The type III secretion system (T3SS) is a protein complex produced by many Gram-negative pathogens. It is capable of injecting effector proteins into host cells that can manipulate cell metabolism and have toxic effects. Understanding how the T3SS is regulated is important in understanding the pathogenesis of bacteria with such systems. Here, we show that RNase E, which is typically thought of as a global regulator of RNA stability, plays a role in regulating the T3SS in Pseudomonas aeruginosa. Depleting RNase E results in the loss of T3SS gene expression as well as a concomitant increase in biofilm formation. These observations are reminiscent of the phenotypes associated with the loss of activity of the posttranscriptional regulator RsmA. However, RNase E-mediated regulation of these systems does not involve changes in the abundance of RsmA and is independent of the known small regulatory RNAs that modulate RsmA activity.
Keywords: GacA, global regulatory networks
ABSTRACT
Pseudomonas aeruginosa is an important opportunistic pathogen that employs a type III secretion system (T3SS) to inject effector proteins into host cells. Using a protein depletion system, we show that the endoribonuclease RNase E positively regulates expression of the T3SS genes. We also present evidence that RNase E antagonizes the expression of genes of the type VI secretion system and limits biofilm production in P. aeruginosa. Thus, RNase E, which is thought to be the principal endoribonuclease involved in the initiation of RNA degradation in P. aeruginosa, plays a key role in controlling the production of factors involved in both acute and chronic stages of infection. Although the posttranscriptional regulator RsmA is also known to positively regulate expression of the T3SS genes, we find that RNase E does not appreciably influence the abundance of RsmA in P. aeruginosa. Moreover, we show that RNase E still exerts its effects on T3SS gene expression in cells lacking all four of the key small regulatory RNAs that function by sequestering RsmA.
IMPORTANCE The type III secretion system (T3SS) is a protein complex produced by many Gram-negative pathogens. It is capable of injecting effector proteins into host cells that can manipulate cell metabolism and have toxic effects. Understanding how the T3SS is regulated is important in understanding the pathogenesis of bacteria with such systems. Here, we show that RNase E, which is typically thought of as a global regulator of RNA stability, plays a role in regulating the T3SS in Pseudomonas aeruginosa. Depleting RNase E results in the loss of T3SS gene expression as well as a concomitant increase in biofilm formation. These observations are reminiscent of the phenotypes associated with the loss of activity of the posttranscriptional regulator RsmA. However, RNase E-mediated regulation of these systems does not involve changes in the abundance of RsmA and is independent of the known small regulatory RNAs that modulate RsmA activity.
INTRODUCTION
Pseudomonas aeruginosa is a pathogenic bacterium capable of causing a number of opportunistic infections. P. aeruginosa infections are a major cause of morbidity and mortality in patients with cystic fibrosis (1). Patients with wounds or burns are also susceptible to P. aeruginosa infection (2). Urinary tract and lung infections with this organism are common in hospitalized patients who require catheters or ventilator-assisted breathing (2). P. aeruginosa infections are especially difficult to treat because these bacteria have a naturally high level of resistance to antibiotic treatment (1–3).
Prominent among those virulence factors that P. aeruginosa uses to infect the host is the type III secretion system (T3SS). P. aeruginosa uses its T3SS to inject effector proteins into host cells (4); the toxic effectors delivered by the T3SSs are critical for the organism to establish an acute infection and are important for the survival of the bacterium inside the host, as well as for the systemic spread of the bacterium within the host (5).
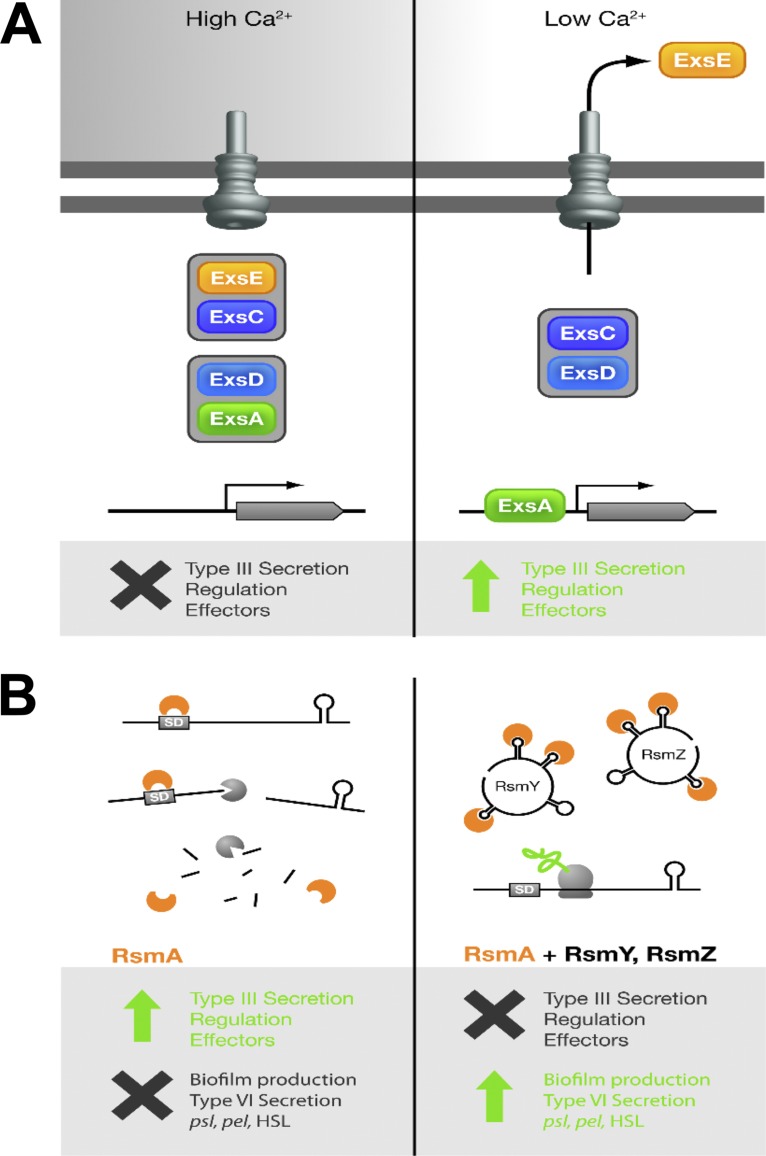
Effector secretion via the T3SS is triggered by host-cell contact or, in vitro, by removing calcium from the growth medium (6, 7). Triggering of effector secretion results in a concomitant upregulation of T3SS gene expression, which results from the activation of the master activator of T3SS gene transcription, ExsA (8). If effector secretion is off, ExsA is sequestered by the antiactivator protein ExsD, allowing only low-level expression of T3SS genes (9). Upon triggering of effector secretion, the sensor protein ExsE is exported via the T3SS, freeing it from the chaperone protein ExsC and allowing ExsC to bind ExsD, thereby releasing ExsA such that it can activate the transcription of the T3SS genes (Fig. 1A) (8, 53).
FIG 1.
Models of the control of type III secretion in P. aeruginosa and the control of RsmA activity. (A) Model of type III secretion system regulation in P. aeruginosa. Under conditions of high calcium (Ca2+) concentrations (left), ExsE forms a complex with ExsC, ensuring that ExsA forms a complex with ExsD, thus preventing expression of the type III secretion genes. In response to low calcium (right), ExsE is secreted, allowing ExsC to form a complex with ExsD, thus allowing ExsA to activate transcription from target promoters. In both panels, the gray lines correspond to the outer membrane (uppermost line) and inner membrane. (B) Reciprocal regulation of acute and chronic virulence determinates by the posttranscriptional regulator RsmA and the sRNAs RsmY and RsmZ. When RsmA (depicted in orange) recognizes GGA target sites within, or in close proximity to, Shine-Dalgarno sequences (SD) on target mRNAs, translation is inhibited, which can result in the degradation of target transcripts. This promotes expression of the T3SS genes (indicated by the arrow) and inhibits biofilm formation as well as expression of T6SS genes (indicated by the cross). Under conditions in which the production of the sRNAs RsmY and RsmZ is upregulated, these sRNAs interact with RsmA, thus sequestering it from target mRNA species, resulting in their translation. This serves to repress T3SS gene expression but enhance biofilm formation and T6SS gene expression. HSL, homoserine lactone.
