (28%), microcephaly burden (15%), and vector competence and ecology (12%) (Table 1). While R0 and RE are not explicitly measurable (e.g. to validate an R0 prediction), studies estimating these values were also included due to their close relationship with predictions about epidemic dynamics [24, 37, 43, 47–49, 53–55, 58, 62, 66, 67, 76, 77, 80–82, 89, 91]. Most of the geographically resolved predictions were concentrated in the Americas (42%) and Asia-Pacific (21%), while few studies were from Africa (4%). Across 73 studies, the most commonly used data were infection case counts, vector data, and demographic data, followed by climate, meteorological, earth science, and transport data (Table 2). Vector data included any vector-related predictor or parameter data, including: spatial distributions and geographic ranges, seasonal/relative abundance, and vector trait data such as biting rate, virus competence, urban preference, endophily, salinity tolerance, extrinsic incubation period, mortality rate, lifespan, and habitat suitability [23, 26, 28, 33, 34, 38, 40–44, 47, 52, 53, 55–58, 67, 68, 71, 72, 84–86, 88, 90, 93, 95, 98]. Genomic data was not used in any of the studies and few studies used novel real-time internet data streams such as those harnessing open access social media and internet search engine platforms.
Fig 1. PRISMA flow-chart indicating the number of studies identified, screened, and confirmed for eligibility into this systematic review.
Table 1. Objectives and study population of eligible studies.
| n | %a | |
|---|---|---|
| Total number of studies | 73 | 100 |
| Zika-related phenomenon forecasted or predictedb | ||
| Predicted microcephaly burdens | 11 | 15 |
| Guillain-Barré syndrome burden | 3 | 4 |
| Epidemic peak size | 4 | 5 |
| Epidemic peak timing | 4 | 5 |
| Epidemic curve trajectory | 8 | 11 |
| Epidemic final size | 5 | 7 |
| Spatial spread | 25 | 34 |
| Force of infection | 7 | 10 |
| Cost-effectiveness | 2 | 3 |
| Intervention impact | 3 | 4 |
| Case fatality ratio | 0 | 0 |
| Ro or REc | 21 | 29 |
| Sexual transmission risk | 3 | 4 |
| Vector competence / ecology | 9 | 12 |
| Otherd | 2 | 3 |
| Geographic region in which predictions madee | ||
| Africa | 3 | 4 |
| Americas (excluding Continental USA) | 31 | 42 |
| Asia–Pacific | 15 | 21 |
| Continental USA | 7 | 10 |
| Europe | 4 | 5 |
| Global | 18 | 24 |
aDenominator excludes those studies where unable or no basis to judge
bSome studies predicted more than one phenomenon
cIncluded estimates of R0
dEcological determinants of vector minimum abundance rate (n = 1); epidemic size and number of infections at time of first microcephaly case detected (n = 1)
eSome studies included >1 geographic category
Table 2. Data sources, methodology and reproducibility of eligible studies.
| N | %a | |
|---|---|---|
| Data types usedb | ||
| Case count | 49 | 67 |
| Demographic | 27 | 37 |
| Genomic sequence data | 0 | 0 |
| Climate, meteorological and earth science | 21 | 29 |
| Transport | 14 | 19 |
| Economic | 7 | 10 |
| Vector | 30 | 41 |
| Internet search engine, social media or news-wire scraping data | 5 | 7 |
| Otherc | 9 | 12 |
| Relevant data made available | ||
| Entirely | 29 | 40 |
| Partially | 27 | 37 |
| Not at all | 16 | 22 |
| Model type(s) used in analysisd | ||
| Stochastic | 21 | 29 |
| Deterministic | 56 | 76 |
| Availability of statistical modeling computational code (e.g. R script provided) | ||
| Entirely | 21 | 29 |
| Partly | 7 | 10 |
| Not at all | 45 | 62 |
| Clear and accurate visual display of the model output | ||
| Entirely | 49 | 67 |
| Partly | 20 | 27 |
| Not at all | 4 | 5 |
| Estimates of prediction uncertainty provided (e.g. confidence intervals) provided | ||
| Entirely | 31 | 43 |
| Partly | 13 | 18 |
| Not at all | 28 | 39 |
| Methods presented with a level of detail that allowed the study to be reproduced | ||
| Entirely | 37 | 54 |
| Partially | 28 | 41 |
| Not at all | 4 | 6 |
aDenominator excludes those studies where unable or no basis to judge
bSome studies used multiple data types
cViremia duration and dynamics (n = 3); sexual contact network (n = 2); semen viral persistence (n = 2), non-human primate demographics (n = 1), mammalian diversity (n = 1)
dSome studies used both stochastic and deterministic models
Only 40% of studies made all relevant source data entirely accessible, while more than 20% of the eligible studies did not make any source data available either directly (e.g. an associated data repository) or indirectly (e.g. a citation or web-link) (Table 2). The visual display of model output was at least partly clear and accurate in 95% of the studies. Over a third of the studies did not present estimates of prediction uncertainty. Approximately half of the studies did not entirely present methods with a level of detail to allow reproducibility. Over 60% of the studies did not provide any computational code used for the analyses. We classified more models as deterministic (76%) as opposed to stochastic. It should be emphasized we only ultimately evaluated whether a model was deterministic versus stochastic.