ExsA expression is positivity regulated by cyclic AMP (cAMP) levels in the cell. The cAMP-dependent DNA binding protein Vfr has been shown to bind to the ExsA promoter and activate its transcription (10). ExsA is also positively regulated at the posttranscriptional level. The RNA helicase DeaD promotes the translation of ExsA by likely reducing secondary structures within the ExsA mRNA (11).
Virulence gene expression in P. aeruginosa is tied into the global regulatory networks of the cell (8, 51). One of the most important of these networks is controlled by the Gac/RsmA system, which reciprocally controls the expression of genes involved in acute and chronic infection (12, 13) (Fig. 1B). In particular, the sensor kinase GacS controls the activity of the response regulator GacA, which in turn positively regulates the expression of the rsmY and rsmZ genes encoding the small RNAs RsmY and RsmZ, respectively (14). These two small RNAs exert their influence on gene expression by binding to and modulating the activity of the RNA-binding protein RsmA, which is a posttranscriptional regulator that typically acts to repress the translation of target transcripts and influences the expression of hundreds of genes (15). For example, RsmA acts to repress type VI secretion system (T6SS) gene expression but activates T3SS gene expression (15, 16). This signaling cascade is subject to further fine control by two hybrid sensor kinases, RetS and LadS, with RetS serving to inhibit the activity of GacS (12, 13, 17) and LadS serving to promote the activity of GacS (18).
In many Gram-negative bacteria, the endonuclease RNase E is highly conserved and is essential for cell survival (19, 20). In Escherichia coli, RNase E is the principal enzyme involved in mRNA degradation and can interact with polynucleotide phosphorylase, as well as the RNA helicase RhlB and the glycolytic enzyme enolase, to form a high-molecular-weight complex referred to as the degradosome (21).
In several pathogenic bacteria, RNase E has been implicated in the control of virulence gene expression. In Escherichia coli O157:H7, RNase E deficiency has been linked to reduced stx2 phage replication (22). In Yersinia spp., RNase E is thought to positively regulate the expression of genes encoding the type III secretion system and influence intracellular survival (23). However, assessing the contribution of RNase E to the control of gene expression in pathogenic bacteria is complicated by the fact that the protein is often essential. Here, we show that depletion of RNase E in P. aeruginosa results in decreased expression of T3SS genes and increased biofilm formation and expression of genes encoding the T6SS. Our findings suggest that RNase E plays a critical role in the control of both acute and chronic virulence factors in P. aeruginosa.
RESULTS
Use of a ClpXP-based protein depletion system to study RNase E in P. aeruginosa.
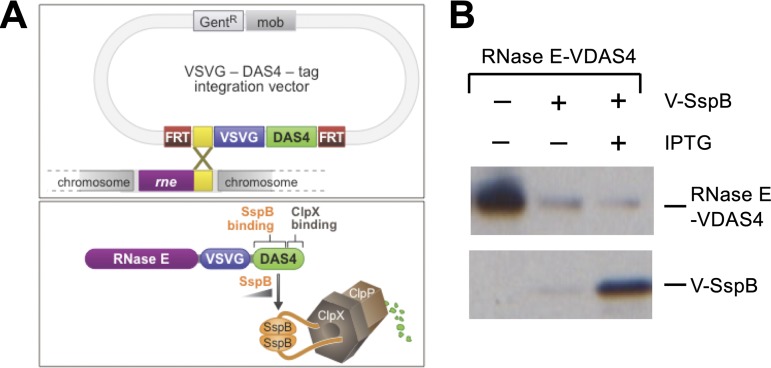
In E. coli, RNase E is essential for viability (19, 20), and transposon insertion sequencing experiments suggest that RNase E is essential in P. aeruginosa strain PAO1 as well as other strains of P. aeruginosa (24, 25). We therefore sought to determine whether we could study the effects of RNase E on the expression of virulence genes in P. aeruginosa using a previously described ClpXP protease-based protein depletion system (26, 27) (Fig. 2A). This system requires the fusion of a small peptide tag, referred to here as DAS4, to the C terminus of a protein of interest, in this case, RNase E. The DAS4 tag contains a low-affinity binding site for ClpX and a high-affinity binding site for the adapter protein SspB. The DAS4-tagged protein is bound by SspB and shuttled to the ClpXP protease for degradation (27). The rate of degradation of the tagged protein by ClpXP is therefore determined by the concentration of SspB in the cell (27). The RNase E depletion strain we constructed (PAO1 ΔsspB rne-VDAS4) carries a deletion of the native sspB gene and contains an RNase E gene that is modified such that it produces RNase E with both a vesicular stomatitis virus glycoprotein (VSV-G) epitope tag and a DAS4 tag at its C terminus (RNase E-VDAS4) (Fig. 2A). The PAO1 ΔsspB rne-VDAS4 depletion strain was first transformed with plasmid pV-SspB, which directs the production of SspB with an N-terminal VSV-G tag (V-SspB) under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. Figure 2B shows that the IPTG-inducible synthesis of V-SspB results in the depletion of RNase E in cells of the PAO1 ΔsspB rne-VDAS4 strain. We note that even in the absence of IPTG, the low level of SspB expressed from the lac promoter on the pPSV38 vector results in appreciable depletion of RNase E; however, the addition of IPTG results in further depletion of RNase E.
FIG 2.
ClpXP-based controllable protein degradation system. (A) Top, diagram of the VSV-G-DAS4 tag integration vector and its use in tagging RNase E with a VSV-G epitope and DAS4 depletion tag. GentR, gentamicin resistant. Bottom, schematic representation of the ClpXP-based protein depletion system. Degradation of the RNase E-VDAS4 protein is depicted. (B) Western blot showing that RNase E depletion is dependent on sspB expression. V corresponds to the VSV-G epitope tag. The DAS4 tag allows for depletion of RNase E-VDAS4 in a manner dependent upon the intracellular concentration of the SspB adapter protein that directs RNase E-VDAS4 to the ClpXP protease complex. SspB synthesis is under the control of an IPTG-inducible promoter. FRT, FLP recombinase.
Depletion of RNase E antagonizes the expression of T3SS genes.
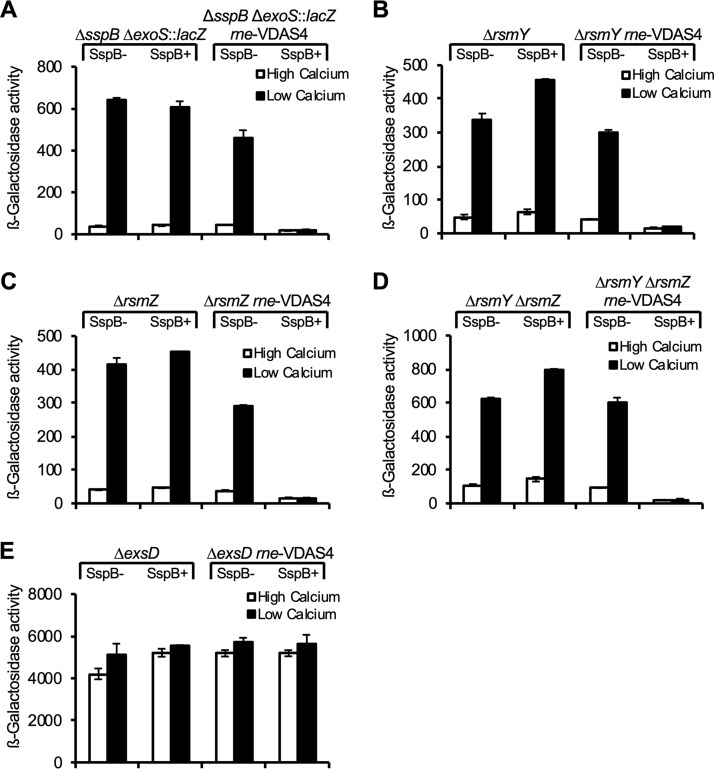
Because RNase E has been implicated in controlling the expression of T3SS genes in Yersinia spp., we sought to determine whether RNase E controlled the expression of T3SS genes in P. aeruginosa (23). Expression of the P. aeruginosa T3SS genes can be induced by growing cells in low-calcium medium (6, 28). To quantify the effect of RNase E depletion on T3SS gene expression, a PAO1 ΔsspB ΔexoS::lacZ reporter strain was generated. This strain has the T3SS effector gene exoS replaced by lacZ, which encodes β-galactosidase. Activation of the T3SS in cells of the PAO1 ΔsspB ΔexoS::lacZ reporter strain results in increased expression of lacZ, suggesting that the lack of SspB did not prevent the induction of exoS expression under conditions of calcium limitation (Fig. 3A). The PAO1 ΔsspB ΔexoS::lacZ reporter strain therefore serves as the control strain for comparison to when RNase E is depleted. PAO1 ΔsspB ΔexoS::lacZ was transformed with either pPSV38 (empty vector) or pPSV38-sspB. The pSV38-sspB plasmid expresses SspB under the control of an IPTG-inducible promoter. The T3SS in the PAO1 ΔsspB ΔexoS::lacZ reporter strain was activated when grown in low-calcium medium in either the absence or presence of SspB (Fig. 3A). However, when SspB was expressed in a PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 reporter strain, lacZ expression decreased 18-fold. This indicated that under conditions of RNase E depletion, T3SS production was significantly reduced (Fig. 3A).