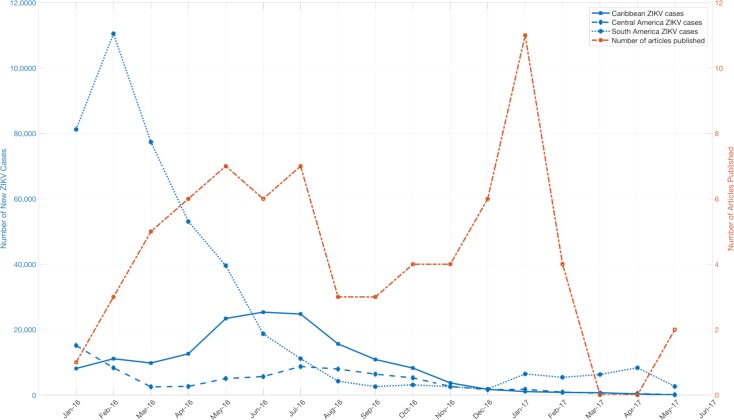
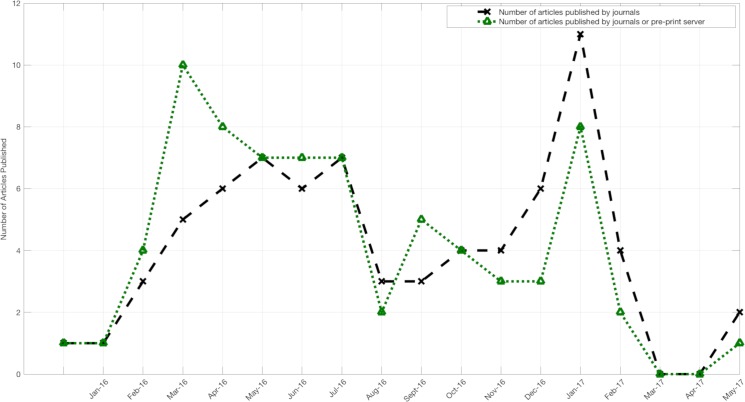
The large majority of published manuscripts were freely accessible (e.g. without a paywall), although 4% were published with paid access only (Table 3). Less than one third of manuscripts were posted on rapid preprint servers (e.g. bioRxiv) [99], prior to publication in a peer-reviewed journal. The median time from journal submission to e-journal publication was 93.5 days, with the maximum time greater than 1 year. This included delays after manuscript acceptance, 25% of the studies had delays of more than 24 days between acceptance and publication (Table 3). Most of the prediction studies were published late in the epidemic, well after the peaks in reported Zika cases (Fig 2, Fig 3). Submitting manuscripts to preprint servers made results available earlier by a median of 119 days (maximum 331 days, IQR 30–177 days) (Table 3). This shift led to more results being available close to the time of the 2016 South America and Central America epidemic peaks and prior to the epidemic peak in the Caribbean and the 2017 peak in Central America (Fig 2, Fig 3). Comparing the impact factor of journals accepting studies which were posted as preprints (versus the impact factor of those journals accepting studies which were not posted as pre-prints), there was no significant difference (median impact factor 4.37 vs. 4.45 respectively; p = 0.84 by Mann-Whitney U test).
Table 3. Accessibility, timeliness and other bibliometrics of eligible studies.
| n | %d | |
|---|---|---|
| Open accessa | 68 | 96 |
| Pre-print accessb | 22 | 30 |
| median | IQR (range) | |
| Journal impact factor | 4.37 | 2.65–7.62 (1.48–79.26) |
| Submission to acceptance time, days | 83 | 44–112 (0–256) |
| Acceptance to publication time, daysc | 15 | 7–24 (-255–279)e |
| Submission to publication time, days | 93.5 | 47–141 (1–389) |
aIncludes non-journal open access websites. Open access defined as able to be viewed without any payment or institutional journal license
bBiorxiv n = 19, ResearchGate n = 1, Bull WHO rapid journal pre-acceptance pre-print n = 2
cNegative values exist as Bull WHO articles published upon receipt (within 24 hrs) and then accepted later
dDenominator may vary in cases where these metrics were unable to be determined
ePublication time based on electronic journal version where available
Fig 2. Comparative trends of reported Zika cases in Latin American and publication times of Zika prediction studies.
Zika case counts were obtained from https://andersen-lab.com/ with permission.
Fig 3. Comparative trends in publication times of ZIKV prediction studies with and without the use of preprints.
Over 90% of the studies included authors with academic affiliations (Table 4). Government affiliated authors participated in a minority of studies, although this may simply reflect “in-house” operational models not being published through journals. Among studies with identifiable funding sources, funding was divided among several sources, though the most common was the United States government, which funded or partially funded 50% of the studies (Table 4). However, many of those studies and others had a variety of funding sources, 85% had at least one non-U.S. government source. Non-governmental organizations were the second most common source, being included in 35% of the studies.
Table 4. Author affiliation and funding source of eligible studies.
| Affiliation of authorsa | n | % |
| Academia | 68 | 93 |
| Govt (US) | 14 | 19 |
| Govt (non-US) | 19 | 26 |
| Industryb | 4 | 5 |
| NGO | 14 | 19 |
| Other type of organizationc | 4 | 5 |
| Funding sourced | n | %e |
| USG | ||
| CDC | 1 | 2 |
| DHS | 2 | 4 |
| DoD | 3 | 6 |
| LANL | 1 | 2 |
| NASA | 1 | 2 |
| NIH | 21 | 39 |
| NSA | 2 | 4 |
| NSF | 12 | 22 |
| USAID | 1 | 2 |
| USDA | 3 | 6 |
| Other USGf | 1 | 2 |
| Any USG | 27 | 50 |
| Any Non-US Govt | 46 | 85 |
| Any Industry | 3 | 6 |
| Any NGO | 19 | 35 |
| Any international normative body | 6 | 11 |
| Otherg | 6 | 11 |
aMultiple affiliations associated with some studies
bScientific contracting/consulting (n = 3), spatial epidemiology software (n = 1)
cWorld Health Organization (n = 2), European Centers for Disease Control (n = 1), HealthMap (n = 1)
dMultiple funding streams associated with some studies
eUnable to be determined or unfunded in a number of studies, denominator = 54
fState Dept of Health (TX)
gAcademic intramural funding (n = 5)
Discussion
Public health agencies, policy-makers, and other stakeholders are carefully examining the response to Zika. Such ‘lessons-learned’ exercises have been fruitful for prior pandemics and outbreaks, including Ebola, SARS, MERS-CoV, pH1N1, and chikungunya viruses. These exercises have included introspection, analysis, and recommended action with respect to research, public health, and policy agendas [100–105]. To date, public health ‘lessons-learned’ activities related to the Zika PHEIC have focused on improved ethics preparedness for rapid research during public health emergencies [106], identification of other high-epidemic-risk pathogens with relatively inadequate countermeasure investment [107], expedited approaches to vaccine and other medical countermeasure development [108], rapid data-sharing and material transfer [109–111], and enhancing the role of media communication during epidemics [112].
In contrast to existing reviews on models developed during the ZIKV pandemic, which described specific contributions of modeling [113] or validated analytical assessment of results [114], this systematic review focused on capturing lessons that could improve the functional use of mathematical modeling in support of future infectious disease outbreaks. Extending an approach used by Chretien et al. in their evaluation of Ebola models, we focused on aspects of the studies that likely are particularly relevant to their usefulness during an outbreak [104]. This included modeling methods and input data, timeliness and accessibility of the publications, reproducibility (e.g. provision of data and code), and the communication of uncertainty.
Our systematic review identified a large number of Zika models that predicted a wide range of epidemiological and ecological phenomena. The most commonly predicted phenomena were spatial spread, R0, epidemic dynamics, microcephaly burden, and vector competence. Notably few of the studies modeled the impact or cost-effectiveness of interventions, sexual transmission risk, or GBS burden. Not surprisingly, the majority of the studies were set in the Americas where most of the cases were reported during the pandemic. Notably one of the global gaps for understanding ZIKV dynamics is Africa, where ZIKV was discovered, is endemic, and poses a risk of future epidemics [115–117].