FIG 3.
RNase E is required for type III secretion gene expression in P. aeruginosa. Shown is the quantification of ΔexoS::lacZ expression in cells of the ΔsspB mutant reporter strain and the rne-VDAS4 mutant derivative containing either empty vector (−SspB) or a SspB expression vector (+SspB). IPTG was added to both the −SspB and +SspB conditions. Various strain genetic backgrounds are as listed above each panel.
RNase E-mediated regulation of the T3SS is not dependent on the small regulatory RNA RsmY, RsmZ, RsmV, or RsmW.
In P. aeruginosa, the small regulatory RNAs RsmY and RsmZ have been shown to inhibit T3SS gene expression (15, 29, 30). We reasoned that depletion of RNase E might inhibit the expression of T3SS genes by increasing the stability of either RsmY, RsmZ, or both of these small RNAs (sRNAs). The T3SS is activated under low-calcium growth conditions in PAO1 ΔsspB ΔexoS::lacZ reporter strains lacking rsmY, rsmZ, or both rsmY and rsmZ in either the absence or presence of SspB (Fig. 3B to D). When SspB was expressed in a PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 reporter strain also lacking rsmY, rsmZ, or rsmYZ, lacZ expression decreased 15- to 18-fold (Fig. 3B to D). RNase E-dependent regulation of the T3SS therefore is not dependent on rsmY or rsmZ.
Recent findings indicate that the activity of RsmA is also subject to control by the sRNAs RsmV and RsmW that, like RsmY and RsmZ, exert their regulatory effects through interaction with RsmA (29, 31). The results depicted in Fig. S1 in the supplemental material indicate that under low-calcium growth conditions, depletion of RNase E results in reduced expression of the exoS-lacZ reporter gene in quadruple-mutant cells lacking rsmY, rsmZ, rsmV, and rsmW. Thus, the regulation of T3SS gene expression is independent of any of the sRNAs that are currently known to limit the activity of RsmA.
RNase E depletion does not influence the expression of T3SS genes in cells lacking exsD.
ExsD represses T3SS gene expression by sequestering the transcription activator ExsA (9, 32, 33) (Fig. 1A). P. aeruginosa cells that lack exsD therefore constitutively express the T3SS genes (Fig. 3E). We found that exoS expression was not affected by RNase E depletion in cells of an ΔexsD mutant (Fig. 3E), indicating that the mechanism by which RNase E regulates the expression of the T3SS genes cannot suppress activation by the liberated ExsA in cells of the ΔexsD mutant.
Depletion of RNase E decreases the abundance of the exsA transcript.
ExsA is the major transcription factor responsible for the activation of T3SS gene expression (8, 32–34). We hypothesized that decreased T3SS gene expression in RNase E-depleted cells is due to decreased expression of exsA. PAO1 ΔsspB ΔexoS::lacZ and PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 cells were grown under low-calcium conditions which should induce the transcription of exsA and activate the expression of T3SS-related genes. The abundance of the exsA transcript was measured by quantitative real-time PCR (qRT-PCR) (Fig. 4A). Under low-calcium conditions, exsA transcripts were detected in both reporter strains. When SspB synthesis is induced by the addition of IPTG to the growth medium in PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 cells, RNase E is depleted, and exsA expression decreases 12-fold (Fig. 4A). This decrease in exsA expression would explain the decrease in exoS expression in RNase E-depleted cells.
FIG 4.
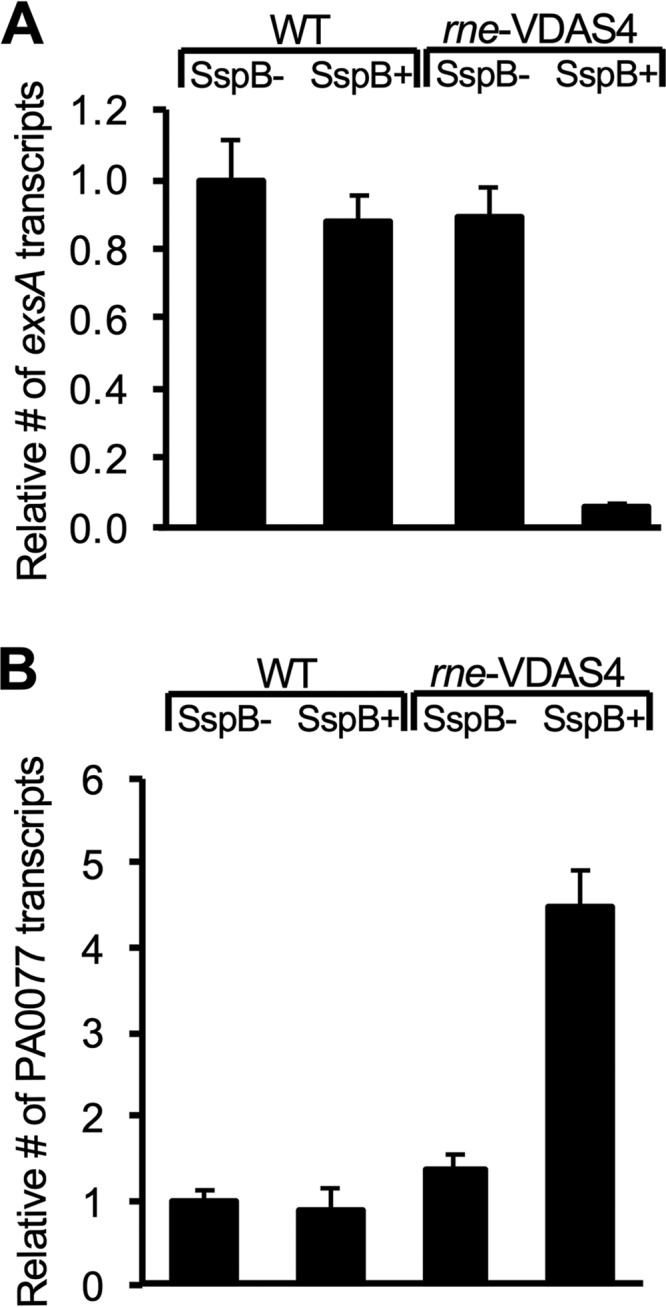
Reciprocal regulation of type III and type VI secretion by RNase E. Effects of RNase E depletion on target gene expression are shown. qRT-PCR was used to measure the indicated transcript abundance relative to the wild-type (WT) strain containing empty vector (SspB−). Error bars represent relative expression values calculated from ±1 standard deviation (SD) from the mean ΔΔCT.
RNase E depletion decreases T3SS protein secretion and cytosolic T3SS-associated regulatory proteins.