The leading data types for the examined studies were conventional case counts, vector, demographic, climate, and transport data. This finding reflects not only the availability but also the importance of such data. Case count data in particular are often hard to access but critical to many modeling approaches. Rapid sharing of case count data during international public health emergencies, as well as open, curated, rapidly accessible baseline demographic, human mobility, climate, and environmental datasets are essential to quickly leverage modeling and forecasting efforts [110]. Our review also identified several relatively underused data streams. First, socioeconomic and behavioral data were conspicuously absent. The lack of behavioral components in these models is concerning given the importance of these factors on disease dynamics [118]. Second, real-time internet-based data-streams, such as social media and internet search engine data, were used in a minority of ZIKV prediction studies identified in this systematic review. The limited use of internet ‘big data’ in the models suggests that either these data are of lower value for epidemic forecasting or that methods have yet to be developed to efficiently extract important information from them. Such data streams may be more commonly used in forecasting in the future as their strengths and weakness become clearer [119].
Genomic data were absent from these published models. During the pandemic, sequencing platforms were employed to generate data critical to diagnostic and countermeasure development [120], but our systematic review revealed that these data were not incorporated into prediction frameworks during the first year of ZIKV pandemic. This may reflect that early molecular epidemiology studies aimed to reconstruct the invasion and evolution of ZIKV rather than forecasting future changes [121, 122]. Some phylodynamic studies were published after the time period of the systematic review, with interesting results highlighting the possibility for phylogenetic data to provide unique insight into epidemic dynamics and possibly forecasting [122–124]. The relative delay of these studies (relative to those using other data sources) echoes a similar time lag of phylogenetic studies during the 2015 Ebola epidemic [104]. The lack of phylogenomic studies captured by this review also suggests that substantial bottlenecks still exist in using these data sources in epidemic response, despite advances in mobile near “real-time” sequencing technologies [120]. In the future, as new methods are developed, and genomic data become more readily available, the use of these data will likely become more common in prospective forecasting frameworks.
Our systematic review did not delve deeply into modeling approaches, but did identify a preponderance of deterministic as opposed to stochastic models. Both categories of models have pros and cons and their use is often informed by the specific question being addressed, in addition to data availability [125]. Deterministic models may generally be easier to produce, but they may have limitations for intrinsically stochastic processes like epidemics, such as underestimating uncertainty, although deterministic models can also be implemented to estimate uncertainty [126]. Uncertainty is particularly important in this context where uncertainties are generated by the epidemic itself, data collection, and analytical approaches. Moreover, forecasts are ideally used to inform the mobilization of resources to save lives, a context in which clearly characterizing uncertainties is paramount. This is also a clear area for improvement in model output reporting; only 43% of studies entirely reported estimates of uncertainty. However, studies which did not present, say a credible interval around a forecasted phenomenon, may still have incorporated uncertainty in other ways which were not captured by the data abstraction tool, for instance a scenario analysis or incorporating uncertainty around parameters in a deterministic model.
Our review also provided a unique evaluation of the more functional aspects of published predictions and forecasts. We determined that the visual clarity of model output was high but indicates room for improvement in publishing datasets used for model fitting and validation, and methodological detail to allow the study to be reproduced.
Our results also identified the need for improved sharing of computational code to permit full result reproducibility. Shared model code could also be adapted by other researchers during time-sensitive epidemics. An important caveat here is that appropriate expertise and rigorous validation should be exercised before the use of model code developed by others, in the same fashion as other biomedical research fields which frequently disseminate code-based research tools in their publications (for instance, pathogen genomics and bioinformatics).
The variable quality in sharing model code and methodological detail shown here does suggest that epidemic model reporting consensus guidelines, which establish a minimum standard for the reporting of epidemic modeling, may be valuable. A recent review of the modeling efforts for the Ebola epidemic also called for standardization of modeling practice [104]. Many other fields of biomedical research have established reporting guidelines to improve research quality and implementation [127–130]. While reporting guidelines have been proposed for population health modeling on a broader scale [131], none have been established for epidemics.
This review also indicated that a majority of studies (60%) did not completely disclose the data they used. To the extent permissible with ethical and privacy constraints, publishing the aggregated data used to fit and validate models is critical. Not only would sharing data support full reproducibility, but sharing would also enable other researchers to use data in their own complementary modeling efforts. Modelers could therefore help answer calls for increased data sharing during public health emergencies [104, 110, 132]. Exploring how data can be shared more openly and quickly during a public health emergency would be useful, as this remains a challenge.
Many studies identified in this review were published on a time-scale that was relevant to the Zika response. However, a large number of predictions were published well after the epidemic peaks, limiting their ability to inform the response. Nonetheless, those studies may well be used to inform other preparedness activities and contributed to the general knowledge of the biology, epidemiology and/or ecology of ZIKV. Further, results may have been informally shared with public health officials or other relevant decision makers prior to publication. Similar delays to publication have also been noted in an analysis of modeling efforts during the 2015 Ebola epidemic, which noted a median publication lag of around three months [103].
We identified two modifiable bottlenecks in the dissemination of results. First, delays from acceptance to journal publication were generally minimal (median 15 days), but a quarter of the evaluated studies had greater than 24 days delay from journal acceptance to publication. Immediate posting of accepted papers, as practiced by many journals, could cut this time down substantially. Second, we found that only 30% of studies were made available as preprints prior to peer review despite endorsements of preprints by major public agencies, funders, and journals. Those posted were available a median of 119 days prior to peer-reviewed publication. An analysis of preprints for all Zika publications over a similar time period found similar publication delays but much lower overall preprint use compared to the studies analyzed here (3.4% versus 30%) [133]. This greater adoption may indicate a changing preprint culture which was also reflected by our finding that preprint posting did not have a demonstrable effect on the impact factor of the journal in which the study was published, and we suggest that pre-prints be more frequently used in future public health emergencies, echoing other similar recent arguments [133].
Our review also provided a unique analysis of the funding sources and author affiliations of the published ZIKV prediction and forecast efforts across the ZIKV pandemic. These results indicated a range of stakeholders, and a diverse source of funding streams, including NGOs. We note that while academia contributed to the greatest volume of published studies, our search strategy would not have captured in-house models developed by US federal agencies or other unpublished models which may have provided direct operational support. Our data also suggested that the Government sector was the leading funder of all Zika prediction and forecasting studies during this period. Despite being a major stakeholder in ZIKV forecasts (e.g. prediction of whether it may have been feasible to plan a Phase III vaccine study in Latin America), we did not find evidence of pharmaceutical industry funded forecasting studies, although our search would not have identified unpublished models.
This systematic review has three important weaknesses. First, due to scale, a completely independent two-reviewer system was not used for abstracting most of the data and for evaluation of aspects such as reproducibility. Second, we did not formally search for preprint manuscripts as part of the literature searching phase of the systematic review, only assessing whether eligible manuscripts had corresponding preprints. We may have therefore missed important research that had been posted but not yet peer-reviewed. Lastly, we had to restrict the time frame for publications to consider in the review. This restriction again led to missing studies, some of which may have already been published but not yet posted in EMBASE or MEDLINE. An additional challenge with our systematic review was that we did not provide a specific definition of “forecasting” and “prediction” to reviewers (to avoid an overly strict eligibility criteria). This led to collection of data on phenomena such as R0, which is an estimate as opposed to a forecast or prediction that could be potentially validated with data.
Overall, the review identified several areas of improvement such as providing data and code, developing reporting standards, posting preprints, and communicating uncertainty. Addressing these areas can improve our understanding of Zika and other outbreaks and ensure that forecasts can inform preparedness and response to future outbreaks, epidemics, and pandemics.
Supporting information
(DOC)
(DOCX)
Acknowledgments
The authors wish to thank members of the Pandemic Prediction and Forecasting Science and Technology Working Group for their helpful feedback. The authors also acknowledge the valuable curated Zika case count data used with permission by the Andersen lab (https://andersen-lab.com/).
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Army, Department of Defense, USUHS, nor the U.S. Government. Several of the authors are US Government Employees. This work was prepared as part of their official duties. Title 17 U.S.C. § 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views, assertions, opinions or policies of the Uniformed Services University, the Department of Defense, the Centers for Disease Control & Prevention, or US Environmental Protection Agency. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government. More than one author is an employee of the U.S. Government and as such under the provisions of 17 U.S.C. 105, copyright protection is not available for this work
Data Availability
All relevant data is available within the manuscript and supporting information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46(5):521–34. 10.1016/0035-9203(52)90043-6 [DOI] [PubMed] [Google Scholar]
- 2.Lee VH, Moore DL. Vectors of the 1969 yellow fever epidemic on the Jos Plateau, Nigeria. Bull World Health Organ. 1972;46(5):669–73. [PMC free article] [PubMed] [Google Scholar]
- 3.Marchette NJ, Garcia R, Rudnick A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg. 1969;18(3):411–5. 10.4269/ajtmh.1969.18.411 [DOI] [PubMed] [Google Scholar]
- 4.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374(10):951–8. 10.1056/NEJMoa1600651 [DOI] [PubMed] [Google Scholar]
- 5.Oliveira Melo AS, Malinger G, Ximenes R, Szejnfeld PO, Alves Sampaio S, Bispo de Filippis AM. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg? Ultrasound in obstetrics & gynecology: the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2016;47(1):6–7. [DOI] [PubMed] [Google Scholar]
- 6.Carteaux G, Maquart M, Bedet A, Contou D, Brugieres P, Fourati S, et al. Zika Virus Associated with Meningoencephalitis. N Engl J Med. 2016;374(16):1595–6. 10.1056/NEJMc1602964 [DOI] [PubMed] [Google Scholar]
- 7.Mécharles S, Herrmann C, Poullain P, Tran TH, Deschamps N, Mathon G, et al. Acute myelitis due to Zika virus infection. Lancet. 2016;387(10026):1481 10.1016/S0140-6736(16)00644-9 [DOI] [PubMed] [Google Scholar]
- 8.Corrêa-Oliveira GE, do Amaral JL, da Fonseca BAL, Del-Ben CM. Zika virus infection followed by a first episode of psychosis: another flavivirus leading to pure psychiatric symptomatology. Rev Bras Psiquiatr. 2017;39(4):381–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Zika Situation Report: Microcephaly and Guillian-Barre Syndrome March 17, 2016. 2016. [Google Scholar]
- 10.DICK GW, KITCHEN SF, HADDOW AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46(5):509–20. 10.1016/0035-9203(52)90042-4 [DOI] [PubMed] [Google Scholar]
- 11.MACNAMARA FN. Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans R Soc Trop Med Hyg. 1954;48(2):139–45. 10.1016/0035-9203(54)90006-1 [DOI] [PubMed] [Google Scholar]
- 12.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet (London, England). 2016;387(10027):1531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360(24):2536–43. 10.1056/NEJMoa0805715 [DOI] [PubMed] [Google Scholar]
- 14.Kwong JC, Druce JD, Leder K. Zika virus infection acquired during brief travel to Indonesia. Am J Trop Med Hyg. 2013;89(3):516–7. 10.4269/ajtmh.13-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkasa A, Yudhaputri F, Haryanto S, Hayati RF, Ma'roef CN, Antonjaya U, et al. Isolation of Zika Virus from Febrile Patient, Indonesia. Emerg Infect Dis. 2016;22(5):924–5. 10.3201/eid2205.151915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alera MT, Hermann L, Tac-An IA, Klungthong C, Rutvisuttinunt W, Manasatienkij W, et al. Zika virus infection, Philippines, 2012. Emerg Infect Dis. 2015;21(4):722–4. 10.3201/eid2104.141707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, et al. Assessing the global threat from Zika virus. Science (New York, NY). 2016;353(6300):aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan American Health Organization, World Health Organization. Zika Cumulative Cases. 2018.
- 19.WHO Director-General summarizes the outcome of the Emergency Committee regarding clusters of microcephaly and Guillain-Barré syndrome [press release]. 2016.