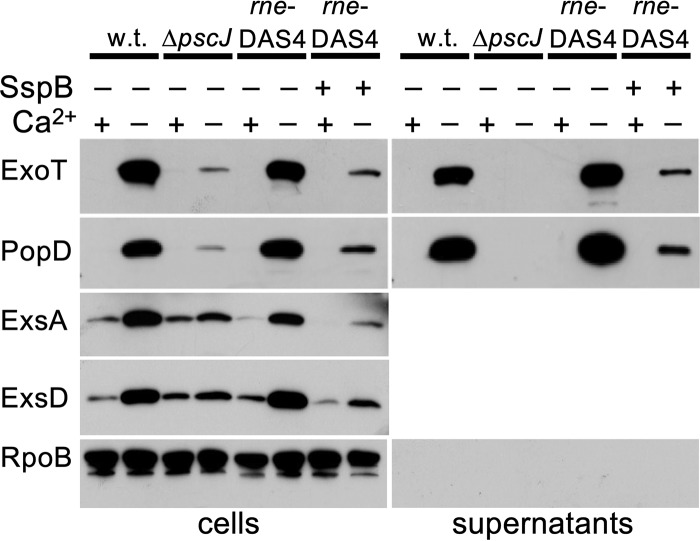
In order to confirm the gene expression phenotype, we monitored secretion of effector and translocator proteins by assaying export of the effector protein ExoT and the translocator protein PopD. Wild-type PAO1 produces PopD and ExoT and secretes both proteins into the culture supernatant in a T3SS-dependent manner when grown under low-calcium conditions (Fig. 5). Depletion of RNase E in the PAO1 strains containing DAS4-tagged RNase E expressing SspB resulted in decreased production of PopD and ExoT in the cells and decreased secretion of both proteins (Fig. 5). We also monitored levels of the regulatory proteins ExsA and ExsD. In wild-type PAO1 cells, PopD, ExsA, and ExsD levels all increase when the cells are grown in low-calcium medium (Fig. 5). When RNase E is depleted in RNase E-VDAS4-tagged strains expressing SspB, ExsA, and ExsD, protein levels decreased (Fig. 5), consistent with the exoS reporter expression data (Fig. 3).
FIG 5.
RNase E depletion decreases type III effector protein secretion. The effects of RNase E depletion on the production of T3SS regulators ExsA and ExsD, as well as production and export of a T3SS translocator protein (PopD), and the effector ExoT were monitored by Western blotting. Amounts of ExsA and ExsD, T3SS regulatory proteins, are also decreased under RNase E depletion conditions. RpoB was detected as a fractionation control. The ΔpscJ mutant strain does not assemble a functional T3SS. w.t., wild type.
Depletion of RNase E results in increased expression of a type 6 secretion system gene.
Studies of RetS and LadS in P. aeruginosa indicate that many virulence factors are reciprocally regulated. For example, the T3SS is RetS activated and LadS repressed, while biofilm formation and the T6SS are LadS activated and RetS repressed (12, 13, 18). We hypothesized that if RNase E depletion results in repression of the T3SS, it might also result in activation of the genes associated with the T6SS. PAO1 ΔsspB ΔexoS::lacZ and PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 cells were grown under low-calcium conditions, which should activate expression of the T3SS. The level of T6SS activation was measured by monitoring the expression of icmF1 (PA0077) by qRT-PCR (Fig. 4B). icmF1 was chosen because it is the last gene of an operon that encodes part of the H1 T6SS. icmF1 also is required for the secretion of Hcp1, which makes up part of the T6SS (35). Under low-calcium conditions, icmF1 transcripts were detected at low levels in both reporter strains. When SspB was expressed in PAO1 ΔsspB ΔexoS::lacZ rne-VDAS4 and RNase E is depleted, icmF1 expression increased 4-fold (Fig. 4B). Taken together, our findings suggest that RNase E plays an important role in T3SS activation while repressing a gene associated with the T6SS. RNase E may therefore reciprocally regulate T3SS and T6SS.
Depletion of RNase E results in increased biofilm formation.
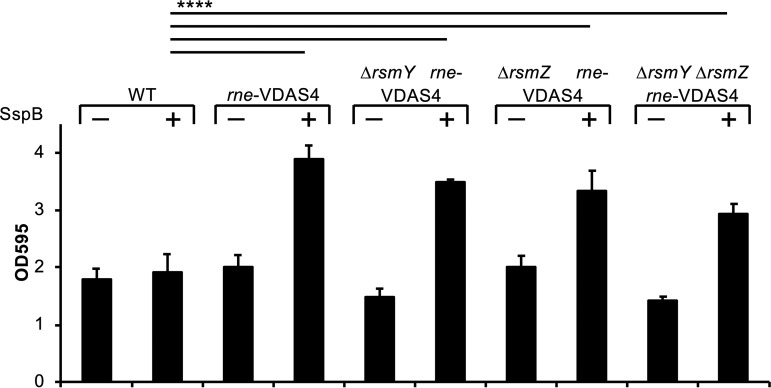
Since RNase E reciprocally regulates the T3SS and the T6SS, we hypothesized that biofilm formation would be upregulated under conditions of RNase E depletion. Twenty-four-well culture plates containing LB with 20 mM IPTG were inoculated with the P. aeruginosa strains indicated in Fig. 6. Following incubation, the culture supernatant was removed, and adherent cells were stained with crystal violet. Biofilm formation was quantitated based on crystal violet retention. Wild-type PAO1 containing pPSV38 or pSspB produced equal amounts of biofilm (Fig. 6). Depletion of RNase E in the PAO1 strains containing DAS4-tagged RNase E expressing SspB resulted in approximately 2-fold increases in biofilm formation (Fig. 6). This increase in biofilm formation was not dependent on rsmY or rsmZ (Fig. 6). The data in Fig. 6 support the idea that RNase E also reciprocally regulates T3S and biofilm formation.
FIG 6.
Depletion of RNase E results in increased biofilm formation. Cells were grown in 24-well plastic tissue culture plates for 16 h at 37°C. After incubation, cells were removed and attached biofilms strained with crystal violet. Biofilm staining was quantified by dissolving the crystal violet in methanol and measuring the OD595. Samples were compared by one-way analysis of variance (ANOVA), using Dunnett’s post hoc test. Significant differences are indicated by a line connecting the data points. ****, P < 0.0001.
RNase E depletion does not influence RsmA abundance.
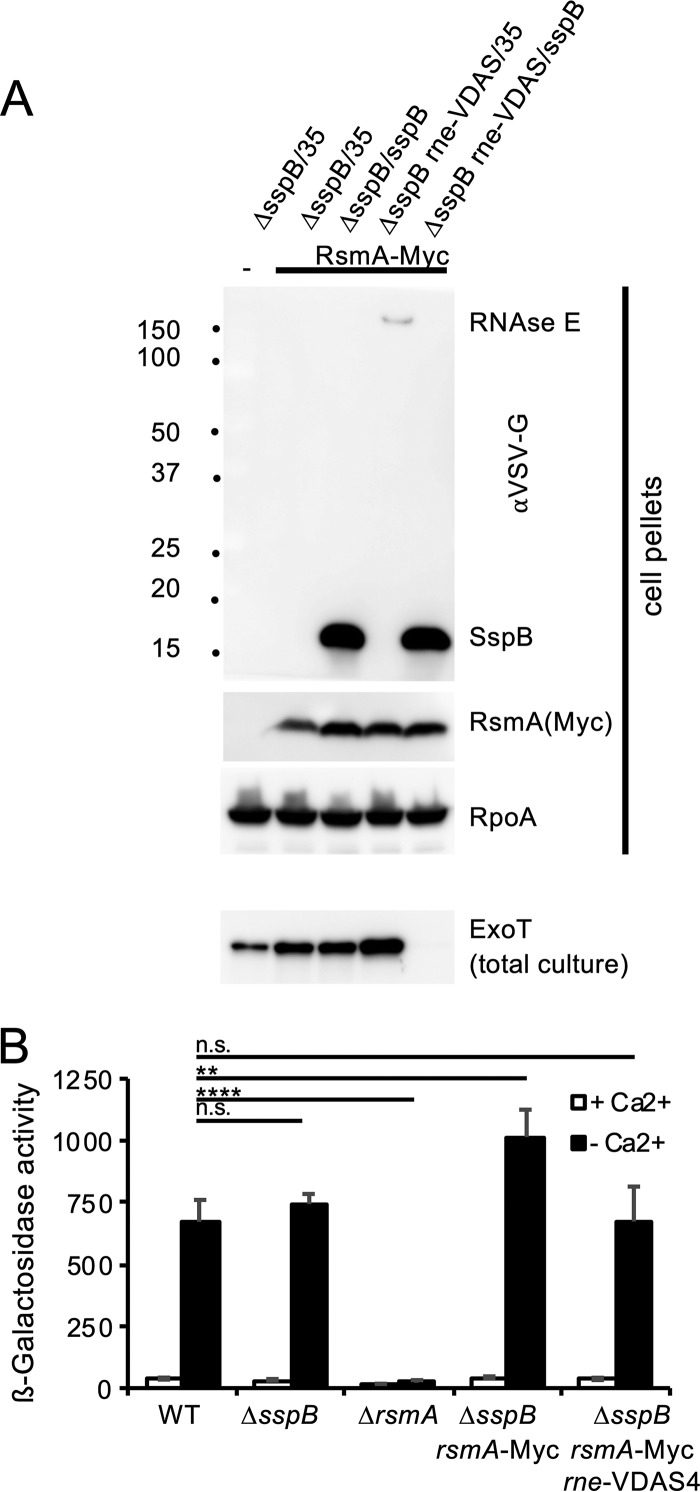
Since the regulatory phenotype of RNase E depletion is consistent with modulation of the Gac/RsmA signal transduction system, we examined whether RNase E depletion interferes with RsmA production. The chromosomal copy of rsmA was modified to encode a version of RsmA that is tagged at the C terminus with 2 repeats of the Myc epitope tag. Tagging of RsmA did not interfere with RsmA function (Fig. 7A). We found that depletion of RNase E did not appreciably alter the abundance of RsmA (Fig. 7B), suggesting that the effect of RNase E depletion on T3SS gene expression cannot be explained through an effect of RNase E on RsmA abundance.
FIG 7.
RNase E depletion does not alter RsmA abundance. (A) RsmA abundance was monitored in strains producing a Myc-tagged version of the protein. The relevant genotype of each strain is indicated above the blot. SspB was supplied from a plasmid, where indicated, or the strain was transformed with the relevant vector control (pPSV35 [denoted “35”]). All strains were grown in the absence of calcium to induce expression of the type III secretion genes. Since this also induces effector secretion, ExoT production was monitored by running total culture samples, rather than cell pellets. RpoA was detected as a loading control. The size standards, indicated at the left, are in kilodaltons. (B) Tagging of RsmA does not interfere with RsmA function. The expression of exoS was monitored in the presence or absence of calcium using a lacZ reporter gene, as described above. The relevant genotype is listed below the columns. β-Galactosidase activity is recorded in Miller units and represents an average of the results from three independent experiments with standard deviation. Samples were compared by one-way ANOVA, using Dunnett’s post hoc test. Specific comparisons are indicated by a line connecting the data points. ****, P < 0.0001; **, P < 0.01; n.s., not significant.
DISCUSSION
Using a ClpXP-based protein depletion system, we have shown that the endoribonuclease RNase E positively regulates expression of the T3SS genes in P. aeruginosa. Depletion of RNase E does not appear to influence the abundance of RsmA, a posttranscriptional regulator that is known to positively regulate T3SS gene expression. Furthermore, the effects of RNase E on T3SS gene expression are independent of the RsmY, RsmZ, RsmV, and RsmW sRNAs that serve to antagonize RsmA activity. We also find that RNase E negatively regulates the expression of a T6SS gene and limits biofilm formation in this organism.
To study the effects of RNase E on the T3SS, we took a targeted protein depletion approach (Fig. 2). By expressing RNase E with a C-terminal VDAS4 tag, we could control ClpXP-dependent degradation of RNase E by IPTG-inducible expression of the SspB adapter protein. When SspB is expressed, it shuttles the VDAS4-tagged RNase E to the ClpXP for degradation (Fig. 2). When RNase E-VDAS4 was expressed in cells in the absence of SspB and grown in calcium-containing medium, the T3SS remained suppressed. Switching these cells to low-calcium medium resulted in the expression of the T3SS. However, depletion of RNase E-VDAS4, by expression of SspB, resulted in cells that maintained suppression of the T3SS, even in low-calcium medium, which should normally trigger expression of the T3SS (Fig. 3).
Since RNase E is predicted to control the degradation of many RNAs in the cell, we considered the possibility that depletion of RNase E leads to increased stability of the sRNAs RsmY and RsmZ. These sRNAs are known to bind and inhibit the activity of RsmA, a key posttranscriptional regulator required for expression of the T3SS genes (36, 37); by depleting RNase E and stabilizing RsmY and RsmZ, free RsmA levels in the cell could in principle be reduced, and the T3SS would be repressed. Indeed, the exoribonuclease polynucleotide phosphorylase (PNPase) has been shown to influence the expression of T3SS gene expression in P. aeruginosa by influencing the stability of RsmY and RsmZ (38). To address this possibility, we depleted RNase E in strains lacking either rsmY, rsmZ, or both. However, depletion of RNase E in these mutant strains still resulted in reduced expression of the T3SS genes in low-calcium medium (Fig. 3A to D). These results establish that RNase E does not regulate the T3SS through a mechanism that is dependent upon rsmY or rsmZ.
RsmV and RsmW are two recently discovered sRNAs that, like RsmY and RsmZ, sequester RsmA and inhibit its activity (31, 39). Depletion of RNase E could potentially stabilize either RsmV, RsmW, or both, thus inhibiting RsmA activity. When RsmV or RsmW was deleted in a strain lacking RsmY and RsmZ, expression of an exoS-lacZ reporter was still reduced upon RNase E depletion (see Fig. S1 in the supplemental material). These results indicate that the RNase E-mediated regulation of the T3SS does not require RsmY, RsmZ, RsmV, or RsmW. However, our findings do not exclude the possibility that RNase E exerts its effects by altering the abundances of other RNA species that act as molecular sponges for RsmA.
We considered the possibility that RNase E regulates T3SS gene expression by influencing the abundance of RsmA which is required for T3SS gene expression. If RNase E depletion leads to decreased RsmA in the cell, that would decrease T3SS gene expression. RNase E depletion did not result in a decrease in the abundance of RsmA in the cell (Fig. 7), suggesting that RNase E does not exert its effects on T3SS gene expression through positive effects on the abundance of RsmA.
RNase E is required for efficient production of the master regulator of T3SS gene expression, ExsA. Both exsA transcript and ExsA protein abundance are reduced when RNase E is depleted (Fig. 4A and 5). This suggests that RNase E influences the expression of the T3SS by controlling the abundance of the ExsA transcription factor. ExsA activity is held in check through sequestration by the antiactivator protein ExsD. The decreased T3SS gene expression we observed in low-calcium medium was reversed in a strain background lacking ExsD (Fig. 3E). These data suggest that the feedforward loop that exists in the absence of ExsD (since ExsA upregulates its own expression as well) may be sufficient to overcome the defect in T3SS gene expression we observed upon depletion of RNase E.
Since the T3SS is often reciprocally regulated with the T6SS and biofilm formation, we measured the effects of RNase E depletion on the expression of icmF1 (PA0077), which is part of the H1 T6SS. While RNase E depletion resulted in decreased expression of exsA, it resulted in an increase in the expression of icmF1 (Fig. 4B). These findings suggest that RNase E participates in a pathway that reciprocally regulates the T3SS and T6SS. Similarly, depletion of RNase E resulted in an approximately 2-fold increase in biofilm formation. Consistent with our observations regarding T3SS gene regulation, the increase in biofilm formation upon RNase E depletion was not dependent on two known regulators of biofilm formation, RsmY and RsmZ (Fig. 5). These data suggest that RNase E has multiple influences on regulating virulence factor gene expression.