- 20.Lewnard JA, Gonsalves G, Ko AI. Low Risk of International Zika Virus Spread due to the 2016 Olympics in Brazil. Annals of internal medicine. 2016;165(4):286–7. 10.7326/M16-1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGough SF, Brownstein JS, Hawkins JB, Santillana M. Forecasting Zika Incidence in the 2016 Latin America Outbreak Combining Traditional Disease Surveillance with Search, Social Media, and News Report Data. PLoS Negl Trop Dis. 2017;11(1):e0005295 10.1371/journal.pntd.0005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogoch II, Brady OJ, Kraemer MU, German M, Creatore MI, Brent S, et al. Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis. 2016;16(11):1237–45. 10.1016/S1473-3099(16)30270-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos J, Meneses BM. An integrated approach for the assessment of the Aedes aegypti and Aedes albopictus global spatial distribution, and determination of the zones susceptible to the development of Zika virus. Acta Trop. 2017;168:80–90. 10.1016/j.actatropica.2017.01.015 [DOI] [PubMed] [Google Scholar]
- 24.Ogden NH, Fazil A, Safronetz D, Drebot MA, Wallace J, Rees EE, et al. Risk of travel-related cases of Zika virus infection is predicted by transmission intensity in outbreak-affected countries. Parasites & vectors. 2017;10(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig AT, Butler MT, Pastore R, Paterson BJ, Durrheim DN. Acute flaccid paralysis incidence and Zika virus surveillance, Pacific Islands. Bull World Health Organ. 2017;95(1):69–75. 10.2471/BLT.16.171892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner LM, Chen N, Sarkar S. Global risk of Zika virus depends critically on vector status of Aedes albopictus. Lancet Infect Dis. 2016;16(5):522–3. 10.1016/S1473-3099(16)00176-6 [DOI] [PubMed] [Google Scholar]
- 27.Pan American Health Organization, World Health Organization. Zika Cumulative Cases. 2018. [Google Scholar]
- 28.Althouse BM, Vasilakis N, Sall AA, Diallo M, Weaver SC, Hanley KA. Potential for Zika Virus to Establish a Sylvatic Transmission Cycle in the Americas. PLoS Negl Trop Dis. 2016;10(12):e0005055 10.1371/journal.pntd.0005055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–12. 10.1016/j.jclinepi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 30.Althaus CL, Low N. How Relevant Is Sexual Transmission of Zika Virus? PLoS medicine. 2016;13(10):e1002157 10.1371/journal.pmed.1002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz-Menendez M, Trigo E, de la Calle-Prieto F, Arsuaga M. Zika virus infection during the Olympic Games in Rio: A fear or an actual risk? Revista clinica espanola. 2016. [DOI] [PubMed] [Google Scholar]
- 32.Iwema CL, LaDue J, Zack A, Chattopadhyay A. search.bioPreprint: a discovery tool for cutting edge, preprint biomedical research articles. F1000Research. 2016;5:1396 10.12688/f1000research.8798.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahrens KA, Hutcheon JA, Gavin L, Moskosky S. Reducing Unintended Pregnancies as a Strategy to Avert Zika-Related Microcephaly Births in the United States: A Simulation Study. Maternal and child health journal. 2017. [DOI] [PubMed] [Google Scholar]
- 34.Perkins TA, Siraj AS, Ruktanonchai CW, Kraemer MU, Tatem AJ. Model-based projections of Zika virus infections in childbearing women in the Americas. Nature microbiology. 2016;1(9):16126 10.1038/nmicrobiol.2016.126 [DOI] [PubMed] [Google Scholar]
- 35.Alfaro-Murillo JA, Parpia AS, Fitzpatrick MC, Tamagnan JA, Medlock J, Ndeffo-Mbah ML, et al. A Cost-Effectiveness Tool for Informing Policies on Zika Virus Control. PLoS Negl Trop Dis. 2016;10(5):e0004743 10.1371/journal.pntd.0004743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Althouse BM, Hanley KA, Diallo M, Sall AA, Ba Y, Faye O, et al. Impact of climate and mosquito vector abundance on sylvatic arbovirus circulation dynamics in Senegal. Am J Trop Med Hyg. 2015;92(1):88–97. 10.4269/ajtmh.13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andronico A, Dorleans F, Ferge JL, Salje H, Ghawche F, Signate A, et al. Real-Time Assessment of Health-Care Requirements During the Zika Virus Epidemic in Martinique. American journal of epidemiology. 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attaway DF, Waters NM, Geraghty EM, Jacobsen KH. Zika virus: Endemic and epidemic ranges of Aedes mosquito transmission. Journal of infection and public health. 2017;10(1):120–3. 10.1016/j.jiph.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 39.Baca-Carrasco D, Velasco-Hernandez JX. Sex, Mosquitoes and Epidemics: An Evaluation of Zika Disease Dynamics. Bulletin of mathematical biology. 2016;78(11):2228–42. 10.1007/s11538-016-0219-4 [DOI] [PubMed] [Google Scholar]
- 40.Bonyah E, Okosun KO. Mathematical Modeling of Zika Virus. Asian Pacific Journal of Tropical Disease. 2016;6(9):673–9. [Google Scholar]
- 41.Burattini MN, Coutinho FA, Lopez LF, Ximenes R, Quam M, Wilder-Smith A, et al. Potential exposure to Zika virus for foreign tourists during the 2016 Carnival and Olympic Games in Rio de Janeiro, Brazil. Epidemiol Infect. 2016;144(9):1904–6. 10.1017/S0950268816000649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butt AM, Siddique S, Gardner LM, Sarkar S, Lancelot R, Qamar R. Zika virus in Pakistan: the tip of the iceberg? The Lancet Global health. 2016;4(12):e913–e4. 10.1016/S2214-109X(16)30246-7 [DOI] [PubMed] [Google Scholar]
- 43.Caminade C, Turner J, Metelmann S, Hesson JC, Blagrove MS, Solomon T, et al. Global risk model for vector-borne transmission of Zika virus reveals the role of El Nino 2015. Proc Natl Acad Sci U S A. 2017;114(1):119–24. 10.1073/pnas.1614303114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson CJ, Dougherty ER, Getz W. An Ecological Assessment of the Pandemic Threat of Zika Virus. PLoS Negl Trop Dis. 2016;10(8):e0004968 10.1371/journal.pntd.0004968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, et al. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet (London, England). 2016;387(10033):2125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cetron M. Revision to CDC's Zika Travel Notices: Minimal Likelihood for Mosquito-Borne Zika Virus Transmission at Elevations Above 2,000 Meters. MMWR Morbidity and mortality weekly report. 2016;65(10):267–8. 10.15585/mmwr.mm6510e1 [DOI] [PubMed] [Google Scholar]
- 47.Champagne C, Salthouse DG, Paul R, Cao-Lormeau VM, Roche B, Cazelles B. Structure in the variability of the basic reproductive number (R0) for Zika epidemics in the Pacific islands. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowell G, Hincapie-Palacio D, Ospina J, Pell B, Tariq A, Dahal S, et al. Using Phenomenological Models to Characterize Transmissibility and Forecast Patterns and Final Burden of Zika Epidemics. PLoS currents. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinh L, Chowell G, Mizumoto K, Nishiura H. Estimating the subcritical transmissibility of the Zika outbreak in the State of Florida, USA, 2016. Theoretical biology & medical modelling. 2016;13(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dirlikov E, Kniss K, Major C, Thomas D, Virgen CA, Mayshack M, et al. Guillain-Barre Syndrome and Healthcare Needs during Zika Virus Transmission, Puerto Rico, 2016. Emerg Infect Dis. 2017;23(1):134–6. 10.3201/eid2301.161290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellington SR, Devine O, Bertolli J, Martinez Quinones A, Shapiro-Mendoza CK, Perez-Padilla J, et al. Estimating the Number of Pregnant Women Infected With Zika Virus and Expected Infants With Microcephaly Following the Zika Virus Outbreak in Puerto Rico, 2016. JAMA pediatrics. 2016;170(10):940–5. 10.1001/jamapediatrics.2016.2974 [DOI] [PubMed] [Google Scholar]