We have established that RNase E plays a role in positively regulating the expression of T3SS genes, but exactly how RNase E exerts its regulatory effects is unclear. It is striking that RNase E appears to have activities reminiscent of RsmA. Although RNase E influences T3SS gene expression independently of sRNAs that are known to limit the activity of RsmA, and although RNase E does not positively influence the abundance of RsmA, our findings raise the possibility that RsmA and RNase E act in concert with one another. Indeed, RsmA is a RNA-binding protein that typically acts to repress the translation of target mRNA species (40). By influencing the translation of target mRNA species, RsmA may render these species accessible to degradation by RNase E. It was shown previously that the metabolic state of P. aeruginosa can control T3SS assembly, presumably through an effect on the envelope (41, 42). We cannot rule out the possibility that the depletion of RNase E influences the metabolic state of the cell, thereby affecting T3SS assembly, and, by extension, the ability to trigger T3SS gene expression by removing calcium from the growth medium.
RNase E is typically thought of as a global regulator of RNA stability in bacterial cells. Here, we have shown that RNase E can influence the expression of genes involved in the virulence of P. aeruginosa. Depletion of RNase E from the cell reduces T3SS gene expression and protein abundance. While further investigation is needed to fully elucidate the mechanism by which RNase E controls T3SS gene expression, our data support recent findings that RNases are important players in the control of virulence gene expression in a variety of pathogenic bacteria (23, 42–46).
MATRIALS AND METHODS
Bacterial strains.
The recipient strain for all plasmid constructions was E. coli DH5αF′IQ (Invitrogen). For strain construction, plasmids were transferred into P. aeruginosa via conjugation utilizing E. coli SM10 λpir. Bacterial cultures were grown in lysogeny broth (LB) or on LB plates containing 15 g/liter agar. For P. aeruginosa strains, gentamicin (Gent; 30 μg/ml), carbenicillin (200 μg/ml), or tetracycline (Tet; 35 μg/ml) was used for selection when required. For P. aeruginosa, cells were grown in liquid LB cultures containing 10 mM MgCl2 and 0.5 mM CaCl2 (LB-MC). Where indicated, type III secretion was triggered by removing calcium from the medium by the addition of EGTA (5 mM final concentration; LB-MC-EGTA). A list of strains and plasmids can be found in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| RP1831 | PAO1F (wild-type PAO1) | 3 |
| RP1868 | PAO1F ΔexoS::GL3 | This study |
| JS1869 | PAO1F ΔexoS::GL3 ΔsspB | This study |
| JS1870 | PAO1F ΔexoS::GL3 ΔsspB rne-VDAS4 | This study |
| JS1871 | PAO1F ΔexoS::GL3 ΔsspB rne-VDAS4 ΔrsmY | This study |
| JS1872 | PAO1F ΔexoS::GL3 ΔsspB rne-VDAS4 ΔrsmZ | This study |
| JS1873 | PAO1F ΔexoS::GL3 ΔsspB rne-VDAS4 ΔrsmY ΔrsmZ | This study |
| RP2093 | PAO1F ΔexoS::GL3 ΔrsmA | This study |
| RP10346 | PAO1F ΔexoS::GL3 ΔsspB rsmA-Myc | This study |
| RP10351 | PAO1F ΔexoS::GL3 ΔsspB rsmA-Myc rne-VDAS4 | This study |
| PAO1F | ΔexoS::GL3 ΔsspB rne-VDAS4 ΔrsmY ΔrsmZ ΔrsmV ΔrsmW | This study |
| Primers | ||
| pPSV35 | Expression plasmid with lacIQ, lacUV5 promoter, Gent resistance, colE1 and Pseudomonas origins of replication, oriT | 43 |
| pEXG2 | Allelic exchange vector, colE1 origin; oriT Gentr sacB | 43 |
| pEXG2-ΔrsmA | pEXG2 with ΔrsmA allele | This study |
| pEXG2-ΔrsmY | pEXG2 with ΔrsmY allele | This study |
| pEXG2-ΔrsmZ | pEXG2 with ΔrsmZ allele | This study |
| pEXG2-ΔrsmV | pEXG2 with ΔrsmV allele | This study |
| pEXG2-ΔrsmW | pEXG2 with ΔrsmW allele | This study |
| pEXG2-rsmA-Myc | pEXG2 with rsmA fused at the 3′ end to 2 copies of the Myc tag | This study |
| pV-sspB | pPSV35 vector directing IPTG-inducible expression of SspB with a N-terminal VSV-G tag | 10 |
| pVDIV | Integration vector to incorporate C-terminal VDAS4 tag | 10 |
| pVDIV-rne | Integration vector to incorporate C-terminal VDAS4 tag to rne | This study |
Construction of depletion strains.
Table 2 contains a list of primers used in this study. The construct for deleting the sspB gene (PA4427) was previously described (26). Briefly, the flanking regions of sspB were amplified by the PCR and spliced together with overlap extension PCR. The resulting PCR product was cloned into plasmid pEXG2 (41), as described previously (26), yielding plasmid pEXG2-ΔsspB. This plasmid, along with recipient strains PAO1 and PAO1 ΔexoS::lacZ, was then used to create the following strains containing in-frame deletions of sspB by allelic exchange: PAO1 ΔsspB and PAO1 ΔexoS::lacZ ΔsspB. Th deletions were confirmed by PCR.
TABLE 2.
Primers used in this study
| Name | Sequence | Description |
|---|---|---|
| rsmA5-1 | AAAAAgaattcCGACACCAACATCGCCAAGGTT | rsmA deletion, removing codons 3–59, outside primer, EcoRI site (lowercase) |
| rsmA5-2 | AACTCGAGCCGCAAGCATGCTGAACAGCATTCCTTTCTCCTCACGCGAA | Pair with rsmA5-1 |
| rsmA3-1 | TTCAGCATGCTTGCGGCTCGAGTTAACCATTAATTTTTATCTAATTTT | Pair with rsmA3-2 |
| rsmA3-2 | AAAAAaagcttTAACGCTTGTTTTACCGTGAAAGA | Outside primer, HindIII site (lowercase) |
| rsmA-Myc-5-2 | CTCCGAGATCAGTTTCTGCTCGCCCAAATCTTCTTCAGAAATCAACTTTTGTTCATGGTTTGGCTCTTGATCTTTCTC | Fuse 2× Myc tag to 3′ end of rsmA, pair with rsmA5-1 |
| rsmA-Myc-3-1 | GAAGAAGATTTGGGCGAGCAGAAACTGATCTCGGAGGAGGACCTGCAATGATTTTTATCTAATTTTCCCTTTGCA | Pair with rsmA3-2 |
| rsmV-5-1 | AAAAAtctagaATGGGTTTCCGGCCAGTTAGC | rsmV deletion 5′ flank outside primer, XbaI site (lowercase) |
| rsmV-5-2 | GGATAGCGGGAGGGGATGAGGTTGGAAATCTACCAAGC | Pair with rsmV-5-1 |
| rsmV-3-1 | GCTTGGTAGATTTCCAACCTCATCCCCTCCCGCTATCC | Pair with rsmV-3-2 |
| rsmV-3-2 | AAAAAaagcttCCGCTGGCGATCAGGTACAGCAC | rsmV deletion, 3′ flank outside primer, HindIII site (lowercase) |
| rsmW-5-1 | AAAAAtctagaGCCCTGGACTATACCGCCAAC | rsmW deletion 5′ flank outside primer, XbaI site (lowercase) |
| rsmW-5-2 | GGAAAGACCGGGACATGAGGGCTCAGCGCAGGGCGCGGGCGTAG | Pair with rsmW-5-1 |
| rsmW-3-1 | CTACGCCCGCGCCCTGCGCTGAGCCCTCATGTCCCGGTCTTTCC | Pair with rsmW-3-2 |
| rsmW-3-2 | AAAAAaagcttGGTCGAACTCTTCGAAGCTGTA | rsmW deletion, 3′ flank outside primer, HindIII site (lowercase) |
| delrsmY F1 | ATATggatccGGTGGCCACGTAGTTCGGGG | rsmY deletion 5′ flank outside primer, BamHI site (lowercase) |
| delrsmY R2 | GCGGTTTTCCTCGGGCAATAAGGTTTGAAGATTACGCATCTCTG | rsmY reverse inner primer |
| delrsmY F3 | TGCGTAATCTTCAAACCTTATTGCCCGAGGAAAACCGCGTCGCT | rsmY forward inner primer |