- 52.Evans MV, Dallas TA, Han BA, Murdock CC, Drake JM. Data-driven identification of potential Zika virus vectors. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferguson NM, Cucunuba ZM, Dorigatti I, Nedjati-Gilani GL, Donnelly CA, Basanez MG, et al. EPIDEMIOLOGY. Countering the Zika epidemic in Latin America. Science (New York, NY). 2016;353(6297):353–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funk S, Kucharski AJ, Camacho A, Eggo RM, Yakob L, Murray LM, et al. Comparative Analysis of Dengue and Zika Outbreaks Reveals Differences by Setting and Virus. PLoS Negl Trop Dis. 2016;10(12):e0005173 10.1371/journal.pntd.0005173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao D, Lou Y, He D, Porco TC, Kuang Y, Chowell G, et al. Prevention and Control of Zika as a Mosquito-Borne and Sexually Transmitted Disease: A Mathematical Modeling Analysis. Sci Rep. 2016;6:28070 10.1038/srep28070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Salazar C, Stephens CR, Sanchez-Cordero V. Predicting the Potential Role of Non-human Hosts in Zika Virus Maintenance. EcoHealth. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grills A, Morrison S, Nelson B, Miniota J, Watts A, Cetron MS. Projected Zika Virus Importation and Subsequent Ongoing Transmission after Travel to the 2016 Olympic and Paralympic Games—Country-Specific Assessment, July 2016. MMWR Morbidity and mortality weekly report. 2016;65(28):711–5. 10.15585/mmwr.mm6528e1 [DOI] [PubMed] [Google Scholar]
- 58.Guzzetta G, Poletti P, Montarsi F, Baldacchino F, Capelli G, Rizzoli A, et al. Assessing the potential risk of Zika virus epidemics in temperate areas with established Aedes albopictus populations. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21(15). [DOI] [PubMed] [Google Scholar]
- 59.Huff A, Allen T, Whiting K, Breit N, Arnold B. FLIRT-ing with Zika: A Web Application to Predict the Movement of Infected Travelers Validated Against the Current Zika Virus Epidemic. PLoS currents. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaenisch T, Rosenberger KD, Brito C, Brady O, Brasil P, Marques ET. Risk of microcephaly after Zika virus infection in Brazil, 2015 to 2016. Bull World Health Organ. 2017;95(3):191–8. 10.2471/BLT.16.178608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johansson MA, Mier-y-Teran-Romero L, Reefhuis J, Gilboa SM, Hills SL. Zika and the Risk of Microcephaly. N Engl J Med. 2016;375(1):1–4. 10.1056/NEJMp1605367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kucharski AJ, Funk S, Eggo RM, Mallet HP, Edmunds WJ, Nilles EJ. Transmission Dynamics of Zika Virus in Island Populations: A Modelling Analysis of the 2013–14 French Polynesia Outbreak. PLoS Negl Trop Dis. 2016;10(5):e0004726 10.1371/journal.pntd.0004726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lessler J, Ott CT, Carcelen AC, Konikoff JM, Williamson J, Bi Q, et al. Times to key events in Zika virus infection and implications for blood donation: a systematic review. Bull World Health Organ. 2016;94(11):841–9. 10.2471/BLT.16.174540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li R, Simmons KB, Bertolli J, Rivera-Garcia B, Cox S, Romero L, et al. Cost-effectiveness of Increasing Access to Contraception during the Zika Virus Outbreak, Puerto Rico, 2016. Emerg Infect Dis. 2017;23(1):74–82. 10.3201/eid2301.161322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Liu T, Lin L, Song T, Du X, Lin H, et al. Application of the analytic hierarchy approach to the risk assessment of Zika virus disease transmission in Guangdong Province, China. BMC Infect Dis. 2017;17(1):65 10.1186/s12879-016-2170-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majumder MS, Santillana M, Mekaru SR, McGinnis DP, Khan K, Brownstein JS. Utilizing Nontraditional Data Sources for Near Real-Time Estimation of Transmission Dynamics During the 2015–2016 Colombian Zika Virus Disease Outbreak. JMIR public health and surveillance. 2016;2(1):e30 10.2196/publichealth.5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manore CA, Ostfeld RS, Agusto FB, Gaff H, LaDeau SL. Defining the Risk of Zika and Chikungunya Virus Transmission in Human Population Centers of the Eastern United States. PLoS Negl Trop Dis. 2017;11(1):e0005255 10.1371/journal.pntd.0005255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez ME. Preventing Zika Virus Infection during Pregnancy Using a Seasonal Window of Opportunity for Conception. PLoS biology. 2016;14(7):e1002520 10.1371/journal.pbio.1002520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Massad E, Tan SH, Khan K, Wilder-Smith A. Estimated Zika virus importations to Europe by travellers from Brazil. Global health action. 2016;9:31669 10.3402/gha.v9.31669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Messina JP, Kraemer MU, Brady OJ, Pigott DM, Shearer FM, Weiss DJ, et al. Mapping global environmental suitability for Zika virus. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monaghan AJ, Morin CW, Steinhoff DF, Wilhelmi O, Hayden M, Quattrochi DA, et al. On the Seasonal Occurrence and Abundance of the Zika Virus Vector Mosquito Aedes Aegypti in the Contiguous United States. PLoS currents. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreno VM, Espinoza B, Bichara D, Holechek SA, Castillo-Chavez C. Role of short-term dispersal on the dynamics of Zika virus in an extreme idealized environment. Infect Dis Model. 2017;2(1):21–34. 10.1016/j.idm.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nah K, Mizumoto K, Miyamatsu Y, Yasuda Y, Kinoshita R, Nishiura H. Estimating risks of importation and local transmission of Zika virus infection. PeerJ. 2016;4:e1904 10.7717/peerj.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ndeffo-Mbah ML, Parpia AS, Galvani AP. Mitigating Prenatal Zika Virus Infection in the Americas. Annals of internal medicine. 2016;165(8):551–9. 10.7326/M16-0919 [DOI] [PubMed] [Google Scholar]
- 75.Nishiura H, Mizumoto K, Rock KS, Yasuda Y, Kinoshita R, Miyamatsu Y. A theoretical estimate of the risk of microcephaly during pregnancy with Zika virus infection. Epidemics. 2016;15:66–70. 10.1016/j.epidem.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 76.Nishiura H, Mizumoto K, Villamil-Gomez WE, Rodriguez-Morales AJ. Preliminary estimation of the basic reproduction number of Zika virus infection during Colombia epidemic, 2015–2016. Travel medicine and infectious disease. 2016;14(3):274–6. 10.1016/j.tmaid.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 77.Nishiura H, Kinoshita R, Mizumoto K, Yasuda Y, Nah K. Transmission potential of Zika virus infection in the South Pacific. Int J Infect Dis. 2016;45:95–7. 10.1016/j.ijid.2016.02.017 [DOI] [PubMed] [Google Scholar]