| delrsmY R4 | ATATctcgagCTGCTCACCGGCAACCTGGA | rsmY deletion 3′ flank outside primer, XhoI site (lowercase) |
| delrsmZ F1 | ATATggatccCGAGCTGCTGCAGGATGACG | rsmZ deletion 5′ flank outside primer, BamHI site (lowercase) |
| delrsmZ R2 | ACGAGTAAAACGGCAGGCAAACAGGAGTGATATTAGCGATTCC | rsmZ reverse inner primer |
| delrsmZ F3 | CGCTAATATCACTCCTGTTTGCCTGCCGTTTTACTCGTCGCCAA | rsmZ forward inner primer |
| delrsmZ R4 | ATATctcgagGCCCGCGGCAAGCTCTCGAT | rsmZ deletion 3′ flank outside primer, XhoI site (lowercase) |
| rneVDAS4-F1 | ATATggatccAGGACGAGCAGGACGATACCGATG | Forward primer to fuse VDAS4 tag to rne 3′ end, BamHI site (lowercase) |
| rneVDAS4-R2 | CCTAGGTCAGCTGGCGTCGGCGTAGTTCTCGCTGTA | rne reverse inner primer to fuse VDAS4 tag to rne 3′ end |
| rneVDAS4-F3 | GCCAGCTGACCTAGGGTGACAGGCTGAAAAAAGGG | rne forward inner primer to fuse VDAS4 tag to rne 3′ end |
| rneVDAS4-R4 | ATATgaattcCTGACCCGTGACGTCGAGGCGCTG | Reverse primer to fuse VDAS4 tag to rne 3′ end, EcoRI site (lowercase) |
| PA0077-RT F1 | ATCGACAGCCTGCTGGAAGACAT | PA0077 (icmF1) forward primer for real-time PCR |
| PA0077-RT R1 | TGGTGGAGTTGACCACGTTCTTCA | PA0077 (icmF1) reverse primer for real-time PCR |
| exsA-RT F1 | ATGCAAGGAGCCAAATCTCTTGG | exsA forward primer for real-time PCR |
| exsA-RT R1 | TCAGTTATTTTTAGCCCGGCATTC | exsA reverse primer for real-time PCR |
The constructs for deleting the genes for rsmW (PA4570.1), rsmY (PA0527.1), and rsmZ (PA3621.1) were made by amplifying 1-kb regions flanking rsmW, rsmY, or rsmZ by PCR and then splicing the flanking regions together by overlap extension PCR. The deletion was in-frame and contained the linker sequence 5′-ATGGCGGCCGCTTAA-3′. The resulting PCR product was cloned on an XbaI/EcoRI fragment into plasmid pEXG2, yielding plasmids pEXG2-ΔrsmW, pEXG2-ΔrsmY, and pEXG2-ΔrsmZ, respectively. These plasmids along with recipient strain PAO1 ΔexoS::lacZ ΔsspB were then used to create the following strains containing in-frame deletions of rsmW, rsmY, and rsmZ by allelic exchange: PAO1 ΔexoS::lacZ ΔsspB ΔrsmY, PAO1 ΔexoS::lacZ ΔsspB ΔrsmZ, PAO1 ΔexoS::lacZ ΔsspB ΔrsmY ΔrsmZ, and PAO1 ΔexoS::lacZ ΔsspB ΔrsmW ΔrsmY ΔrsmZ. The deletions were confirmed by PCR.
The VSV-G-DAS4 tag (VDAS4 tag) integration vector pVDIV was described previously (26). Plasmid pVDIV-rne was constructed by amplifying approximately 300 bp of the 3′ end of the rne (PA2976) containing a 5′ HindIII site and a 3′ NotI site. This allowed cloning of the 3′ end of rne into pVDIV cut with HindIII and NotI; the portion of the rne gene was cloned such that they were in-frame with the DNA specifying the VDAS4 tag. This plasmid together with recipient strains PAO1 ΔsspB, PAO1 ΔexoS::lacZ ΔsspB, PAO1 ΔexoS::lacZ ΔsspB ΔrsmY, PAO1 ΔexoS::lacZ ΔsspB ΔrsmZ, PAO1 ΔexoS::lacZ ΔsspB ΔrsmY ΔrsmZ, and PAO1 ΔexoS::lacZ ΔsspB ΔrsmW ΔrsmY ΔrsmZ were used to create strains PAO1 ΔsspB rne-VDAS4, PAO1 ΔexoS::lacZ ΔsspB rne-VDAS4, PAO1 ΔexoS::lacZ ΔsspB ΔrsmY rne-VDAS4, PAO1 ΔexoS::lacZ ΔsspB ΔrsmZ rne-VDAS4, PAO1 ΔexoS::lacZ ΔsspB ΔrsmY ΔrsmZ rne-VDAS4, and PAO1 ΔexoS::lacZ ΔsspB ΔrsmW ΔrsmY ΔrsmZ rne-VDAS4 by integration of plasmid into the chromosome. The plasmid backbone was excised by the expression FLP recombinase from pFLP2.
SspB expression vectors.
Plasmid pV-SspB directs IPTG-inducible expression of SspB with an N-terminal VSV-G tag and was previously described (26).
Depletion of RNase E.
Strains containing VSV-G-DAS4-tagged RNase E were transformed with pV-SspB. IPTG (1 mM final concentration) was added to liquid cultures to induce the expression of SspB and subsequent degradation of RNase E by the ClpXP protease.
β-Galactosidase assays.
P. aeruginosa cultures were grown at 37°C to log phase in LB-MC medium. One milliliter of that initial culture was then added to 1 ml LB-MC medium (+Ca) and 1 ml of LB-MC medium containing 5 mM EGTA (−Ca). These cultures were incubated, and β-galactosidase assays were performed essentially as described in references 47 and 52.
RNA isolation and cDNA synthesis.
P. aeruginosa strains were grown with aeration at 37°C in LB-MC (+Ca) or LB-MC-EGTA (−Ca) medium containing 30 μg/ml gentamicin and 1 mM IPTG. Triplicate cultures of each strain were inoculated to a starting optical density at 600 nm (OD600) of 0.01 and grown to a final OD600 of ∼0.5. RNA isolation and cDNA synthesis were performed essentially as described previously (48).
Quantitative real-time PCR.
The abundance of target transcripts relative to that of the clpX transcript was determined by quantitative real-time reverse transcription-PCR (RT-PCR) using the iTaq SYBR green kit (Bio-Rad). cDNAs were amplified by real-time PCR utilizing an ABI Prism 7000 system (Applied Biosystems). PCR primer specificities were confirmed by melting curve analyses (Table 2). Relative transcript abundance was determined using the comparative threshold cycle (CT) method (ΔΔCT), as described previously (49). The values presented in Fig. 4 are the averages of 3 real-time RT-PCR amplifications from three independent RNA isolations. Error bars represent the relative expression values calculated from ±1 standard deviation from the mean ΔΔCT.
Biofilm assays.
Twenty-four-well culture plates (VWR) containing 10 ml LB broth with 20 mM IPTG and appropriate antibiotic selection were inoculated with the P. aeruginosa strains to a starting OD600 of 0.03. Plates were incubated at 37°C for 16 h without shaking. Following incubation, the culture supernatant was removed, and adherent cells were stained with 0.1% crystal violet for 15 min. Biofilm assays and quantitation were performed essentially as described in reference 50.
Western blots.
Overnight cultures were grown in high-salt LB medium with Gent and, if needed (rne-VDAS), 100 μg/ml tetracycline and then diluted 1:200 (rne-VDAS and sspB 1:100) into high-salt LB medium with 15 μg/ml Gent, 1 mM IPTG, and 200 μg/ml Tet for rne-VDAS strains. Cultures were grown for 1.5 h, at which point EGTA was added to 5 mM and incubated for another 2.5 h. For total culture samples, 75 μl of culture was removed and mixed with 25 μl of 4× SDS sample buffer. Cell pellets from 1 ml of culture were also collected, resuspended in 1× sample buffer to an OD of 5, and lysed. Supernatant samples were collected by removing 500 μl of culture supernatant after pelleting cells and precipitating secreted proteins by added trichloroacetic acid to 10%. Precipitates were collected by centrifugation and washed with acetone to remove residual trichloroacetic acid before being resuspended in 1× SDS sample buffer, also normalized to a culture with an OD of 5.