- 78.Quam MB, Wilder-Smith A. Estimated global exportations of Zika virus infections via travellers from Brazil from 2014 to 2015. Journal of travel medicine. 2016;23(6). [DOI] [PubMed] [Google Scholar]
- 79.Reefhuis J, Gilboa SM, Johansson MA, Valencia D, Simeone RM, Hills SL, et al. Projecting Month of Birth for At-Risk Infants after Zika Virus Disease Outbreaks. Emerg Infect Dis. 2016;22(5):828–32. 10.3201/eid2205.160290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riou J, Poletto C, Boelle PY. A comparative analysis of Chikungunya and Zika transmission. Epidemics. 2017. [DOI] [PubMed] [Google Scholar]
- 81.Rocklov J, Quam MB, Sudre B, German M, Kraemer MU, Brady O, et al. Assessing Seasonal Risks for the Introduction and Mosquito-borne Spread of Zika Virus in Europe. EBioMedicine. 2016;9:250–6. 10.1016/j.ebiom.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rojas DP, Dean NE, Yang Y, Kenah E, Quintero J, Tomasi S, et al. The epidemiology and transmissibility of Zika virus in Girardot and San Andres island, Colombia, September 2015 to January 2016. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21(28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saad-Roy CM, van den Driessche P, Ma J. Estimation of Zika virus prevalence by appearance of microcephaly. BMC Infect Dis. 2016;16(1):754 10.1186/s12879-016-2076-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samy AM, Thomas SM, Wahed AA, Cohoon KP, Peterson AT. Mapping the global geographic potential of Zika virus spread. Memorias do Instituto Oswaldo Cruz. 2016;111(9):559–60. 10.1590/0074-02760160149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scata M, Di Stefano A, Lio P, La Corte A. The Impact of Heterogeneity and Awareness in Modeling Epidemic Spreading on Multiplex Networks. Sci Rep. 2016;6:37105 10.1038/srep37105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang B, Xiao Y, Wu J. Implication of vaccination against dengue for Zika outbreak. Sci Rep. 2016;6:35623 10.1038/srep35623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teng Y, Bi D, Xie G, Jin Y, Huang Y, Lin B, et al. Dynamic Forecasting of Zika Epidemics Using Google Trends. PLoS One. 2017;12(1):e0165085 10.1371/journal.pone.0165085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teng Y, Bi D, Xie G, Jin Y, Huang Y, Lin B, et al. Model-informed risk assessment for Zika virus outbreaks in the Asia-Pacific regions. The Journal of infection. 2017. [DOI] [PubMed] [Google Scholar]
- 89.Towers S, Brauer F, Castillo-Chavez C, Falconar AK, Mubayi A, Romero-Vivas CM. Estimate of the reproduction number of the 2015 Zika virus outbreak in Barranquilla, Colombia, and estimation of the relative role of sexual transmission. Epidemics. 2016;17:50–5. 10.1016/j.epidem.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 90.Viennet E, Mincham G, Frentiu FD, Jansen CC, Montgomery BL, Harley D, et al. Epidemic Potential for Local Transmission of Zika Virus in 2015 and 2016 in Queensland, Australia. PLoS currents. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Villela DA, Bastos LS, LM DEC, Cruz OG, Gomes MF, Durovni B, et al. Zika in Rio de Janeiro: Assessment of basic reproduction number and comparison with dengue outbreaks. Epidemiol Infect. 2017:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiwanitkit S, Wiwanitkit V. Predicted pattern of Zika virus infection distribution with reference to rainfall in Thailand. Asian Pacific journal of tropical medicine. 2016;9(7):719–20. 10.1016/j.apjtm.2016.05.014 [DOI] [PubMed] [Google Scholar]
- 93.Yakob L, Kucharski A, Hue S, Edmunds WJ. Low risk of a sexually-transmitted Zika virus outbreak. Lancet Infect Dis. 2016;16(10):1100–2. 10.1016/S1473-3099(16)30324-3 [DOI] [PubMed] [Google Scholar]
- 94.Zinszer K, Morrison K, Brownstein JS, Marinho F, Santos AF, Nsoesie EO. Reconstruction of Zika Virus Introduction in Brazil. Emerg Infect Dis. 2017;23(1):91–4. 10.3201/eid2301.161274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bogoch II, Brady OJ, Kraemer MUG, German M, Creatore MI, Kulkarni MA, et al. Anticipating the international spread of Zika virus from Brazil. Lancet. 2016;387(10016):335–6. 10.1016/S0140-6736(16)00080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castro LA, Fox SJ, Chen X, Liu K, Bellan SE, Dimitrov NB, et al. Assessing real-time Zika risk in the United States. BMC Infect Dis. 2017;17(1):284 10.1186/s12879-017-2394-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodriguez-Barraquer I, Salje H, Lessler J, Cummings DA. Predicting intensities of Zika infection and microcephaly using transmission intensities of other arboviruses. bioRxiv. 2016. [Google Scholar]
- 98.Bogoch II, Brady OJ, Kraemer MUG, German M, Creatore MI, Brent S, et al. Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis. 2016;16(11):1237–45. 10.1016/S1473-3099(16)30270-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.bioRxiv: The Preprint Server for Biology: Cold Spring Harbor Laboratory; [Available from: https://www.biorxiv.org/.
- 100.Chowell G, Viboud C, Simonsen L, Merler S, Vespignani A. Perspectives on model forecasts of the 2014–2015 Ebola epidemic in West Africa: lessons and the way forward. BMC Med. 2017;15(1):42 10.1186/s12916-017-0811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng VC, Chan JF, To KK, Yuen KY. Clinical management and infection control of SARS: lessons learned. Antiviral research. 2013;100(2):407–19. 10.1016/j.antiviral.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johansson MA, Powers AM, Pesik N, Cohen NJ, Staples JE. Nowcasting the spread of chikungunya virus in the Americas. PLoS One. 2014;9(8):e104915 10.1371/journal.pone.0104915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zumla A, Alagaili AN, Cotten M, Azhar EI. Infectious diseases epidemic threats and mass gatherings: refocusing global attention on the continuing spread of the Middle East Respiratory syndrome coronavirus (MERS-CoV). BMC Med. 14 England2016. p. 132 10.1186/s12916-016-0686-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chretien JP, Riley S, George DB. Mathematical modeling of the West Africa Ebola epidemic. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.World Health Organization. Preparing for the second wave: lessons from current outbreaks. Geneva; 2009. August 28, 2009. Contract No.: Briefing Note 9. [Google Scholar]
- 106.Saenz C. Zika virus: ethics preparedness for old and new challenges. The Lancet Global health. 2016;4(10):e686 10.1016/S2214-109X(16)30222-4 [DOI] [PubMed] [Google Scholar]
- 107.World Health Organization. Methodology for Prioritizing Severe Emerging Diseases for Research and Development. Geneva; 2017 February.