Four percent to 12% Bis-Tris NuPAGE gels (Invitrogen) were utilized to separate purified proteins and cell lysates. Western blotting was performed as previously described (28). Polyclonal rabbit anti-VSV-G (Sigma-Aldrich) and peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich) antibodies were used to detect the VSV-G tag. In some experiments, samples were separated on 12% TGX protein gels (Bio-Rad), transferred to polyvinylidene difluoride (PVDF) membranes using a Pierce Power Station semidry transfer apparatus (Thermo), and blocked using 5% nonfat milk resuspended in Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 150 mM NaCl) with 0.05% Tween 20 (TBS-T). All antibody incubations were performed in the same buffer. Primary antibodies to the Myc tag and VSV-G tag were purchased commercially (Sigma-Aldrich), as well as the antibody directed against the RNA polymerase alpha subunit (BioLegend). Affinity-purified antibodies to PopD, ExoT, ExsA, and ExsD were raised against purified His-tagged proteins in rabbits and affinity purified (Covance). Primary antibodies were detected with species-specific secondary antibodies conjugated to horseradish peroxidase (Sigma-Aldrich), using the Advansta Quantum chemiluminescent substrate.
Statistical analysis.
Statistical analysis was performed using the Prism software package, version 6 (GraphPad).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by National Institutes of Health grants AI069007 and AI118955 to S.L.D., NIH grant EY022052 to A.R., and NIH grant T32-AI07061-32 to J.S.S.
We thank Bryan McGuffie for designing the artwork for Fig. 1 and Renate Hellmiss for the artwork in Fig. 2.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00336-19.
REFERENCES
- 1.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerr KG, Snelling AM. 2009. P. aeruginosa: a formidable and ever-present adversary. J Hosp Infect 73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N. 1993. Antibiotic therapy for chronic infection of Pseudomonas in the lung. Annu Rev Med 44:1–10. doi: 10.1146/annurev.me.44.020193.000245. [DOI] [PubMed] [Google Scholar]
- 4.Yahr TL, Goranson J, Frank DW. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol 22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 5.Galle M, Carpentier I, Beyaert R. 2012. Structure and function of the type III secretion system of Pseudomonas aeruginosa. Curr Protein Pept Sci 13:831–842. doi: 10.2174/138920312804871210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornef MW, Roggenkamp A, Geiger AM, Hogardt M, Jacobi CA, Heesemann J. 2000. Triggering the ExoS regulon of Pseudomonas aeruginosa: a GFP-reporter analysis of exoenzyme (Exo) S, ExoT and ExoU synthesis. Microb Pathog 29:329–343. doi: 10.1006/mpat.2000.0398. [DOI] [PubMed] [Google Scholar]
- 7.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67:914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams McMackin EA, Djapgne L, Corley JM, Yahr TL. 1999. Fitting pieces into the puzzle of Pseudomonas aeruginosa type III secretion system gene expression. J Bacteriol 201:e00209-19. doi: 10.1128/JB.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCaw ML, Lykken GL, Singh PK, Yahr TL. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol 46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 10.Marsden AE, Intile PJ, Schulmeyer KH, Simmons-Patterson ER, Urbanowski ML, Wolfgang MC, Yahr TL. 2016. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 198:1442–1450. doi: 10.1128/JB.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. 2015. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 197:2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 17.Francis VI, Waters EM, Finton-James SE, Gori A, Kadioglu A, Brown AR, Porter SL. 2018. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat Commun 9:2219. doi: 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apirion D. 1978. Isolation, genetic mapping, and some characterization of a mutation in Escherichia coli that affects the processing of ribonucleic acids. Genetics 90:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono M, Kuwano M. 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol 129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 21.Mackie GA. 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 22.Thuraisamy T, Lodato PB. 2018. Influence of RNase E deficiency on the production of stx2-bearing phages and Shiga toxin in an RNase E-inducible strain of enterohaemorrhagic Escherichia coli (EHEC) O157:H7. J Med Microbiol 67:724–732. doi: 10.1099/jmm.0.000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Jain C, Schesser K. 2008. RNase E regulates the Yersinia type 3 secretion system. J Bacteriol 190:3774–3778. doi: 10.1128/JB.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulsen BE, Yang R, Clatworthy AE, White T, Osmulski SJ, Li L, Penaranda C, Lander ES, Shoresh N, Hung DT. 2019. Defining the core essential genome of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castang S, McManus HR, Turner KH, Dove SL. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A 105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinness K, Baker T, Sauer R. 2006. Engineering controllable protein degradation. Mol Cell 22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen KH, Diaz MR, Golden M, Graham JW, Sanders W, Wolfgang MC, Yahr TL. 2018. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J Bacteriol 200:e00736-17. doi: 10.1128/JB.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Callaghan J, Reen FJ, Adams C, O’Gara F. 2011. Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology 157:3417–3428. doi: 10.1099/mic.0.052050-0. [DOI] [PubMed] [Google Scholar]
- 31.Miller CL, Romero M, Karna SL, Chen T, Heeb S, Leung KP. 2016. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol 16:155. doi: 10.1186/s12866-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brutinel ED, Vakulskas CA, Yahr TL. 2009. Functional domains of ExsA, the transcriptional activator of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 191:3811–3821. doi: 10.1128/JB.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brutinel ED, Vakulskas CA, Yahr TL. 2010. ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol 192:1479–1486. doi: 10.1128/JB.01457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vakulskas CA, Brady KM, Yahr TL. 2009. Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J Bacteriol 191:6654–6664. doi: 10.1128/JB.00902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burrowes E, Baysse C, Adams C, O’Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 37.Mulcahy H, O’Callaghan J, O’Grady EP, Adams C, O’Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen R, Weng Y, Zhu F, Jin Y, Liu C, Pan X, Xia B, Cheng Z, Jin S, Wu W. 2016. Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front Microbiol 7:247. doi: 10.3389/fmicb.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janssen KH, Diaz MR, Gode CJ, Wolfgang MC, Yahr TL. 2018. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J Bacteriol 200:e00277-18. doi: 10.1128/JB.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 41.Rietsch A, Mekalanos JJ. 2006. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol 59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rietsch A, Wolfgang MC, Mekalanos JJ. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun 72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bearson SM, Bearson BL, Lee IS, Kich JD. 2013. Polynucleotide phosphorylase (PNPase) is required for Salmonella enterica serovar Typhimurium colonization in swine. Microb Pathog 65:63–66. doi: 10.1016/j.micpath.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Casinhas J, Matos RG, Haddad N, Arraiano CM. 2018. Biochemical characterization of Campylobacter jejuni PNPase, an exoribonuclease important for bacterial pathogenicity. Biochimie 147:70–79. doi: 10.1016/j.biochi.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Engman J, Negrea A, Sigurlásdóttir S, Geörg M, Eriksson J, Eriksson OS, Kuwae A, Sjölinder H, Jonsson AB. 2016. Neisseria meningitidis polynucleotide phosphorylase affects aggregation, adhesion, and virulence. Infect Immun 84:1501–1513. doi: 10.1128/IAI.01463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haddad N, Matos RG, Pinto T, Rannou P, Cappelier JM, Prévost H, Arraiano CM. 2014. The RNase R from Campylobacter jejuni has unique features and is involved in the first steps of infection. J Biol Chem 289:27814–27824. doi: 10.1074/jbc.M114.561795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee PC, Zmina SE, Stopford CM, Toska J, Rietsch A. 2014. Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc Natl Acad Sci U S A 111:E2027–E2036. doi: 10.1073/pnas.1402658111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.O’Toole G, Kolter R. 2002. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 51.Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. 2005. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol 187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dove SL, Hochschild A. 2004. A bacterial two-hybrid system based on transcription activation. Methods Mol Biol 261:231–246. [DOI] [PubMed] [Google Scholar]
- 53.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.