- 108.Malone RW, Homan J, Callahan MV, Glasspool-Malone J, Damodaran L, Schneider Ade B, et al. Zika Virus: Medical Countermeasure Development Challenges. PLoS Negl Trop Dis. 2016;10(3):e0004530 10.1371/journal.pntd.0004530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wellcome Trust. Statement on data sharing in public health emergencies 2016. [Available from: https://wellcome.ac.uk/what-we-do/our-work/statement-data-sharing-public-health-emergencies. [Google Scholar]
- 110.Chretien JP, Rivers CM, Johansson MA. Make Data Sharing Routine to Prepare for Public Health Emergencies. PLoS medicine. 2016;13(8):e1002109 10.1371/journal.pmed.1002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yozwiak NL, Schaffner SF, Sabeti PC. Data sharing: Make outbreak research open access. Nature. 2015;518(7540):477–9. 10.1038/518477a [DOI] [PubMed] [Google Scholar]
- 112.United Nations Educational Scientific, and Cultural Organization,. Inform, engage, investigate: Lessons learned from Zika outbreak 2016. November 30, 2017. Available from: http://www.unesco.org/new/en/media-services/single-view/news/inform_engage_investigate_lessons_learned_from_zika_outbr/. [Google Scholar]
- 113.Keegan LT, Lessler J, Johansson MA. Quantifying Zika: Advancing the Epidemiology of Zika With Quantitative Models. J Infect Dis. 2017;216(suppl_10):S884–S90. 10.1093/infdis/jix437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carlson CJ, Dougherty E, Boots M, Getz W, Ryan SJ. Consensus and conflict among ecological forecasts of Zika virus outbreaks in the United States. Sci Rep. 2018;8(1):4921 10.1038/s41598-018-22989-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pollett S, Melendrez MC, Maljkovic Berry I, Duchêne S, Salje H, Cummings DAT, et al. Understanding dengue virus evolution to support epidemic surveillance and counter-measure development. Infect Genet Evol. 2018;62:279–95. 10.1016/j.meegid.2018.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lourenço J, de Lourdes Monteiro M, Valdez T, Monteiro Rodrigues J, Pybus O, Rodrigues Faria N. Epidemiology of the Zika Virus Outbreak in the Cabo Verde Islands, West Africa. PLoS Curr. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kraemer MUG, Brady OJ, Watts A, German M, Hay SI, Khan K, et al. Zika virus transmission in Angola and the potential for further spread to other African settings. Trans R Soc Trop Med Hyg. 2017;111(11):527–9. 10.1093/trstmh/try001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ajelli M, Moise IK, Hutchings T, Brown SC, Kumar N, Johnson NF, et al. Host outdoor exposure variability affects the transmission and spread of Zika virus: Insights for epidemic control. PLoS Negl Trop Dis. 2017;11(9):e0005851 10.1371/journal.pntd.0005851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pollett S, Althouse BM, Forshey B, Rutherford GW, Jarman RG. Internet-based biosurveillance methods for vector-borne diseases: Are they novel public health tools or just novelties? PLoS Negl Trop Dis. 2017;11(11):e0005871 10.1371/journal.pntd.0005871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Faria NR, Sabino EC, Nunes MR, Alcantara LC, Loman NJ, Pybus OG. Mobile real-time surveillance of Zika virus in Brazil. Genome medicine. 2016;8(1):97 10.1186/s13073-016-0356-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Faria NR, Azevedo Rdo S, Kraemer MU, Souza R, Cunha MS, Hill SC, et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science (New York, NY). 2016;352(6283):345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Faria NR, Quick J, Claro IM, Thézé J, de Jesus JG, Giovanetti M, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546(7658):406–10. 10.1038/nature22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grubaugh ND, Ladner JT, Kraemer MUG, Dudas G, Tan AL, Gangavarapu K, et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature. 2017;546(7658):401–5. 10.1038/nature22400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thézé J, Li T, du Plessis L, Bouquet J, Kraemer MUG, Somasekar S, et al. Genomic Epidemiology Reconstructs the Introduction and Spread of Zika Virus in Central America and Mexico. Cell Host Microbe. 2018;23(6):855–64.e7. 10.1016/j.chom.2018.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Del Valle SY, McMahon BH, Asher J, Hatchett R, Lega JC, Brown HE, et al. Summary results of the 2014–2015 DARPA Chikungunya challenge. BMC Infectious Diseases. 2018;18(1):245 10.1186/s12879-018-3124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.King AA, Domenech de Cellès M, Magpantay FM, Rohani P. Avoidable errors in the modelling of outbreaks of emerging pathogens, with special reference to Ebola. Proc Biol Sci. 2015;282(1806):20150347 10.1098/rspb.2015.0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6(11):e012799 10.1136/bmjopen-2016-012799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schulz KF, Altman DG, Moher D, Group C. CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. Int J Surg. 2011;9(8):672–7. 10.1016/j.ijsu.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 129.White RG, Hakim AJ, Salganik MJ, Spiller MW, Johnston LG, Kerr L, et al. Strengthening the Reporting of Observational Studies in Epidemiology for respondent-driven sampling studies: "STROBE-RDS" statement. J Clin Epidemiol. 2015;68(12):1463–71. 10.1016/j.jclinepi.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1 10.1186/2046-4053-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bennett C, Manuel DG. Reporting guidelines for modelling studies. BMC Med Res Methodol. 2012;12:168 10.1186/1471-2288-12-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Littler K, Boon WM, Carson G, Depoortere E, Mathewson S, Mietchen D, et al. Progress in promoting data sharing in public health emergencies. Bull World Health Organ. 2017;95(4):243 10.2471/BLT.17.192096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Johansson MA, Reich NG, Meyers LA, Lipsitch M. Preprints: An underutilized mechanism to accelerate outbreak science. PLoS Med. 2018;15(4):e1002549 10.1371/journal.pmed.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
Data Availability Statement
All relevant data is available within the manuscript and supporting information files.